Summary
Dendritic cells (DC) are a class of bone‐marrow‐derived cells arising from lympho‐myeloid haematopoiesis that form an essential interface between the innate sensing of pathogens and the activation of adaptive immunity. This task requires a wide range of mechanisms and responses, which are divided between three major DC subsets: plasmacytoid DC (pDC), myeloid/conventional DC1 (cDC1) and myeloid/conventional DC2 (cDC2). Each DC subset develops under the control of a specific repertoire of transcription factors involving differential levels of IRF8 and IRF4 in collaboration with PU.1, ID2, E2‐2, ZEB2, KLF4, IKZF1 and BATF3. DC haematopoiesis is conserved between mammalian species and is distinct from monocyte development. Although monocytes can differentiate into DC, especially during inflammation, most quiescent tissues contain significant resident populations of DC lineage cells. An extended range of surface markers facilitates the identification of specific DC subsets although it remains difficult to dissociate cDC2 from monocyte‐derived DC in some settings. Recent studies based on an increasing level of resolution of phenotype and gene expression have identified pre‐DC in human blood and heterogeneity among cDC2. These advances facilitate the integration of mouse and human immunology, support efforts to unravel human DC function in vivo and continue to present new translational opportunities to medicine.
Keywords: antigen presentation/processing, dendritic cell, transcriptomics
Introduction
The initiation and control of immune responses depends upon dendritic cells (DC), a class of bone‐marrow‐derived cells found in blood, tissues and lymphoid organs. In a broad sense, the function of DC is to bridge the innate and adaptive immune systems. DC are innate immune cells because they recognize and respond to pathogen‐associated and danger‐associated signals, shaping the acute inflammatory response. Their defining role in adaptive immunity is to process extracellular and intracellular proteins and to present antigens in the context of MHC molecules to prime naive T cells. Previously, DC have been characterized as universal ‘all purpose’ antigen‐presenting cells, but an important aspect of the control of immune responses, is the existence of several different types of DC, each specialized to respond to particular pathogens and to interact with specific subsets of T cells. This expands the flexibility of the immune system to react appropriately to a wide range of different pathogens and danger signals (Fig. 1a).
Figure 1.

Ontological overview and functional specialization of human dendritic cells (DC). (a) DC are often depicted as a single ‘all purpose’ cell in diagrams of T‐cell differentiation but each subset is specialized to make specific responses to pathogen or danger signals. Depending on the context, many different responses may be observed and selected principal functions of human plasmacytoid DC (pDC), conventional DC1 (cDC1) and cDC2 are depicted. (b) Ontological basis of DC, monocyte and macrophage classification. Haematopoietic stem cells (HSC) give rise to DC and monocyte‐derived cells by distinct routes marked by differences in the relative expression of interferon regulatory factor 8 (IRF8) and IRF4 as shown in schematic bivariate plots beneath. Monocytes are IRF4/8 low but can be induced to differentiate into monocyte‐derived DC (mo‐DC). Monocyte‐derived macrophages are also ontologically distinct from many populations of long‐lived resident macrophages derived from early myeloid progenitors (EMP).
The unified classification of mammalian DC
The functional and anatomical classification of DC as ‘migratory tissue DC’ or ‘lymph node resident DC’ remains useful to define context. However, comparative gene expression studies have driven a robust classification of DC based primarily on lineage, that correlates with the differential expression of key transcription factors such as interferon regulatory factors 8 and 4 (IRF8 and IRF4) and performs well across all mammalian species.1, 2, 3, 4 This recognizes plasmacytoid DC (pDC) and two types of ‘conventional’ or ‘classical’ DC (cDC) corresponding to the two subsets of myeloid DC previously defined by CD141 and CD1c expression in humans.5, 6, 7 These DC lineage cells are treated as distinct entities to monocyte‐derived cells and monocyte‐derived DC (mo‐DC) and macrophage populations derived from primitive myeloid progenitors arising in the yolk sac (Fig. 1b).
The term ‘myeloid’, introduced into human DC biology around two decades ago,5, 6, 7 remains valid in defining a common set of antigens found on cDC and for clarity, ‘myeloid’ and ‘conventional’ will be used together to describe the human DC of this class. The original markers, CD141 and CD1c have limitations as both are induced on cDC and monocyte‐derived cells in tissues and in culture. Expression profiling has now provided a suite of more consistent markers that perform well across species, such as CLEC9A, CADM1, BTLA and CD26 for CD141+ myeloid cDC1, and CD2, FcεR1 and SIRPA for CD1c+ myeloid cDC22, 3, 4 (Table 1). Additional complexity discussed below is the emergence of subsets of cDC2 and the realization that conventional pDC gates include myeloid cDC precursors that also express CD123 and CD303.8, 9, 10
Table 1.
Human dendritic cell subset characterization
| Unified classification | Differential TFs | Conventional markers | Extended markers | Notes |
|---|---|---|---|---|
| Plasmacytoid DC | E2‐2 | CD123, | FCER1 | DC6 9 |
| ZEB2 | CD303/CLEC4C/BDCA‐2 | ILT3, ILT7 | ||
| IRF8 | CD304/NRP1/BDCA‐4 | DR6 | ||
| IRF4 | ||||
| Myeloid cDC1 | ID2 | CD141/BDCA‐1 | CLEC9A | DC1 9 |
| IRF8 | CADM1 | No antibody for XCR1 in human | ||
| BATF3 | XCR1 | |||
| BTLA | ||||
| CD26 | ||||
| DNAM‐1/CD226 | ||||
| Myeloid cDC2 | ID2 | CD1c/BDCA‐1 | CD2 | DC2/DC3 9 |
| ZEB2 | CD11c | FCER1 | DCIR clone specific [26] | |
| IRF4 | CD11b | SIRPA | ||
| Notch2/KLF4 | ILT1 | |||
| DCIR/CLEC4A | ||||
| CLEC10A | ||||
| Langerhans cell | ID2 | CD207 | EpCAM | |
| RUNX3 | CD1a | TROP2 | ||
| E‐Cadherin | ||||
| Pre‐DC | ZEB2 | CD123, CD303 | AXL | DC5 ‘AS’ DC 9 |
| IRF4 | SIGLEC 6 | |||
| KLF4 | CX3CR1 | |||
| CD169 (SIGLEC 1) | ||||
| CD22 (SIGLEC 2) | ||||
| CD33 (SIGLEC 3) | ||||
| Mo‐DC | MAFB | CD11c | SIRPA | |
| KLF4 | CD1c/BDCA‐1 | S100A8/A9 | ||
| CD1a | CD206 | |||
| DC‐SIGN/CD209 | ||||
| Non‐classical monocyte | CD16 | DC4 9 | ||
| CX3CR1 | SLAN DC? | |||
| +/‐SLAN |
cDC, conventional DC; DC, dendritic cell; Mo‐DC, monocyte‐derived DC; pDC, plasmacytoid DC; TF, transcription factor. IRF4 and IRF8 are highlighted in bold.
Analysis of human dendritic cells, monocytes and macrophages
Fluorescence flow cytometry is the most commonly used platform for identifying human DC.2, 3, 4 This has been expanded from the analysis of blood, to single‐cell suspensions of tissues including skin,11, 12, 13 lung14, 15, 16 intestine17 and liver of humans2, 18, 19 and to lymphoid tissue3, 4, 20, 21, 22 fetal tissues23 and other body fluids.24
The Tenth Human Leucocyte Differentiation Antigen workshop has recently reported a range of new antibodies characterized on human DC populations (Table 1).25, 26 Advances through cytometry by time‐of‐flight now enable 30–40 antigens to be analysed simultaneously, facilitating the dissection of complex populations of leucocytes or ‘deep phenotyping’ of selected lineages. A number of computational flow cytometry tools have been developed to scale and represent high dimensional data such as FlowSOM, tSNE, oneSENSE2, 23 and ISOMAP.10 This approach, rather than sequential manual gating, aids the unbiased mapping and discovery of new cell phenotypes from multiparameter data, including that generated by conventional fluorescence flow cytometry (reviewed in ref. 27).
In recent years, transcriptomics has moved rapidly from expression arrays of bulk populations to single cell RNAseq.9, 10 These studies provide novel surface markers, reveal heterogeneity within subsets and identify small but critical DC precursor populations.9, 10 It is noteworthy that such unbiased approaches support the major classification of known populations of pDC, cDC1 and cDC2 and confirm that empirically defined markers such as CLEC9A and CD1c are the most highly discriminatory according to formal computation.9, 10 Using oligonucleotide tags, it is also possible to combine antibody‐based phenotyping with transcriptomics.28
New models of haematopoiesis and the origin of human DC
Dendritic cells have a finite lifespan of days to weeks after entering the periphery and must be continually replenished by haematopoiesis. The use of media containing Flt3 ligand, stem cell factor and granulocyte–macrophage colony‐stimulating factor (GM‐CSF; ‘FSG’) with murine stromal cells such as MS5 permits the generation of pDC and cDC1 that are easy to identify as genuine DC with no relationship to mo‐DC.29, 30, 31, 32 It is more challenging to distinguish between myeloid cDC2 and DC derived from monocytes, that form naturally in these cultures, but retention of CD14 has been used as a de facto marker of likely monocyte origin.9, 10, 32
Recent conceptual revolutions in haematopoiesis have had a profound impact upon models of DC ontogeny. First, the existence of a hierarchy of multipotent progenitors that make a series of dichotomous fate decisions (Fig. 2a), has been replaced by the notion that each progenitor follows a predestined pathway according to lineage priming that occurs at early stages in development (Fig. 2b). In experimental terms, this means that a phenotypically defined population does not contain a homogeneous population of multi‐potent cells, but rather, a cross‐section of cells primed by related but distinct developmental pathways that share a common, transient phenotype.33, 34, 35, 36 Entities such as the macrophage–dendritic cell progenitor (MDP) and common dendritic cell progenitor (CDP) are evanescent. Although bi‐potential and tri‐potential cells exist, profiling of > 2000 clonal outputs from the entire range of human progenitors does not find any significant populations corresponding to human MDP or CDP.32 Regions thought to contain such multi‐potent cells mostly comprise phenotypically related cells with a single potential.
Figure 2.
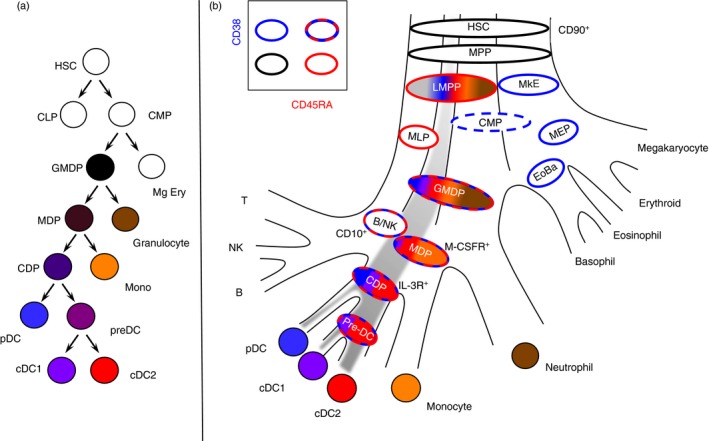
Classical and revised models of human haematopoiesis. (a) In classical models of haematopoiesis, cell potential partitions by successive bifurcations descending from the apex where common lymphoid and common myeloid progenitors (CLP; CMP) arise from the haematopietic stem cell (HSC). Each progenitor population has homogeneous differentiation potential such that every cell has an equal probability of two mutually exclusive fates. Hence, dendritic cells (DC) were proposed to arise in the sequence: CMPs, granulocyte–macrophage DC progenitor (GMDP), macrophage DC progenitor (MDP), common DC progenitor (CDP) with a final pre‐DC stage leading to conventional DC1 (cDC1) and cDC2. Each population is given a uniform colour to indicate homogeneous potential. (b) Experimental data support several revisions to the classical model. First lineage is primed in early progenitors so that most populations contain only cells with a single potential. Second, lymphoid and myeloid potential run together originating as the lymphoid primed multi‐potent progenitor (LMPP) that separates from megakaryocyte and erythroid potential (MkE) at the apex. Hence the gates defined by CD38 (blue borders) and CD45RA (red borders) contain phenotypically related cells but with restricted potentials, indicated by bands of colour each corresponding to a discrete lineage.
Second, the classical dichotomy between lymphoid and myeloid lineages, placed at the apex of haematopoiesis, has been thoroughly revised. Common myeloid progenitors are mixtures of mega‐erythroid and myeloid precursors and the most significant early partitioning of cell fate occurs when megakaryocyte and erythroid potentials separate from lympho‐myeloid potential.33, 34, 37 In contemporary models, lymphoid‐primed multipotent progenitors are at the apex of all myeloid and lymphoid lineages.34, 36 The important consequence of this is that it is no longer necessary to puzzle over the apparent ‘dual’ lymphoid and myeloid origin of DC, because DC are a product of the core lympho‐myeloid pathway in which both traits may be expressed by emerging progeny.
Hence pDC, cDC1 and cDC2 potential can be traced through all the previously defined human progenitor compartments from haematopoietic stem cells, through lymphoid‐primed multipotent progenitors to portions of the granulocyte macrophage DC progenitor (GMDP) with either high CD115 expression (MDP‐like) or high CD123 expression (CDP‐like) that contain mainly uni‐potent progenitors for each DC lineage32 (Fig. 3). Where DC are derived from two different regions of the CD34+ compartment, they emerge transcriptionally homogeneous, illustrating the importance of intrinsic regulatory circuits in defining lineage and the limitations of phenotyping in identifying discrete potentials.31
Figure 3.
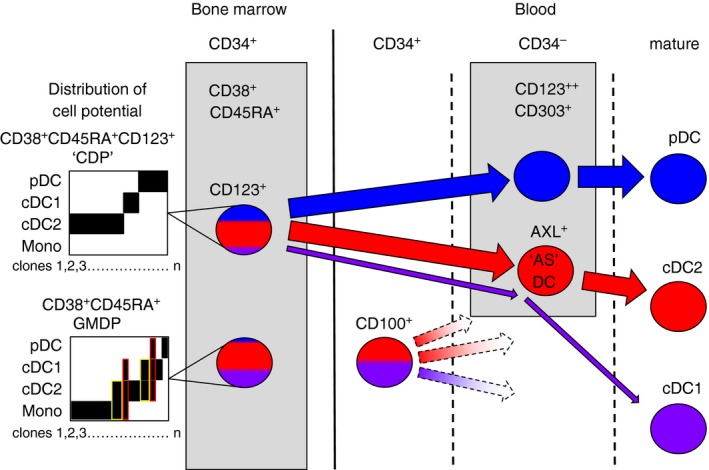
Segregation of human dendritic cell (DC) potential in late precursor compartments. The CD34+ CD38+ CD45RA+ human granulocyte–macrophage DC progenitor (GMDP) contains only a minority of progenitor cells with bi‐ or tri‐potential indicated in yellow and red, respectively in the diagrams of cell potential of several hundred individual progenitors differentiated in vitro (schematic redrawn from data of Lee et al.32). A small minority of progenitors in the GMDP qualify as MDP [except without plasmacytoid DC (pDC) potential] or common DC progenitor (CDP; all three DC potentials). The CD123+ fraction of GMDP has been described as a CDP; although it does not contain monocyte potential, trilineage common DC progenitors are not found in this gate. In the blood, a CD34+ CD123− precursor fraction is found to contain conventional DC1 (cDC1) and cDC2 potential9 together with a CD34– CD123+ AXL+ SIGLEC6+ pre‐DC with mainly cDC2 potential.9, 10 Traces of cDC1 potential are also found in CD123+ cells. It is not known how the CD34+ CD123− blood precursor relates to GMDP, if it is a physiological route of cDC differentiation, or whether this occurs via AXL+ precursors. Broken lines indicate unconfirmed relationships.
Another aspect of the lympho‐myeloid model of haematopoiesis is that DC can be ordered in a spectrum of phenotypes from lymphoid to myelo‐monocytic that mirrors their dominant developmental pathway.32 It has been known for many years that pDC are most ‘lymphoid’ harbouring RAG and rearranged IgH loci38, 39 and expressing many markers that are distinct from monocytes. Myeloid cDC1 still express a mixture of unique markers that separate them from monocytes in addition to myeloid antigens that connect them with myeloid lineages. At the other end of the spectrum, myeloid cDC2 share the most markers with monocytes and can be difficult to dissociate entirely from mo‐DC.
Human pre‐dendritic cells and AXL+SIGLEC6+ DC
Between CD34+ progenitors and mature DC, there exist potential pre‐DC defined as DC‐restricted precursors that do not yet express the full phenotype of mature DC. Several groups have reported human pre‐DC that fulfil these criteria although it is not yet completely clear how they relate to one another. Breton and colleagues focused on CD34‐negative myeloid cDC precursors, excluding pDC with CD123 and CD303, cDC1 with CD141 and cDC2 with CD1c40 before selecting cells with expression of the growth factor receptors CD117 (c‐kit), CD116 (GM‐CSF receptor) and CD135 (Flt‐3). Other investigators have found more numerous populations that fit the definition of pre‐DC by examining every lineage‐negative HLA‐DR‐positive cell using single‐cell transcriptomics9 or a combination of deep phenotyping and single‐cell trancriptomics.10 The critical advantage of these recent studies is that they did not begin by excluding mature DC and so found many pre‐DC among the CD123+ populations that had been excluded as mature pDC by Breton and colleagues. CD123 has long been used as a marker to identify pDC but both new studies emphasize the critical point that myeloid cDC‐like cells are also captured in CD123+ populations (Fig. 3). Moreover, the inadvertent inclusion of these cells has confounded many previous studies of pDC and explains the observations that pDC can apparently differentiate into myeloid cDC in vitro.41 A re‐interpretation of these results would suggest that in vitro culture causes short‐lived mature pDC to decline, while differentiating myeloid cDC come to dominate the preparation. This conclusion had been anticipated to a degree by a number of reports describing subsets of CD123+ pDC marked by CD2 or CD56 that show myeloid DC characteristics of inferior type I interferon production, higher interleukin‐12 (IL‐12) production and greater allo‐stimulatory capacity.42, 43, 44, 45, 46
CD303/CLEC4C and CD304/NRP1 overlap in expression between the myeloid cDC precursor component and pDC and cannot be used to separate the two populations completely, although the highest expressing cells will include only pDC.9 The non‐pDC cells begin to express myeloid cDC antigens such as CD11c, CD33 (SIGLEC 3) and CX3CR1.10 AXL and SIGLEC 6 (CD327) emerge as particularly useful, leading Villani and colleagues to define a new ‘AS’ DC subset (DC5 in their classification) (Table 1). In favour of AS DC being a distinct functional entity they stimulate T cells vigorously and are found in lymphoid tissue. As potential pre‐DC, the authors note that they ‘exist in a continuum of states in vivo with the potential to transition towards cDC2’.9 Intrinsic DC function or precursor status are not mutually exclusive roles and AS DC may be immediately recruited for function by some stimuli or undergo maturation to cDC2 in response to others (as yet to be defined). See et al.10 prefer to characterize a population containing AXL+ SIGLEC6+ cells primarily as precursors, describing them as ‘early pre‐DC’ with the ability to develop into cDC1 and cDC2. In support of this, See et al. observe that the ratio of cDC1:cDC2 production by their pre‐DC is in proportion with that of mature cells in the blood. Taking a slightly wider population of CD33+ CD45RA+ CD123+ cells, these authors also describe lineage priming in pre‐DC beginning to express CADM1 (pre‐cDC1) and CD1c (pre‐cDC2).
By further relaxing the gating to include CD34+ cells, Villani and colleagues identified another DC‐restricted precursor in human blood described as CD34+ CD100+ (Fig. 3). Intriguingly, this cell has lower expression of CD123 than AXL+ SIGLEC 6+ pre‐cDC and appears to be more primitive, owing to its lack of CD116 (GM‐CSF receptor) and higher proliferative capacity.9 It has approximately equal cDC1 and cDC2 potential in vitro and has the highest enrichment of pre‐cDC1 potential in human blood so far described. CD34+ CD100+ cells potentially emerge directly from a compartment of the GMDP but further experiments will be required to test this and also to map any relationship with the AXL+ pre‐cDC.
Plasmacytoid dendritic cells
Phenotype and distribution
Plasmacytoid dendritic cells have an eccentric nucleus and prominent endoplasmic reticulum and golgi (resembling a plasma cell) for the production of type I interferon (Fig. 4a). They were first identified in human blood and tonsil.41, 47, 48 Unlike myeloid cDC, they do not express the myeloid antigens CD11c, CD33, CD11b or CD13.5, 6, 49 They retain expression of the GMDP markers CD123 (IL‐3R) and CD45RA, which are down‐regulated when DC progenitors differentiate into myeloid cDC. Like all human DC they express CD4, at a higher level than myeloid cDC.50 In addition, pDC have an array of surface receptors that are intimately involved in the regulation of their major physiological function, the production of type I interferon. These include the well‐known human pDC markers CD303 (CLEC4C; BDCA‐2), CD304 (neuropilin; BDCA‐4) CD85k (ILT3) and CD85g (ILT7) together with more recently characterized antigens FcεR1, BTLA, DR6 (TNFRSF21/CD358) and CD300A.51, 52 Transciptional profiling has also added the markers FAM129C, CUX2 and GZMB.3 Several recent papers have described a small subset of CD123+ pDC that express CD2+,42, 43 CD56+ 44, 45 or CD5.46 These cells have a distinct gene expression pattern that overlaps with myeloid cDC and are now known to contain AXL+ SIGLEC 6+ myeloid pre‐cDC as described above. The two populations do not completely overlap; some AXL‐negative pDC appear to express CD2 or CD543, 46 and CD56 cannot be evaluated in the single‐cell studies because it was excluded in lineage. However, studies seeking to define the characteristics of ‘pure’ pDC should exclude contamination with myeloid pre‐DC using AXL and other markers.9, 10
Figure 4.

Features of the principal human dendritic cell (DC) subsets. Diagrams of the main surface markers, pathogen sensors and responses of (a) plasmacytoid DC (pDC); (b) conventional DC1 (cDC1) and (c) cDC2. Data are principally drawn from observations on freshly isolated blood DC and do not capture the full range of responses possible following inflammatory activation.
Development
Dendritic cell development in mammals is dependent on the coordinated action of many transcription factors that facilitate lympho‐myeloid differentiation including GATA2, PU.1, GFI1, IKZF1 and IRF853, 54, 55, 56, 57 (reviewed in refs. 58, 59, 60). Heterozygous GATA2 mutation and bi‐allelic IRF8 mutation abrogate pDC development in humans.61, 62 Humans with an IKZF1 mutation have a selective pDC deficiency EcDC1 production.63
A key axis in regulating the balance of pDC and myeloid cDC development is the antagonism between ID2, an inhibitor of DNA binding, and E2‐2, a basic hemophagocytic lymphohistiocytosis protein.58, 60 E2‐2 is a lineage‐determining factor for pDC that is negatively regulated by ID2.64 Several recent reports describe transcription factors that influence the relative production of pDC and cDC, through interaction with this axis. The ETO family protein MTG16 and the transcription factor ZEB2 are reported to repress ID2, so increasing pDC production,65, 66, 67 while NFIL3 acts to reduce pDC in favour of cDC1 production.68 Exogenous growth factors GM‐CSF (acting through STAT5) and Flt3L (acting through STAT3) respectively inhibit or enhance pDC development by modulating the expression of ID2 and E2‐2. SPIB and BCL11A also positively regulate the pDC lineage through early commitment and enhanced survival. Downstream targets of E2‐2 include SPIB and BCL11A, so stabilizing the lineage through positive feedback. E2‐2 also promotes many factors critical to the function of pDC including IRF7 and IRF8, RUNX2 and CIITA (reviewed in refs. 58, 59, 60). In humans, heterozygous mutation or loss of E2‐2 causes Pitt–Hopkins syndrome in which mature interferon‐α (IFN‐α)‐secreting pDC are reduced in number. Accurate exclusion of pre‐cDC in the analysis of this syndrome reveals a greater pDC deficiency than previously suspected,10 although this does not cause overt immunodeficiency.
Functions and role in immunity
Plasmacytoid DC are specialized to sense and respond to viral infection through several mechanisms by the rapid production of high quantities of type I and type III interferons and secretion of cytokines.52, 59 Toll‐like receptor 7 (TLR7) and TLR9 are the key endosomal pattern recognition receptors that sense single‐stranded RNA and double‐stranded DNA, respectively.52 STING has also been reported as playing a role in DNA sensing.69 Depending upon the nature of the nucleic acid cargo and mode of delivery, interferon and cytokine production may be differentially regulated.70 IRF7 is the major transducer of type I interferon production in pDC,71 whereas production of tumour necrosis factor and IL‐6 is dependent upon the nuclear factor‐κB pathway. Many other signalling molecules participate in and regulate this process including MyD88 and DOCK2 (reviewed in refs. 52, 59).
Ligation of surface receptors modulates activation or tolerance through the regulation of the IRF7 and nuclear factor‐κB pathways. CD300A transmits enhancing signals through an ITIM domain51 while ligation of FcεR1, ILT7 and CD303 (BDCA2) inhibits IFN‐α production via ITAM signalling (reviewed in ref. 52). Death Receptor 6 (DR6; TNFRSF21/CD358) is a specific marker of human pDC and knock‐down in pDC cell lines impairs IRF7 translocation to the nucleus.72 Sphingosine‐1‐phosphate signalling interacts with ILT7 to limit TLR‐induced interferon production73 and with IFNAR to terminate the IFN‐α response.74
The importance of IRF7 in regulating IFN‐α production in humans was underscored by the severe susceptibility to influenza and demonstrable lack of IFN‐α production by pDC in a patient with compound heterozygous IRF7 mutation.75 Deficiency of MyD88 and IRAK4 are predicted to affect pDC whereas DOCK8 deficiency is known to be associated with decreased pDC number and function.76 In addition to acute viral infection, pDC have been studied in chronic infections. Early production of IFN‐α by gut pDC appears to be beneficial in HIV elite controllers77 but in chronic hepatitis, persistence may be promoted by attenuated pDC responses and pDC‐mediated induction of T‐cell tolerance.78, 79 There is scope for more detailed analysis of the role of pDC in viral infections and even more so for bacterial and fungal pathogens. The prominent role of type I interferon production and signalling and potential of pDC to sense self‐nucleic acids80 have implicated pDC in the pathogenesis of psoriasis,81 systemic lupus erythematosus82 and other autoimmune diseases. Many potential therapeutic targets are presented by the factors that modulate IFN‐α release by pDC.
Direct targeting of antigens to pDC through CD303 (CLEC4C; BDCA‐2)49 or CD367 (CLEC4A, DCIR)83, 84 suggests that pDC are capable of priming CD4 T cells. In vitro experiments have also measured cross‐presentation to CD8 T cells.85, 86 However, some pDC preparations used in functional studies may have been contaminated with myeloid cDC precursors with superior ability to process and present antigens to T cells9, 10 and further evaluations of the antigen‐presenting capacity of pDC are warranted.
Conflicting roles for pDC have been reported in allergy.87, 88 Tolerogenic pDC under the influence of GM‐CSF have also been proposed to contribute to tumour progression.89
IRF8/BATF3‐dependent myeloid cDC1
Phenotype and distribution
Human myeloid cDC1 are present at approximately one‐tenth the frequency of cDC2 in steady‐state blood and tissues.2, 3, 4, 7, 12 They were originally described as a subset of blood DC with high expression of CD141+ (BDCA‐3, thrombomodulin)5, 6 (Fig. 4b). In common with myeloid cDC2, they express CD13 and CD33, but differ by low CD11c and little CD11b or SIRPα (CD172). Care must be taken not to exclude them by selecting myeloid cDC with only high CD11c expression. CD141 alone is not a reliable marker because monocytes and CD1c+ cDC2 acquire a moderate level of expression in tissues and in vitro.12, 90 Other markers should be used for confirmation: CLEC9A, the receptor for actin exposed during cell necrosis,91, 92 the cell adhesion molecule CADM1 (NECL2) and the antigen BTLA considerably increase the accuracy of identification. Indoleamine 2,3‐dioxygenase is also highly expressed.3 Lack of expression of monocyte and cDC2 markers such as CD14, CD11b and SIRPα is also important. XCR1 is also a conserved marker in many species, as identified by several gene expression studies.93 Monoclonal antibodies have been harder to develop because the structure is highly conserved, but hybrid cytokine‐Fc reagents have been successfully used.2 Intracellular detection of IRF8 may be considered a reference standard for identifying this lineage because unopposed expression of IRF8 (without IRF4) defines the lineage.2 However, intracellular staining prevents many subsequent transcriptomic and functional tests from being performed.
Human cDC1 are found in blood and among resident DC of lymph node, tonsil, spleen and bone marrow3, 4, 20, 29, 94, 95 and non‐lymphoid tissues, skin, lung, intestine and liver.2, 3, 4, 14, 15, 16, 17, 18, 19 There are suggestions that they are more enriched in tissues than in the blood, although this has not always been backed up by sufficiently accurate characterization.
Development
Myeloid cDC1 development is dependent upon GATA2, PU.1, GFI1, Id2, IRF8 and Basic leucine zipper transcription factor (BATF3)53, 54, 55, 56, 96, 97, 98 (reviewed in refs. 58 and 60). IKZF1 deficiency in humans ablates pDC but results in an increase in cDC1.63 IRF8 acts to preserve DC potential at several points in haematopoiesis by direct or indirect competition with a series of transcription factors that promote other lineages: (i) IRF8 limits CEBPA‐mediated granulocytic differentiation; (ii) with PU.1 it interacts with KLF4 to modulate the balance of DC to monocyte differentiation; (iii) it competes with IRF4 to control cDC1:cDC2 output and; (iv) a BATF3 ‘switch’ ensures that unopposed IRF8 maintains cDC1 maturation.99 Gene dosage is a critical determinant in understanding the effect of IRF8 variants upon DC development. This has been thoroughly explored in the mouse through targeted hemizygous and homozygous deletion100 but becomes even more complex when amino acid substitutions are considered, as in human examples of IRF8 variation.61, 62 Generally speaking, more severe losses of IRF8 activity incur earlier defects in haematopoiesis. Hence homozygous deletion causes excess production of neutrophils and loss of monocytes and all DC.61, 62 At the other extreme, subtle losses of IRF8 activity may only affect the production of cDC1, as documented in BXH2 mice carrying the IRF8 hypomorphic allele R294C and IRF8 hemizygous mice.100, 101 Other lineages are also affected by IRF8 mutation including B cells and natural killer cells, and human phenotypes show variant‐specific idiosyncracies.61, 62, 102 IRF8 is expressed at a low level in haematopoietic stem cells and early progenitors. It has been suggested that the ratio of IRF8 to PU.1 determines DC lineage potential from primitive stages of haematopoiesis.32 The multi‐level regulation of DC‐poiesis by IRF8, in concert with interferon‐mediated signalling and other pathways such as wnt/β‐catenin and notch, is likely to modulate cellular output during inflammatory stress.103 In humans, short hairpin RNA knock‐down of BATF3 prevents cDC1 maturation in vitro.104 Immune activation of other BATFs is able to bypass BATF3 deficiency in mice, suggesting another pathway in which inflammation may increase the flux of cDC1 development.105
Functions and role in immunity
Myeloid cDC1 have been characterized as a subset of DC that have a high intrinsic capacity to cross‐present antigens via MHC class I to activate CD8+ T cells and to promote T helper type 1 (Th1) and natural killer responses through IL‐12.12, 29, 94, 106 Cross‐presenting capacity per se is less restricted to the cDC1 subset in humans than in mouse,20, 21, 107, 108, 109 as reflected by lower enrichment for MHC class I presentation pathway genes than in murine cDC1.110, 111
Human cDC1 secrete surprisingly low IL‐12 compared with appropriately activated cDC2 or mo‐DC.12, 20, 107 This has been a subject of some controversy but is in line with the observation that human cDC2 and mo‐DC also have significant ability to interact and present antigens to Th1 cells.107, 109, 112 It appears that both cross‐presentation to CD8+ T cells and Th1 activation are less restricted to the cDC1 lineage in humans than in mice.
These differences aside, human cDC1 possess several conserved mechanisms that mediate efficient recognition of viral and intracellular antigens, transport of antigen to the appropriate endosomal compartments108 and production of type III interferon. They are also intrinsically resistant to productive viral infection.113 CLEC9A, a key marker of cDC1, is a unique receptor that recognizes bare actin filaments exposed upon necrotic cell death91, 92 and directs cell‐associated antigens into the cross‐presentation pathway.114, 115 Myeloid cDC1 express TLR3, TLR9 and TLR10 and among DC, TRL3 and TLR10 are selectively expressed.9, 116, 117 TLR3 plays an important role in the recognition of dsRNA and production of type I interferons via IRF3.118 Defects in the TLR3/IRF3 axis in humans are associated with specific susceptibility to HSV1 encephalitis through attenuated responses to viral RNA, although not necessarily uniquely mediated by cDC1.119 TLR9 is less well studied in cDC1 but is potentially also important in interferon responses to DNA, as in pDC. Myeloid cDC1 are also major producers of type III interferons IFNλ1–3120 and their accumulation in hepatitis C virus infection has been linked to the beneficial role of type III interferon in viral clearance.18
Expression of the XCR1 chemokine receptor is another conserved feature of cDC1 that enables close interaction with XCL‐producing activated T cells and natural killer cells. Several recent studies indicate the importance of this axis in coordinating peripheral Th1 and cytotoxic responses121, 122 in reciprocity with the action of DC‐derived CXCL9/10 upon activated T cells.123
In mice, cDC1 have also been characterized as cross‐priming tolerogenic cells but this potential is not well documented in humans.124, 125 In this respect, the restricted expression of TLR10 by cDC1 is intriguing. It has no known ligand and no mouse homologue but has recently been shown to be a negative regulator of TLR signalling.126 CD141 is also thought to transmit negative regulation.127
IRF4/KLF4/NOTCH2‐dependent myeloid cDC2
Phenotype and distribution
The major population of myeloid cDC in human blood, tissues and lymphoid organs are characterized as myeloid cDC2 expressing CD1c, CD2, FcεR1, SIRPA and the myeloid antigens CD11b, CD11c, CD13 and CD33 (Fig. 4c). Recent transcriptional profiling has identified CLEC10A (CD301a), VEGFA and FCGR2A (CD32A) as consistent cDC2 markers, together with the lack of cDC1 markers.3 In tissues, dermal cDC2 were first characterized as migratory CD1a+ CD1c+ DC.128, 129 CD1a is easily acquired by cDC2 but neither CD1a nor CD1c are completely restricted to cDC2 and may be expressed by cDC1 and mo‐DC isolated from tissues or in culture.12, 130
In the skin, cDC2 may be distinguished from Langerhans cells (LC) by higher CD11c and CD11b but lower CD1a, Langerin and EpCAM.22, 131 Notably, tissue cDC2 spontaneously express low langerin,22, 131, 132 in contrast to mouse in which cDC1 express langerin. Furthermore, blood cDC2 can be induced to express high langerin and Birbeck granules in response to transforming growth factor‐β (TGF‐β) in vitro 133, 134 although it remains unknown whether cDC2 have any potential to form LC in vivo. Myeloid cDC2 have also been described in the lung,14, 15, 16 intestine17 and liver of humans.2, 4, 19 In the lymph node, most interdigitating cells of the T‐cell areas have a phenotype compatible with cDC2 lineage.3, 4, 21, 135, 136, 137 Tonsil and spleen also contain CD1c+ DC.20, 29, 138 As these tissues do not receive afferent lymph, it is concluded that some CD1c+ DC are ‘resident DC’ originating directly from the blood.
Detailed phenotyping and single‐cell gene expression studies have recently characterized two subsets of cDC2 in human blood, one ‘DC‐like’ with higher expression of CD5, CD1c, HLA‐DQ and IRF4 and the other more ‘monocyte‐like’ showing CD14, CD32, CD36, CD163 and proportionately higher MAFB expression.8, 9 CD14+ CD1c+ cells have previously been detected and characterized as CD1c+ monocytes139 but by gene expression, most cluster with cDC2.8, 9 In tissues, especially during inflammation, and in humans affected by cancer, it is relatively easy to detect dual positive CD1c+ CD14+ cells12, 13, 140 but the origin of these may be difficult to ascertain precisely because as mo‐DC, they converge towards the monocyte‐like cDC2 phenotype.30, 141 Antibodies to CD2, CD5, FcεR1, CLEC4A (DCIR/CD367) and CLEC10A (CD301) may be useful but are still inducible and labile in inflammation.
Development
Myeloid cDC2 are dependent on GATA2, PU.1, GFI1, ID2, ZEB2, RELB, IRF4, NOTCH2 and KLF4, but unlike pDC and cDC1, no single transcription factor has exclusive control over their development.53, 60, 66, 142 In mice, ZEB2 has recently been identified as a factor influencing the fate of pre‐DC towards the cDC2 lineage66 and IRF4 is considered to be a lineage‐defining factor.2 Depending upon the tissue site, subsets of murine IRF4+ cDC2 show variable dependence upon RELB, NOTCH2/RBPJ and lymphotoxin‐β or a requirement for KLF4.60 In humans, much less is known about the differential regulation of cDC2 production. Heterozygous GATA2 deficiency leads to eventual loss of all cDC2.53 Bi‐allelic IRF8 deficiency also abrogates cDC2 development because the entire monocyte‐DC tract of lympho‐myeloid development is lost.61, 62 In contrast, cDC2 are IRF8‐independent in mice,100 although great care is required to avoid inadvertently counting expanded primitive myeloid cells as cDC2 when both alleles of IRF8 are deleted. This may at least partly explain the phenotype of humans with heterozygous IRF8 T80A mutation, in which an abnormal population of CD11c+ CD1c– cells appears in place of CD1c+ cDC2.61 Transcription factor zbtb46 is not required for development of cDC2 but is up‐regulated during differentiation and also appears on mo‐DC.143, 144
The heterogeneity of transcription factor dependence observed in mice is consistent with multiple subsets of cDC2, as recently described in humans.8, 9 IRF4, KLF4, LTBR and MAFB are slightly differentially expressed by cDC2 subsets but RELB and NOTCH2 are uniform across cDC2. There is no obvious correlation with pre‐DC or monocyte origin; pre‐cDC have high expression of both IRF4 and KLF4 whereas monocytes express comparable levels with DC of KLF4, MAFB, RELB and NOTCH2 (data from ref. 9).
Functions and role in immunity
Myeloid cDC2 are equipped with a wide range of lectins, TLRs, NOD‐like receptors and RIG‐I‐like receptors similar in range to that expressed by monocytes. Human blood cDC2 respond well to lipopolysaccharide, flagellin, poly IC and R848.145 The potential of CD1c and CD1a to present the glycolipid antigens of mycobacteria and other pathogens is often overlooked.146 Among the lectins, CLEC4A (DCIR/CD367), CLEC10A (CD301) CLEC12A (CD371) and the asialoglycoprotein receptor are highly expressed. In common with monocytes, TLRs 2, 4, 5, 6 and 8 are present with notable expression of NOD2, NLRP1, NLRP3 and NAIP (data from ref. 9). Dectin‐1 (CLEC7A) and Dectin‐2 (CLEC6A) are highly expressed in tissue cDC2, suggesting a role in fungal recognition.147, 148 DEC205 (CD205; CLEC13B) and macrophage mannose receptor (CD206; CLEC13D) are variable.95
In contrast with mice, human cDC2 can be stimulated to become high producers of IL‐12 and excellent cross‐presenting cells.20, 21, 107, 108 Their ability to synthesize IL‐12 is greater than that of cDC1 in most conditions analysed. They secrete IL‐23, IL‐1, tumour necrosis factor‐α (TNF‐α), IL‐8 and IL‐10 but are consistently low in the secretion of type III interferon.109, 149 In vitro, human cDC2 are potent in the activation of Th1, Th2, Th17 and CD8+ T cells,109, 149, 150 suggesting the capacity to promote a wide range of immune responses in vivo. Subsets of cDC2 defined by CD5 and other markers differ in their production of TNF‐α, IL‐6, IL‐10 and IL‐23 in response to TLR ligation.8, 140 CD5‐high ‘DC‐like’ cDC2 are more active in CCR7‐dependent migration, stimulate high naive T‐cell proliferation and preferential priming of Th2, Th17, Th22 and regulatory T cells. ‘Monocyte‐like’ cDC2 with lower CD5 are less active in proliferation assays and produce mainly Th1 cells.8 Subsets of cDC2 specializing in Th2 or Th17 responses are dependent upon IRF4 or KLF4, respectively,58, 60 but this has not been corroborated in humans.
Langerhans cells
Phenotype and distribution
Langerhans cells are specialized DC that inhabit the basal epidermis and other stratified squamous epithelia. They express the C‐type lectin langerin and the invariant MHC class I molecule CD1a. The close integration of LC within the epithelial layer is mediated by E‐Cadherin, EpCAM (TROP1) TROP2, AXL and tight junction proteins claudin, occludin and ZO‐1.151, 152 In common with myeloid cDC2 they express high levels of FcεR1 and CD39 (ATPase). Myeloid cDC2 spontaneously express a low level of langerin in many sites, but the high expression of langerin, CD1a and EpCAM, together with lower expression of CD11c, CD11b and CD13 by LC, is usually sufficient to separate them clearly.22, 131 Care must be taken not to include cDC1 and LC together as both show lower CD11c expression than cDC2. This has led to controversy about the relationship of LC with the cDC1 lineage.111, 153 Although LC are distinct from cDC1 ontogenetically,111 it is clear that they do have functional cross‐presentation capacity154, 155 and high MHC class I‐related gene expression in humans.111 LC migrate to skin‐draining lymph nodes, where they appear in the T‐cell areas expressing langerin, CD1a and CD1c. EpCAM and other epithelial markers are down‐regulated, making it more difficult to distinguish LC from cDC2 by microscopy, although langerin is still expressed. Differences between inflamed and non‐inflamed skin‐draining nodes156 and between skin‐draining nodes and tonsil22 have been used to highlight the contribution of migratory LC to nodal DC populations.
Development
Langerhans cells are phylogenetically ancient and share a primitive origin with tissue macrophages and microglia of the brain.157 This has led to their classification as ‘macrophages’,1 a controversial position given that they epitomize the function of myeloid DC that capture antigen, mature and migrate to lymph nodes.158 Recent lineage tracing experiments in mice indicate that LC have an equal claim to DC and macrophage heritage by virtue of unique dual expression of ZBTB46 and MAFB.159 Their development is unique among DC in that once established by primitive and fetal liver haematopoiesis, LC are capable of local self‐renewal, independently of the bone marrow.160 Proliferating human LC were described many years ago161 and self‐renewal may be demonstrated in humans lacking DC‐poiesis due to mutations in GATA2 or IRF862 ,162 or recipients of limb transplantation.163 Recent data indicate that mucosal LC are more dependent upon bone‐marrow‐derived precursors.164
In mice, the establishment of a self‐renewing network is dependent upon PU.1 RUNX3 and ID2 in combination with locally produced cytokines IL‐34, TFG‐β and bone morphogenetic protein 7 (reviewed in ref. 165). The sequential formation of human fetal LC from myeloid precursors has been observed in detailed microscopic studies and ascribed to non‐monocyte precursors.166 In the steady state, the influence of TGF‐β, E‐cadherin/β‐catenin and the binding of Axl to gas 6 maintains LC in a quiescent state.151, 152 ,167,168
In postnatal life, local self‐renewal restores LC numbers following chronic or low‐grade inflammatory insults.169 However, studies in mice show that severe inflammation recruits de novo bone‐marrow‐derived precursors in two waves; a transient population of classical monocytes followed by uncharacterized myeloid precursors that form a stable self‐renewing LC network as inflammation subsides.170,171 Bone marrow transplantation in humans also results in replacement by donor cells even after non‐myeloablative treatment and in the absence of overt graft‐versus‐host disease.172,173
In vitro models of LC development from CD34+ progenitors, cDC2 and monocytes offer potential routes to explain the repopulation of LC in vivo, following severe inflammation.174 Purified monocytes, contrary to early reports, do not make LC efficiently with GM‐CSF and TGF‐β,175 but require additional notch signals to down‐regulate a KLF4‐dependent pathway of differentiation.176–178 Myeloid cDC2, however, rapidly up‐regulate langerin to high levels and form Birbeck granules with GM‐CSF or TSLP and TGF‐β or BMP7.133, 134 ,179 In vivo evidence for these two pathways has recently been reported in the homeostasis of mucosal LC164 but direct evidence is lacking in humans.
Functions and role in immunity
When the skin becomes inflamed, local production of TNF‐α and IL‐1β stimulate LC to lose their connections with the surrounding epithelium and migrate across the basement membrane into the afferent lymphatics. Although LC were the primary model of migratory myeloid DC, their non‐redundant function in immunity has been surprisingly difficult to pin down. In humans, they can be matured into potent cross‐presenting DC with high IL‐15 production and the ability to present mycobacterial glycolipid antigens and stimulate CD8 T cells.154, 155 Transgenic expression of human CD1a on murine LC licensed them for presentation of lipid antigens to Th17 and Th22 cells. resulting in skin inflammation.180 However, it has also been reported that LC lack critical TLRs145 and can induce regulatory T cells and IL‐22 production through CD1a‐restricted antigens to autologous T cells.181 Overall, the role of LC has been summarized as maintaining epidermal health and tolerance to commensals, while retaining the ability to respond to selected intracellular pathogens and viruses under inflammatory conditions.182
Monocyte‐derived inflammatory DC
Phenotype and distribution
The term ‘inflammatory dendritic cell’ has come to be defined as monocyte‐derived DC (mo‐DC) that appear in inflammation. Ontogeny‐based nomenclature would require that the term monocyte‐derived is retained, since DC lineage cells may also be recruited in inflammation.183 In mice, inflammatory mo‐DC were originally demonstrated in leishmaniasis but have now been recognized in other infections and inflammatory settings (reviewed in refs. 184,185). In humans, inflammatory myeloid cells have been reported in several settings including eczema,186 psoriasis,187,188 skin sensitization,189 allergic rhinitis,190 coeliac disease,191 inflammatory bowel disease,192,193 synovitis and peritonitis.24 ,194 Kinetics, surface markers, gene expression and even direct labelling suggest that monocyte‐derived cells dominate these populations. Considerable heterogeneity is observed with no clear consensus on the use of the terms monocyte, macrophage or DC to describe inflammatory monocyte‐derived cells (Table 2). Inflammatory monocytes retain expression of CD13, CD33, CD11b, CD11c and CD172a and may show recent evidence of recruitment from monocytes by their expression of S100A8/9 and CCR2.190,194 In humans, monocytes express CD11c and MHC class II so these markers are not helpful in separating monocytes from DC. Evidence of DC differentiation is supported by the expression of CD1c, CD1a, FcεR1, IRF4 and ZBTB46. Inflammatory mo‐DC have also been described to express CD206 and CD209, but retained expression of CD14 and co‐expression of CD16, CD163 and FXIIIA, together with lack of CD1c, CD1a and FceR1, are more consistent with a phenotype usually described as macrophage‐like. In two studies, where it was possible to characterize dual populations of DC‐like and macrophage‐like cells, the key properties linked to DC phenotype were CD11c, CD1c, FcεR1, CD206, IRF4 and allo‐stimulation. CD14 was variable but CD16 and CD163 were negative.24 ,194 Although gene expression showed subset‐restricted patterns, there were many shared transcripts between populations designated as mo‐DC and those described as monocyte‐derived macrophages.
Table 2.
Features of monocyte‐derived dendritic cells
Development and functions in immunity
Populations of monocyte‐derived cells exist in human steady‐state tissues, including the skin,13 lung14, 15, 16 and intestine.17 It is not known if this is a distinct mechanism of recruitment or one that is accelerated and activated in inflammation. During inflammation, monocyte‐derived cells expand resident populations many‐fold.189–194 Prevailing models suggest that monocyte‐derived cells function mainly at a site of inflammation rather than migrating to lymph nodes.195 However, mo‐DC studied ex vivo secrete IL‐1, TNF‐α, IL‐12 and IL‐23, stimulate CD4 and CD8 T cells and express CCR7.24 ,188,194 Although it is possible to derive potent mo‐DC in vitro with GM‐CSF and IL‐4 followed by activating stimuli such as lipopolysaccharide or prostaglandin E2, the use of such preparations in immunotherapy is declining in favour of ‘naturally‐occurring’ DC such as pDC or cDC2 (reviewed in ref. 196). The disappointing performance of mo‐DC in cancer immunotherapy is being interpreted as due to an intrinsic lack of biological potency of monocyte‐derived cells.141 However, the in vivo conditions under which mo‐DC form are still poorly replicated by in vitro experiments.
CD16+ non‐classical monocytes, SLAN+ DC and DC4
The CD16+ non‐classical monocyte is still considered as a DC by some authors. In particular, those that express a carbohydrate modification of the P‐selectin glycoprotein ligand 1, SLAN (recognized by the antibody M‐DC8) have been identified as ‘SLAN DC’.197 The nomenclature is confusing however, as others have since used SLAN to identify a ‘true’ non‐classical monocyte population (distinct from intermediate CD14+ CD16+ cells).198 The simple facts are that CD16+ monocytes are heterogeneous and that SLAN expression identifies a subpopulation with lower CD11b, CD14 and CD36 but higher expression of CD16. Concerning the question of whether SLAN+ cells are monocytes or DC, their gene expression is clearly monocytic.13, 198, 199, 200 Furthermore, recent human in vivo and xenograft experiments support the hypothesis that non‐classical monocytes differentiate from classical monocytes by down‐regulating inflammatory pathways and adopting a CX3CR+ CCR2– CD11chi CD11blo phenotype.201 However, it should be noted that gene expression alone does not absolutely exclude an independent origin of SLAN+ cells from the remainder of CD16+ monocytes and that SLAN itself was not measured on the output of cells derived from human classical monocytes in the adoptive transfer experiments.201
Most recently, single cell RNAseq studies found CD16+ cells among HLA‐DR+ lineage‐negative (CD3, CD19, CD56) CD14‐negative cells and classified them as ‘DC4’.9 DC4 was described as transcriptomically distinct from non‐classical monocytes, yet it was obtained from the same CD16+ population that contains non‐classical monocytes, by a slightly different gating route. One of the drawbacks of transcriptomic clustering is that it is difficult to relate population frequency back to input cells. The most likely explanation for DC4 is that is signifies a subset of CD16+ monocytes. Although it is possible to ‘index’ single cells back to their flow parameter space, the position of DC4 within the non‐classical monocyte gate was not shown. It is apparent that DC4 has a transcriptional profile reminiscent of SLAN+ cells with lower CD11b, CD14 and CD36 but higher expression of CD16. The expression of SLAN itself, a post‐translational modification, was naturally invisible to transcriptomic analysis. Recent experiments conclude that DC4 and SLAN+ cells are indeed identical202 but further studies are required to discover their non‐redundant roles in immunity.
Conclusion
In summary, human DC arise by a dedicated pathway of lympho‐myeloid haematopoiesis and differentiate into specialized subsets under the influence of lineage‐specific transcription factors, notably IRF8 and IRF4. Plasmacytoid DC and cDC1 are the most specialized with relatively restricted roles in sensing nucleic acid and responding by interferon production. Cross‐presentation to CD8 cells is not restricted to cDC1 but can be performed by cDC2 and mo‐DC with appropriate activation. The wide range of CD4+ T‐cell priming capacity by cDC2 is likely to reflect heterogeneity within this population. Single‐cell experiments have clarified that pDC and pre‐cDC have often been previously analysed as a mixed population. DC subset specialization increases the range and flexibility of immune responses through mechanisms that are relatively conserved across mammalian species. This knowledge has become essential in translating murine immunology to human pathology and continues to expand the therapeutic horizon of DC in medicine.
Disclosures
The authors have no competing interests to declare.
Acknowledgement
The authors acknowledge funding from CRUK (MC; grant: C30484/A21025) and the Wellcome Trust (VB; grant: 101155/Z/13/Z).
Dedicated to the memory of Derek NJ Hart, friend and mentor in human dendritic cells.
References
- 1. Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Donna N, Schraml BU et al Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol 2014; 14:571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guilliams M, Dutertre CA, Scott CL, McGovern N, Sichien D, Chakarov S et al Unsupervised high‐dimensional analysis aligns dendritic cells across tissues and species. Immunity 2016; 45:669–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Heidkamp GF, Sander J, Lehmann CHK, Heger L, Eissing N, Baranska A et al Human lymphoid organ dendritic cell identity is predominantly dictated by ontogeny, not tissue microenvironment. Sci Immunol 2016; 1:eaai7677. [DOI] [PubMed] [Google Scholar]
- 4. Granot T, Senda T, Carpenter DJ, Matsuoka N, Weiner J, Gordon CL et al Dendritic cells display subset and tissue‐specific maturation dynamics over human life. Immunity 2017; 46:504–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dzionek A, Fuchs A, Schmidt P, Cremer S, Zysk M, Miltenyi S et al BDCA‐2, BDCA‐3, and BDCA‐4: three markers for distinct subsets of dendritic cells in human peripheral blood. J Immunol 2000; 165:6037–46. [DOI] [PubMed] [Google Scholar]
- 6. MacDonald KP, Munster DJ, Clark GJ, Dzionek A, Schmitz J, Hart DN. Characterization of human blood dendritic cell subsets. Blood 2002; 100:4512–20. [DOI] [PubMed] [Google Scholar]
- 7. Ziegler‐Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN et al Nomenclature of monocytes and dendritic cells in blood. Blood 2010; 116:e74–80. [DOI] [PubMed] [Google Scholar]
- 8. Yin X, Yu H, Jin X, Li J, Guo H, Shi Q et al Human blood CD1c+ dendritic cells encompass CD5high and CD5low subsets that differ significantly in phenotype, gene expression, and functions. J Immunol 2017; 198:1553–64. [DOI] [PubMed] [Google Scholar]
- 9. Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J et al Single‐cell RNA‐seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 2017; 356:eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. See P, Dutertre CA, Chen J, Günther P, McGovern N, Irac SE et al Mapping the human DC lineage through the integration of high‐dimensional techniques. Science 2017; 356:eaag3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haniffa M, Ginhoux F, Wang XN, Bigley V, Abel M, Dimmick I et al Differential rates of replacement of human dermal dendritic cells and macrophages during hematopoietic stem cell transplantation. J Exp Med 2009; 206:371–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haniffa M, Shin A, Bigley V, McGovern N, Teo P, See P et al Human tissues contain CD141hi cross‐presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity 2012; 37:60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McGovern N, Schlitzer A, Gunawan M, Jardine L, Shin A, Poyner E et al Human dermal CD14⁺ cells are a transient population of monocyte‐derived macrophages. Immunity 2014; 41:465–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Desch AN, Gibbings SL, Goyal R, Kolde R, Bednarek J, Bruno T et al Flow cytometric analysis of mononuclear phagocytes in non‐diseased human lung and lung‐draining lymph nodes. Am J Respir Crit Care Med 2016; 193:614–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baharom F, Thomas S, Rankin G, Lepzien R, Pourazar J, Behndig AF et al Dendritic cells and monocytes with distinct inflammatory responses reside in lung mucosa of healthy humans. J Immunol 2016; 196:4498–509. [DOI] [PubMed] [Google Scholar]
- 16. Patel VI, Booth JL, Duggan ES, Cate S, White VL, Hutchings D et al Transcriptional classification and functional characterization of human airway macrophage and dendritic cell subsets. J Immunol 2017; 198:1183–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Watchmaker PB, Lahl K, Lee M, Baumjohann D, Morton J, Kim SJ et al Comparative transcriptional and functional profiling defines conserved programs of intestinal DC differentiation in humans and mice. Nat Immunol 2014; 15:98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yoshio S, Kanto T, Kuroda S, Matsubara T, Higashitani K, Kakita N et al Human blood dendritic cell antigen 3 (BDCA3)(+) dendritic cells are a potent producer of interferon‐λ in response to hepatitis C virus. Hepatology 2013; 57:1705–15. [DOI] [PubMed] [Google Scholar]
- 19. Kelly A, Fahey R, Fletcher JM, Keogh C, Carroll AG, Siddachari R et al CD141⁺ myeloid dendritic cells are enriched in healthy human liver. J Hepatol 2014; 60:135–42. [DOI] [PubMed] [Google Scholar]
- 20. Mittag D, Proietto AI, Loudovaris T, Mannering SI, Vremec D, Shortman K et al Human dendritic cell subsets from spleen and blood are similar in phenotype and function but modified by donor health status. J Immunol 2011; 186:6207–17. [DOI] [PubMed] [Google Scholar]
- 21. Segura E, Durand M, Amigorena S. Similar antigen cross‐presentation capacity and phagocytic functions in all freshly isolated human lymphoid organ‐resident dendritic cells. J Exp Med 2013; 210:1035–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. De Monte A, Olivieri CV, Vitale S, Bailleux S, Castillo L, Giordanengo V et al CD1c‐related DCs that express CD207/langerin, but are distinguishable from Langerhans cells, are consistently present in human tonsils. Front Immunol 2016; 7:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McGovern N, Shin A, Low G, Duan K, Yao LJ et al Human fetal dendritic cells promote prenatal T‐cell immune suppression through arginase‐2. Nature 2017; 546:662–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Segura E, Touzot M, Bohineust A, Cappuccio A, Chiocchia G, Hosmalin A et al Human inflammatory dendritic cells induce Th17 cell differentiation. Immunity 2013; 38:336–48. [DOI] [PubMed] [Google Scholar]
- 25. Autenrieth SE, Grimm S, Rittig SM, Grünebach F, Gouttefangeas C, Bühring HJ. Profiling of primary peripheral blood‐ and monocyte‐derived dendritic cells using monoclonal antibodies from the HLDA10 Workshop in Wollongong, Australia. Clin Transl Immunol 2015; 4:e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fromm PD, Kupresanin F, Brooks AE, Dunbar PR, Haniffa M, Hart DN et al A multi‐laboratory comparison of blood dendritic cell populations. Clin Transl Immunol 2016; 5:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saeys Y, Gassen SV, Lambrecht BN. Computational flow cytometry: helping to make sense of high‐dimensional immunology data. Nat Rev Immunol 2016; 16:449–62. [DOI] [PubMed] [Google Scholar]
- 28. Stoeckius M, Hafemeister C, Stephenson W, Houck‐Loomis B, Chattopadhyay PK, Swerdlow H et al Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017; 14:865–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Poulin LF, Salio M, Griessinger E, Anjos‐Afonso F, Craciun L, Chen JL et al Characterization of human DNGR‐1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J Exp Med 2010; 207:1261–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Balan S, Ollion V, Colletti N, Chelbi R, Montanana‐Sanchis F, Liu H et al Human XCR1+ dendritic cells derived in vitro from CD34+ progenitors closely resemble blood dendritic cells, including their adjuvant responsiveness, contrary to monocyte‐derived dendritic cells. J Immunol 2014; 193:1622–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Helft J, Anjos‐Afonso F, van der Veen AG, Chakravarty P, Bonnet D, Reis E et al Dendritic cell lineage potential in human early hematopoietic progenitors. Cell Rep 2017; 20:529–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee J, Zhou YJ, Ma W, Zhang W, Aljoufi A, Luh T et al Lineage specification of human dendritic cells is marked by IRF8 expression in hematopoietic stem cells and multipotent progenitors. Nat Immunol 2017; 18:877–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paul F, Arkin Y, Giladi A, Jaitin DA, Kenigsberg E, Keren‐Shaul H et al Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell 2015; 163:1663–77. [DOI] [PubMed] [Google Scholar]
- 34. Notta F, Zandi S, Takayama N, Dobson S, Gan OI, Wilson G et al Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 2016; 351:aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Velten L, Haas SF, Raffel S, Blaszkiewicz S, Islam S, Hennig BP et al Human haematopoietic stem cell lineage commitment is a continuous process. Nat Cell Biol 2017; 19:271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Karamitros D, Stoilova B, Aboukhalil Z, Hamey F, Reinisch A, Samitsch M et al Single‐cell analysis reveals the continuum of human lympho‐myeloid progenitor cells. Nat Immunol 2017; 19:85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kawamoto H, Ikawa T, Masuda K, Wada H, Katsura Y. A map for lineage restriction of progenitors during hematopoiesis: the essence of the myeloid‐based model. Immunol Rev 2010; 238:23–36. [DOI] [PubMed] [Google Scholar]
- 38. Pelayo R, Hirose J, Huang J, Garrett KP, Delogu A, Busslinger M et al Derivation of 2 categories of plasmacytoid dendritic cells in murine bone marrow. Blood 2005; 105:4407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Corcoran L, Ferrero I, Vremec D, Lucas K, Waithman J, O'Keeffe M et al The lymphoid past of mouse plasmacytoid cells and thymic dendritic cells. J Immunol 2003; 170:4926–32. [DOI] [PubMed] [Google Scholar]
- 40. Breton G, Zheng S, Valieris R, Tojal da Silva I, Satija R, Nussenzweig MC. Human dendritic cells (DCs) are derived from distinct circulating precursors that are precommitted to become CD1c+ or CD141+ DCs. J Exp Med 2016; 213:2861–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grouard G, Rissoan MC, Filgueira L, Durand I, Banchereau J, Liu YJ. The enigmatic plasmacytoid T cells develop into dendritic cells with interleukin (IL)‐3 and CD40‐ligand. J Exp Med 1997; 185:1101–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matsui T, Connolly JE, Michnevitz M, Chaussabel D, Yu CI, Glaser C et al CD2 distinguishes two subsets of human plasmacytoid dendritic cells with distinct phenotype and functions. J Immunol 2009; 182:6815–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bryant C, Fromm PD, Kupresanin F, Clark G, Lee K, Clarke C et al A CD2 high‐expressing stress‐resistant human plasmacytoid dendritic‐cell subset. Immunol Cell Biol 2016; 94:447–57. [DOI] [PubMed] [Google Scholar]
- 44. Osaki Y, Yokohama A, Saito A, Tahara K, Yanagisawa K, Ogawa Y et al Characterization of CD56+ dendritic‐like cells: a normal counterpart of blastic plasmacytoid dendritic cell neoplasm. PLoS ONE 2013; 8:e81722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yu H, Zhang P, Yin X, Yin Z, Shi Q, Cui Y et al Human BDCA2+CD123+CD56+ dendritic cells (DCs) related to blastic plasmacytoid dendritic cell neoplasm represent a unique myeloid DC subset. Protein Cell 2015; 6:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang H, Gregorio JD, Iwahori T, Zhang X, Choi O, Tolentino LL et al A distinct subset of plasmacytoid dendritic cells induces activation and differentiation of B and T lymphocytes. Proc Natl Acad Sci U S A 2017; 114:1988–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cella M, Jarrossay D, Facchetti F, Alebardi O, Nakajima H, Lanzavecchia A et al Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat Med 1999; 5:919–23. [DOI] [PubMed] [Google Scholar]
- 48. Siegal FP, Kadowaki N, Shodell M, Fitzgerald‐Bocarsly PA, Shah K, Ho S et al The nature of the principal type 1 interferon‐producing cells in human blood. Science 1999; 284:1835–7. [DOI] [PubMed] [Google Scholar]
- 49. Dzionek A, Sohma Y, Nagafune J, Cella M, Colonna M, Facchetti F et al BDCA‐2, a novel plasmacytoid dendritic cell‐specific type II C‐type lectin, mediates antigen capture and is a potent inhibitor of interferon α/β induction. J Exp Med 2001; 194:1823–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jardine L, Barge D, Ames‐Draycott A, Pagan S, Cookson S, Spickett G et al Rapid detection of dendritic cell and monocyte disorders using CD4 as a lineage marker of the human peripheral blood antigen‐presenting cell compartment. Front Immunol 2013; 4:495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ju X, Zenke M, Hart DN, Clark GJ. CD300a/c regulate type I interferon and TNF‐alpha secretion by human plasmacytoid dendritic cells stimulated with TLR7 and TLR9 ligands. Blood 2008; 112:1184–94. [DOI] [PubMed] [Google Scholar]
- 52. Bao M, Liu YJ. Regulation of TLR7/9 signaling in plasmacytoid dendritic cells. Protein Cell 2013; 4:40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Collin M, Dickinson R, Bigley V. Haematopoietic and immune defects associated with GATA2 mutation. Br J Haematol 2015; 169:173–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Onodera K, Fujiwara T, Onishi Y, Itoh‐Nakadai A, Okitsu Y, Fukuhara N et al GATA2 regulates dendritic cell differentiation. Blood 2016; 128:508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Carotta S, Dakic A, D'Amico A, Pang SH, Greig KT, Nutt SL et al The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose‐dependent manner. Immunity 2010; 32:628–41. [DOI] [PubMed] [Google Scholar]
- 56. Rathinam C, Geffers R, Yucel R, Buer J, Welte K, Moroy T et al The transcriptional repressor Gfi1 controls STAT3‐dependent dendritic cell development and function. Immunity 2005; 22:717–28. [DOI] [PubMed] [Google Scholar]
- 57. Allman D, Dalod M, Asselin‐Paturel C, Delale T, Robbins SH, Trinchieri G et al Ikaros is required for plasmacytoid dendritic cell differentiation. Blood 2006; 108:4025–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tussiwand R, Gautier EL. Transcriptional regulation of mononuclear phagocyte development. Front Immunol 2015; 6:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Swiecki M, Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol 2015; 15:471–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Murphy TL, Grajales‐Reyes GE, Wu X, Tussiwand R, Briseño CG, Iwata A et al Transcriptional control of dendritic cell development. Annu Rev Immunol 2016; 34:93–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hambleton S, Salem S, Bustamante J, Bigley V, Boisson‐Dupuis S, Azevedo J et al IRF8 mutations and human dendritic‐cell immunodeficiency. N Engl J Med 2011; 365:127–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bigley V, Maisuria S, Cytlak U, Jardine L, Care MA, Green K et al Biallelic interferon regulatory factor 8 mutation: a complex immunodeficiency syndrome with dendritic cell deficiency, monocytopenia, and immune dysregulation. J Allergy Clin Immunol 2017; doi: 10.1016/j.jaci.2017.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cytlak U, Resteu AB. D, Kuehn HS, Altmann TG, A. Ikaros family zinc finger 1 regulates dendritic cell development and function in humans. Nature . Communications 2018; In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Spits H, Couwenberg F, Bakker AQ, Weijer K, Uittenbogaart CH. Id2 and Id3 inhibit development of CD34(+) stem cells into predendritic cell (pre‐DC)2 but not into pre‐DC1. Evidence for a lymphoid origin of pre‐DC2. J Exp Med 2000; 192:1775–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ghosh HS, Ceribelli M, Matos I, Lazarovici A, Bussemaker HJ, Lasorella A et al ETO family protein Mtg16 regulates the balance of dendritic cell subsets by repressing Id2. J Exp Med 2014; 211:1623–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Scott CL, Soen B, Martens L, Skrypek N, Saelens W, Taminau J et al The transcription factor Zeb2 regulates development of conventional and plasmacytoid DCs by repressing Id2. J Exp Med 2016; 213:897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wu X, Briseño CG, Grajales‐Reyes GE, Haldar M, Iwata A, Kretzer NM et al Transcription factor Zeb2 regulates commitment to plasmacytoid dendritic cell and monocyte fate. Proc Natl Acad Sci U S A 2016; 113:14775–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kashiwada M, Pham NL, Pewe LL, Harty JT, Rothman PB. NFIL3/E4BP4 is a key transcription factor for CD8alpha dendritic cell development. Blood 2011; 117:6193–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bode C, Fox M, Tewary P, Steinhagen A, Ellerkmann RK, Klinman D et al Human plasmacytoid dentritic cells elicit a Type I Interferon response by sensing DNA via the cGAS‐STING signaling pathway. Eur J Immunol 2016; 46:1615–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bruni D, Chazal M, Sinigaglia L, Chauveau L, Schwartz O, Després P et al Viral entry route determines how human plasmacytoid dendritic cells produce type I interferons. Sci Signal 2015; 8:ra25. [DOI] [PubMed] [Google Scholar]
- 71. Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T et al IRF‐7 is the master regulator of type‐I interferon‐dependent immune responses. Nature 2005; 434:772–7. [DOI] [PubMed] [Google Scholar]
- 72. Li J, Du Q, Hu R, Wang Y, Yin X, Yu H et al Death receptor 6 is a novel plasmacytoid dendritic cell‐specific receptor and modulates type I interferon production. Protein Cell 2016; 7:291–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dillmann C, Ringel C, Ringleb J, Mora J, Olesch C, Fink AF et al S1PR4 signaling attenuates ILT 7 internalization to limit IFN‐α production by human plasmacytoid dendritic cells. J Immunol 2016; 196:1579–90. [DOI] [PubMed] [Google Scholar]
- 74. Teijaro JR, Studer S, Leaf N, Kiosses WB, Nguyen N, Matsuki K et al S1PR1‐mediated IFNAR1 degradation modulates plasmacytoid dendritic cell interferon‐α autoamplification. Proc Natl Acad Sci USA 2016; 113:1351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ciancanelli MJ, Huang SX, Luthra P, Garner H, Itan Y, Volpi S et al Infectious disease. Life‐threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science 2015; 348:448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Keles S, Jabara HH, Reisli I, McDonald DR, Barlan I, Hanna‐Wakim R et al Plasmacytoid dendritic cell depletion in DOCK8 deficiency: rescue of severe herpetic infections with IFN‐α 2b therapy. J Allergy Clin Immunol 2014; 133:1753–5.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Li H, Goepfert P, Reeves RK. Short communication: plasmacytoid dendritic cells from HIV‐1 elite controllers maintain a gut‐homing phenotype associated with immune activation. AIDS Res Hum Retroviruses 2014; 30:1213–5. [DOI] [PubMed] [Google Scholar]
- 78. Yonkers NL, Rodriguez B, Milkovich KA, Asaad R, Lederman MM, Heeger PS et al TLR ligand‐dependent activation of naive CD4 T cells by plasmacytoid dendritic cells is impaired in hepatitis C virus infection. J Immunol 2007; 178:4436–44. [DOI] [PubMed] [Google Scholar]
- 79. Woltman AM, Op den Brouw ML, Biesta PJ, Shi CC, Janssen HL. Hepatitis B virus lacks immune activating capacity, but actively inhibits plasmacytoid dendritic cell function. PLoS ONE 2011; 6:e15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lande R, Ganguly D, Facchinetti V, Frasca L, Conrad C, Gregorio J et al Neutrophils activate plasmacytoid dendritic cells by releasing self‐DNA‐peptide complexes in systemic lupus erythematosus. Sci Transl Med 2011; 3:73ra19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ganguly D, Chamilos G, Lande R, Gregorio J, Meller S, Facchinetti V et al Self‐RNA‐antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. J Exp Med 2009; 206:1983–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Berggren O, Alexsson A, Morris DL, Tandre K, Weber G, Vyse TJ et al IFN‐α production by plasmacytoid dendritic cell associations with polymorphisms in gene loci related to autoimmune and inflammatory diseases. Hum Mol Genet 2015; 24:3571–81. [DOI] [PubMed] [Google Scholar]
- 83. Meyer‐Wentrup F, Benitez‐Ribas D, Tacken PJ, Punt CJ, Figdor CG, de Vries IJ et al Targeting DCIR on human plasmacytoid dendritic cells results in antigen presentation and inhibits IFN‐α production. Blood 2008; 111:4245–53. [DOI] [PubMed] [Google Scholar]
- 84. Klechevsky E, Flamar AL, Cao Y, Blanck JP, Liu M, O'Bar A et al Cross‐priming CD8+ T cells by targeting antigens to human dendritic cells through DCIR. Blood 2010; 116:1685–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tel J, Sittig SP, Blom RA, Cruz LJ, Schreibelt G, Figdor CG et al Targeting uptake receptors on human plasmacytoid dendritic cells triggers antigen cross‐presentation and robust type I IFN secretion. J Immunol 2013; 191:5005–12. [DOI] [PubMed] [Google Scholar]
- 86. Tel J, Schreibelt G, Sittig SP, Mathan TS, Buschow SI, Cruz LJ et al Human plasmacytoid dendritic cells efficiently cross‐present exogenous Ags to CD8+ T cells despite lower Ag uptake than myeloid dendritic cell subsets. Blood 2013; 121:459–67. [DOI] [PubMed] [Google Scholar]
- 87. Froidure A, Vandenplas O, D'Alpaos V, Evrard G, Pilette C. Defects in plasmacytoid dendritic cell expression of inducible costimulator ligand and IFN‐α are associated in asthma with disease persistence. Am J Respir Crit Care Med 2015; 192:392–5. [DOI] [PubMed] [Google Scholar]
- 88. Pritchard AL, Carroll ML, Burel JG, White OJ, Phipps S, Upham JW. Innate IFNs and plasmacytoid dendritic cells constrain Th2 cytokine responses to rhinovirus: a regulatory mechanism with relevance to asthma. J Immunol 2012; 188:5898–905. [DOI] [PubMed] [Google Scholar]
- 89. Ghirelli C, Reyal F, Jeanmougin M, Zollinger R, Sirven P, Michea P et al Breast cancer cell‐derived GM‐CSF licenses regulatory Th2 induction by plasmacytoid predendritic cells in aggressive disease subtypes. Cancer Res 2015; 75:2775–87. [DOI] [PubMed] [Google Scholar]
- 90. Chu CC, Ali N, Karagiannis P, Di Meglio P, Skowera A, Napolitano L et al Resident CD141 (BDCA3)+ dendritic cells in human skin produce IL‐10 and induce regulatory T cells that suppress skin inflammation. J Exp Med 2012; 209:935–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ahrens S, Zelenay S, Sancho D, Hanc P, Kjaer S, Feest C et al F‐actin is an evolutionarily conserved damage‐associated molecular pattern recognized by DNGR‐1, a receptor for dead cells. Immunity 2012; 36:635–45. [DOI] [PubMed] [Google Scholar]
- 92. Zhang JG, Czabotar PE, Policheni AN, Caminschi I, Wan SS, Kitsoulis S et al The dendritic cell receptor Clec9A binds damaged cells via exposed actin filaments. Immunity 2012; 36:646–57. [DOI] [PubMed] [Google Scholar]
- 93. Crozat K, Guiton R, Contreras V, Feuillet V, Dutertre CA, Ventre E et al The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J Exp Med 2010; 207:1283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jongbloed SL, Kassianos AJ, McDonald KJ, Clark GJ, Ju X, Angel CE et al Human CD141+ (BDCA−3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross‐presents necrotic cell antigens. J Exp Med 2010; 207:1247–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Segura E, Valladeau‐Guilemond J, Donnadieu MH, Sastre‐Garau X, Soumelis V, Amigorena S. Characterization of resident and migratory dendritic cells in human lymph nodes. J Exp Med 2012; 209:653–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hacker C, Kirsch RD, Ju XS, Hieronymus T, Gust TC, Kuhl C et al Transcriptional profiling identifies Id2 function in dendritic cell development. Nat Immunol 2003; 4:380–6. [DOI] [PubMed] [Google Scholar]
- 97. Tsujimura H, Tamura T, Ozato K. Cutting edge: IFN consensus sequence binding protein/IFN regulatory factor 8 drives the development of type I IFN‐producing plasmacytoid dendritic cells. J Immunol 2003; 170:1131–5. [DOI] [PubMed] [Google Scholar]
- 98. Grajales‐Reyes GE, Iwata A, Albring J, Wu X, Tussiwand R, Kc W et al Batf3 maintains autoactivation of Irf8 for commitment of a CD8α+ conventional DC clonogenic progenitor. Nat Immunol 2015; 16:708–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Tamura T, Kurotaki D, Koizumi SI. Regulation of myelopoiesis by the transcription factor IRF8. Int J Hematol 2015; 101:342–51. [DOI] [PubMed] [Google Scholar]
- 100. Sichien D, Scott CL, Martens L, Vanderkerken M, Van Gassen S, Plantinga M et al IRF8 transcription factor controls survival and function of terminally differentiated conventional and plasmacytoid dendritic cells, respectively. Immunity 2016; 45:626–40. [DOI] [PubMed] [Google Scholar]
- 101. Tailor P, Tamura T, Morse HC, Ozato K. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood 2008; 111:1942–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Mace EM, Bigley V, Gunesch JT, Chinn IK, Angelo LS, Care MA et al Biallelic mutations in IRF8 impair human NK cell maturation and function. J Clin Invest 2017; 127:306–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Cohen SB, Smith NL, McDougal C, Pepper M, Shah S, Yap GS et al β‐catenin signaling drives differentiation and proinflammatory function of IRF8‐dependent dendritic cells. J Immunol 2015; 194:210–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Poulin LF, Reyal Y, Uronen‐Hansson H, Schraml BU, Sancho D, Murphy KM et al DNGR‐1 is a specific and universal marker of mouse and human Batf3‐dependent dendritic cells in lymphoid and nonlymphoid tissues. Blood 2012; 119:6052–62. [DOI] [PubMed] [Google Scholar]
- 105. Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, Wumesh KC, Albring JC et al Compensatory dendritic cell development mediated by BATF‐IRF interactions. Nature 2012; 490:502–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Bachem A, Guttler S, Hartung E, Ebstein F, Schaefer M, Tannert A et al Superior antigen cross‐presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J Exp Med 2010; 207:1273–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Nizzoli G, Krietsch J, Weick A, Steinfelder S, Facciotti F, Gruarin P et al Human CD1c+ dendritic cells secrete high levels of IL‐12 and potently prime cytotoxic T‐cell responses. Blood 2013; 122:932–42. [DOI] [PubMed] [Google Scholar]
- 108. Cohn L, Chatterjee B, Esselborn F, Smed‐Sorensen A, Nakamura N, Chalouni C et al Antigen delivery to early endosomes eliminates the superiority of human blood BDCA3+ dendritic cells at cross presentation. J Exp Med 2013; 210:1049–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sittig SP, Bakdash G, Weiden J, Sköld AE, Tel J, Figdor CG et al A comparative study of the T cell stimulatory and polarizing capacity of human primary blood dendritic cell subsets. Mediators Inflamm 2016; 2016:3605643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Dalod M, Chelbi R, Malissen B, Lawrence T. Dendritic cell maturation: functional specialization through signaling specificity and transcriptional programming. EMBO J 2014; 33:1104–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Carpentier S, Vu Manh TP, Chelbi R, Henri S, Malissen B, Haniffa M et al Comparative genomics analysis of mononuclear phagocyte subsets confirms homology between lymphoid tissue‐resident and dermal XCR1+ DCs in mouse and human and distinguishes them from Langerhans cells. J Immunol Methods 2016; 432:35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Geginat J, Nizzoli G, Paroni M, Maglie S, Larghi P, Pascolo S et al Immunity to pathogens taught by specialized human dendritic cell subsets. Front Immunol 2015; 6:527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Silvin A, Yu CI, Lahaye X, Imperatore F, Brault JB, Cardinaud S et al Constitutive resistance to viral infection in human CD141+ dendritic cells. Sci Immunol 2017; 2:eaai8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Schreibelt G, Klinkenberg LJ, Cruz LJ, Tacken PJ, Tel J, Kreutz M et al The C‐type lectin receptor CLEC9A mediates antigen uptake and (cross‐)presentation by human blood BDCA3+ myeloid dendritic cells. Blood 2012; 119:2284–92. [DOI] [PubMed] [Google Scholar]
- 115. Li J, Ahmet F, Sullivan LC, Brooks AG, Kent SJ, De Rose R et al Antibodies targeting Clec9A promote strong humoral immunity without adjuvant in mice and non‐human primates. Eur J Immunol 2015; 45:854–64. [DOI] [PubMed] [Google Scholar]
- 116. Hemont C, Neel A, Heslan M, Braudeau C, Josien R. Human blood mDC subsets exhibit distinct TLR repertoire and responsiveness. J Leukoc Biol 2013; 93:599–609. [DOI] [PubMed] [Google Scholar]
- 117. Colletti NJ, Liu H, Gower AC, Alekseyev YO, Arendt CW, Shaw MH. TLR3 signaling promotes the induction of unique human BDCA‐3 dendritic cell populations. Front Immunol 2016; 7:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Liu S, Cai X, Wu J, Cong Q, Chen X, Li T et al Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015; 347:aaa2630. [DOI] [PubMed] [Google Scholar]
- 119. Lafaille FG, Pessach IM, Zhang SY, Ciancanelli MJ, Herman M, Abhyankar A et al Impaired intrinsic immunity to HSV‐1 in human iPSC‐derived TLR3‐deficient CNS cells. Nature 2012; 491:769–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lauterbach H, Bathke B, Gilles S, Traidl‐Hoffmann C, Luber CA, Fejer G et al Mouse CD8α+ DCs and human BDCA3+ DCs are major producers of IFN‐λ in response to poly IC. J Exp Med 2010; 207:2703–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Brewitz A, Eickhoff S, Dähling S, Quast T, Bedoui S, Kroczek RA et al CD8+ T Cells Orchestrate pDC‐XCR1+ dendritic cell spatial and functional cooperativity to optimize priming. Immunity 2017; 46:205–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ohta T, Sugiyama M, Hemmi H, Yamazaki C, Okura S, Sasaki I et al Crucial roles of XCR1‐expressing dendritic cells and the XCR1‐XCL1 chemokine axis in intestinal immune homeostasis. Sci Rep 2016; 6:23505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Alexandre YO, Ghilas S, Sanchez C, Le Bon A, Crozat K, Dalod M. XCR1+ dendritic cells promote memory CD8+ T cell recall upon secondary infections with Listeria monocytogenes or certain viruses. J Exp Med 2016; 213:75–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Guilliams M, Crozat K, Henri S, Tamoutounour S, Grenot P, Devilard E et al Skin‐draining lymph nodes contain dermis‐derived CD103− dendritic cells that constitutively produce retinoic acid and induce Foxp3+ regulatory T cells. Blood 2010; 115:1958–68. [DOI] [PubMed] [Google Scholar]
- 125. Idoyaga J, Fiorese C, Zbytnuik L, Lubkin A, Miller J, Malissen B et al Specialized role of migratory dendritic cells in peripheral tolerance induction. J Clin Invest 2013; 123:844–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Jiang S, Li X, Hess NJ, Guan Y, Tapping RI. TLR10 is a negative regulator of both MyD88‐dependent and ‐independent TLR signaling. J Immunol 2016; 196:3834–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Li YH, Kuo CH, Shi GY, Wu HL. The role of thrombomodulin lectin‐like domain in inflammation. J Biomed Sci 2012; 19:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Lenz A, Heine M, Schuler G, Romani N. Human and murine dermis contain dendritic cells. Isolation by means of a novel method and phenotypical and functional characterization. J Clin Invest 1993; 92:2587–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Nestle FO, Zheng XG, Thompson CB, Turka LA, Nickoloff BJ. Characterization of dermal dendritic cells obtained from normal human skin reveals phenotypic and functionally distinctive subsets. J Immunol 1993; 151:6535–45. [PubMed] [Google Scholar]
- 130. Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony‐stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor α. J Exp Med 1994; 179:1109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Bigley V, McGovern N, Milne P, Dickinson R, Pagan S, Cookson S et al Langerin‐expressing dendritic cells in human tissues are related to CD1c+ dendritic cells and distinct from Langerhans cells and CD141high XCR1+ dendritic cells. J Leukoc Biol 2015; 97:627–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Melum GR, Farkas L, Scheel C, Van Dieren B, Gran E, Liu YJ et al A thymic stromal lymphopoietin‐responsive dendritic cell subset mediates allergic responses in the upper airway mucosa. J Allergy Clin Immunol 2014; 134:613–21.e7. [DOI] [PubMed] [Google Scholar]
- 133. Martinez‐Cingolani C, Grandclaudon M, Jeanmougin M, Jouve M, Zollinger R, Soumelis V. Human blood BDCA‐1 dendritic cells differentiate into Langerhans‐like cells with thymic stromal lymphopoietin and TGF‐beta. Blood 2014; 124:2411–20. [DOI] [PubMed] [Google Scholar]
- 134. Milne P, Bigley V, Gunawan M, Haniffa M, Collin M. CD1c+ blood dendritic cells have Langerhans cell potential. Blood 2015; 125:470–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Takahashi K, Asagoe K, Zaishun J, Yanai H, Yoshino T, Hayashi K et al Heterogeneity of dendritic cells in human superficial lymph node: in vitro maturation of immature dendritic cells into mature or activated interdigitating reticulum cells. Am J Pathol 1998; 153:745–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Angel CE, Chen CJ, Horlacher OC, Winkler S, John T, Browning J et al Distinctive localization of antigen‐presenting cells in human lymph nodes. Blood 2009; 113:1257–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. van de Ven R, van den Hout MF, Lindenberg JJ, Sluijter BJ, van Leeuwen PA, Lougheed SM et al Characterization of four conventional dendritic cell subsets in human skin‐draining lymph nodes in relation to T‐cell activation. Blood 2011; 118:2502–10. [DOI] [PubMed] [Google Scholar]
- 138. Summers KL, Hock BD, McKenzie JL, Hart DN. Phenotypic characterization of five dendritic cell subsets in human tonsils. Am J Pathol 2001; 159:285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Schrøder M, Melum GR, Landsverk OJ, Bujko A, Yaqub S, Gran E et al CD1c‐expression by monocytes ‐ implications for the use of commercial CD1c+ dendritic cell isolation kits. PLoS ONE 2016; 11:e0157387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Bakdash G, Buschow SI, Gorris MA, Halilovic A, Hato SV, Sköld AE et al Expansion of a BDCA1+CD14+ myeloid cell population in melanoma patients may attenuate the efficacy of dendritic cell vaccines. Cancer Res 2016; 76:4332–46. [DOI] [PubMed] [Google Scholar]
- 141. Osugi Y, Vuckovic S, Hart DN. Myeloid blood CD11c+ dendritic cells and monocyte‐derived dendritic cells differ in their ability to stimulate T lymphocytes. Blood 2002; 100:2858–66. [DOI] [PubMed] [Google Scholar]
- 142. Briseño CG, Gargaro M, Durai V, Davidson JT, Theisen DJ, Anderson DA et al Deficiency of transcription factor RelB perturbs myeloid and DC development by hematopoietic‐extrinsic mechanisms. Proc Natl Acad Sci USA 2017; 114:3957–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Meredith MM, Liu K, Darrasse‐Jeze G, Kamphorst AO, Schreiber HA, Guermonprez P et al Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J Exp Med 2012; 209:1153–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Satpathy AT, Wumesh KC, Albring JC, Edelson BT, Kretzer NM, Bhattacharya D et al Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J Exp Med 2012; 209:1135–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. van der Aar AM, Sylva‐Steenland RM, Bos JD, Kapsenberg ML, de Jong EC, Teunissen MB. Loss of TLR2, TLR4, and TLR5 on Langerhans cells abolishes bacterial recognition. J Immunol 2007; 178:1986–90. [DOI] [PubMed] [Google Scholar]
- 146. Van Rhijn I, Ly D, Moody DB. CD1a, CD1b, and CD1c in immunity against mycobacteria. Adv Exp Med Biol 2013; 783:181–97. [DOI] [PubMed] [Google Scholar]
- 147. Lundberg K, Albrekt AS, Nelissen I, Santegoets S, de Gruijl TD, Gibbs S et al Transcriptional profiling of human dendritic cell populations and models ‐ unique profiles of in vitro dendritic cells and implications on functionality and applicability. PLoS ONE 2013; 8:e52875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Harman AN, Bye CR, Nasr N, Sandgren KJ, Kim M, Mercier SK et al Identification of lineage relationships and novel markers of blood and skin human dendritic cells. J Immunol 2013; 190:66–79. [DOI] [PubMed] [Google Scholar]
- 149. Nizzoli G, Larghi P, Paroni M, Crosti MC, Moro M, Neddermann P et al IL‐10 promotes homeostatic proliferation of human CD8(+) memory T cells and when produced by CD1c(+) DCs, shapes naive CD8(+) T‐cell priming. Eur J Immunol 2016; 46:1622–32. [DOI] [PubMed] [Google Scholar]
- 150. Di Blasio S, Wortel IM, van Bladel DA, de Vries LE, Duiveman‐de Boer T, Worah K et al Human CD1c(+) DCs are critical cellular mediators of immune responses induced by immunogenic cell death. Oncoimmunology 2016; 5:e1192739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Bauer T, Zagorska A, Jurkin J, Yasmin N, Köffel R, Richter S et al Identification of Axl as a downstream effector of TGF‐beta1 during Langerhans cell differentiation and epidermal homeostasis. J Exp Med 2012; 209:2033–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Hieronymus T, Zenke M, Baek JH, Sere K. The clash of Langerhans cell homeostasis in skin: should I stay or should I go? Semin Cell Dev Biol 2015; 41:30–8. [DOI] [PubMed] [Google Scholar]
- 153. Artyomov MN, Munk A, Gorvel L, Korenfeld D, Cella M, Tung T et al Modular expression analysis reveals functional conservation between human Langerhans cells and mouse cross‐priming dendritic cells. J Exp Med 2015; 212:743–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Romano E, Cotari JW, Barreira da Silva R, Betts BC, Chung DJ, Avogadri F et al Human Langerhans cells use an IL‐15R‐α/IL‐15/pSTAT5‐dependent mechanism to break T‐cell tolerance against the self‐differentiation tumor antigen WT1. Blood 2012; 119:5182–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Banchereau J, Thompson‐Snipes L, Zurawski S, Blanck JP, Cao Y, Clayton S et al The differential production of cytokines by human Langerhans cells and dermal CD14(+) DCs controls CTL priming. Blood 2012; 119:5742–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Geissmann F, Dieu‐Nosjean MC, Dezutter C, Valladeau J, Kayal S, Leborgne M et al Accumulation of immature Langerhans cells in human lymph nodes draining chronically inflamed skin. J Exp Med 2002; 196:417–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Mueller CG, Voisin B. Of skin and bone: did Langerhans cells and osteoclasts evolve from a common ancestor. J Anat 2016. doi: 10.1111/joa.12543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Romani N, Brunner PM, Stingl G. Changing views of the role of Langerhans cells. J Invest Dermatol 2012; 132:872–81. [DOI] [PubMed] [Google Scholar]
- 159. Wu X, Briseño CG, Durai V, Albring JC, Haldar M, Bagadia P et al Mafb lineage tracing to distinguish macrophages from other immune lineages reveals dual identity of Langerhans cells. J Exp Med 2016; 213:2553–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Price JG, Idoyaga J, Salmon H, Hogstad B, Bigarella CL, Ghaffari S et al CDKN1A regulates Langerhans cell survival and promotes Treg cell generation upon exposure to ionizing irradiation. Nat Immunol 2015; 16:1060–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Czernielewski JM, Demarchez M. Further evidence for the self‐reproducing capacity of Langerhans cells in human skin. J Invest Dermatol 1987; 88:17–20. [DOI] [PubMed] [Google Scholar]
- 162. Bigley V, Haniffa M, Doulatov S, Wang XN, Dickinson R, McGovern N et al The human syndrome of dendritic cell, monocyte, B and NK lymphoid deficiency. J Exp Med 2011; 208:227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Kanitakis J, Morelon E, Petruzzo P, Badet L, Dubernard JM. Self‐renewal capacity of human epidermal Langerhans cells: observations made on a composite tissue allograft. Exp Dermatol 2011; 20:145–6. [DOI] [PubMed] [Google Scholar]
- 164. Capucha T, Mizraji G, Segev H, Blecher‐Gonen R, Winter D, Khalaileh A et al Distinct Murine Mucosal Langerhans Cell Subsets Develop from Pre‐dendritic Cells and Monocytes. Immunity 2015; 43:369–81. [DOI] [PubMed] [Google Scholar]
- 165. Collin M, Milne P. Langerhans cell origin and regulation. Curr Opin Hematol 2016; 23:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Schuster C, Mildner M, Mairhofer M, Bauer W, Fiala C, Prior M et al Human embryonic epidermis contains a diverse Langerhans cell precursor pool. Development 2014; 141:807–15. [DOI] [PubMed] [Google Scholar]
- 167. Riedl E, Stockl J, Majdic O, Scheinecker C, Knapp W, Strobl H. Ligation of E‐cadherin on in vitro‐generated immature Langerhans‐type dendritic cells inhibits their maturation. Blood 2000; 96:4276–84. [PubMed] [Google Scholar]
- 168. Yasmin N, Konradi S, Eisenwort G, Schichl YM, Seyerl M, Bauer T et al β‐Catenin promotes the differentiation of epidermal langerhans dendritic cells. J Invest Dermatol 2013; 133:1250–9. [DOI] [PubMed] [Google Scholar]
- 169. Chorro L, Sarde A, Li M, Woollard KJ, Chambon P, Malissen B et al Langerhans cell (LC) proliferation mediates neonatal development, homeostasis, and inflammation‐associated expansion of the epidermal LC network. J Exp Med 2009; 206:3089–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Seré K, Baek JH, Ober‐Blöbaum J, Müller‐Newen G, Tacke F, Yokota Y et al Two distinct types of Langerhans cells populate the skin during steady state and inflammation. Immunity 2012; 37:905–16. [DOI] [PubMed] [Google Scholar]
- 171. Nagao K, Kobayashi T, Moro K, Ohyama M, Adachi T, Kitashima DY et al Stress‐induced production of chemokines by hair follicles regulates the trafficking of dendritic cells in skin. Nat Immunol 2012; 13:744–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Collin MP, Hart DN, Jackson GH, Cook G, Cavet J, Mackinnon S et al The fate of human Langerhans cells in hematopoietic stem cell transplantation. J Exp Med 2006; 203:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Mielcarek M, Kirkorian AY, Hackman RC, Price J, Storer BE, Wood BL et al Langerhans cell homeostasis and turnover after nonmyeloablative and myeloablative allogeneic hematopoietic cell transplantation. Transplantation 2014; 98:563–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Strobl H, Bello‐Fernandez C, Riedl E, Pickl WF, Majdic O, Lyman SD et al flt3 ligand in cooperation with transforming growth factor‐beta1 potentiates in vitro development of Langerhans‐type dendritic cells and allows single‐cell dendritic cell cluster formation under serum‐free conditions. Blood 1997; 90:1425–34. [PubMed] [Google Scholar]
- 175. Geissmann F, Prost C, Monnet JP, Dy M, Brousse N, Hermine O. Transforming growth factor beta1, in the presence of granulocyte/macrophage colony‐stimulating factor and interleukin 4, induces differentiation of human peripheral blood monocytes into dendritic Langerhans cells. J Exp Med 1998; 187:961–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Hoshino N, Katayama N, Shibasaki T, Ohishi K, Nishioka J, Masuya M et al A novel role for Notch ligand Delta‐1 as a regulator of human Langerhans cell development from blood monocytes. J Leukoc Biol 2005; 78:921–9. [DOI] [PubMed] [Google Scholar]
- 177. Hutter C, Kauer M, Simonitsch‐Klupp I, Jug G, Schwentner R, Leitner J et al Notch is active in Langerhans cell histiocytosis and confers pathognomonic features on dendritic cells. Blood 2012; 120:5199–208. [DOI] [PubMed] [Google Scholar]
- 178. Jurkin J, Krump C, Köffel R, Fieber C, Schuster C, Brunner PM et al Human skin dendritic cell fate is differentially regulated by the monocyte identity factor KLF4 during steady state and inflammation. J Allergy Clin Immunol 2016; 139:1873–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Milne P, Bigley V, Bacon CM, Néel A, McGovern N, Bomken S et al Hematopoietic origin of Langerhans cell histiocytosis and Erdheim‐Chester disease in adults. Blood 2017; 130:167–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Kim JH, Hu Y, Yongqing T, Kim J, Hughes VA, Le Nours J et al CD1a on Langerhans cells controls inflammatory skin disease. Nat Immunol 2016; 17:1159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Seneschal J, Clark RA, Gehad A, Baecher‐Allan CM, Kupper TS. Human epidermal Langerhans cells maintain immune homeostasis in skin by activating skin resident regulatory T cells. Immunity 2012; 36:873–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Kashem SW, Igyarto BZ, Gerami‐Nejad M, Kumamoto Y, Mohammed JA, Jarrett E et al Candida albicans morphology and dendritic cell subsets determine T helper cell differentiation. Immunity 2015; 42:356–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Langlet C, Tamoutounour S, Henri S, Luche H, Ardouin L, Grégoire C et al CD64 expression distinguishes monocyte‐derived and conventional dendritic cells and reveals their distinct role during intramuscular immunization. J Immunol 2012; 188:1751–60. [DOI] [PubMed] [Google Scholar]
- 184. Segura E, Amigorena S. Inflammatory dendritic cells in mice and humans. Trends Immunol 2013; 34:440–5. [DOI] [PubMed] [Google Scholar]
- 185. Schlitzer A, McGovern N, Ginhoux F. Dendritic cells and monocyte‐derived cells: Two complementary and integrated functional systems. Semin Cell Dev Biol 2015; 41:9–22. [DOI] [PubMed] [Google Scholar]
- 186. Wollenberg A, Kraft S, Hanau D, Bieber T. Immunomorphological and ultrastructural characterization of Langerhans cells and a novel, inflammatory dendritic epidermal cell (IDEC) population in lesional skin of atopic eczema. J Invest Dermatol 1996; 106:446–53. [DOI] [PubMed] [Google Scholar]
- 187. Wollenberg A, Mommaas M, Oppel T, Schottdorf EM, Gunther S, Moderer M. Expression and function of the mannose receptor CD206 on epidermal dendritic cells in inflammatory skin diseases. J Invest Dermatol 2002; 118:327–34. [DOI] [PubMed] [Google Scholar]
- 188. Zaba LC, Fuentes‐Duculan J, Eungdamrong NJ, Abello MV, Novitskaya I, Pierson KC et al Psoriasis is characterized by accumulation of immunostimulatory and Th1/Th17 cell‐polarizing myeloid dendritic cells. J Invest Dermatol 2009; 129:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Jenner W, Motwani M, Veighey K, Newson J, Audzevich T, Nicolaou A et al Characterisation of leukocytes in a human skin blister model of acute inflammation and resolution. PLoS ONE 2014; 9:e89375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Eguíluz‐Gracia I, Bosco A, Dollner R, Melum GR, Lexberg MH, Jones AC et al Rapid recruitment of CD14(+) monocytes in experimentally induced allergic rhinitis in human subjects. J Allergy Clin Immunol 2016; 137:1872–81.e12. [DOI] [PubMed] [Google Scholar]
- 191. Beitnes AC, Raki M, Brottveit M, Lundin KE, Jahnsen FL, Sollid LM. Rapid accumulation of CD14+CD11c+ dendritic cells in gut mucosa of celiac disease after in vivo gluten challenge. PLoS ONE 2012; 7:e33556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Grimm MC, Pullman WE, Bennett GM, Sullivan PJ, Pavli P, Doe WF. Direct evidence of monocyte recruitment to inflammatory bowel disease mucosa. J Gastroenterol Hepatol 1995; 10:387–95. [DOI] [PubMed] [Google Scholar]
- 193. Bain CC, Scott CL, Uronen‐Hansson H, Gudjonsson S, Jansson O, Grip O et al Resident and pro‐inflammatory macrophages in the colon represent alternative context‐dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol 2013; 6:498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Liao CT, Andrews R, Wallace LE, Khan MW, Kift‐Morgan A, Topley N et al Peritoneal macrophage heterogeneity is associated with different peritoneal dialysis outcomes. Kidney Int 2017; 91:1088–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Guilliams M, van de Laar L. A Hitchhiker's guide to myeloid cell subsets: practical implementation of a novel mononuclear phagocyte classification system. Front Immunol 2015; 6:406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Wimmers F, Schreibelt G, Skold AE, Figdor CG, De Vries IJ. Paradigm shift in dendritic cell‐based immunotherapy: from generated monocyte‐derived DCs to naturally circulating DC subsets. Front Immunol 2014; 5:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Schakel K, von Kietzell M, Hansel A, Ebling A, Schulze L, Haase M et al Human 6‐sulfo LacNAc‐expressing dendritic cells are principal producers of early interleukin‐12 and are controlled by erythrocytes. Immunity 2006; 24:767–77. [DOI] [PubMed] [Google Scholar]
- 198. Hofer TP, Zawada AM, Frankenberger M, Skokann K, Satzl AA, Gesierich W et al slan‐defined subsets of CD16‐positive monocytes: impact of granulomatous inflammation and M‐CSF receptor mutation. Blood 2015; 126:2601–10. [DOI] [PubMed] [Google Scholar]
- 199. Cros J, Cagnard N, Woollard K, Patey N, Zhang SY, Senechal B et al Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity 2010; 33:375–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. van Leeuwen‐Kerkhoff N, Lundberg K, Westers TM, Kordasti S, Bontkes HJ, de Gruijl TD et al Transcriptional profiling reveals functional dichotomy between human slan(+) non‐classical monocytes and myeloid dendritic cells. J Leukoc Biol 2017; 102:1055–68. [DOI] [PubMed] [Google Scholar]
- 201. Patel AA, Zhang Y, Fullerton JN, Boelen L, Rongvaux A, Maini AA et al The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J Exp Med 2017; 214:1913–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Calzetti F, Tamassia N, Micheletti A, Finotti G, Bianchetto‐Aguilera F, Cassatella MA. Human dendritic cell subset 4 (DC4) correlate to a subset of CD14dim/‐ CD16++ monocytes. J Allergy Clin Immunol 2018. doi: 10.1016/j.jaci.2017.12.988 [DOI] [PubMed] [Google Scholar]


