Summary
The few initial formative studies describing non‐specific and apparently spontaneous activity of natural killer (NK) cells have since multiplied into thousands of scientific reports defining their unique capacities and means of regulation. Characterization of the array of receptors that govern NK cell education and activation revealed an unexpected relationship with the major histocompatibility molecules that NK cells originally became well known for ignoring. Proceeding true to form, NK cells continue to up‐end archetypal understanding of their ever‐expanding capabilities. Discovery that the NK cell repertoire is extremely diverse and can be reshaped by particular viruses into unique subsets of adaptive NK cells challenges, or at least broadens, the definition of immunological memory. This review provides an overview of studies identifying adaptive NK cells, addressing the origins of NK cell memory and introducing the heretical concept of NK cells with extensive antigenic specificity. Whether these newly apparent properties reflect adaptive utilization of known NK cell attributes and receptors or a specially creative allocation from an undefined receptor array remains to be fully determined.
Keywords: Fc receptors, memory, major histocompatibility complex/histocompatibility‐linked antigen, natural killer cell, virus
Introduction
Our perception of natural killer (NK) cells has transformed dramatically since their discovery just over 40 years ago.1, 2, 3, 4, 5 Once viewed as a monolithic fraction of peripheral blood lymphocytes with an innate ability to find and kill aberrant cells, NK cells have since been revealed as a highly diverse lymphocyte population within which idiosyncratic education, regulation and activation processes govern subset function and prominence. Subsets distinguished by variable constellations of regulatory receptors diverge further along functional scales delineated by the presence and affinity of polymorphic ligands. Genetic, stochastic, deterministic and epistatic factors interact to produce up to 30 000 discrete NK cell subsets.6 We now know that the ontogenetic NK cell repertoire is dynamically modified by environmental influences (Fig. 1). As with the responses of B and T lymphocytes, NK cell engagement in an immune response reshapes the baseline repertoire, in part by introducing an adapted, preferentially selected, memory‐like population with distinct phenotypic and functional features. Selective expansion, together with acquisition of novel functional and phenotypic characteristics, is common to memory B, T and NK cells alike. However, to our knowledge, only NK cells mature into immune memory cells independently of antigen recognition through somatically generated, clonotypic receptors. Therefore, the selection mechanisms underlying dynamic remodelling of NK cell repertoires and NK cell maturation into memory cells remain mostly obscure.
Figure 1.
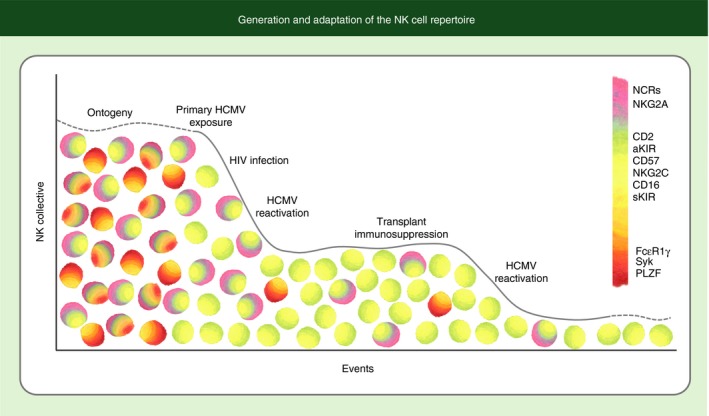
Generation and adaptation of the natural killer (NK) cell repertoire. A diverse NK cell repertoire is generated through variable expression of a number of germline‐encoded receptors, creating a myriad collection of NK cells with different receptor constellations. Genetic and environmental influences generally lead to modest selection of NK cells with self‐specific and activating killer cell immunoglobulin‐like receptors (KIRs). Infection with human cytomegalovirus (HCMV) drives a more stringent selection leading to a highly expanded population of mature CD57pos NK cells expressing NKG2C and high levels of CD16. This selection process appears to be exaggerated in the setting of immunodeficiency or immunosuppression with increasingly narrow bottlenecks favouring selective expansion of these cells, which are specialized to mediate antibody‐dependent cell‐mediated cytotoxicity, into a dominant fraction of the overall repertoire. Narrowing of the repertoire with additional events on the x‐axis represents a diminution of the frequency of certain NK cell subsets, not of total NK cell numbers.
This review will address accumulating evidence for antigen‐dependent and ‐independent NK cell maturation and memory formation. The response against murine cytomegalovirus (MCMV) is the first and best characterized example of antigen‐driven activation and maturation of NK cells into memory cells. We will review aspects of the NK cell response to MCMV, followed by a series of as yet mechanistically unexplained examples of NK cell memory and antigen specificity in mice and primates, followed by a description of human memory‐like NK cell development, primarily in response to human CMV (HCMV) infection. Of note, the process by which NK cell memory forms in the case of HCMV infection appears to have less in common with MCMV infection than initially expected. Finally, we will speculate on how features of NK cells, viruses and other antigens might interact to confer a semblance of antigen specificity independent of somatically rearranged clonotypic receptors.
Murine cytomegalovirus infection
The role that NK cells play against MCMV in certain strains of mice illustrates the two key aspects of immunological memory previously attributed exclusively to lymphocytes bearing clonotypic receptors. There is selective expansion of NK cells bearing antigen‐specific receptors and a more efficient secondary response against the same antigen. Appreciation of the importance of murine NK cell responses to MCMV began with the observation that BALB/c mice are more susceptible to MCMV infection than C57BL/6 mice. Classical genetics mapped resistance to Ly49H, a gene encoding an activating receptor on murine NK cells.7 This receptor is now known to directly interact with the MCMV‐encoded m157 glycoprotein, thereby stimulating expansion of Ly49H‐bearing NK cells that help to control primary infection and provide enhanced protection against subsequent challenge.7, 8, 9 Intensive study of this system over the last two decades revealed distinct phases necessary for the propagation of memory NK cells. The first phase occurs during early infection and probably precedes physical interactions between receptor–ligand pairs. During this time, pro‐inflammatory cytokines such as type‐I interferon (IFN‐α/β) and interleukin‐12 (IL‐12) provide non‐specific signals to prime NK cells. These cytokines, initially produced by myeloid cells in response to MCMV infection, stimulate NK cell antiviral cytokine production, induce their activation and initiate non‐specific NK cell proliferation.10, 11, 12 Interferon‐α/β‐induced IL‐15 is also thought to contribute to non‐specific NK cell proliferation during the early stages of MCMV infection and provide stimulation pertinent to NK cell survival.13, 14 These cytokines sensitize NK cells to IL‐18 and IL‐33, which are required for optimal responses, but dispensable for forming memory NK cells.15 Interactions between cytokines and their receptors encourage NK cell adaptation to MCMV by transmitting signals through signal transducer and activator of transcription (STAT) mediators. Engaging IFN‐α and IL‐12 receptors stimulates STAT1 and STAT4, crucial regulators of NK cell IFN‐γ expression and cytolytic activity.16, 17, 18, 19, 20 Although the Yokoyama laboratory initially described the prolonged effects of IL‐12 on NK cell activation, the central role for IL‐12 in NK cell memory was definitively illustrated by the inability of NK cells from IL‐12Rk/o mice to expand and provide protection against MCMV challenge.21, 22, 23 No correlations between NK cell inflammatory cytokine stimulation and antigen‐specific recall responses have been noted, but IL‐12 modulates NK cell adaption, enhances NK cell responsiveness and causes inheritable modifications in NK cell progeny.23, 24 Exposure to IFN‐α/β and IL‐12 during the later stages of NK cell adaptation also enables the proliferative burst observed after MCMV challenge, which is incited by a cytokine‐specific increase in expression of zinc finger transcription factor Zbtb32.25, 26
This early generic NK cell response to cytokines may be critical for priming NK cells to respond more effectively to virus‐specific interactions during subsequent stages of MCMV infection. The initial NK cell response to cytokines is independent of activating receptor expression; however, NK cells lacking Ly49H fail to proliferate in response to MCMV during the later stages of infection.27 The key feature in NK cell memory responses to MCMV infection is the specific interaction between the MCMV m157 glycoprotein and Ly49H activating receptors that trigger selective proliferation and differentiation into a long‐lived memory cell population with enhanced protective capacity.8, 9, 28, 29 The interaction between Ly49H and m157 induces a two to threefold increase in NK cell numbers during the first week after MCMV infection, with this expansion remaining detectable at least 70 days after infection.27, 30 The importance of this interaction in NK cell adaptation is illustrated by adoptive transfer studies in which Ly49Hpos NK cells vigorously proliferate in recipients lacking Ly49H after challenge with MCMV.30 Infection with an MCMV construct lacking m157 also fails to induce proliferation or expansion of this protective pool of NK cells, reiterating the importance of direct interaction between Ly49H and m157.29, 30
More recent studies show that the activating receptor Ly49P recognizes an MCMV m04‐derived peptide in the context of murine H‐2D class I major histocompatibility complex (MHC) molecules and acts similarly to Ly49H in conferring protection against MCMV in MA/MyJ mice.31 With the exception of clonal selection, C57BL/6 and MA/MyJ NK cell responses against MCMV display the key defining features of adaptive immune memory displayed by B and T lymphocytes. Recognition of MCMV by NK cell activating receptors may have evolved under pressure from the numerous MCMV evasion strategies targeting NK cell inhibitory receptors. Polymorphism within MCMV m157 and m04 genes and the interacting Ly49 alleles supports this possibility.31, 32, 33, 34 The evolution of both direct and, in the case of m04, MHC‐restricted recognition processes illustrates multiple NK cell adaptation strategies arising against a single virus. Other similar as yet uncharted events may have advanced murine NK cell evolution and driven diversification of the analogous human killer cell immunoglobulin‐like receptor (KIR) genes, however, specific recognition of MCMV by NK cells from C57BL/6 and MA/MyJ mice remain the only definitive examples.
Antigen‐specific NK cell expansion initiated by m157/Ly49H interactions relies on directed signalling through adaptor proteins.35 Although Ly49H receptors transmit signals through either DAP10 or DAP12, Orr et al. showed that signalling through both adaptor proteins is necessary for optimal NK cell adaptation.35 In the absence of either DAP10 or DAP12 expression, NK cells still mediate resistance to MCMV infection; however, deleting either of these two proteins reduces surface Ly49H expression, NK cell proliferation and IFN‐γ production in response to MCMV.35 Signal transmission through either of these adaptor proteins after m157/Ly49H interactions may represent a redundancy mechanism whereby adequate memory‐like NK cell responses are maintained in the absence of either DAP10 or DAP12 expression. However, if both DAP10 and DAP12 are absent, Ly49H expression is completely suppressed and mice are susceptible to MCMV challenge.35
Analogous to the activation of naive B and T cells, co‐stimulatory receptor engagement is a necessary component of NK cell adaptation. Upon infection, MCMV induces CD155 and/or CD112 expression on monocytes or dendritic cells.36 Engagement of NK cell‐expressed DNAX accessory molecule‐1 (DNAM‐1) by CD112 and/or CD155 drives signal transduction through Fyn and PKCƞ, which promotes NK cell differentiation after initial and subsequent MCMV challenges.37 This process generates a pool of memory NK cells with high expression levels of maturation markers (CD11b, Ly6C, and KLRG1) and of Ly49H.37 It is unknown whether other co‐stimulatory molecules are involved in the activation, proliferation or persistence of MCMV‐specific memory NK cells.
During the first week of MCMV infection, NK cells are exposed to non‐specific cytokines, viral proteins or virus‐induced host ligands that support NK cell proliferation and expansion. Following activation and differentiation towards enhanced effector function, the next phase of memory NK cell generation begins as the NK cell pool contracts into an elite collection of highly functional mature NK cells.38 Pro‐apoptotic Bcl‐2‐like protein 11 (Bim) is a key regulator in this contraction phase.39 After MCMV challenge, mice lacking Bim accumulate NK cells with a less matured phenotype (KLRG1lo) that respond poorly to secondary m157 stimulation.39 In wild‐type mice, most of these cells undergo apoptosis, but a repair process controlled by mitochondrial‐associated proteins, BNIP3 and BNIP3L, spares a small proportion of NK cells by selectively removing dysfunctional mitochondria.40 The cells that survive mitophagy persist as the long‐lived memory NK cells that proliferate, expand and contract in response to repeated MCMV challenge and provide greater protection against successive MCMV infection than their naive counterparts.30 The C57BL/6 MCMV model draws parallels with T‐cell memory development, extending from the need for cytokine stimulation and co‐stimulatory receptor engagement, to the critical contraction phase following antigen‐driven NK cell expansion. This model has provided detailed information on the role of cytokine‐priming, accessory interactions and signalling pathways underlying formation of antigen‐specific NK cell memory populations, ultimately broadening our perspective on immunological memory to include cells restricted to recognition of antigens with germ‐line receptors.
Antigen‐specific NK cells in mice
Other adaptive properties that were previously thought to be restricted to lymphocytes bearing somatically rearranged antigen‐specific receptors were demonstrated for NK cells by O'Leary et al.41 In classic crossover control style experiments, they showed that rag k/o mice lacking B and T cells can be sensitized against a particular hapten and mount hapten‐specific contact hypersensitivity reactions for at least 4 weeks afterwards.41 This phenomenon reflects selective expansion and differentiation of hapten‐specific cells during sensitization, persistence of some fraction of the sensitized cells as memory cells, recruitment of memory cells to the site of secondary challenge and their in situ antigen‐specific reactivation. Adoptive transfer of NK cells from sensitized mice enabled naive mice to mount contact hypersensitivity responses specific for the sensitizing hapten, recapitulating a phenomenon previously associated exclusively with adoptive transfer of antigen‐specific T cells.41 The haptens used to induce contact hypersensitivity (dinitrofluorobenzene and oxalazone) fall within a special category of antigens that trigger immune responses by modifying self‐proteins, but investigators have since demonstrated similar NK cell‐mediated responses following immunization with viral proteins. Human immunodeficiency virus (HIV) gag or env proteins, or influenza M1 matrix protein delivered in virus‐like particles or immunization with ultraviolet‐light‐killed vesicular stomatitis virus also elicited NK‐dependent immune memory.42 Although it lacks mechanistic explanation, this form of apparent antigen‐specific NK cell reactivity has now been generalized across a range of antigens. Characterization of the murine NK cell subset responsible for antigen specificity indicates CXCR6‐dependent liver residency, expression of Ly49C, DX5 (in most cases) and NKG2D with dependence on IL‐12, IFN‐α and IFN‐γ priming for their initial development.43, 44 A notable feature of the contact hypersensitivity mediated by sensitized NK cells is the speed with which the reaction occurs following secondary exposure to the sensitizing antigen. Unlike T‐cell‐dependent contact hypersensitivity, which takes 24–48 hr to develop, NK cell‐dependent responses are detectable within 30 min.43 Presumably, this reflects the lack of any need for antigen processing and presentation and possibly faster recruitment or enhanced potency on a per cell basis. If so, this might either be an intrinsic property of the NK cells active in this context or related to the absence of (regulatory) T cells in the rag k/o models. Further studies have extended the phenomenon of antigen‐specific NK cell memory to recognition of vaccinia virus and Mycobacterium tuberculosis.45, 46 In contrast to the case of influenza M1 matrix protein, where the memory NK cells induced selectively reside in the lungs, respiratory influenza virus infection induces memory‐like NK cells resident in the liver.42, 47 Although M. tuberculosis‐induced memory NK cells can develop in rag k/o mice, they require T‐cell‐derived IL‐21 for induction.46
These studies in mice have been rigorously conducted with a general consensus emerging from multiple laboratories as to the phenotype, effector functions and homing behaviour of the antigen‐specific NK cells detected. There is preliminary evidence of a genetic basis for the murine NK cell specificity, but the nature of any receptor genes responsible remains mysterious.48 Adaptation to ancient, chronic pathogens like MCMV is a logical explanation for pressure to evolve activating receptors that counter viral decoy proteins, but it is difficult to envision something similar for adaptation to the synthetic haptens that specifically sensitize NK cells. More research is clearly required to discern the full antigenic range of this phenomenon and whether specific recognition of synthetic haptens and microbial products reflects their inclusion in some form of pre‐existing or inducible antigen receptor repertoire unique to NK cells.
Antigen‐specific NK cells in primates
Demonstration of antigen‐specific memory NK cells in mice raises the question of whether similar phenomena occur in primates. Subsets of human NK cells respond selectively to certain viruses, bacteria and fungi with proliferation and augmented effector functions.49, 50, 51, 52, 53, 54, 55 Expanded numbers of these cells can persist after initial exposure and mount memory‐like reactions upon secondary exposure. A number of associations between protection against viral infection or disease progression and co‐ordinate inheritance of NK cell receptor/ligand gene pairings indicate that particular subsets of NK cells have pathogen‐selective, if not pathogen‐specific, activity in humans and other primates. One of the more prominent associations with enhanced NK cell function, protection from viral infection or protection from disease progression, involves inheritance of KIR3DS1 together with or without class I human histocompatibility‐linked antigen (HLA) molecules bearing the HLA‐Bw4*80I motif.53, 54, 56, 57, 58, 59, 60 The human KIR locus, analogous to murine Ly49, encodes inhibitory and activating receptors that interact selectively with class I HLA molecules. In HIV infection, NK cells bearing particular KIR molecules increase in prominence, suggesting that they might play a role in pathogen recognition.54, 60 Interaction between class I HLA molecules and KIR proteins can be modulated by the nature of the HLA‐bound peptide, as can the interaction between C‐type lectin‐like receptors and HLA‐E.61, 62, 63, 64, 65, 66, 67 It is possible, therefore, that viral infection alters the peptide composition of expressed class I HLA molecules in ways that tilt the balance between NK cell inhibition and activation. Selective expansion of NK cells bearing KIR3DS1 occurs during acute HIV infection, and with co‐ordinate expression of HLA‐Bw4*80I, NK cells bearing KIR3DS1 more effectively inhibit HIV replication in vitro.53, 54 There is epidemiological evidence of a co‐ordinate role for KIR3DS1 expression together with HLA genotypes expressing the HLA‐Bw4*80I epitope in protection from HIV disease progression as well as some indirect evidence for enhanced NK cell function with this combination, but the nature of this effect remains controversial. No functional interaction between KIR3DS1 and HLA molecules bearing the Bw4*80I epitope was ever demonstrated and in fact, it was recently shown that KIR3DS1 binds open conformers of the relatively non‐polymorphic HLA‐F molecule.68, 69 In light of this finding, the data indicating epistatic interaction between KIR3DS1 and HLA‐B molecules bears re‐assessment or re‐analysis. A study of inhibitory and activating KIR expression on NK cells from monozygotic twins in comparison with unrelated individuals suggested that expression of inhibitory receptors is highly influenced by genetics, but that environmental factors have more influence on expression of activating KIR than does co‐ordinate expression of their cognate ligands.6 As none of the subjects in this study were infected with HIV, this implies that a number of other infections might select for NK cells expressing activating KIR. Conversely, there is also evidence for HIV escape mutations selective for individuals expressing KIR2DL2 that work by enhancing its inhibitory function.70 While fine antigenic specificity is not implied by any of these examples, expression of various KIR molecules with or without their ligands clearly affects, and is affected by, the interface between NK cells and multiple pathogens.
Two published studies describe antigen‐specific NK cell behaviour in primates. Splenic NK cells from rhesus macaques, either infected with simian immunodeficiency virus (SIV) constructs or vaccinated with recombinant adenoviruses expressing SIV antigens, selectively killed dendritic cells pulsed with relevant retroviral Env or Gag antigens.71 Specialized longer‐term killing assays were used in these cases, suggesting either a low frequency or low cytotoxic potency of the relevant NK cell population. However, SIV‐specific NK cell activity detected by this assay persisted up to 5 years after vaccination with the recombinant adenoviruses, indicating a robust and durable response.71 Blocking either NKG2C or NKG2A reduced killing, but it is unclear if or how either receptor is directly involved. In similar assays, circulating human NK cells were reported to selectively kill autologous B cells pulsed with viral peptides, most notably those pulsed with HCMV pp65 peptides.72 Significant NK cell killing was reported with cells from 9 of 35 donors tested, but the representative data shown in the publication indicated weak killing barely above background and these findings have not since been corroborated by other published research.
Cytokine‐dependent NK cell memory
Another interesting feature of NK cells is their ability to manifest a form of immunological memory simply from exposure to pro‐inflammatory cytokines in the absence of virus, hapten or other specific receptor/ligand engagement. The first indication that cytokines invoke NK cell adaptation followed an adoptive transfer study of murine splenic NK cells that were briefly activated in vitro with IL‐12, IL‐15 and IL‐18 before administration to naive recipients.23 Cytokine‐primed NK cells proliferated in their new host and generated a stable population with augmented IFN‐γ responses upon re‐exposure to IL‐12, IL‐15 and IL‐18, or upon activating receptor engagement.23 Heightened IFN‐γ responses were maintained for up to 12 weeks, and cytokine‐induced NK cell memory was demonstrable as a heritable property.23, 24 Similar effects were noted with human NK cells, where ex vivo IL‐12, IL‐15 and IL‐18 re‐stimulation induced robust IFN‐γ production in NK cells previously exposed to the same cytokine milieu in vitro.73 In contrast with the phenotype associated with HCMV‐induced NK cell adaptation, namely acquisition of the maturation marker CD57 and loss of NKG2A and natural cytotoxicity receptors, the cytokine‐induced IFN‐γ‐producing adaptive NK cell population lacks CD57, while retaining NKp46 and NKG2A.73 It remains to be established whether epigenetic modification of the IFNG locus occurs, and if the cytokine‐induced alterations to NK cell functions are as durable as those seen in HCMV‐driven NK cell adaptation. Another intriguing question is whether immunomodulatory cytokines produced during viral infections contribute to in vivo cytokine‐induced NK cell adaptation. Commonly produced cytokines, such as IFN‐α/β, or in the case of HCMV, a viral homologue to IL‐10, could act alone or in concert with signalling through various NK cell receptors to promote adaptation.
The NK cell response to HCMV
There is now clear appreciation that diversity within the NK cell repertoire at the population and individual levels affects interactions with a multitude of pathogens and that in turn, these pathogens effect enrichment of certain NK subsets. Diversity within NK cells is thought to have arisen in part under pressure from what has been described as an evolving ‘arms race’ between pathogens and the host immune system.74 If longevity and antagonistic recidivism in this arms race are important factors, then the influence of human herpesviruses on the NK cell repertoire should be prominently reflected. Despite evidence of marked susceptibility to multiple herpesvirus infections in NK‐deficient humans, only CMV infection is known to have a dramatic effect on the composition of the NK cell repertoire.75 This is true in the case of both MCMV and HCMV, but despite superficial similarities, the operative mechanism for NK cell adaptation to murine and human CMV appears mostly unrelated.
In 2004, Guma et al.76 first reported that infection with HCMV leaves a durable imprint on the human NK cell repertoire. This imprint is reflected in an increased frequency of NK cells expressing CD57 and C‐type lectin‐like activating receptor NKG2C, together with high levels of the IgG Fc receptor CD16 (Fig. 2). Expression of activating and self‐specific KIRs is higher on these NK cells, whereas levels of natural cytotoxicity receptors and NKG2A are reduced.76, 77, 78, 79, 80, 81, 82, 83 Functionally, the cells are especially efficient at mediating antibody‐dependent cell‐mediated cytotoxicity, perhaps in adaptation to down‐modulated natural cytotoxicity receptor signalling. Recently, data from two independent laboratories demonstrated substantial overlap of the population of NK cells co‐expressing CD57 and NKG2C with NK cells that have reduced expression of the FcεR1γ signalling protein, Syk kinase, promyelocytic leukaemia zinc finger protein (PLZF) transcription factor and Ewing's sarcoma's/FLI‐1 activated transcript‐2 (EAT‐2).84, 85 Although accumulation of these cells was initially associated with a number of other viral infections, it later became clear that HCMV infection is the one constant required for marked expansion of CD57pos NK cells expressing NKG2C. This association between selective expansion of NK cells bearing the activating receptor NKG2C and HCMV infection inspired consideration that NKG2C might be analogous to Ly49H with its specific recognition of an HCMV protein or peptide driving expansion of NKG2Cpos NK cells. Lending credence to this idea, in some in vitro HCMV infection/NK cell co‐culture systems, CD57pos NKG2Cpos NK cell expansion is blocked by antibodies against either HLA‐E or NKG2C.86 Both NKG2C and its inhibitory counterpart NKG2A bind to HLA‐E, which in turn is modulated by HCMV infection.64 Expression of HLA‐E is generally stabilized by peptides derived from class I molecules and its function as an inhibitory or activating ligand for NKG2A or NKG2C receptors, respectively, is critically dependent on the peptide presented.61, 62, 63, 87 Although peptides derived from HCMV UL40 protein (i.e. VMAPRTLIL) bind and stabilize HLA‐E, expression of HCMV UL16, UL18 and UL40 is dispensable for NKG2Cpos NK cell expansion in vitro.77, 88 In contrast, the US2–US11 genes crucially contribute to in vitro NKG2C‐driven NK cell expansion, indicating a role for class I HLA molecules.77, 89
Figure 2.
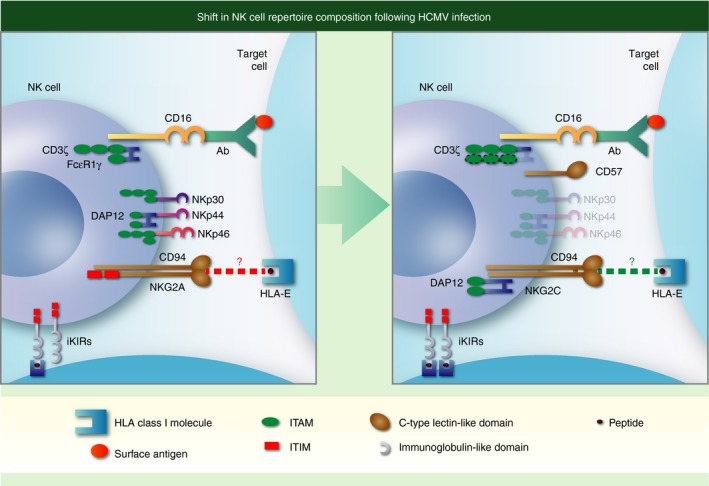
Adaptation of natural killer (NK) cells in response to human cytomegalovirus (HCMV) infection. Individuals infected with HCMV (right hand panel) have an increased fraction of circulating NK cells that express CD57 and the activating C‐type lectin‐like receptor NKG2C with reduced levels of natural cytotoxicity receptors NKp30, NKp44 and NKp46. These NK cells are skewed towards expression of activating KIR and self‐specific inhibitory killer cell immunoglobulin‐like receptors (iKIR) with cognate HLA‐I ligands present in the host, express higher levels of CD16 and are more effective at mediating antibody‐dependent cell‐mediated cytotoxicity. Enhanced triggering through CD16 is associated with down‐regulation of the FcεR1γ adaptor protein [one immunoreceptor tyrosine‐based activation motif (ITAM) each] and potential replacement with the CD3ζ adaptor protein (three ITAMS each). Additional changes not shown include epigenetic modifications (demethylation) activating the interferon‐γ locus and down‐regulation of Syk, Ewing's sarcoma's/FLI‐1 activated transcript (EAT) and move as indicated to end of sentence promyelocytic leukaemia zinc finger protein (PLZF) transcription factor. Although NK cells expressing NKG2C and CD57 have undergone differentiation and are considered adaptive NK in the setting of HCMV infection, specificity for HCMV has not been demonstrated. Both NKG2A and NKG2C interact with HLA‐E and CMV infection can increase expression of HLA‐E, but the role that either interaction plays in shaping the NK cell repertoire in CMV infection is unknown, hence a ‘?’ punctuates the dotted lines denoting receptor–ligand interactions.
The largely NKG2Cpos phenotype of adapted NK cells in HCMV infection supports the possibility that NKG2C plays a direct role. However, in co‐culture systems without exogenous cytokines, NK cells from HCMV‐infected individuals selectively proliferate and release IFN‐γ in response to HCMV‐infected fibroblasts with little apparent role for NKG2C.90 In addition, NK cells from individuals lacking the NKG2C gene respond similarly to HCMV infection with accelerated maturation, and co‐express activating KIRs (KIR2DS1, KIR2DS2, KIR2DS4 and KIR3DS1) to a similar extent to NK cells from NKG2Cpos donors.91 Therefore, at the very least, NKG2C‐mediated interactions are not the only driving force behind HCMV's impact on the NK cell repertoire.92 An extensive study of adaptive NK cell responses in NKG2C‐bearing and NKG2Cnull individuals suggests that CD2 co‐stimulation may be a critical component in effector potency, irrespective of NKG2C expression.92 Paralleling the ‘second signal’ or DNAM‐1 co‐activation of adaptive murine NK cells, human CD2 on NK cells interacts with CD58 on target cells.93 Increased CD2 expression on in vitro‐expanded adaptive NK cells favours increased IFN‐γ and TNF‐α production, and is critical for robust antibody‐dependent responses.92, 93 However, the extent to which CD2 engagement contributes to adaptive NK cell formation in vivo, or whether other ligands for CD2 provoke the same response, remains unresolved.
Elucidation of intracellular mechanisms accompanying NK cell adaptation to HCMV infection was bolstered by identification of NK cells deficient for FcεR1γ, as emergence of adaptive NK cells in HCMV‐infected individuals is closely associated with loss of this transmembrane signalling adaptor protein.94, 95 Although CD57 and NKG2C are mostly co‐expressed on adapted NK cells, some FcεR1γ neg NK cells lack NKG2C and retain low levels of NKG2A.85, 95 The FcεR1γ adaptor protein associates with NKp30, NKp46 and/or CD16 receptors as a homodimer or heterodimer with CD3ζ to signal through immunoreceptor tyrosine‐based activation motifs.96, 97 Although CD3ζ expression remains unchanged, loss of NK cell FcεRIγ expression in response to HCMV parallels the development of adaptive NK cells with low levels of NKp30 and NKp46 and impaired cytokine and cytotoxic responses against classical targets.94, 95 In contrast to the reduced functional and phenotypic natural cytotoxicity receptor expression, loss of FcεR1γ has limited impact on CD16 expression and is associated with broadly enhanced antibody‐dependent cytokine production and cytotoxic potential.95 Further, HCMV‐dependent loss of NK cell FcεR1γ relates to markedly decreased expression of the PLZF transcription factor and DAB2.84, 85 Loss of PLZF, in concert with epigenetic hypermethylation of the FcεR1γ, Syk and EAT‐2 promoter regions to which PLZF binds, reduces FcεR1γ, and in some instances Syk and EAT‐2 transcription, producing a pattern that reliably identifies adaptive NK cells.84, 85 The differential methylation pattern observed in adaptive NK cells represents a shift from the epigenetic profile of conventional NK cells towards that of CD8pos cytotoxic T cells.84
Ancillary to the epigenetic modifications that create a population of NK cells highly specialized for antibody‐dependent cell‐mediated cytotoxicity, partial demethylation of the IFNG locus gives rise to daughter cells with heritable DNA modifications and an enhanced capacity for IFN‐γ production.98, 99 Epigenetic imprinting of both the IFNG promoter and conserved non‐coding sequence 1 (CNS‐1), located 4 kbp upstream of the human IFNG promoter, fixes NKG2Chi NK cells with strong and stable IFN‐γ responses.98, 99, 100, 101 During NK cell differentiation, hypomethylated CNS‐1 encourages binding of T‐bet, STAT4, nuclear factor‐κB and nuclear factor of activated T cells to enhance downstream IFNG transcription after stimulation through activating NK cell receptors, particularly NKG2C.99, 100, 101, 102 The epigenetic imprint that HCMV leaves on the NK cell repertoire contributes to a highly specialized collective readily able to produce IFN‐γ in response to appropriate stimuli.
In the absence of demonstrable specific interactions between activating NK cell receptors and HCMV proteins, the question remains as to why HCMV in particular has such a dramatic effect on the NK cell repertoire. Latency and reactivation are common features of all herpesviruses, but the apparently special relationship between HCMV and the immune system is also reflected in conventional adaptive immunity, wherein phenotypically and functionally altered T cells with clonotypic receptors specific for HCMV expand into an unusually large fraction of the T‐cell repertoire.103 This process of T‐cell memory inflation parallels phenotypic adaptation of the NK cell repertoire, suggesting that similar stressors impact on both responses.104 Like other herpesviruses, a long period of co‐existence with the human immune system infers an extended arms race influencing both host and virus evolution. Accordingly, a large number of HCMV genes encode proteins that function to promote evasion of T or NK cells (Table 1). Through the nature of its latent reservoir and mode of reactivation, HCMV may disseminate more broadly than other herpesviruses and adopt a wider cellular host range. Hence, the adapted lymphocyte population emerging following primary infection or persisting following viral reactivation should be selectively configured to resist HCMV immune evasion mechanisms more effectively than the naive population. Possibly a number of as yet undefined features of the NK cells that expand following HCMV infection serve to counter one or more of the multiple NK cell evasion mechanisms enacted by HCMV. Although NKG2C is an effective surrogate marker for NK cell adaptation to HCMV infection, its role in either in vivo selection or protection against HCMV has not been confirmed. In fact, there is only one reported example of a T‐cell‐deficient child where HCMV replication was reduced in vivo coincident with emergence of a predominantly NKG2Cpos NK cell subset with high CD16 expression.105 The most notable functional feature attributed to the NK cells responding to HCMV infection is an enhanced capacity for antibody‐dependent activation.95 Whether CD16, NKG2C, activating KIR, self‐specific KIR, the absence of inhibitory receptors such as NKG2A, epigenetic remodelling, or all of the above endow NK cells with a selective advantage for expansion and persistence in HCMV‐infected individuals remains to be clarified. More effective suppression of HCMV by the adapted NK cell population through antibody‐dependent or other mechanisms has also not been proven, but the consistency and robustness of the response suggests either a strong adaptive advantage to the host or that HCMV has evolved to shape the NK cell repertoire to its own advantage. Many individuals infected with HCMV do not have large CD57pos NKG2Cpos NK cell populations and low numbers of these cells are not associated with reduced containment, at least not symptomatically. Conversely, the NK cell response to HCMV infection is markedly accentuated in conditions of immune deficiency, immune suppression or ongoing immune reconstitution, such as organ transplantation, bone marrow transplantation and antiretroviral suppression of HIV infection.81, 83, 104, 105 Whatever selection criteria are generally active appear to operate with greater stringency in these settings, creating a narrow bottleneck strongly favouring eminence of NK cells phenotypically and functionally adapted in response to CMV infection (Fig. 1). Either the frequency or magnitude of viral reactivation in these instances may be an important factor underlying the more pronounced NK cell adaptive responses that occur. The impact of viral variation or superinfection has not been addressed.
Table 1.
Human cytomegalovirus‐encoded immune evasion genes
| HCMV immune evasion gene | Description of interaction | References |
|---|---|---|
| UL16 | Blocks surface expression of NKG2D ligands MICB and ULBP1/2 | 106, 107, 108 |
| UL18 |
(i) Class I HLA homologue (ii) Binds NK cell receptor CD85j (LIR‐1) (iii) Weakly binds NK cell activating receptor NKG2C |
109, 110, 111 |
| UL40 |
(i) Class I HLA homologue (ii) Leader sequence peptide (VMAPRTLIL) binds and stabilizes both UL18 (see above) and HLA‐E (NKG2A ligand) |
88, 112, 113, 114 |
| UL83 (pp65) | Interacts with NKp30 to dissociate CD3ζ | 115 |
| UL111a | hIL‐10 homologue with immunoregulatory properties | 116, 117, 118, 119 |
| miR‐UL112‐1 | Down‐modulates NKG2D ligand, MICB | 120 |
| UL135 | Remodels actin cytoskeleton to disrupt immune synapse formation | 121 |
| UL141 |
(i) TRAIL ligand (ii) Sequesters CD155 (PVR) in the ER (iii) Decreases CD112 expression in concert with US2 to reduce NK cell co‐stimulation through DNAM‐1 |
122, 123, 124, 125 |
| UL142 | Class I HLA homologue that retains NKG2D ligands, MICA and ULBP3, in Golgi | 126, 127, 128 |
| UL146 | Chemoattractant virokine (vCXCL1) that binds NK cell and neutrophil IL‐8R | 129, 130 |
| gp34 & gp68 | HCMV‐encoded FcγR that impairs CD16‐mediated NK cell activation | 131, 132, 133, 134 |
| US2 | Prevents HLA‐A2 and B27 expression by translocating HLA complexes from ER lumen to cytosol | 135, 136, 137, 138 |
| US3 | Blocks tapasin and retains class I HLA in ER | 137, 138, 139 |
| miR‐US4‐1 | Inhibits ERAP1, prevents peptide loading into class I HLA complexes | 140 |
| US6 | Inhibits TAP, prevents peptide loading into class I HLA complexes | 138, 141, 142 |
| US9 | Targets MICA*008 for proteasomal degradation | 143, 144 |
| US10 | Induces degradation of HLA‐G; classical HLA complexes resist degradation | 145, 146 |
| US11 | Degrades HLA‐A2 by transporting to cytoplasmic proteases; HLA‐C and HLA‐G are resistant | 135, 138, 147, 148 |
| US18 & US20 |
(i) Targets MICA for lysosomal degradation (ii) Down‐modulates B7‐H6 expression |
149, 150, 151 |
Abbreviations: ER, endoplasmic reticulum; HCMV, human cytomegalovirus; HLA, human histocompatibility‐linked antigen; NK, natural killer.
Concluding remarks
Ample evidence has accumulated that NK cells proliferate, mature and differentiate in response to various environmental cues. These responses transform the naive NK cell repertoire, which already varies with host genetic background, into a mature repertoire adapted in terms of activating and self‐specific KIR expression, C‐type lectin‐like receptor expression, CD16 expression, maturation markers, intracellular signalling molecules and transcription factors. Although this constitutes a memory population of immune effector cells, only in the case of MCMV is there definitive evidence of a mechanism for selective expansion of NK cells with specificity for a particular antigen. A limited number of examples in mice and macaques hint at the possibility of antigen specificity involving an NK cell activating receptor repertoire beyond that currently known, but evidence for the nature of a receptor repertoire specially created for NK cell adaptive responses is lacking. Hence, some ambivalence still exists as to the perceived nature of NK cell memory. In the clearest examples, there is monochromatic recognition of select foreign proteins and non‐specific cytokines that enhance secondary effector function. Still to be fully explained is the NK cell antigen specificity similar to that bestowed by clonotypic receptors reported in mice and peptide or other modification of NK cell receptor ligands that tilt the balance towards activation of NK cells with particular receptor constellations. In the latter case, memory for any particular pathogen would reflect collective recognition of multiple ligands with signal integration triggering a select set of NK cells to proliferate and differentiate in a manner that lowered the threshold for activation upon secondary exposure to the same set of environmental cues. This possibility requires only subtle extension of what is already accepted about integration of positive and negative signals to control NK cell activation. A more speculative possibility would be collective memory of specific environmental cues distributed among a network of NK cells, rather than attributable to single antigen‐specific cells. In this scenario, NK cells would have evolved to provide context‐dependent help through intercellular communications in a manner similar to the establishment of memory with neuronal networks. Much remains to be explained regarding the unanticipated behaviour of NK cells revealed over the last two decades. The massive implications for basic and translational science will continue to drive research on NK cell memory that promises more surprises.
Disclosure
The authors declare no conflict of interest.
Acknowledgements
The manuscript was written and edited by KH, MG and EC. Figures created by KH and EC were adapted for publication by Alison Schrooer. Research by the authors related to material in this review was supported by the Canadian Institutes for Health Research (CIHR) bridge operating grant (HBF‐131552), Memorial University Faculty of Medicine Dean's Innovation award and Canadian Foundation for AIDS Research innovation grant awarded to MG. KH is a trainee in the Cancer Research Training Programme of the Beatrice Hunter Cancer Research Institute, with funds provided by the Terry Fox Research Institute with matching funds from the Memorial University School of Graduate Studies. EC is supported by CANFAR and a Memorial University Faculty of Medicine Dean's fellowship.
References
- 1. Greenberg AH, Hudson L, Shen L, Roitt IM. Antibody‐dependent cell‐mediated cytotoxicity due to a “null” lymphoid cell. Nat New Biol 1973; 242:111–3. [DOI] [PubMed] [Google Scholar]
- 2. Herberman RB, Nunn ME, Holden HT, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors. II. Characterization of effector cells. Int J Cancer 1975; 16:230–9. [DOI] [PubMed] [Google Scholar]
- 3. Kiessling R, Klein E, Wigzell H. “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol 1975; 5:112–7. [DOI] [PubMed] [Google Scholar]
- 4. Sendo F, Aoki T, Boyse EA, Buafo CK. Natural occurrence of lymphocytes showing cytotoxic activity to BALB/c radiation‐induced leukemia RL male 1 cells. J Natl Cancer Inst 1975; 55:603–9. [DOI] [PubMed] [Google Scholar]
- 5. Zarling JM, Nowinski RC, Bach FH. Lysis of leukemia cells by spleen cells of normal mice. Proc Natl Acad Sci USA 1975; 72:2780–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Horowitz A, Strauss‐Albee DM, Leipold M, Kubo J, Nemat‐Gorgani N, Dogan OC et al Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med 2013; 5:208ra145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown MG, Dokun AO, Heusel JW, Smith HR, Beckman DL, Blattenberger EA et al Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science 2001; 292:934–7. [DOI] [PubMed] [Google Scholar]
- 8. Smith HR, Heusel JW, Mehta IK, Kim S, Dorner BG, Naidenko OV et al Recognition of a virus‐encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci USA 2002; 99:8826–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 2002; 296:1323–6. [DOI] [PubMed] [Google Scholar]
- 10. Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar‐Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol 1999; 17:189–220. [DOI] [PubMed] [Google Scholar]
- 11. Welsh RM, O'Donnell CL, Shultz LD. Antiviral activity of NK 1.1+ natural killer cells in C57BL/6 scid mice infected with murine cytomegalovirus. Nat Immun 1994; 13:239–45. [PubMed] [Google Scholar]
- 12. Wang LL, Chu DT, Dokun AO, Yokoyama WM. Inducible expression of the gp49B inhibitory receptor on NK cells. J Immunol 2000; 164:5215–20. [DOI] [PubMed] [Google Scholar]
- 13. Orange JS, Biron CA. Characterization of early IL‐12, IFN‐αβ, and TNF effects on antiviral state and NK cell responses during murine cytomegalovirus infection. J Immunol 1996; 156:4746–56. [PubMed] [Google Scholar]
- 14. Nguyen KB, Salazar‐Mather TP, Dalod MY, Van Deusen JB, Wei XQ, Liew FY et al Coordinated and distinct roles for IFN‐αβ, IL‐12, and IL‐15 regulation of NK cell responses to viral infection. J Immunol 2002; 169:4279–87. [DOI] [PubMed] [Google Scholar]
- 15. Madera S, Sun JC. Cutting edge: stage‐specific requirement of IL‐18 for antiviral NK cell expansion. J Immunol 2015; 194:1408–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Au‐Yeung N, Mandhana R, Horvath CM. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK‐STAT pathway. JAK‐STAT 2013; 2:e23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miyagi T, Gil MP, Wang X, Louten J, Chu WM, Biron CA. High basal STAT4 balanced by STAT1 induction to control type 1 interferon effects in natural killer cells. J Exp Med 2007; 204:2383–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liang S, Wei H, Sun R, Tian Z. IFNα regulates NK cell cytotoxicity through STAT1 pathway. Cytokine 2003; 23:190–9. [DOI] [PubMed] [Google Scholar]
- 19. Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA et al T‐bet regulates the terminal maturation and homeostasis of NK and Vα14i NKT cells. Immunity 2004; 20:477–94. [DOI] [PubMed] [Google Scholar]
- 20. Wang KS, Frank DA, Ritz J. Interleukin‐2 enhances the response of natural killer cells to interleukin‐12 through up‐regulation of the interleukin‐12 receptor and STAT4. Blood 2000; 95:3183–90. [PubMed] [Google Scholar]
- 21. Madera S, Rapp M, Firth MA, Beilke JN, Lanier LL, Sun JC. Type I IFN promotes NK cell expansion during viral infection by protecting NK cells against fratricide. J Exp Med 2016; 213:225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun JC, Madera S, Bezman NA, Beilke JN, Kaplan MH, Lanier LL. Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J Exp Med 2012; 209:947–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cooper MA, Elliott JM, Keyel PA, Yang L, Carrero JA, Yokoyama WM. Cytokine‐induced memory‐like natural killer cells. Proc Natl Acad Sci USA 2009; 106:1915–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keppel MP, Yang L, Cooper MA. Murine NK cell intrinsic cytokine‐induced memory‐like responses are maintained following homeostatic proliferation. J Immunol 2013; 190:4754–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beaulieu AM, Zawislak CL, Nakayama T, Sun JC. The transcription factor Zbtb32 controls the proliferative burst of virus‐specific natural killer cells responding to infection. Nat Immunol 2014; 15:546–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu D, Holko M, Sadler AJ, Scott B, Higashiyama S, Berkofsky‐Fessler W et al Promyelocytic leukemia zinc finger protein regulates interferon‐mediated innate immunity. Immunity 2009; 30:802–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dokun AO, Kim S, Smith HR, Kang HS, Chu DT, Yokoyama WM. Specific and nonspecific NK cell activation during virus infection. Nat Immunol 2001; 2:951–6. [DOI] [PubMed] [Google Scholar]
- 28. Tripathy SK, Smith HR, Holroyd EA, Pingel JT, Yokoyama WM. Expression of m157, a murine cytomegalovirus‐encoded putative major histocompatibility class I (MHC‐I)‐like protein, is independent of viral regulation of host MHC‐I. J Virol 2006; 80:545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bubic I, Wagner M, Krmpotic A, Saulig T, Kim S, Yokoyama WM et al Gain of virulence caused by loss of a gene in murine cytomegalovirus. J Virol 2004; 78:7536–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature 2009; 457:557–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kielczewska A, Pyzik M, Sun T, Krmpotic A, Lodoen MB, Munks MW et al Ly49P recognition of cytomegalovirus‐infected cells expressing H2‐Dk and CMV‐encoded m04 correlates with the NK cell antiviral response. J Exp Med 2009; 206:515–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Voigt V, Forbes CA, Tonkin JN, Degli‐Esposti MA, Smith HR, Yokoyama WM et al Murine cytomegalovirus m157 mutation and variation leads to immune evasion of natural killer cells. Proc Natl Acad Sci USA 2003; 100:13483–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kielczewska A, Kim HS, Lanier LL, Dimasi N, Vidal SM. Critical residues at the Ly49 natural killer receptor's homodimer interface determine functional recognition of m157, a mouse cytomegalovirus MHC class I‐like protein. J Immunol 2007; 178:369–77. [DOI] [PubMed] [Google Scholar]
- 34. Corbett AJ, Coudert JD, Forbes CA, Scalzo AA. Functional consequences of natural sequence variation of murine cytomegalovirus m157 for Ly49 receptor specificity and NK cell activation. J Immunol 2011; 186:1713–22. [DOI] [PubMed] [Google Scholar]
- 35. Orr MT, Sun JC, Hesslein DG, Arase H, Phillips JH, Takai T et al Ly49H signaling through DAP10 is essential for optimal natural killer cell responses to mouse cytomegalovirus infection. J Exp Med 2009; 206:807–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pende D, Castriconi R, Romagnani P, Spaggiari GM, Marcenaro S, Dondero A et al Expression of the DNAM‐1 ligands, Nectin‐2 (CD112) and poliovirus receptor (CD155), on dendritic cells: relevance for natural killer‐dendritic cell interaction. Blood 2006; 107:2030–6. [DOI] [PubMed] [Google Scholar]
- 37. Nabekura T, Kanaya M, Shibuya A, Fu G, Gascoigne NR, Lanier LL. Costimulatory molecule DNAM‐1 is essential for optimal differentiation of memory natural killer cells during mouse cytomegalovirus infection. Immunity 2014; 40:225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robbins SH, Tessmer MS, Mikayama T, Brossay L. Expansion and contraction of the NK cell compartment in response to murine cytomegalovirus infection. J Immunol 2004; 173:259–66. [DOI] [PubMed] [Google Scholar]
- 39. Min‐Oo G, Bezman NA, Madera S, Sun JC, Lanier LL. Proapoptotic Bim regulates antigen‐specific NK cell contraction and the generation of the memory NK cell pool after cytomegalovirus infection. J Exp Med 2014; 211:1289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O'Sullivan TE, Johnson LR, Kang HH, Sun JC. BNIP3‐ and BNIP3L‐mediated mitophagy promotes the generation of natural killer cell memory. Immunity 2015; 43:331–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. O'Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell‐ and B cell‐independent adaptive immunity mediated by natural killer cells. Nat Immunol 2006; 7:507–16. [DOI] [PubMed] [Google Scholar]
- 42. Paust S, Gill HS, Wang BZ, Flynn MP, Moseman EA, Senman B et al Critical role for the chemokine receptor CXCR6 in NK cell‐mediated antigen‐specific memory of haptens and viruses. Nat Immunol 2010; 11:1127–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Majewska‐Szczepanik M, Paust S, von Andrian UH, Askenase PW, Szczepanik M. Natural killer cell‐mediated contact sensitivity develops rapidly and depends on interferon‐α, interferon‐γ and interleukin‐12. Immunology 2013; 140:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Paust S, von Andrian UH. Natural killer cell memory. Nat Immunol 2011; 12:500–8. [DOI] [PubMed] [Google Scholar]
- 45. Gillard GO, Bivas‐Benita M, Hovav AH, Grandpre LE, Panas MW, Seaman MS et al Thy1+ NK [corrected] cells from vaccinia virus‐primed mice confer protection against vaccinia virus challenge in the absence of adaptive lymphocytes. PLoS Pathog 2011; 7:e1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Venkatasubramanian S, Cheekatla S, Paidipally P, Tripathi D, Welch E, Tvinnereim AR et al IL‐21‐dependent expansion of memory‐like NK cells enhances protective immune responses against Mycobacterium tuberculosis . Mucosal Immunol 2017; 10:1031–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li T, Wang J, Wang Y, Chen Y, Wei H, Sun R et al Respiratory influenza virus infection induces memory‐like liver NK cells in mice. J Immunol 2017; 198:1242–52. [DOI] [PubMed] [Google Scholar]
- 48. Paust S, Blish CA, Reeves RK. Redefining memory: building the case for adaptive NK cells. J Virol 2017; 91:pii: e00169‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lunemann A, Vanoaica LD, Azzi T, Nadal D, Munz C. A distinct subpopulation of human NK cells restricts B cell transformation by EBV. J Immunol 2013; 191:4989–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Suliman S, Geldenhuys H, Johnson JL, Hughes JE, Smit E, Murphy M et al Bacillus Calmette–Guérin (BCG) revaccination of adults with latent Mycobacterium tuberculosis infection induces long‐lived BCG‐reactive NK cell responses. J Immunol 2016; 197:1100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ma LL, Wang CL, Neely GG, Epelman S, Krensky AM, Mody CH. NK cells use perforin rather than granulysin for anticryptococcal activity. J Immunol 2004; 173:3357–65. [DOI] [PubMed] [Google Scholar]
- 52. Alter G, Teigen N, Davis BT, Addo MM, Suscovich TJ, Waring MT et al Sequential deregulation of NK cell subset distribution and function starting in acute HIV‐1 infection. Blood 2005; 106:3366–9. [DOI] [PubMed] [Google Scholar]
- 53. Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A et al Differential natural killer cell‐mediated inhibition of HIV‐1 replication based on distinct KIR/HLA subtypes. J Exp Med 2007; 204:3027–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Alter G, Rihn S, Walter K, Nolting A, Martin M, Rosenberg ES et al HLA class I subtype‐dependent expansion of KIR3DS1+ and KIR3DL1+ NK cells during acute human immunodeficiency virus type 1 infection. J Virol 2009; 83:6798–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Song R, Lisovsky I, Lebouche B, Routy JP, Bruneau J, Bernard NF. HIV protective KIR3DL1/S1‐HLA‐B genotypes influence NK cell‐mediated inhibition of HIV replication in autologous CD4 targets. PLoS Pathog 2014; 10:e1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ et al Epistatic interaction between KIR3DS1 and HLA‐B delays the progression to AIDS. Nat Genet 2002; 31:429–34. [DOI] [PubMed] [Google Scholar]
- 57. Boulet S, Sharafi S, Simic N, Bruneau J, Routy JP, Tsoukas CM et al Increased proportion of KIR3DS1 homozygotes in HIV‐exposed uninfected individuals. AIDS 2008; 22:595–9. [DOI] [PubMed] [Google Scholar]
- 58. Long BR, Ndhlovu LC, Oksenberg JR, Lanier LL, Hecht FM, Nixon DF et al Conferral of enhanced natural killer cell function by KIR3DS1 in early human immunodeficiency virus type 1 infection. J Virol 2008; 82:4785–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Boulet S, Song R, Kamya P, Bruneau J, Shoukry NH, Tsoukas CM et al HIV protective KIR3DL1 and HLA‐B genotypes influence NK cell function following stimulation with HLA‐devoid cells. J Immunol 2010; 184:2057–64. [DOI] [PubMed] [Google Scholar]
- 60. Jiang Y, Chen O, Cui C, Zhao B, Han X, Zhang Z et al KIR3DS1/L1 and HLA‐Bw4‐80I are associated with HIV disease progression among HIV typical progressors and long‐term nonprogressors. BMC Infect Dis 2013; 13:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vales‐Gomez M, Reyburn HT, Erskine RA, Lopez‐Botet M, Strominger JL. Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2‐A and the activating receptor CD94/NKG2‐C to HLA‐E. EMBO J 1999; 18:4250–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wada H, Matsumoto N, Maenaka K, Suzuki K, Yamamoto K. The inhibitory NK cell receptor CD94/NKG2A and the activating receptor CD94/NKG2C bind the top of HLA‐E through mostly shared but partly distinct sets of HLA‐E residues. Eur J Immunol 2004; 34:81–90. [DOI] [PubMed] [Google Scholar]
- 63. Kraemer T, Celik AA, Huyton T, Kunze‐Schumacher H, Blasczyk R, Bade‐Doding C. HLA‐E: Presentation of a broader peptide repertoire impacts the cellular immune response‐implications on HSCT outcome. Stem Cells Int 2015; 2015:346714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Braud VM, Allan DS, O'Callaghan CA, Soderstrom K, D'Andrea A, Ogg GS et al HLA‐E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998; 391:795–9. [DOI] [PubMed] [Google Scholar]
- 65. Fadda L, Borhis G, Ahmed P, Cheent K, Pageon SV, Cazaly A et al Peptide antagonism as a mechanism for NK cell activation. Proc Natl Acad Sci USA 2010; 107:10160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fadda L, Korner C, Kumar S, van Teijlingen NH, Piechocka‐Trocha A, Carrington M et al HLA‐Cw*0102‐restricted HIV‐1 p24 epitope variants can modulate the binding of the inhibitory KIR2DL2 receptor and primary NK cell function. PLoS Pathog 2012; 8:e1002805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fadda L, O'Connor GM, Kumar S, Piechocka‐Trocha A, Gardiner CM, Carrington M et al Common HIV‐1 peptide variants mediate differential binding of KIR3DL1 to HLA‐Bw4 molecules. J Virol 2011; 85:5970–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Garcia‐Beltran WF, Holzemer A, Martrus G, Chung AW, Pacheco Y, Simoneau CR et al Open conformers of HLA‐F are high‐affinity ligands of the activating NK‐cell receptor KIR3DS1. Nat Immunol 2016; 17:1067–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Burian A, Wang KL, Finton KA, Lee N, Ishitani A, Strong RK et al HLA‐F and MHC‐I open conformers bind natural killer Cell Ig‐like receptor KIR3DS1. PLoS ONE 2016; 11:e0163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM et al HIV‐1 adaptation to NK‐cell‐mediated immune pressure. Nature 2011; 476:96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Reeves RK, Li H, Jost S, Blass E, Li H, Schafer JL et al Antigen‐specific NK cell memory in rhesus macaques. Nat Immunol 2015; 16:927–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tong L, Assenmacher M, Zanker KS, Jahn P. Virus‐specific peptide dependent NK cell cytotoxicity. Inflamm Allergy Drug Targets 2014; 13:128–33. [DOI] [PubMed] [Google Scholar]
- 73. Romee R, Schneider SE, Leong JW, Chase JM, Keppel CR, Sullivan RP et al Cytokine activation induces human memory‐like NK cells. Blood 2012; 120:4751–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Miletic A, Krmpotic A, Jonjic S. The evolutionary arms race between NK cells and viruses: who gets the short end of the stick? Eur J Immunol 2013; 43:867–77. [DOI] [PubMed] [Google Scholar]
- 75. Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med 1989; 320:1731–5. [DOI] [PubMed] [Google Scholar]
- 76. Guma M, Angulo A, Vilches C, Gomez‐Lozano N, Malats N, Lopez‐Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 2004; 104:3664–71. [DOI] [PubMed] [Google Scholar]
- 77. Guma M, Cabrera C, Erkizia I, Bofill M, Clotet B, Ruiz L et al Human cytomegalovirus infection is associated with increased proportions of NK cells that express the CD94/NKG2C receptor in aviremic HIV‐1‐positive patients. J Infect Dis 2006; 194:38–41. [DOI] [PubMed] [Google Scholar]
- 78. Monsivais‐Urenda A, Noyola‐Cherpitel D, Hernandez‐Salinas A, Garcia‐Sepulveda C, Romo N, Baranda L et al Influence of human cytomegalovirus infection on the NK cell receptor repertoire in children. Eur J Immunol 2010; 40:1418–27. [DOI] [PubMed] [Google Scholar]
- 79. Muntasell A, Vilches C, Angulo A, Lopez‐Botet M. Adaptive reconfiguration of the human NK‐cell compartment in response to cytomegalovirus: a different perspective of the host‐pathogen interaction. Eur J Immunol 2013; 43:1133–41. [DOI] [PubMed] [Google Scholar]
- 80. Noyola DE, Fortuny C, Muntasell A, Noguera‐Julian A, Munoz‐Almagro C, Alarcon A et al Influence of congenital human cytomegalovirus infection and the NKG2C genotype on NK‐cell subset distribution in children. Eur J Immunol 2012; 42:3256–66. [DOI] [PubMed] [Google Scholar]
- 81. Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X et al Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood 2012; 119:2665–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lopez‐Verges S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA et al Expansion of a unique CD57+ NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci USA 2011; 108:14725–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Della Chiesa M, Falco M, Podesta M, Locatelli F, Moretta L, Frassoni F et al Phenotypic and functional heterogeneity of human NK cells developing after umbilical cord blood transplantation: a role for human cytomegalovirus? Blood 2012; 119:399–410. [DOI] [PubMed] [Google Scholar]
- 84. Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD et al Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 2015; 42:443–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M et al Epigenetic modification and antibody‐dependent expansion of memory‐like NK cells in human cytomegalovirus‐infected individuals. Immunity 2015; 42:431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Rolle A, Pollmann J, Ewen EM, Le VT, Halenius A, Hengel H et al IL‐12‐producing monocytes and HLA‐E control HCMV‐driven NKG2C+ NK cell expansion. J Clin Invest 2014; 124:5305–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kaiser BK, Pizarro JC, Kerns J, Strong RK. Structural basis for NKG2A/CD94 recognition of HLA‐E. Proc Natl Acad Sci USA 2008; 105:6696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Prod'homme V, Tomasec P, Cunningham C, Lemberg MK, Stanton RJ, McSharry BP et al Human cytomegalovirus UL40 signal peptide regulates cell surface expression of the NK cell ligands HLA‐E and gpUL18. J Immunol 2012; 188:2794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Falk CS, Mach M, Schendel DJ, Weiss EH, Hilgert I, Hahn G. NK cell activity during human cytomegalovirus infection is dominated by US2‐11‐mediated HLA class I down‐regulation. J Immunol 2002; 169:3257–66. [DOI] [PubMed] [Google Scholar]
- 90. Newhook N, Fudge N, Grant M. NK cells generate memory‐type responses to human cytomegalovirus‐infected fibroblasts. Eur J Immunol 2017; 47:1032–9. [DOI] [PubMed] [Google Scholar]
- 91. Beziat V, Liu LL, Malmberg JA, Ivarsson MA, Sohlberg E, Bjorklund AT et al NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 2013; 121:2678–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Liu LL, Landskron J, Ask EH, Enqvist M, Sohlberg E, Traherne JA et al Critical role of CD2 co‐stimulation in adaptive natural killer cell responses revealed in NKG2C‐deficient humans. Cell Rep 2016; 15:1088–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rolle A, Halenius A, Ewen EM, Cerwenka A, Hengel H, Momburg F. CD2‐CD58 interactions are pivotal for the activation and function of adaptive natural killer cells in human cytomegalovirus infection. Eur J Immunol 2016; 46:2420–5. [DOI] [PubMed] [Google Scholar]
- 94. Hwang I, Zhang T, Scott JM, Kim AR, Lee T, Kakarla T et al Identification of human NK cells that are deficient for signaling adaptor FcRγ and specialized for antibody‐dependent immune functions. Int Immunol 2012; 24:793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zhang T, Scott JM, Hwang I, Kim S. Cutting edge: antibody‐dependent memory‐like NK cells distinguished by FcRγ deficiency. J Immunol 2013; 190:1402–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol 2008; 9:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Watzl C, Long EO. Signal transduction during activation and inhibition of natural killer cells. Curr Protoc Immunol 2010; Chapter 11:Unit 11 9B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Luetke‐Eversloh M, Hammer Q, Durek P, Nordstrom K, Gasparoni G, Pink M et al Human cytomegalovirus drives epigenetic imprinting of the IFNG locus in NKG2Chi natural killer cells. PLoS Pathog 2014; 10:e1004441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Luetke‐Eversloh M, Cicek BB, Siracusa F, Thom JT, Hamann A, Frischbutter S et al NK cells gain higher IFN‐γ competence during terminal differentiation. Eur J Immunol 2014; 44:2074–84. [DOI] [PubMed] [Google Scholar]
- 100. Shnyreva M, Weaver WM, Blanchette M, Taylor SL, Tompa M, Fitzpatrick DR et al Evolutionarily conserved sequence elements that positively regulate IFN‐γ expression in T cells. Proc Natl Acad Sci USA 2004; 101:12622–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Dong J, Chang HD, Ivascu C, Qian Y, Rezai S, Okhrimenko A et al Loss of methylation at the IFNG promoter and CNS‐1 is associated with the development of functional IFN‐γ memory in human CD4+ T lymphocytes. Eur J Immunol 2013; 43:793–804. [DOI] [PubMed] [Google Scholar]
- 102. Lee DU, Avni O, Chen L, Rao A. A distal enhancer in the interferon‐γ (IFN‐γ) locus revealed by genome sequence comparison. J Biol Chem 2004; 279:4802–10. [DOI] [PubMed] [Google Scholar]
- 103. Wikby A, Johansson B, Olsson J, Lofgren S, Nilsson BO, Ferguson F. Expansions of peripheral blood CD8 T‐lymphocyte subpopulations and an association with cytomegalovirus seropositivity in the elderly: the Swedish NONA immune study. Exp Gerontol 2002; 37:445–53. [DOI] [PubMed] [Google Scholar]
- 104. Heath J, Newhook N, Comeau E, Gallant M, Fudge N, Grant M. NKG2C+CD57+ natural killer cell expansion parallels cytomegalovirus‐specific CD8+ T cell evolution towards senescence. J Immunol Res 2016; 2016:7470124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kuijpers TW, Baars PA, Dantin C, van den Burg M, van Lier RA, Roosnek E. Human NK cells can control CMV infection in the absence of T cells. Blood 2008; 112:914–5. [DOI] [PubMed] [Google Scholar]
- 106. Vales‐Gomez M, Winterhalter A, Roda‐Navarro P, Zimmermann A, Boyle L, Hengel H et al The human cytomegalovirus glycoprotein UL16 traffics through the plasma membrane and the nuclear envelope. Cell Microbiol 2006; 8:581–90. [DOI] [PubMed] [Google Scholar]
- 107. Vales‐Gomez M, Browne H, Reyburn HT. Expression of the UL16 glycoprotein of human cytomegalovirus protects the virus‐infected cell from attack by natural killer cells. BMC Immunol 2003; 4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Dunn C, Chalupny NJ, Sutherland CL, Dosch S, Sivakumar PV, Johnson DC et al Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J Exp Med 2003; 197:1427–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Leong CC, Chapman TL, Bjorkman PJ, Formankova D, Mocarski ES, Phillips JH et al Modulation of natural killer cell cytotoxicity in human cytomegalovirus infection: the role of endogenous class I major histocompatibility complex and a viral class I homolog. J Exp Med 1998; 187:1681–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Kim Y, Park B, Cho S, Shin J, Cho K, Jun Y et al Human cytomegalovirus UL18 utilizes US6 for evading the NK and T‐cell responses. PLoS Pathog 2008; 4:e1000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Chen KC, Stanton RJ, Banat JJ, Wills MR. Leukocyte immunoglobulin‐like receptor 1‐expressing human natural killer cell subsets differentially recognize isolates of human cytomegalovirus through the viral major histocompatibility complex class I homolog UL18. J Virol 2016; 90:3123–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Heatley SL, Pietra G, Lin J, Widjaja JM, Harpur CM, Lester S et al Polymorphism in human cytomegalovirus UL40 impacts on recognition of human leukocyte antigen‐E (HLA‐E) by natural killer cells. J Biol Chem 2013; 288:8679–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Tomasec P, Braud VM, Rickards C, Powell MB, McSharry BP, Gadola S et al Surface expression of HLA‐E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 2000; 287:1031. [DOI] [PubMed] [Google Scholar]
- 114. Ulbrecht M, Martinozzi S, Grzeschik M, Hengel H, Ellwart JW, Pla M et al Cutting edge: the human cytomegalovirus UL40 gene product contains a ligand for HLA‐E and prevents NK cell‐mediated lysis. J Immunol 2000; 164:5019–22. [DOI] [PubMed] [Google Scholar]
- 115. Arnon TI, Achdout H, Levi O, Markel G, Saleh N, Katz G et al Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat Immunol 2005; 6:515–23. [DOI] [PubMed] [Google Scholar]
- 116. Spencer JV, Cadaoas J, Castillo PR, Saini V, Slobedman B. Stimulation of B lymphocytes by cmvIL‐10 but not LAcmvIL‐10. Virology 2008; 374:164–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Jenkins C, Abendroth A, Slobedman B. A novel viral transcript with homology to human interleukin‐10 is expressed during latent human cytomegalovirus infection. J Virol 2004; 78:1440–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Jenkins C, Garcia W, Godwin MJ, Spencer JV, Stern JL, Abendroth A et al Immunomodulatory properties of a viral homolog of human interleukin‐10 expressed by human cytomegalovirus during the latent phase of infection. J Virol 2008; 82:3736–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. Human cytomegalovirus harbors its own unique IL‐10 homolog (cmvIL‐10). Proc Natl Acad Sci USA 2000; 97:1695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Nachmani D, Lankry D, Wolf DG, Mandelboim O. The human cytomegalovirus microRNA miR‐UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat Immunol 2010; 11:806–13. [DOI] [PubMed] [Google Scholar]
- 121. Stanton RJ, Prod'homme V, Purbhoo MA, Moore M, Aicheler RJ, Heinzmann M et al HCMV pUL135 remodels the actin cytoskeleton to impair immune recognition of infected cells. Cell Host Microbe 2014; 16:201–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Smith W, Tomasec P, Aicheler R, Loewendorf A, Nemcovicova I, Wang EC et al Human cytomegalovirus glycoprotein UL141 targets the TRAIL death receptors to thwart host innate antiviral defenses. Cell Host Microbe 2013; 13:324–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Prod'homme V, Sugrue DM, Stanton RJ, Nomoto A, Davies J, Rickards CR et al Human cytomegalovirus UL141 promotes efficient downregulation of the natural killer cell activating ligand CD112. J Gen Virol 2010; 91:2034–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Tomasec P, Wang EC, Davison AJ, Vojtesek B, Armstrong M, Griffin C et al Downregulation of natural killer cell‐activating ligand CD155 by human cytomegalovirus UL141. Nat Immunol 2005; 6:181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Hsu JL, van den Boomen DJ, Tomasec P, Weekes MP, Antrobus R, Stanton RJ et al Plasma membrane profiling defines an expanded class of cell surface proteins selectively targeted for degradation by HCMV US2 in cooperation with UL141. PLoS Pathog 2015; 11:e1004811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ashiru O, Bennett NJ, Boyle LH, Thomas M, Trowsdale J, Wills MR. NKG2D ligand MICA is retained in the cis‐Golgi apparatus by human cytomegalovirus protein UL142. J Virol 2009; 83:12345–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Chalupny NJ, Rein‐Weston A, Dosch S, Cosman D. Down‐regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem Biophys Res Commun 2006; 346:175–81. [DOI] [PubMed] [Google Scholar]
- 128. Wills MR, Ashiru O, Reeves MB, Okecha G, Trowsdale J, Tomasec P et al Human cytomegalovirus encodes an MHC class I‐like molecule (UL142) that functions to inhibit NK cell lysis. J Immunol 2005; 175:7457–65. [DOI] [PubMed] [Google Scholar]
- 129. Yamin R, Lecker LSM, Weisblum Y, Vitenshtein A, Le‐Trilling VTK, Wolf DG et al HCMV vCXCL1 binds several chemokine receptors and preferentially attracts neutrophils over NK cells by interacting with CXCR2. Cell Rep 2016; 15:1542–53. [DOI] [PubMed] [Google Scholar]
- 130. Luttichau HR. The cytomegalovirus UL146 gene product vCXCL1 targets both CXCR1 and CXCR2 as an agonist. J Biol Chem 2010; 285:9137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Corrales‐Aguilar E, Trilling M, Hunold K, Fiedler M, Le VT, Reinhard H et al Human cytomegalovirus Fcγ binding proteins gp34 and gp68 antagonize Fcγ receptors I. II and III. PLoS Pathog 2014; 10:e1004131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Sprague ER, Reinhard H, Cheung EJ, Farley AH, Trujillo RD, Hengel H et al The human cytomegalovirus Fc receptor gp68 binds the Fc CH2‐CH3 interface of immunoglobulin G. J Virol 2008; 82:3490–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Atalay R, Zimmermann A, Wagner M, Borst E, Benz C, Messerle M et al Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcγ receptor homologs. J Virol 2002; 76:8596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Ndjamen B, Joshi DS, Fraser SE, Bjorkman PJ. Characterization of antibody bipolar bridging mediated by the human cytomegalovirus Fc receptor gp68. J Virol 2016; 90:3262–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Schust DJ, Tortorella D, Seebach J, Phan C, Ploegh HL. Trophoblast class I major histocompatibility complex (MHC) products are resistant to rapid degradation imposed by the human cytomegalovirus (HCMV) gene products US2 and US11. J Exp Med 1998; 188:497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Hegde NR, Johnson DC. Human cytomegalovirus US2 causes similar effects on both major histocompatibility complex class I and II proteins in epithelial and glial cells. J Virol 2003; 77:9287–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Jones TR, Sun L. Human cytomegalovirus US2 destabilizes major histocompatibility complex class I heavy chains. J Virol 1997; 71:2970–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Park B, Oh H, Lee S, Song Y, Shin J, Sung YC et al The MHC class I homolog of human cytomegalovirus is resistant to down‐regulation mediated by the unique short region protein (US)2, US3, US6, and US11 gene products. J Immunol 2002; 168:3464–9. [DOI] [PubMed] [Google Scholar]
- 139. Noriega VM, Hesse J, Gardner TJ, Besold K, Plachter B, Tortorella D. Human cytomegalovirus US3 modulates destruction of MHC class I molecules. Mol Immunol 2012; 51:245–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Kim S, Lee S, Shin J, Kim Y, Evnouchidou I, Kim D et al Human cytomegalovirus microRNA miR‐US4‐1 inhibits CD8+ T cell responses by targeting the aminopeptidase ERAP1. Nat Immunol 2011; 12:984–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Lehner PJ, Karttunen JT, Wilkinson GW, Cresswell P. The human cytomegalovirus US6 glycoprotein inhibits transporter associated with antigen processing‐dependent peptide translocation. Proc Natl Acad Sci USA 1997; 94:6904–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Hewitt EW, Gupta SS, Lehner PJ. The human cytomegalovirus gene product US6 inhibits ATP binding by TAP. EMBO J 2001; 20:387–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Ashiru O, Boutet P, Fernandez‐Messina L, Aguera‐Gonzalez S, Skepper JN, Vales‐Gomez M et al Natural killer cell cytotoxicity is suppressed by exposure to the human NKG2D ligand MICA*008 that is shed by tumor cells in exosomes. Cancer Res 2010; 70:481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Seidel E, Le VT, Bar‐On Y, Tsukerman P, Enk J, Yamin R et al Dynamic co‐evolution of host and pathogen: HCMV downregulates the prevalent allele MICA*008 to escape elimination by NK cells. Cell Rep 2015; 10:968–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Furman MH, Dey N, Tortorella D, Ploegh HL. The human cytomegalovirus US10 gene product delays trafficking of major histocompatibility complex class I molecules. J Virol 2002; 76:11753–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Park B, Spooner E, Houser BL, Strominger JL, Ploegh HL. The HCMV membrane glycoprotein US10 selectively targets HLA‐G for degradation. J Exp Med 2010; 207:2033–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Tirosh B, Iwakoshi NN, Lilley BN, Lee AH, Glimcher LH, Ploegh HL. Human cytomegalovirus protein US11 provokes an unfolded protein response that may facilitate the degradation of class I major histocompatibility complex products. J Virol 2005; 79:2768–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 1996; 84:769–79. [DOI] [PubMed] [Google Scholar]
- 149. Charpak‐Amikam Y, Kubsch T, Seidel E, Oiknine‐Djian E, Cavaletto N, Yamin R et al Human cytomegalovirus escapes immune recognition by NK cells through the downregulation of B7‐H6 by the viral genes US18 and US20. Sci Rep 2017; 7:8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Fielding CA, Aicheler R, Stanton RJ, Wang EC, Han S, Seirafian S et al Two novel human cytomegalovirus NK cell evasion functions target MICA for lysosomal degradation. PLoS Pathog 2014; 10:e1004058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Fielding CA, Weekes MP, Nobre LV, Ruckova E, Wilkie GS, Paulo JA et al Control of immune ligands by members of a cytomegalovirus gene expansion suppresses natural killer cell activation. Elife 2017; 6:pii: e22206. [DOI] [PMC free article] [PubMed] [Google Scholar]


