Highlights
-
•
Florida Lanai is a tomato variety suitable for virus-host interaction studies.
-
•
Florida-Lanai was infected by geminiviruses delivered by different methods.
-
•
Florida-Lanai shows distinct measurable symptoms for different geminiviruses.
-
•
Florida-Lanai has a small size, rapid growth and is easy to maintain.
-
•
Florida-Lanai is an excellent choice for comparing geminivirus infections.
Keywords: Florida lanai, Tomato, Geminiviruses, Symptoms, qPCR, Ploidy, Seed transmission
Abstract
Geminiviruses are devastating single-stranded DNA viruses that infect a wide variety of crops in tropical and subtropical areas of the world. Tomato, which is a host for more than 100 geminiviruses, is one of the most affected crops. Developing plant models to study geminivirus-host interaction is important for the design of virus management strategies. In this study, “Florida Lanai” tomato was broadly characterized using three begomoviruses (Tomato yellow leaf curl virus, TYLCV; Tomato mottle virus, ToMoV; Tomato golden mosaic virus, TGMV) and a curtovirus (Beet curly top virus, BCTV). Infection rates of 100% were achieved by agroinoculation of TYLCV, ToMoV or BCTV. Mechanical inoculation of ToMoV or TGMV using a microsprayer as well as whitefly transmission of TYLCV or ToMoV also resulted in 100% infection frequencies. Symptoms appeared as early as four days post inoculation when agroinoculation or bombardment was used. Symptoms were distinct for each virus and a range of features, including plant height, flower number, fruit number, fruit weight and ploidy, was characterized. Due to its small size, rapid growth, ease of characterization and maintenance, and distinct responses to different geminiviruses, “Florida Lanai” is an excellent choice for comparing geminivirus infection in a common host.
1. Introduction
Geminiviruses belong to a large, diverse family of plant infecting viruses (Geminiviridae) that are transmitted by insects and cause economically significant diseases worldwide (Zhang et al., 2001; Rojas et al., 2005; Hanley-Bowdoin et al., 2013). Geminiviruses are among the most economically important pathogens in a variety of crops including vegetables, fruits, root crops, cereals, spices and legumes (Morales and Anderson, 2001; Mansoor et al., 2003; Seal et al., 2006). The genomes of geminiviruses consist of either one (monopartite) or two (bipartite) circular, single-stranded DNA molecules, with the components of bipartite viruses known as DNA-A and DNA-B (Zhang et al., 2001; Brown et al., 2012; Hanley-Bowdoin et al., 2013). Geminiviruses are classified in nine genera according to their genome, host and insect vector (Zerbini et al., 2017).
Management of plant viruses is of vital importance to reduce the damage (Sastry and Zitter, 2014), especially in areas where food security is at risk due to high viral diversity and the emergence of more virulent strains (Damsteegt, 1999; Mansoor et al., 2003; Sastry and Zitter, 2014). In 2009, Rodrigues et al. (Rodrigues et al., 2009) concluded that disease management strategies need extensive knowledge of the virus infection, transmission, spread and their effects on host plants to select the best control measures. Studying viruses can be simplified if a tractable host system is available. The suitability of a host for studying the infection process is determined by its ability to become infected and to allow the virus to replicate and induce typical symptoms (Scholthof et al., 1996).
Geminiviruses, have been studied using model plant systems such as Arabidopsis thaliana (Muangsan et al., 2004; Ascencio-Ibáñez et al., 2008; Hanley-Bowdoin et al., 2013; Raja et al., 2014), Nicotiana benthamiana (Goodin et al., 2008), Solanum nigrum (Urbino et al., 2008), and Datura stramonium (Chen et al., 2013). These model plants have many advantages including small size, short life cycles, high seed germination rates and ease of genetic analysis (Meissner et al., 1997; Meinke et al., 1998; Matsukura et al., 2008). For example, Arabidopsis has one of the smallest genomes, making it useful for genetic manipulation (Bevan and Walsh, 2006). Model plants are also usually inexpensive to study and readily accessible. However, information obtained using model plants may not always accurately reflect viral interactions or processes that occur in a non-model crop or reservoir plants in nature and disease can be the result of specific interactions between a virus and a host (Dawson and Hilf, 1992; Pallas and Garcia, 2011).
Of the 322 begomoviruses recognized by the International Committee on Taxonomy of Viruses, more than a third infect tomato and probably many others can infect solanaceous plants, underscoring the importance of having a suitable tomato variety for virus testing. Tomato (Solanum lycopersicum L., Solanaceae) is an herbaceous plant with hundreds of varieties that differ in size and generation time. Tomato has long been the preferred system for studying plant-pathogen interactions involving plants from the Solanaceae family (Arie et al., 2007; Meissner et al. 1997; Emmanuel and Levy, 2002). Tomato is susceptible to a wide range of viral diseases, many of which are associated with significant agronomic losses (Hanssen et al., 2010; Inoue-Nagata et al., 2016). As an example, tomato yellow leaf curl disease is caused by begomoviruses and has spread worldwide to become one of the most important viral diseases of tomato (Lefeuvre et al., 2010).
There is considerable physiological and genetic variation among tomato varieties that affects their suitability for laboratory studies. Among tomato varieties, Micro-Tom (TGRC accession # LA3911, UC Davis, Department of Plant Sciences, USA), a dwarf tomato cultivar derived from crossing cv. Florida Basket and Ohio 4013-3 (Scott and Harbaugh, 1989), is widely used in laboratory studies due to its small size (15–20 cm in height), rapid life cycle (70–90 days), and because it can be readily and efficiently transformed (Emmanuel and Levy, 2002; Meissner et al., 1997, Martí et al., 2006; Carvalho et al., 2011; Okabe et al., 2011; Sun et al., 2006). Studies require less time to complete because of its rapid life cycle that can accommodate up to four generations per year. Even though Micro-Tom has been widely adopted, its potential for molecular studies is limited because of its mutant genetic background, which results in brassinosteroid deficiency and deep green rugose leaves induced by the presence of the dwarf (d) and miniature (mnt) recessive genes (Bishop et al., 1996; Pnueli et al., 1998; Martí et al., 2006). The brassinosteroid pathway has been implicated in viral disease and symptom development, and alterations in the pathway may interfere with virus-plant interaction studies in Micro-Tom (Campos et al., 2010). Moreover, the gibberellin response is altered in Micro-Tom (Martí et al., 2006) and further interferes with data interpretation. In addition, Micro-Tom has a mutation in the self-pruning (sp) gene, which controls the regularity of the vegetative-reproductive switch along the compound shoot of tomato. This mutation is responsible for its determinate phenotype (Pnueli et al., 1998). Thus, it is important to look for new model systems that are either alternative or complementary to those currently used.
S. lycopersicum ‘Florida Lanai’ is also a small tomato variety that was developed for home gardens (Augustine et al., 1981). It has regular leaves and determinate growth, reaching a height of 60–90 cm. Flowers are open pollinated and produce a medium sized fruit (under 450 g) maturing approximately 60 days after transplanting or 90 days from seeding. Seed germination rate ranges from 82% to 96%. Even though ‘Florida Lanai’ plants are small and have a short generation time, they do not carry the recessive genes that place the use of Micro-Tom in doubt. ‘Florida Lanai’ has been used previously to characterize a new begomovirus species (Tomato yellow margin leaf curl virus) using biolistics to inoculate infectious clones (Nava et al., 2013). It has also been used to study geminivirus-insect interactions (McKenzie, 2002), although there has been no systematic characterization of its suitability as a model system for geminiviruses. In this study, we used three inoculation methods to examine ‘Florida Lanai’ as a model system for studying diverse geminiviruses that naturally infect tomato.
2. Materials and methods
2.1. Plant growth conditions and inoculation protocols
Florida Lanai seeds were kindly supplied by J. Scott (University of Florida, USA). ‘Florida Lanai’ plants were grown in sterile soil from seeds in a walk-in growth chamber at 25 °C, 80% humidity and a 16:8 light/dark (LD) cycle. After one week, the seedlings were transplanted into pots and propagated for two more weeks before inoculation. Virus inoculation was done by either Agrobacterium-mediated inoculation, low-pressure particle acceleration DNA delivery using a microdrop sprayer (Venganza, Inc.) or whitefly transmission from infected to healthy plant. The infectious clones corresponding to Beet curly top virus (BCTV), Tomato yellow leaf curl virus (TYLCV), Tomato mottle virus (ToMoV DNA-A and DNA-B), Tomato golden mosaic virus (TGMV DNA-A and DNA-B), Cabbage leaf curl virus (CaLCuV DNA-A and DNA-B), are described in Table 1. E. coli cultures for TYLCV, ToMoV, TGMV and CaLCuV DNA A and DNA B were prepared in LB broth containing 0.1 μg/ml carbenicillin, subsequently grown overnight at 37 °C with vigorous shaking. Similarly their corresponding Agrobacterium clones were prepared in LB broth containing 0.075 μg/ml Spectinomycin grown at 30 °C. For BCTV, E. coli and Agrobacterium clones were prepared in 0.05 μg/ml kanamycin LB broth cultured overnight at their respective temperatures. All experiments were repeated three times.
Table 1.
Infectious viral clones used to inoculate ‘Florida Lanai’ plants by agroinoculation or biolistics.
| Virusa | Plasmid used for biolistics | Plasmid used for agroinoculation | References and comments |
|---|---|---|---|
| BCTV | BCTV in pMON521 | BCTV in pMON521 | Beet curly top virus (BCTV; strain Logan), a pMON525-based plasmid containing a BCTV DNA containing a partial tandem copy (provided by D. M. Bisaro of Ohio State University, Stenger et al., 1992). |
| TYLCV | pTYLCV2 | pNSB1736 | Partial tandem copy of Tomato yellow leaf curl virus (TYLCV; Dominican Republic isolate) cloned into pMON721 (Settlage et al., 2005; Reyes et al., 2013), from Acc. number AF024715. |
| ToMoV DNA A | pNSB1906 | pNSB1906 | Partial tandem copy of Tomato mottle virus (ToMoV) DNA-A cloned into pMON721 (Abouzid et al., 1992, Reyes et al., 2013) |
| ToMoV DNA B | pNSB1877 | pNSB1877 | Partial tandem copy of Tomato mottle virus (ToMoV) DNA-B cloned into pMON721 (Abouzid et al., 1992, Reyes et al., 2013) |
| TGMV DNA A | pMON1565 | pMON337 | Partial tandem copy of Tomato golden mosaic virus (TGMV) DNA-A (Fontes et al., 1994; Orozco and Hanley-Bowdoin, 1996; Elmer et al., 1988). |
| TGMV DNA B | pTG1.4B | pMON393 | Partial tandem copy of Tomato golden mosaic virus (TGMV) DNA-B cloned in pTG1.4B (Fontes et al., 1994; Orozco and Hanley-Bowdoin, 1996). |
| CaLCuV DNA A | pCpCLCV A.003 | pNSB1090 | Cabbage leaf curl virus (CaLCuV) with a partial tandem copy (Turnage et al., 2002; Egelkrout et al., 2002). |
| CaLCuV DNA B | pCpCLCV B.003 | pNSB1091 | Cabbage leaf curl virus (CaLCuV) with a partial tandem copy (Turnage et al., 2002; Egelkrout et al., 2002). |
All clones have been designed to contain two viral origins of replication which allow the vector to release a functional viral monomer circularized by Rep and identical to wild-type viral DNA.
2.1.1. Agrobacterium-mediated inoculation
Agrobacterium cultures containing infectious clones in binary vectors were grown in LB broth with their corresponding antibiotics at 30 °C overnight. The bacterial cultures were diluted 10-fold with LB media and used to inoculate ten plants for each treatment. For bipartite viruses, equal amounts of cultures corresponding to DNA-A and DNA-B genomes were mixed prior to inoculation. An Agrobacterium strain carrying an empty T-DNA vector was used for mock inoculation. Plants were then returned to the growth chamber. Agroinoculation procedures were described previously by Reyes et al, (2013).
2.1.2. Biolistics
Plasmid DNA (5 μg) carrying infectious clones was coated onto 1 μm gold (Au) particle suspensions as described in Cabrera-Ponce et al. (1997). The final pellet was resuspended in 65 μL of absolute ethanol and used to spray 6 plants (10 μL/plant) at 40 psi. For the bipartite geminiviruses, 5 μg of each viral DNA component were mixed prior to coating the gold particles. The sprayer was positioned 2.5 cm from the plant apex. Empty plasmid DNA was used for the mock controls.
2.1.3. Whitefly transmission
Experiments were carried out in whitefly proof cages using Bemisia tabaci MEAM1 adults from a colony maintained on ‘Florida Lanai’ tomato at 27 °C and a 16:8 LD cycle in an environmental chamber. Approximately 100 adult whiteflies between 2 and 10 days post-eclosion were allowed to acquire virus by caging for 72 hr with a symptomatic ‘Florida Lanai’ plant infected with either TYLCV or ToMoV. The whiteflies were transferred to new cages containing healthy ‘Florida Lanai’ plants and allowed to feed continuously. The mock treatment was done by feeding the whiteflies on healthy plants. The plants were inspected for symptoms at 28 days post inoculation (dpi) and leaf samples collected for PCR analysis.
2.1.4. Seed transmission
Seeds were harvested from plants showing typical symptoms of TYLCV, ToMoV, BCTV or TGMV. Harvested seeds were washed, dried and sown in new pots. Samples were taken for DNA isolation from one leaflet of the fourth compound leaf (counted from the top of the plant) at 3 and 6 weeks after planting from 6 plants per treatment. Equal amounts of DNA from 6 plants were pooled for each treatment. For BCTV-infected plants, which do not produce fruit if infected early, healthy plants were inoculated with BCTV after initial fruit-setting. Seeds were harvested and analyzed as described above. All pooled samples were analysed by conventional PCR using virus-specific primers (Table 2).
Table 2.
List of primers used for PCR amplification of viruses in this study.
| Primer name | Sequence (5′ → 3′) | Virus species | Expected size (nt) |
|---|---|---|---|
| BCTV15-fora | CGTTACTGTGACGAAGCATTTG | BCTV | 283 |
| BCTV15-reva | CTCCTTCCCTCCATATCCAGTA | BCTV | |
| TYLCV15-forb | CCTCTGGCTGTGTTCTGTTATC | TYLCV | 257 |
| TYLCV15-revb | GCAATCTTCGTCACCCTCTAC | TYLCV | |
| ToMoV pNSB1c | GTCCAATACTCTCTCGTCCAATC | ToMoV | 239 |
| ToMoV pNSB2c | CAGCGGCCTTGTTAATTCTTG | ToMoV | |
| Sal-Ncod | CGACAAAGACGGAGATACTCT | TGMV | 397 |
| AL1 RTd | GCCTAGTGAACGAGCCCACA | TGMV | |
| CaLCuV1990-Fe | ACATACATCAGAGTCGCAAGAG | CaLCuV | 223 |
| CaLCuV1990-Rd | ACTGCCCGGATTCACAATAA | CaLCuV |
2.2. Disease, growth and yield monitoring
Plants were inspected weekly from 1 dpi to record disease symptoms and plant height. Disease symptoms were recorded by photography using a digital camera (Panasonic Lumix DMC-FZ28). Plant height (cm) was measured from the base to the tip of the main shoot for each plant (Olaniyi et al., 2010). The measurements were recorded as height increase by subtracting initial height of a plant at the time of inoculation from the height measured at the time of data recording. Data were also recorded on yield parameters (number of flowers, number of fruit and fresh fruit weight). The number of flowers was recorded 60 days after planting. The number of fruit and the fresh fruit weight was recorded at harvest (95 days from planting).
2.3. DNA extraction and virus detection
Samples were collected from the fourth compound leaf from the top of individual plants, and consisted of a single base leaflet. Independent samples were placed in 2-mL cryovials at 14, 17, 21 and 31 dpi from 10 plants for each treatment and frozen immediately in liquid nitrogen. DNA was extracted using the CTAB DNA extraction method (Doyle and Doyle, 1987). DNA concentrations and quality were assessed using a Nanodrop (Thermo Scientific™). For plants infected with ToMoV, which showed a recovery phenotype, DNA was prepared from the first, second and third compound leaves from the apex.
A convergent primer pair that amplifies a short DNA fragment (≤300 bp) was designed for each virus (Table 2). Primers were first tested in conventional PCR to establish optimum annealing temperature and amplification efficiency before being used in quantitative real-time PCR (qPCR). Viral DNA was quantified using a qPCR standard curve generated by amplification of known amounts of plasmid DNA containing viral sequences (Table 1) that was 10-fold serially diluted from 10−1 to 10−4 range. The concentration of the template DNA in the reaction mix was converted from ng/μL to copy number/μL using the following formula: (C × 10−9/MW) × NA where C = template concentration ng/μL, MW = template molecular weight in Daltons and NA = Avogadro's constant, 6.022 × 1023. MW was obtained by multiplying the number of base pairs of a plasmid by the average molecular mass of one base pair (660 g/mol). A base 10 logarithmic graph of copy number versus the threshold cycle (Ct) for the dilution factor was plotted and used as a standard curve to determine the amount of viral DNA (copy number) in each μL of total DNA in a reaction mix.
The qPCR analyses were performed with the MX300P real-time thermocycler (Stratagene, La Jolla, CA) using Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). The amplification reactions were performed in 50 μL containing 0.2 μM forward and reverse primers, ultrapure water and the optimum amount of DNA template as determined in titration experiments for the respective viruses (data not shown). Each virus was tested in a separate 96-well plate in which the first row contained the 10-fold serially diluted plasmid DNA for the standard curve.
2.4. DNA ploidy levels
To determine DNA ploidy levels of ‘Florida Lanai’ infected with different geminiviruses, leaf samples were taken from plants showing symptoms of TYLCV, ToMoV and BCTV as well as from mock-inoculated and healthy plants for comparison. Three biological replicas were collected for each treatment. Ploidy levels were determined using an Accuri™ C6 Flow Cytometer (BD Biosciences). Nuclei suspensions were prepared by chopping ca. 200 mg of fresh leaf tissue with a sharp razor blade in chopping buffer (3 mL Galbraith buffer + 10 μL β-ME and 2 μL RNase A) for 5 min on ice. Buffer preparation and other processes were done according to the BD Accuri™ C6 Flow Cytometer user manual. Data were plotted using internal BD Accuri C6 software, and peak positions and relative ploidy indices determined.
2.5. Statistical analysis
Statistical analysis was performed using Microsoft Excel (Office 2013). Analysis was performed using paired, two-tailed Student's t-test and p < 0.05 as the statistically significant cutoff. One-way analysis of variance was used to establish differences among group means and the Least significant difference (LSD) test was used in pairwise comparison to analyze differences between means.
3. Results
Three inoculation protocols were used: agroinoculation, particle bombardment and whitefly transmission, to inoculate ‘Florida Lanai’ plants with 5 diverse geminiviruses. A characterization of the effects of inoculation with each virus onto ‘Florida Lanai’ was performed. Also, seed transmission was determined for four of the viruses. A comparison between 'Florida Lanai' and Micro-Tom is shown for healthy plants (Fig. 1).
Fig. 1.
Comparison at 45 day-old A: Florida Lanai and B: Micro-Tom tomato varieties.
3.1. Symptom expression
Agroinoculation was a very efficient method for inoculating ‘Florida Lanai’ with TYLCV, ToMoV and BCTV resulting in 100% infection. Typical symptoms were observed in plants inoculated with these three viruses (Fig. 2A–D). Symptoms started to appear as early as 4 dpi for TYLCV and ToMoV and 7 dpi for BCTV. There were no observable symptoms in plants inoculated with TGMV or CaLCuV (data not shown) and no virus was detected by PCR. The failure of the TGMV to induce symptoms in tomato is well documented and we may have some issues with our agrobacterium inoculum (Wyant et al., 2012).
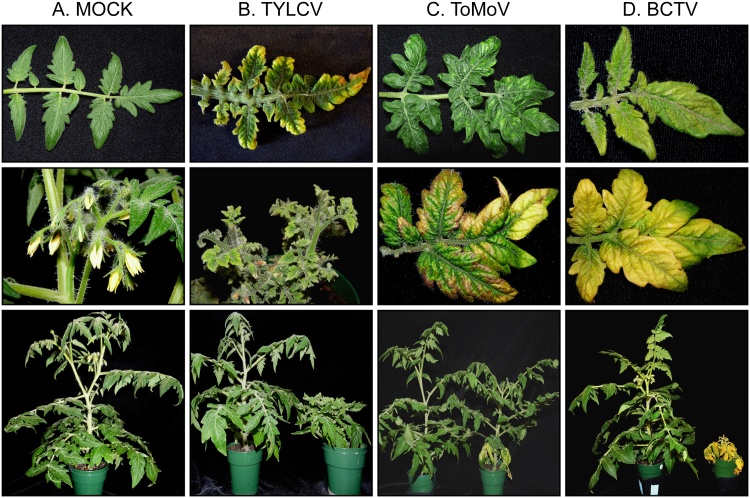
Fig. 2.
Symptoms observed on Florida Lanai plants mock- and agro-inoculated with TYLCV, ToMoV and BCTV.A: Mock-inoculated plant showing a healthy leaf, healthy flowers and a healthy plant (top to bottom). B: TYLCV inoculation showing chlorotic leaf margins, severe leaf size reduction, flower abscission and severe height reduction. C: ToMoV inoculation displaying bright yellow mottling in upper leaves, severe yellowing of lower leaves and medium plant height reduction. D: BCTV inoculation with general yellowing with mixed shades of green at early stages of infection, deep yellowing of the whole plant and very severe stunting at late stages of infection’.
When young plants were agroinoculated with TYLCV (28 days after planting), the plants showed chlorotic leaf margins, upward leaf curling, severe leaf size reduction and flower abscission (Fig. 2B). When older plants were inoculated with TYLCV (45 days after planting), symptoms were limited to middle and upper leaves and ca. 85% of the floral buds were lost by abscission. Other symptoms included swelling of veins and severe stunting.
Plants agroinoculated with BCTV developed a general yellowing mixed with green at early stages of infection that progressed to deep yellow at advanced stages (Fig. 2D). Leaves were stunted, thicker and crisp with swollen veins. BCTV-infected plants generally exhibited severe stunting (Table 3 and Fig. 5). Approximately 25% of the plants infected at an early growth stage (28 days after planting) exhibited root decay and were dead by 45 dpi (data not shown), while the remaining plants did not recover or produce flowers. Plants infected later (45 days after planting) produced a few flowers, which did not open and dropped before fruit set.
Table 3.
Comparison between infected and healthy plants for the change in height at different days after inoculation.
| Mean (cm)a | P-valueb | % of height reduction | |
|---|---|---|---|
| Mock | |||
| 7 dpi | 2.65 ± 0.66 | ||
| 14 dp1 | 5.27 ± 1.28 | ||
| 21 dpi | 7.68 ± 1.56 | ||
| 28 dpi | 8.58 ± 1.31 | ||
| 35 dpi | 11.1 ± 1.26 | ||
| TYLCV | |||
| 7 dpi | 1.05 ± 0.38 | ≤0.001 | 60.4 |
| 14 dpi | 2.09 ± 0.54 | ≤0.001 | 60.6 |
| 21 dpi | 2.52 ± 0.60 | ≤0.001 | 67.2 |
| 28 dpi | 2.99 ± 0.62 | ≤0.001 | 65.2 |
| 35 dpi | 4.31 ± 0.65 | ≤0.001 | 61.0 |
| ToMoV | |||
| 7 dpi | 1.79 ± 0.63 | 0.008 | 32.5 |
| 14 dpi | 3.94 ± 1.04 | 0.02 | 25.8 |
| 21 dpi | 6.15 ± 1.64 | 0.05 | 19.9 |
| 28 dpi | 8.00 ± 0.99 | 0.28 | 6.76 |
| 35 dpi | 10.4 ± 1.42 | 0.27 | 6.15 |
| BCTV | |||
| 7 dpi | 1.93 ± 0.64 | 0.008 | 28.3 |
| 14 dp1 | 2.07 ± 0.72 | 0.002 | 62.1 |
| 21 dpi | 2.16 ± 0.73 | ≤0.001 | 72.5 |
| 28 dpi | 2.36 ± 0.87 | ≤0.001 | 72.8 |
| 35 dpi | 2.88 ± 0.15 | ≤0.001 | 73.3 |
Mean±S.D, n = 10.
Significance level (P ≤ 0.05).
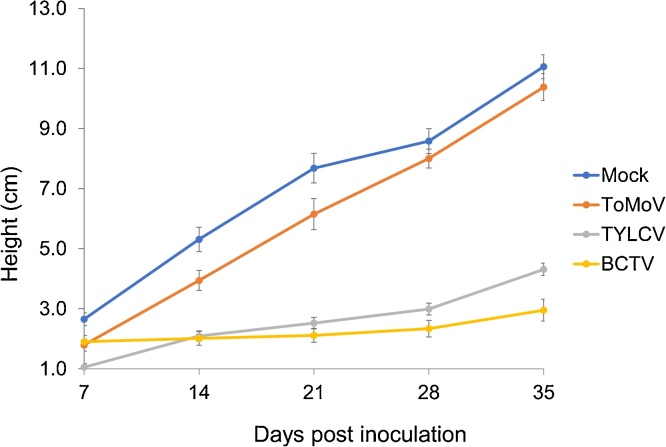
Fig. 5.
Change in plant height for Florida-Lanai tomato plants infected with TYLCV, ToMoV, BCTV and mock at different days post inoculation. Vertical bars represent the standard error (SE) of the means. N = 10 for all treatments.
Plants agroinoculated with ToMoV typically developed a bright yellow chlorotic mottling on younger leaves and severe yellowing, leaf deformation and upward curling on lower leaves (Fig. 2C). Compared to plants infected with TYLCV or BCTV, ToMoV-infected plants showed only moderate stunting, less flower abscission and a smaller reduction in fruit (Figs. 2, 3, and Table 3). During ToMoV infection, the yellow chlorotic symptoms observed from 5 to 14 dpi changed to a recovery phenotype in which new leaf growth was symptomless and the plant grew faster producing many flowers and fruit (Fig. 3 and Table 3). ToMoV DNA was detected by PCR in leaves showing the recovery phenotype.
Fig. 3.
Florida Lanai recovering from ToMoV infection. A: Infection at 14 dpi, B: Infection at 28 dpi, C: Impact of recovery on yield (i) ToMoV, (ii) TYLCV and (iii) BCTV.
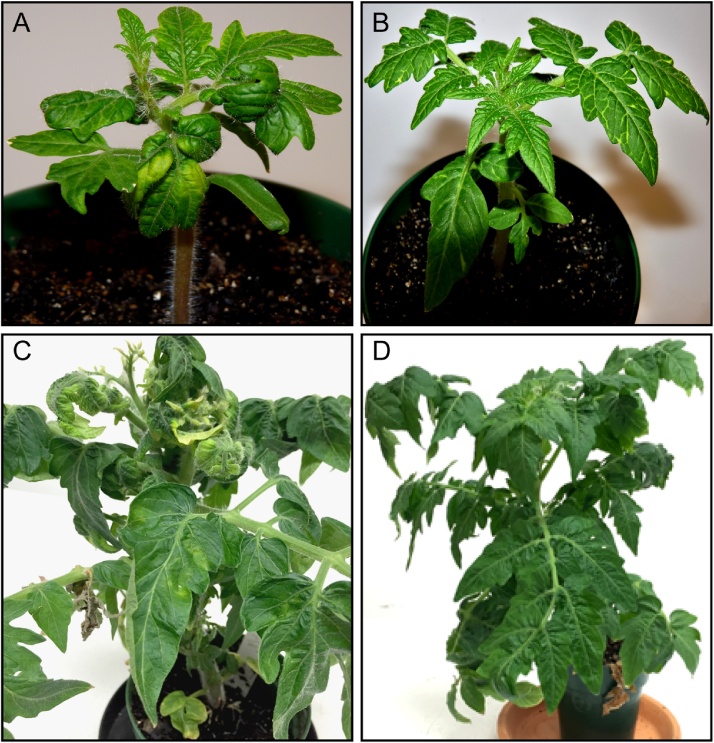
Particle bombardment led to the infection of two of the five viruses used in this study. Virus symptoms were observed in 100% of the ‘Florida Lanai’ plants inoculated with ToMoV or TGMV by bombardment. No infected plants were observed in equivalent experiments using plasmids corresponding to TYLCV, BCTV or CaLCuV. Plants bombarded with ToMoV developed symptoms indistinguishable from those in agroinoculation experiments (Fig. 4A). Bombardment of TGMV DNA resulted in bright yellow coloration along veins (Fig. 4B). In comparison, TGMV inoculated plants (N. benthamiana) exhibited chlorotic mottling, leaf curling or spiral distortion, which was not observed in Florida Lanai (data not shown).
Fig. 4.
Symptoms on inoculated Florida Lanai by biolistics using A: ToMoV and B: TGMV. Bottom row: Florida Lanai infected by whitefly transmission using C: TYLCV and D: ToMoV, showing severe and very mild symptoms respectively.
We tested TYLCV and ToMoV in whitefly transmission assays. Based on symptoms and PCR analysis, TYLCV was successively transmitted by viruliferous whiteflies from a TYLCV-infected source plant to a healthy Florida Lanai. By 30 days after introduction of viruliferous whiteflies, the target plants exhibited chlorotic leaf margins, upward curling of leaves, reduced leaf size and others symptoms characteristic of TYLCV infection described above (Fig. 4C). Whitefly transmission of ToMoV resulted in a very mild mottling on leaves (Fig. 4D).
A recent study (Kil et al., 2016) reported that geminiviruses can be transmitted through seed collected from TYLCV-infected plants. We produced seed from fruit collected from plants infected with TYLCV, ToMoV, BCTV or TGMV. After washing carefully with water, the seeds were planted and F1 and F2 progeny plants were examined for symptoms and viral DNA. None of the plants developed symptoms, and PCR assays did not detect viral DNA in any of the plants. These results showed that the geminiviruses we tested are not transmitted through ‘Florida Lanai’ seed.
3.2. Virus titer
Analyses of virus titer by conventional PCR or qPCR used total DNA extracted from leaves of ‘Florida Lanai’ plants. Primer pairs (Table 2) were optimized to amplify viral DNA at an annealing temperature of 58 °C. The TYLCV, ToMoV and BCTV standard curves for qPCR were linear in the range of 50 (1:10 dilution) to 5 × 10−6 ng (1:1 × 106 dilution) per reaction (r2 > 0.99). We used 5 ng/reaction of total DNA for qPCR analysis of unknown viral DNA titers. This amount (5 ng) can be easily measured using a spectrophotometer.
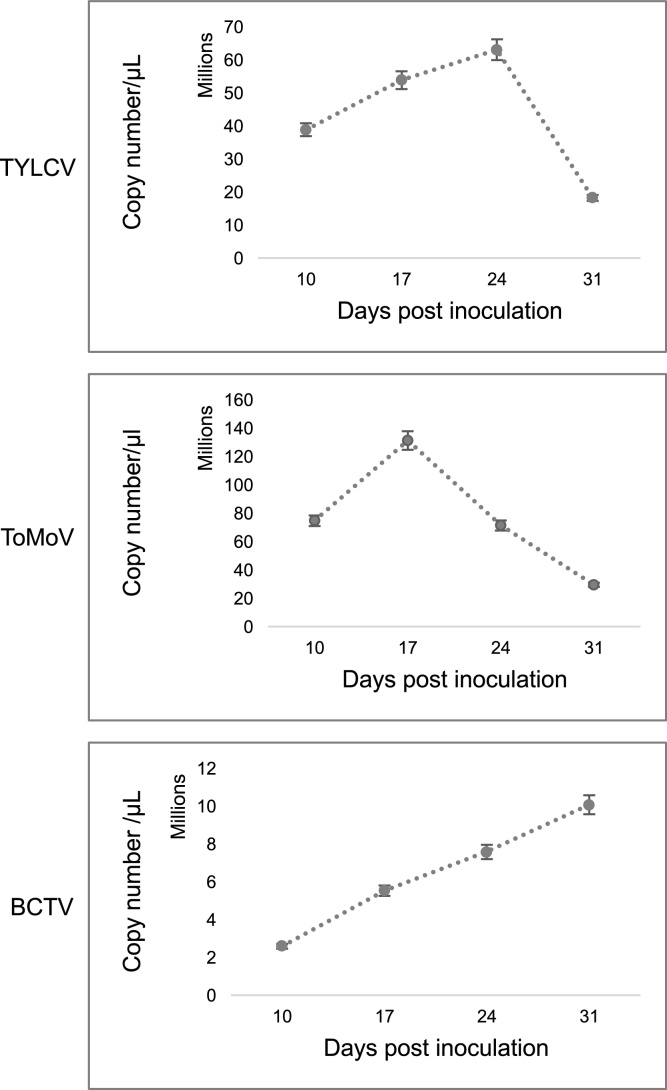
Analysis of variance (ANOVA) showed that virus levels changed over time in all treatments (Fig. 7). There was a significant change up to 31 dpi in the means of viral load in plants infected with TYLCV (F3,24 = 5.30, p < 0.05), ToMoV (F3,24 = 7.28, p < 0.05) or BCTV (F3,24 = 3.08, p < 0.05) (Table 5A). Mean separation by a LSD test (Table 5B) showed that virus titer in plants infected with TYLCV increased significantly (α = 0.01) at 10, 17 and 24 dpi and then decreased at 31 dpi to a level similar to 10 dpi. Viral DNA increased in ToMoV-infected plants over a shorter window of time between 10 to 17 dpi (α = 0.05) and then declined (31 dpi, α = 0.01) consistent with the recovery phenotype. BCTV infected plants showed a continuous increase in virus titer from 10 to 31 dpi, with a significant increase at 31 dpi (α = 0.01). This correlates with observed continuous increase in symptom severity over time.
Fig. 7.
Changes in viral load over time for Florida Lanai infected with A: TYLVC, B: ToMoV and C: BCTV. Vertical bars represent the standard error (SE) of the means. N = 7 for all treatments.
Table 5A.
One-way analysis of variance (ANOVA) for means of virus titer (copy number) for TYLCV, ToMoV and BCTV.
| Source of Variation | SS | df | MS | F | P-value | F crit | |
|---|---|---|---|---|---|---|---|
| TYLCV | Between Groups | 8.05E + 15 | 3 | 2.68E + 15 | 5.29 | 0.00602 | 3.01 |
| Within Groups | 1.21E + 16 | 24 | 5.07E + 14 | ||||
| Total | 2.02E + 16 | 27 | |||||
| ToMoV | Between Groups | 3.66E + 16 | 3 | 1.22E + 16 | 7.27 | 0.00123 | 3.01 |
| Within Groups | 4.03E + 16 | 24 | 1.68E + 15 | ||||
| Total | 7.69E + 16 | 27 | |||||
| BCTV | Between Groups | 2.11E + 14 | 3 | 7.03E + 13 | 3.08 | 0.04649 | 3.01 |
| Within Groups | 5.48E + 14 | 24 | 2.28E + 13 | ||||
| Total | 7.59E + 14 | 27 | |||||
Table 5B.
Difference between means and significance of pairwise comparison (LSD) for means of virus copy number for TYLCV, ToMoV and BCTV at different days post inoculation. Differences indicated by * are significant at the α < 0.05 level and ** are significant at the α < 0.01 level.
| 10dpi | 17dpi | 24dpi | 31dpi | ||
|---|---|---|---|---|---|
| TYLCV | 10dpi | 0 | 4.99E + 7** | 2.42E + 7 ns | 2.06E + 7 ns |
| 17dpi | 0 | 4.75E + 8** | 5.20E + 8** | ||
| 24dpi | 0 | 4.48E + 7** | |||
| 31dpi | 0 | ||||
| ToMoV | 10dpi | 0 | 5.66E + 7* | 3.36E + 6 ns | 4.52E + 7 ns |
| 17dpi | 0 | 6.00E + 7* | 1.02E + 8** | ||
| 24dpi | 0 | 4.18E + 7 ns | |||
| 31dpi | 0 | ||||
| BCTV | 10dpi | 0 | 2.94E + 6 ns | 4.99E + 6 ns | 7.48E + 6** |
| 17dpi | 0 | 2.05E + 6 ns | 4.54E + 6 ns | ||
| 24dpi | 0 | 2.50E + 6 ns | |||
| 31dpi | 00 | ||||
dpi = days post inoculation.
ns = not significant.
3.3. Plant height
Plants infected with TYLCV, ToMoV or BCTV were shorter than the mock-inoculated controls (Table 3 and Fig. 5). The reduction in height was highly significant (P < 0.05) for plants infected with BCTV or TYLCV at all sampling times. In contrast, ToMoV infection resulted in a significant height reduction during the initial stages of infection (7 and 14 dpi). During the later stages (21 and 28 dpi), ToMoV-infected plants underwent recovery and the heights of infected and mock-inoculated plants were not statistically different.
The establishment of BCTV infection was initially delayed (Table 3), but it ultimately caused the most severe disease symptoms. BCTV caused the largest reduction in the mean plant height (73.3%) at 35 dpi followed by TYLCV (67.2%) at 21 dpi. ToMoV had the smallest effect on Lanai growth. It recorded only 32.5% reduction in plant height at the initial stage of infection (7 dpi) before the plants recovered (Table 3).
3.4. Yield
Plants infected with TYLCV, BCTV or ToMoV showed reduced yields (Table 4). Reductions were most pronounced for TYLCV and BCTV, which were reduced for mean flower number, fruit number and fruit weight (g) per plant by 69.3, 93.5, and 95.3% respectively for TYLCV and 87.8, 100 and 100% respectively for BCTV. In contrast, ToMoV reduced the yield metrics by 8.5, 27.4 and 29.8%, respectively. The reductions were significant (P < 0.05) for numbers of flowers and fruit and fruit weight for plants infected with TYLCV and BCTV. The reductions in number of fruit and fruit weight were also significant for ToMoV-infected plants, but the reduction in number of flowers was not. From these results, it appears that TYLCV reduces the number of flowers and the proportion of flowers resulting in fruit due to excessive abscission, while ToMoV does not change the number of flowers produced by plants but increases flower abscission and causes a smaller reduction in fruit size. BCTV impairs the ability of plants to produce viable flowers and had a greater effect on yield than TYLCV. Plants infected early (21 days old) with BCTV produced very few flowers and none of them set fruit (Table 4). When older plants (at flowering, 45 days old) were infected with BCTV they formed flower buds that failed to open and eventually died (data not shown). Generally, all plants including the mock-inoculated controls produced many more flowers that set and produced fruit.
Table 4.
Effect of TYLCV, ToMoV and BCTV on yield.
| Meana | P-valueb | |
|---|---|---|
| Mock | ||
| Mean flower number per plant | 18.9 ± 4.15 | |
| Mean fruit number per plant | 6.20 ± 1.62 | |
| Mean fruit weight per plant (g) | 61.3 ± 14.4 | |
| TYLCV | ||
| Mean flower number per plant | 5.80 ± 2.57 | ≤0.001 |
| Mean fruit number per plant | 0.40 ± 0.70 | ≤0.001 |
| Mean fruit weight per plant (g) | 2.91 ± 8.63 | ≤0.001 |
| ToMoV | ||
| Mean flower number per plant | 17.3 ± 5.71 | 0.48 |
| Mean fruit number per plant | 4.50 ± 1.90 | 0.045 |
| Mean fruit weight per plant (g) | 43.0 ± 22.2 | 0.045 |
| BCTV | ||
| Mean flower number per plant | 2.30 ± 0.2.21 | ≤0.001 |
| Mean fruit number per plant | 0 ± 0.00 | ≤0.001 |
| Mean fruit weight per plant (g) | 0 ± 0.00 | ≤0.001 |
Mean±S.D, n = 10.
Significance level (P ≤ 0.05).
3.5. DNA ploidy
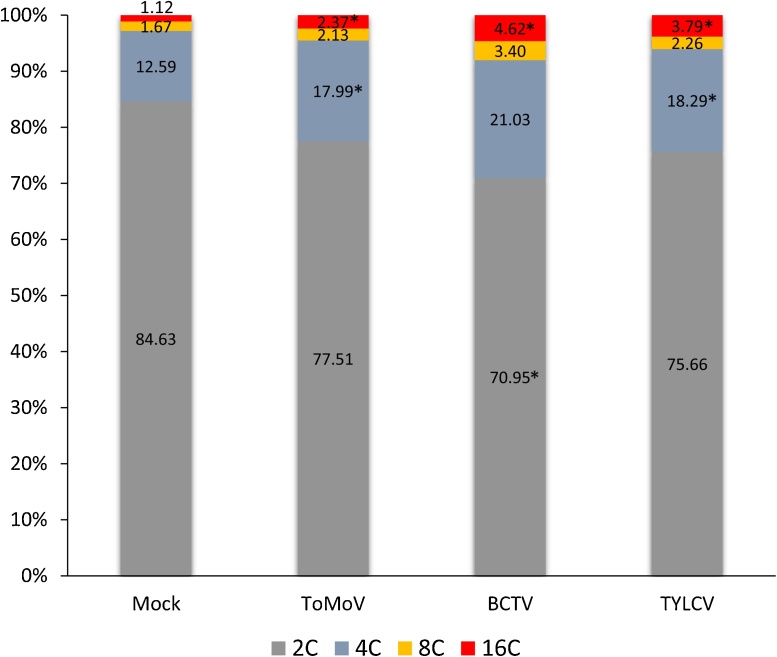
Geminivirus infection modifies plant cell cycle controls to support replication of both viral DNA and plant chromosomes leading to increase genome ploidy (Ascencio-Ibáñez et al., 2008). Flow cytometry analysis of leaf nuclei of Lanai plants infected with TYLCV, ToMoV or BCTV and uninfected leaf controls showed four peaks corresponding to nuclei with 2C, 4C, 8C and 16C ploidy (Fig. 6). Virus infection changed the distribution of the peaks. A reduction in cells with lower ploidy (2C) and enrichment in cells with higher ploidy (4C, 8C and 16C) was observed during infection, with BCTV-infected plants displaying the largest changes in ploidy. The differences were found to be statistically significant for 4C and 16C for ToMoV and TYLCV infected plants, as well as for 16C for BCTV infected plants (Fig. 6).
Fig. 6.
Histogram of the relative fluorescence intensity of nuclei isolated from leaves of Lanai plants either mock-inoculated or inoculated with ToMoV, BCTV or TYLCV. The bars represent ploidy percentages for each treatment. Values indicated by * are statistically significant (P < 0.05).
4. Discussion
Other studies have highlighted the facility of virus transmission and ability to allow rapid replication as the most important characteristics of a good model system (Gergerich and Dolja, 2006; MacLean et al., 2011). TYLCV is an Old World monopartite begomovirus. ToMoV and TGMV are New World bipartite begomoviruses. Two of these viruses were identified and isolated from tomato, whereas TGMV was identified in tomato but propagated in and isolated from N. benthamiana (Cohen and Nitzany, 1966; Matyis et al., 1975; Costa, 1976; Buck and Coutts, 1985; Bisaro et al., 1982; Abouzid et al., 1992; Crespi et al., 1995). CaLCuV is a bipartite begomovirus in the Squash leaf curl clade from the New World, (Nawaz-ul-Rehman et al., 2009). CaLCuV was not cloned from or considered to be a pathogen of tomato. BCTV, which has a single-component genome, is a curtovirus with a broad host range that includes tomato (Bennett, 1971; Chen et al., 2010). ‘Florida Lanai’ plants were readily infected (100% success rate) by TYLCV, ToMoV and BCTV using agroinoculation, regardless of plant growth stage. ToMoV and TGMV were transmitted mechanically by a microdrop-sprayer, while ToMoV and TYLCV were transmitted by whiteflies (whitefly transmission of TGMV was not tested). Together, these results established that ‘Florida Lanai’ is a versatile model for studying geminivirus infection in tomato. The ability to infect the variety using more than one method of inoculation provides important alternatives when facilities or expertise to carry out other methods are lacking. The inability to inoculate TYLCV and BCTV by bombardment most likely reflects that they are largely phloem limited. With a few exceptions, phloem-limited viruses are not mechanically transmitted (Schneider, 1973; Esau, 1977, Rojas et al., 2001; Wyant, et al., 2012; Miozzi et al., 2014). Although recent reports showed geminivirus seed transmission (Kil et al., 2016), no seed transmission was detected for the viruses infecting Florida Lanai. Seeds were washed extensively prior to planting to minimize any potential contamination of the seed coat from surrounding fruit tissue, which may contain virus.
Another interesting observation is the ability of TGMV to infect Florida Lanai. Tomato is thought to be a non-host for TGMV even though the virus was originally found in tomato but maintained and cloned from N. benthamiana (Stenger et al., 1992; Matyis et al., 1975). Florida Lanai was readily infected with TGMV by biolistics with a 100% success rate. A previous study (Wyant et al., 2012) inoculated three tomato cultivars, including var. Moneymaker, with 25% efficiency.
The observation that ‘Florida Lanai’ plants displayed typical disease symptoms as early as 4 dpi, is an important characteristic of a good model plant. The short incubation period of the pathogen depends not only on the infectious agent but also on host susceptibility and ability to express symptoms (Dmitry and Van den Ackerveken, 2013). ‘Florida Lanai’ plants developed viral symptoms quickly, producing typical and distinct symptoms for different geminiviruses and enabling a systematic evaluation of the impact of different viruses in a common host. Walkey (1991) stated that good indicator plants respond to viral infections consistently and distinctively. These are important requirements for a model plant, especially when making a disease diagnosis, fulfilling Koch’s postulates or characterizing virus-host interactions. Quantifiable effects of virus infection on symptoms, leaf deformation, plant height, flower number, fruit number, fruit weight, effect on roots, and DNA ploidy were detected.
We used flow cytometry to examine the effect of virus infection on plant ploidy. TYLCV, ToMoV or BCTV infection increased the number of cells with higher ploidy levels (4C, 8C and 16C) and reduced the number of cells with lower ploidy levels (2C). These results confirm previous reports of increases in ploidy in mature leaves during geminivirus infection (Ascencio-Ibáñez et al., 2008). The earlier study detected ploidy changes in CaLCuV which is not confinced to the phloem. Thus, it was surprising to detect significant ploidy changes for BCTV and TYLCV, both of which have been reported to be phloem-limited in tomato (Schneider, 1973; Esau, 1977, Rojas et al., 2001; Miozzi et al., 2014), and it will be interesting to characterize further the interactions of these two viruses with ‘Florida Lanai’.
The patterns of virus accumulation in ‘Florida Lanai’ plants infected with TYLCV, ToMoV and BCTV provide more evidence of its suitability as a model system. The patterns related clearly with the severity of symptoms exhibited by the plants, fitting the general concept that higher virus titer leads to more plant damage (Ponz and Bruening, 1986). The kinetics of virus accumulation for TYLCV and BCTV followed general virus infection patterns (Rom et al., 1993) and corresponded well with the development and maintenance of severe symptoms throughout the time course of infection. In contrast, ToMoV plants showed significant rise in viral load early in infection followed by a rapid decrease. This decline was associated with the disappearance of symptoms. Reduced virus accumulation and a recovery phenotype are thought to be the consequence of host defenses overcoming the virus (Covey et al., 1997; Ratcliff et al., 1999; Zhou et al., 2008; Ma et al., 2015). One of the main factors in the success of infection is the ability of a given virus to suppress plant silencing pathways (Qu and Morris, 2005; Hanley-Bowdoin et al., 2013; Pumplin and Voinnet, 2013). Our results suggested that TYLCV and BCTV have stronger silencing suppressing activities than ToMoV. The use of a common plant host provides an excellent system for studying these differences because it eliminates any effects due to potential differences between host silencing factors across plant species and varieties (Nie and Molen, 2015).
Conclusions: The Florida Lanai tomato variety is an excellent model system for studying and comparing tomato infecting geminivirus-host interactions. Using this model system, researchers can obtain reliable results quickly even when space is limited. ‘Florida Lanai’ plants are readily infected by different viruses, delivered using different methods, to produce distinct measurable symptoms. More than 60 geminivirus species infect tomato (Inoue-Nagata et al., 2016), and we tested only four here. Hence, there is a need for further studies to determine if more geminiviruses can infect Florida Lanai. We recommend Florida Lanai as an excellent tomato variety for use as a model system for agroinoculation studies of TYLCV, ToMoV and BCTV, for mechanical bombardment of ToMoV and TGMV, and for whitefly transmission for TYLCV and ToMoV. Researchers may find it useful to use Florida Lanai in virus transmission studies, disease epidemiology studies and when investigating various physiological phenomena.
Conflicts of interest
The authors have no conflict of interest to declare.
Acknowledgements
This study was supported by grants from the Bill & Melinda Gates Foundation to JTA-I and LHB (OPP1 149990). We thank MSc student Jonatan Isaksson and undergraduate students, Maxie G. Jollie, Daniel Bayha, Yamilex Rosado, Ananya Talikoti and Vanessa Ly, for their assistance with the flow cytometry analysis. We thank Wayne Curtis (PSU) for critical reading of the manuscript. We also thank Mary Beth Dallas and Wei Shen (NCSU) for their technical advice.
References
- Abouzid A.M., Polston J.A., Hiebert E. The nucleotide sequence of tomato mottle virus, a new geminivirus isolated from tomatoes in Florida. J. Gen. Virol. 1992;73:3225–3229. doi: 10.1099/0022-1317-73-12-3225. [DOI] [PubMed] [Google Scholar]
- Arie T., Takahashi H., Kodama M., Teraoka T. Tomato as a model plant for plant-pathogen interactions. Plant Biotechnol. 2007;24:135–147. [Google Scholar]
- Ascencio-Ibáñez J.T., Sozzani R., Lee T.J., Chu T.M., Wolfinger R.D., Cella R., Hanley-Bowdoin L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008;148:436–454. doi: 10.1104/pp.108.121038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine J.J., Harbaugh B.K., Crill J.P. Agricultural Experiment Stations, Institute of Food and Agricultural Sciences, University of Florida; Gainesville [Fla.]: 1981. Florida Lanai: A Dwarf Tomato for the Patio. [Google Scholar]
- Bennett C.W. American Phytopathological Society; St. Paul, MN: 1971. The Curly Top Disease of Sugarbeet and Other Plants. [Google Scholar]
- Bevan M., Walsh S. The Arabidopsis genome: a foundation for plant research. Genome Res. 2006;15:1632–1642. doi: 10.1101/gr.3723405. [DOI] [PubMed] [Google Scholar]
- Bisaro D.M., Hamilton W.D.O., Coutts R.H.A., Buck K.W. Molecular cloning and characterization of the two DNA components of tomato golden mosaic virus. Nucleic Acids Res. 1982;10(16):4913–4922. doi: 10.1093/nar/10.16.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop G.J., Harrison K., Jones J.D.G. The tomato Dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell. 1996;8:959–969. doi: 10.1105/tpc.8.6.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.K., Fauquet C.M., Briddon R.W., Zerbini M., Moriones E., Navas-Castillo J. Family Geminiviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy – Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; 2012. pp. 351–373. [Google Scholar]
- Buck K.W., Coutts R.H.A. AAB Descriptions of Plant Viruses No. 303. Spottiswode Balantyne Printers Ltd.; Warwick, U.K: 1985. Tomato golden mosaic virus. [Google Scholar]
- Cabrera-Ponce J.L., López L., Assad-García N., Medina-Arevalo C., Bailey A.M., Herrera-Estrella L. An efficient particle bombardment system for the genetic transformation of asparagus (Asparagus officinalis L.) Plant Cell Rep. 1997;16:255–260. doi: 10.1007/BF01088276. [DOI] [PubMed] [Google Scholar]
- Campos M.L., Carvalho R.F., Benedito V.A., Peres L.E.P. Small and remarkable: the Micro-Tom model system as a tool to discover novel hormonal functions and interactions. Plant Signal. Behav. 2010;5:267–270. doi: 10.4161/psb.5.3.10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho R.F., Campos M.L., Pino L.E., Crestana S.L., Zsögön A., Lima J.E., Benedito V.A., Peres L.E.P. Convergence of developmental mutants into a single tomato model system: ‘Micro-Tom' as an effective toolkit for plant development research. Plant Methods. 2011;7:18. doi: 10.1186/1746-4811-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Pan H., Xie W., Wang S., Wu Q., Fang Y., Shi X., Zhang Y. Virus infection of a weed increases vector attraction to and vector fitness on the weed. Sci. Rep. 2013;3:2253. doi: 10.1038/srep02253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.F., Brannigan K., Clark R., Gilbertson R.L. Characterization of curtoviruses associated with curly top disease of tomato in California and monitoring for these viruses in beet leafhoppers. Plant Dis. 2010;94:99–108. doi: 10.1094/PDIS-94-1-0099. [DOI] [PubMed] [Google Scholar]
- Cohen S., Nitzany F.E. Transmission and host range of the tomato yellow leaf curl virus. Phytopath. 1966;56:1127–1131. [Google Scholar]
- Costa A.S. Whitefly transmitted plant diseases. Annu. Rev. Phytopath. 1976;14:429–449. [Google Scholar]
- Covey S.N., Al-Kaff N.S., Langara A., Turner D.S. Plants combat infection by gene silencing. Nature. 1997;385:781–782. [Google Scholar]
- Crespi S., Noris E., Vaira A.M., Accotto G.P. Molecular characterization of cloned DNA from tomato yellow leaf curl virus isolate from Sicily. Phytopathol. Mediterr. 1995;34:93–99. [Google Scholar]
- Damsteegt V. APSnet Features; 1999. New and Emerging Plant Viruses. Online. [Google Scholar]
- Dawson W.O., Hilf M.E. Host-range determinants of plant viruses. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992;43:527–555. [Google Scholar]
- Dmitry L., Van den Ackerveken G. Susceptibility to plant disease: more than a failure of host immunity. Trends Plant Sci. 2013;18:546–554. doi: 10.1016/j.tplants.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Doyle J.J., Doyle J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- Egelkrout E.M., Mariconti L., Settlage S.B., Cella R., Robertson D., Hanley-Bowdoin L. Two E2F elements regulate the proliferating cell nuclear antigen promoter differently during leaf development. Plant Cell. 2002;14:3225–3236. doi: 10.1105/tpc.006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer J.S., Brand L., Sunter G., Gardiner W.E., Bisaro D.M., Rogers S.G. Genetic analysis of the tomato golden mosaic virus. II. The product of the AL1 coding sequence is required for replication. Nucleic Acids Res. 1988;16:7043–7060. doi: 10.1093/nar/16.14.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanuel E., Levy A.A. Tomato mutants as tools for functional genomics. Curr. Biol. 2002;5:112–117. doi: 10.1016/s1369-5266(02)00237-6. [DOI] [PubMed] [Google Scholar]
- Esau K. Virus-like particles in nuclei of phloem cells in spinach leaves infected with the curly top virus. J. Ultrastruct. Res. 1977;61:78–88. doi: 10.1016/s0022-5320(77)90007-7. [DOI] [PubMed] [Google Scholar]
- Fontes E.P.B., Gladfelter H.J., Schaffer R.L., Petty I.T.D., Hanley-Bowdoin L.K. Geminivirus replication origins have a modular organization. Plant Cell. 1994;6:405–416. doi: 10.1105/tpc.6.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergerich R.C., Dolja V.V. Introduction to plant viruses, the invisible foe. Plant Health Instruct. 2006 https://www.apsnet.org/edcenter/intropp/PathogenGroups/Pages/PlantViruses.aspx [Google Scholar]
- Goodin M.M., Zaitlin D., Naidu R.A., Lommel S.A. Nicotiana benthamiana: Its history and future as a model for plant-pathogen interactions. Mol. Plant-Microbe Interact. 2008;21:1015–1026. doi: 10.1094/MPMI-21-8-1015. [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin L., Bejarano E.R., Robertson D., Mansoor S. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microb. 2013;11:777–788. doi: 10.1038/nrmicro3117. [DOI] [PubMed] [Google Scholar]
- Hanssen I.M., Lapidot M., Thomma B.P.H.J. Emerging viral diseases of tomato crops. Mol. Plant-Microbe Interact. 2010;23:539–548. doi: 10.1094/MPMI-23-5-0539. [DOI] [PubMed] [Google Scholar]
- Inoue-Nagata A.K., Lima M.F., Gilbertson R.L. A review of geminivirus diseases in vegetables and other crops in Brazil: current status and approaches for management. Hortic. Bras. 2016;34:8–18. [Google Scholar]
- Kil E., Kim S., Lee Y., Byun H., Park J., Seo H., Kim C., Shim J., Lee J., Kim J., Lee K., Choi H., Lee S. Tomato yellow leaf curl virus (TYLCV-IL): a seed-transmissible geminivirus in tomatoes. Sci. Rep. 2016;6:19013. doi: 10.1038/srep19013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefeuvre P., Martin D.P., Harkins G., Lemey P., Gray A.J., Meredith S., Lakay F., Monjane A., Lett J.M., Varsani A., Heydarnejad J. The spread of tomato yellow leaf curl virus from the Middle East to the world. PLoS Pathog. 2010;6:e1001164. doi: 10.1371/journal.ppat.1001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Nicole M.C., Meteignier L.V., Hong N., Wang G., Moffett P. Different roles for RNA silencing and RNA processing components in virus recovery and virus-induced gene silencing in plants. J. Exp. Bot. 2015;66:919–932. doi: 10.1093/jxb/eru447. [DOI] [PubMed] [Google Scholar]
- MacLean A.M., Sugio A., Kingdom H.N., Grieve V.M., Hogenhout S.A. Arabidopsis thaliana as a model plant for understanding phytoplasmas interactions with plant and insect hosts. Annu. Rev. Phytopathol. 2011;49:175–195. doi: 10.1146/annurev-phyto-072910-095323. [DOI] [PubMed] [Google Scholar]
- Mansoor S., Briddon R.W., Zafar Y., Stanley J. Geminivirus disease complexes: An emerging threat. Trends Plant Sci. 2003;8:128–134. doi: 10.1016/S1360-1385(03)00007-4. [DOI] [PubMed] [Google Scholar]
- Martí E., Gisbert C., Bishop G.J., Dixon M.S., Garcia-Martinez J.L. Genetic and physiological characterization of tomato cv. Micro-Tom. J. Exp. Bot. 2006;57:2037–2047. doi: 10.1093/jxb/erj154. [DOI] [PubMed] [Google Scholar]
- Matsukura C., Aoki K., Fukuda N., Mizoguchi T., Asamizu E., Saito T., Shibata D., Ezura H. Comprehensive resources for tomato functional genomics based on the miniature model tomato Micro-Tom. Curr. Genom. 2008;9:436–443. doi: 10.2174/138920208786241225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyis J.C., Silva D.M., Oliveira A.R., Costa A.S. Purification and morphology of tomato golden mosaic virus. Summa Phytopathol. 1975;1:267–275. [Google Scholar]
- McKenzie C.L. Effect of tomato mottle virus (ToMoV) on Bemisia tabaci biotype B (Homoptera: Aleyrodidae) oviposition and adult survivorship on healthy tomato. Fla. Entomol. 2002;85:367–368. [Google Scholar]
- Meinke D.W., Cherry J.M., Dean C., Rounsley S.D., Koornneef M. Arabidopsis thaliana: a model plant for genome analysis. Science. 1998;282:662–682. doi: 10.1126/science.282.5389.662. [DOI] [PubMed] [Google Scholar]
- Meissner R., Jacobson Y., Melamed S., Levyatuv S., Shalev G., Ashri A., Elkind Y., Levy A. A new model system for tomato genetics. Plant J. 1997;12:1465–1472. [Google Scholar]
- Miozzi L., Napoli C., Sardo L., Accotto G.P. Transcriptomics of the interaction between the monopartite phloem-limited Geminivirus Tomato yellow leaf curl Sardinia virus and Solanum lycopersicum highlights a role for plant hormones, autophagy and plant immune system fine tuning during infection. PLoS One. 2014;9:e89951. doi: 10.1371/journal.pone.0089951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales F.J., Anderson P.K. The emergence and dissemination of whitefly-transmitted geminiviruses in Latin America. Arch. Virol. 2001;146:415–441. doi: 10.1007/s007050170153. [DOI] [PubMed] [Google Scholar]
- Muangsan N., Beclin C., Vaucheret H., Robertson D. Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants. Plant J. 2004;38:1004–1014. doi: 10.1111/j.1365-313X.2004.02103.x. [DOI] [PubMed] [Google Scholar]
- Nava A., Londono A., Polston J.E. Characterization and distribution of tomato yellow margin leaf curl virus, a begomovirus from Venezuela. Arch. Virol. 2013;158:399–406. doi: 10.1007/s00705-012-1501-x. [DOI] [PubMed] [Google Scholar]
- Nawaz-ul-Rehman M.S., Mansoor S., Briddon R.W., Fauquet C.M. Maintenance of an old world betasatellite by a new world helper begomovirus and possible rapid adaptation of the betasatellite. J. Virol. 2009;83:9347–9355. doi: 10.1128/JVI.00795-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X., Molen T.A. Host recovery and reduced virus level in the upper leaves after potato virus Y infection occur in tobacco and tomato but not in potato plants. Viruses. 2015;7:680–698. doi: 10.3390/v7020680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe Y., Asamizu E., Saito T., Matsukura C., Ariizumi T., Bres C., Rothan C., Mizoguchi T., Ezura H. Tomato TILLING technology: development of a reverse genetics tool for the efficient isolation of mutants from Micro-Tom mutant libraries. Plant Cell Physiol. 2011;52:1994–2005. doi: 10.1093/pcp/pcr134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaniyi J.O., Akanbi W.B., Adejumo T.A., Akande O.G. Growth, fruit yield and nutritional quality of tomato varieties. Afr. J. Food Sci. 2010;4:398–402. [Google Scholar]
- Orozco B.M., Hanley-Bowdoin L. A DNA structure is required for geminivirus origin function. J. Virol. 1996;270:148–158. doi: 10.1128/jvi.70.1.148-158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallas V., Garcia J.A. How do plant viruses induce disease? Interactions and interference with host components. J. Gen. Virol. 2011;92:2691–2705. doi: 10.1099/vir.0.034603-0. [DOI] [PubMed] [Google Scholar]
- Pnueli L., Carmel-Goren L., Hareven D., Gutfinger T., Alvarez J., Ganal M., Zamir D., Lifschitz E. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development. 1998;125:1979–1989. doi: 10.1242/dev.125.11.1979. [DOI] [PubMed] [Google Scholar]
- Ponz F., Bruening G. Mechanism of resistance to plant viruses. Annu. Rev. Phytopathol. 1986;24:355–381. [Google Scholar]
- Pumplin N., Voinnet O. RNA silencing suppression by plant pathogens: defense, counter-defence and counter-counter-defence. Nat. Rev. Microb. 2013;11:745–760. doi: 10.1038/nrmicro3120. [DOI] [PubMed] [Google Scholar]
- Qu F., Morris J.T. Suppressors of RNA silencing encoded by plant viruses and their role in viral infections. FEBS Lett. 2005;579:5958–5964. doi: 10.1016/j.febslet.2005.08.041. [DOI] [PubMed] [Google Scholar]
- Raja P., Jackel J.N., Li S., Heard I.M., Bisaro D.M. Arabidopsis double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against geminiviruses. J. Virol. 2014;88:2611–2622. doi: 10.1128/JVI.02305-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F.G., MacFarlane S.A., Baulcombe D.C. Gene silencing without DNA: RNA-mediated cross-protection between viruses. Plant Cell. 1999;11:1207–1216. doi: 10.1105/tpc.11.7.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes M.I., Nash T.E., Dallas M.M., Ascencio-Ibanez J.T., Hanley-Bowdoin L.J. Peptide aptamers that bind to geminivirus replication proteins confer a resistance phenotype to tomato yellow leaf curl virus and tomato mottle virus infection in tomato. J. Virol. 2013;87:9691–9706. doi: 10.1128/JVI.01095-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues S.P., Lindsey G.G., Fernandes P.M.B. Biotechnological approaches for plant viruses resistance: from general to the modern RNA silencing pathway. Braz. Arch. Biol. Technol. 2009;52:795–808. [Google Scholar]
- Rojas M.R., Hagen C., Lucas W.J., Gilbertson R.L. Exploiting chinks in the plant's armor: evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 2005;43:361–394. doi: 10.1146/annurev.phyto.43.040204.135939. [DOI] [PubMed] [Google Scholar]
- Rojas M.R., Jiang H., Salati R., Xoconostle-Cázares B., Sudarshana M.R., Lucas W.J., Gilbertson R.L. Functional analysis of proteins involved in movement of the monopartite begomovirus, Tomato yellow leaf curl virus. Virology. 2001;291:110–125. doi: 10.1006/viro.2001.1194. [DOI] [PubMed] [Google Scholar]
- Rom M., Antignus Y., Gidoni D., Pilowsky M., Cohen S. Accumulation of tomato yellow leaf curl virus DNA in tolerant and susceptible tomato lines. Plant Dis. 1993;77:253–257. [Google Scholar]
- Sastry K.S., Zitter T.A. Management of virus and viroid diseases of crops in the tropics. In: Sastry K.S., Zitter T.A., editors. vol. 2. Springer; Netherlands: 2014. pp. 149–480. (Plant Virus and Viroid Diseases in the Tropics: Epidemiology and Management). [Google Scholar]
- Schneider H. Cytological and histological aberrations in woody plants following infection with viruses, mycoplasmas, rickettsias, and flagellates. Annu. Rev. Phytopathol. 1973;11:119–146. [Google Scholar]
- Scholthof H.B., Scholthof K.B., Jackson A.O. Plant virus gene vectors for transient expression of foreign proteins in plants. Annu. Rev. Phytopathol. 1996;34:299–323. doi: 10.1146/annurev.phyto.34.1.299. [DOI] [PubMed] [Google Scholar]
- Scott J.W., Harbaugh B.K. Agricultural Experiment Station, Institute of Food and Agricultural Sciences, University of Florida; Gainesville, Fla: 1989. Micro-Tom: a Miniature Dwarf Tomato; pp. 1–6. Circular 1989. [Google Scholar]
- Seal S.E., van denBosch F., Jeger M.J. Factors influencing begomovirus evolution and their increasing global significance: implications for sustainable control. Crit. Rev. Plant Sci. 2006;25:23–46. [Google Scholar]
- Settlage S.B., See R.G., Hanley-Bowdoin L. Geminivirus C3 protein: replication enhancement and protein interactions. J. Virol. 2005;79:9885–9895. doi: 10.1128/JVI.79.15.9885-9895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger D.C., Davis K.R., Bisaro D.M. Limited replication of tomato golden mosaic virus DNA in explants of nonhost species. Mol. Plant-Microbe Interact. 1992;5:525–527. [Google Scholar]
- Sun H.J., Uchii S., Watanabe S., Ezura H. A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol. 2006;47:426–431. doi: 10.1093/pcp/pci251. [DOI] [PubMed] [Google Scholar]
- Turnage M.A., Muangsan N., Peele C.G., Robertson D. Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J. 2002;30:107–114. doi: 10.1046/j.1365-313x.2002.01261.x. [DOI] [PubMed] [Google Scholar]
- Urbino C., Thébaud G., Granier M., Blanc S., Peterschmitt M. A novel cloning strategy for isolating, genotyping and phenotyping genetic variants of geminiviruses. Virol. J. 2008;5 doi: 10.1186/1743-422X-5-135. 135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkey D. 2nd edition. Chapman and Hall; London: 1991. Applied Plant Virology. [Google Scholar]
- Wyant P.S., Kober S., Schwierzok A., Kocher C., Schäfer B., Jeske H., Wege C. Cloned tomato golden mosaic virus back in tomatoes. Virus Res. 2012;167:397–403. doi: 10.1016/j.virusres.2012.05.021. [DOI] [PubMed] [Google Scholar]
- Zerbini F.M., Briddon R.W., Idris A., Martin D.P., Moriones E., Navas-Castillo J., Rivera-Bustamante R., Varsani A., Consortium I.R. ICTV virus taxonomy profile: Geminiviridae. J. Gen. Virol. 2017;98:131–133. doi: 10.1099/jgv.0.000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Olson N.H., Baker T.S., Faulkner L., Agbandje-McKenna M., Boulton M.I., Davies J.W., McKenna R. Structure of the maize streak virus geminate particle. Virology. 2001;279:471–477. doi: 10.1006/viro.2000.0739. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Ryabov E., Zhang X., Hong Y. Influence of viral genes on the cell-to-cell spread of RNA silencing. J. Exp. Bot. 2008;59:2803–2813. doi: 10.1093/jxb/ern141. [DOI] [PMC free article] [PubMed] [Google Scholar]