Abstract
Hereditary nonsyndromic hearing loss is extremely heterogeneous. Mutations in the POU class 4 transcription factor 3 (POU4F3) are known to cause autosomal dominant nonsyndromic hearing loss linked to the loci of DFNA15. In this study, we describe a pathogenic missense mutation in POU4F3 in a four-generation Chinese family (6126) with midfrequency, progressive, and postlingual autosomal dominant nonsyndromic hearing loss (ADNSHL). By combining targeted capture of 129 known deafness genes, next-generation sequencing, and bioinformatic analysis, we identified POU4F3 c.602T>C (p.Leu201Pro) as the disease-causing variant. This variant cosegregated with hearing loss in other family members but was not detected in 580 normal controls or the ExAC database and could be classified as a “pathogenic variant” according to the American College of Medical Genetics and Genomics guidelines. We conclude that POU4F3 c.602T>C (p.Leu201Pro) is related to midfrequency hearing loss in this family. Routine examination of POU4F3 is necessary for the genetic diagnosis of midfrequency hearing loss.
1. Introduction
Hearing loss is a common sensory defect in humans. Nonsyndromic hereditary forms, in which hearing loss is the only clinical sign, are known to be genetically heterogeneous. So far, more than 30 genes responsible for autosomal dominant nonsyndromic hearing loss (ADNSHL) have been identified (Hereditary Hearing Loss homepage, http://hereditaryhearingloss.org). Most patients with ADNSHL show large variations in age of onset and degrees of variety. According to the affected frequency, the phenotypes are divided into low-frequency, midfrequency, high-frequency, and all-frequency hearing loss. Midfrequency hearing loss (i.e., a U-shape audiogram) is a rare form of hearing loss, and six associated genes have been reported to date: EYA4, TECTA, COL11A2, CCDC50, POU4F3, and SLC44A4 [1–6]. Thus, in clinical molecular diagnosis, the hearing loss phenotype in a patient can aid the selection of a limited number of genes for mutational analysis.
This study identified one Chinese family with ADNSHL (number 6126), in which affected individuals showed typical midfrequency hearing loss. Using next-generation sequencing, we performed large-scale mutational screening of 129 known deafness-related genes and identified one novel dominant disease-segregating mutation, c.602T>C (p.Leu201Pro) in the POU4F3 gene, as the causative mutation that led to the midfrequency hearing loss phenotype in this family.
2. Materials and Methods
2.1. Clinical Data
Family 6126 is a four-generation Chinese family with autosomal dominant, late onset, progressive, nonsyndromic sensorineural hearing loss. Fully informed written consent was attained from each subject or their guardians. The study was approved by the Chinese PLA General Hospital's ethics research committees. Clinical information was gathered through multiple interviews with all participating members of the family. Medical history collection, otoscopy, physical examination, pure tone audiometric examination, and vestibular function were performed as previously described [7]. CT scans of the temporal bone in the index patients were performed.
2.2. DNA Preparation
Genomic DNA was extracted from peripheral blood using a blood DNA extraction kit according to the manufacturer's instructions (TianGen, Beijing, China).
2.3. Deafness Gene Capture and Illumina Library Preparation
Three prevalent deafness-associated genes, GJB2, SLC26A4, and mtDNA12SrRNA, were first screened for mutations in all participating cases and controls. Two affected individuals (II:1 and III:1) and one unaffected individual (II:3) of family 6126 were subjected to a gene panel containing 129 deafness genes. Capture and NGS of the coding exons for the 129 deafness genes and their flanking 100 bps (Supplemental Table 1) were performed on an Illumina HiSeq 2000 by Otogenetics Corporation (Norcross, GA, USA).
The details of the deafness gene capture, sequencing, and bioinformatics analysis methods have been described in detail previously [8]. According to the autosomal dominant pattern of inheritance, only variants that were heterozygous in the affected siblings were selected as candidates.
Segregation of the POU4F3 c.602T>C (p.Leu201Pro) variant was tested in seven family members (II:1, II:3, II:4, II:5, III:1, III:2, and IV:1), including the three whose gDNA had been subjected to NGS screening, using polymerase chain reaction (PCR) (primer sequences available on request) followed by bidirectional Sanger sequencing. Sequence alterations were identified by alignment with the POU4F3 GenBank sequence (NM_002700.2 and NP_002691.1) using the GeneTools software. To identify pathogenic mutations, cosegregation analysis was performed with the family members and an in-house database of 481 Chinese controls with normal hearing.
2.4. Multiple Sequence Alignment
Multiple sequence alignment was performed according to a Homologene program with default settings and the sequences NP_002691.1 (H. sapiens), XP_001100319.1 (M. mulatta), XP_527063.1 (P. troglodytes), XP_544328.1 (C. lupus), NP_001178964.1 (B. taurus), NP_620395.2 (M. musculus), NP_001102359.1 (R. norvegicus), NP_990090.1 (G. gallus), NP_571353.1 (D. rerio), NP_524876.1 (D. melanogaster), XP_308015.5 (A. gambiae), and XP_002935313.1 (X. tropicalis) (https://www.ncbi.nlm.nih.gov/homologene?cmd=Retrieve&dopt=MultipleAlignment&list_uids=2023).
3. Results
3.1. Family and Clinical Evaluations
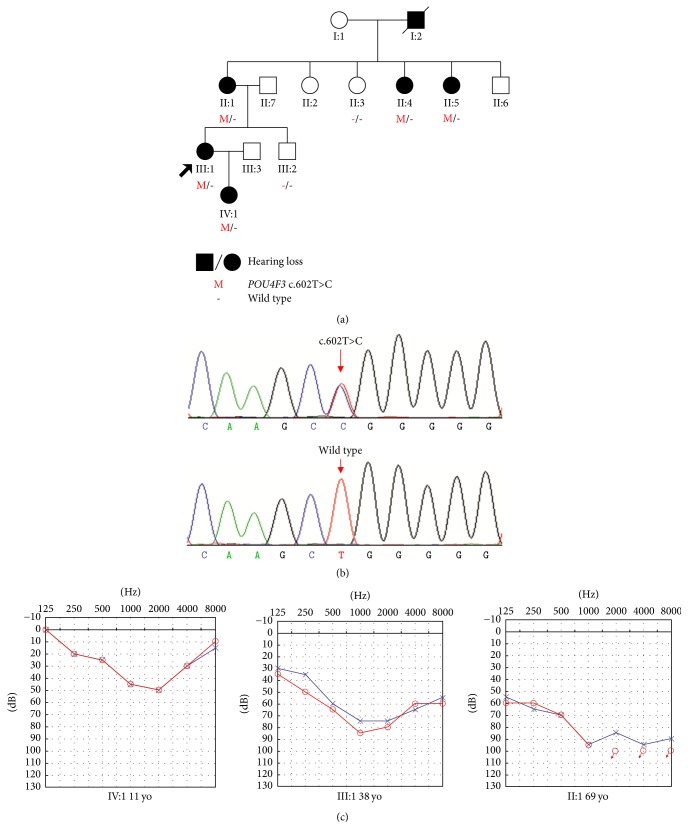
Family 6126 is a four-generation Chinese family with ADNSHL (Figure 1) and includes six affected patients: II:1 (female, 69 years old), II:4 (female, 60 years old), II:5 (female, 57 years old), III:1 (female, 42 years old), and IV:1 (female, 11 years old). For this family, hearing impairment was postlingual, late onset (after 10 years old), and progressive. Their hearing loss progressed gradually with advancing age. Audiograms show that although low-frequency and high-frequency hearing were normal in the beginning, their hearing would ultimately deteriorate at all frequencies. Flatter audiogram configurations were observed at 69 years of age (II:1), whereas the audiograms of III:1 and IV:1 were U-shaped (Figure 1(c)). For affected subjects II:4 and II:5, audiograms were unavailable.
Figure 1.
Pedigree, mutational analysis, and audiogram of Chinese family 6126 with ADNSHI. (a) The proband is indicated by an arrow. Subjects II:1, II:3 and III:1 were tested by NGS. (b) DNA sequencing profile showing the POU4F3 c.602T>C cosegregated with the hearing loss. (c) Audiogram showed bilateral sensorineural hearing impairment of affected subjects II:1, III:1, and IV:1 (red: right ear; blue: left ear).
Detailed vestibular analysis was performed in III:1, who did not complain about dizziness, vertigo, or imbalance. Vestibular tests revealed normal vestibular function via caloric tests. All position tests produced no nystagmus without vertigo sensation. Affected individuals did not have obvious delayed gross motor development. The physical examinations of all participating members revealed no signs of systemic illness or dysmorphic features. High-resolution computed tomography of the temporal bone in III:1 was normal, excluding inner ear malformations. This phenotype was consistent with that reported for DFNA15.
3.2. Mutation Detection and Analysis
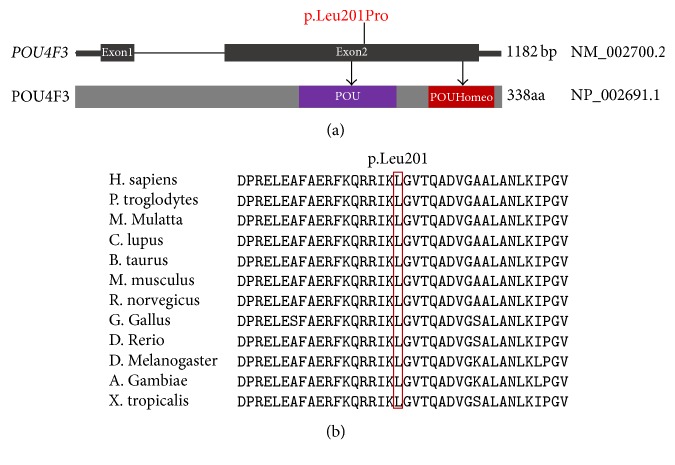
We sequenced all the coding exons plus ~100 bp of the flanking intronic sequence of 129 deafness genes in one unaffected (II:4) individual and two affected (II:1 and III:1) individuals of family 6126. One variant leading to amino acid change was detected in POU4F3: c.602T>C (p.Leu201Pro), which is located within exon 2 (Figure 2(a)). This variant has not been reported previously nor found in the ExAC database (http://exac.broadinstitute.org/) and was not observed in the 481 Chinese controls with normal hearing. The substitution occurred in an evolutionarily conserved region across different species in the POU domain (Figure 2(b)) and is predicted to be damaging by SIFT, Polyphen2, and CADD.
Figure 2.
Conservation analysis and genomic structure of POU4F3 based on the open reading frame (NM_002700.2) containing 2 exons (black rectangles). (a) The position of POU4F3 c.602T>C (p.Leu201Pro) is highlighted in red and shown both at the gene (top) and the protein level (bottom). The protein diagram depicts the predicted functional domains and sequence motifs. (b) Protein alignment showing POU4F3 p.Leu201 occurred at evolutionarily conserved amino acids (in red box) across twelve species.
Using Sanger sequencing, seven participating family members (five affected and two unaffected) in family 6126 were genotyped to identify the mutation. POU4F3 heterozygous variant p.Leu201Pro was found in five patients (Figure 1), consistent with autosomal dominant inheritance.
These data, together with the clinical presentation of the affected siblings and consistent autosomal dominant inheritance of the mutations in the affected and unaffected members, indicate that POU4F3 c.602T>C (p.Leu201Pro) is the cause of hearing impairment in this family.
4. Discussion
Inner ear hair cells play a crucial role in the mechanical transmission of sound and stimulation of the auditory nerve. A defect in hair cells in the cochlea can be a major reason for sensorineural hearing loss. POU4F3, a POU domain class IV protein, has two exons and encodes a protein of 338 amino acids that belongs to the POU domain family of transcription factors, which are expressed specifically in inner ear hair cells and play a critical role in the maturation, differentiation, and maintenance of inner ear hair cells [9]. POU4F3 contains two conserved DNA-binding domains (a POU-specific domain and a POU homeodomain; amino acids 179–256 and 274–333, resp.), which are the main functional parts [10].
In 1998, POU4F3 was first described as a disease-causing gene within the DFNA15 locus in an Israeli Jewish family [11]. So far, 27 variants in POU4F3 (13 missense variants, 8 frameshift variants, 4 nonsense variants, and 1 splice-site variant) (Table 1) and a whole deletion of POU4F3 [12] have been reported to cause ADNSHL with variable ages of onset and degrees of severity in various ethnic groups, including Chinese, Japanese, Dutch, Korean, and Brazilian populations [6, 10, 13–18]. Recently, Kitano et al. reported that POU4F3 variants represent the third largest cause of ADNSHL (2.5%, 15/602) in Japan and usually presented with mid- or high-frequency hearing loss. They also noticed that patients with truncating variants showed earlier onset and slower progression of hearing loss compared to those with nontruncating variants [6]. Through next-generation sequencing, He et al. reported that mutation in POU4F3 is a relatively common (3/18) cause of ADNSHL among Han Chinese people. Notably, most causative variants were located within or close to the POU-specific domain or the POU homeodomain, the two conserved DNA-binding domains of POU4F3 encoded in exon 2. The age of onset of hearing loss, ranging from the first to fourth decade of life, differs among POU4F3 mutations. In the present study, the earliest age of onset of hearing loss for affected family members was recorded at 11 years old (IV:1). Although the pathogenic mechanisms underlying hearing impairment of patients with POU4F3 variants remain unclear, the mechanism of haploinsufficiency has been supported by several studies [12, 19, 20].
Table 1.
Overview of POU4F3 mutations described in DFNA15.
| Number | Nucleotide change | Protein change | Exon | Domain | Origin | Audiometric configuration | Reference |
|---|---|---|---|---|---|---|---|
| (1) | Whole deletion of POU4F3 | Brazil | Flat and HF | Freitas et al., 2014 | |||
| (2) | c.74dupA | p.His25fs∗18 | 1 | Japan | HF | Kitano et al., 2017 | |
| (3) | c.120+1G>C | 1 | China | Flat | He et al., 2016 | ||
| (4) | c.191A>T | p.Asp64Val | 2 | Japan | HF | Kitano et al., 2017 | |
| (5) | c.337C>T | p.Gln113Ter | 2 | China | Zhang et al., 2016 | ||
| (6) | c.367delA | p.Ile123fs∗3 | 2 | Japan | MF | Kitano et al., 2017 | |
| (7) | c.427C>T | p.Gln143Ter | 2 | Japan | MF | Kitano et al., 2017 | |
| (8) | c.491C>G | p.Pro164Arg | 2 | POU | China | Flat and HF | Wei et al., 2014 |
| (9) | c.574G>T | p.Glu192Ter | 2 | POU | Japan | HF | Kitano et al., 2017 |
| (10) | c.581T>A | p.Phe194Tyr | 2 | POU | Japan | HF | Kitano et al., 2017 |
| (11) | c.602T>C | p.Leu201Pro | 2 | POU | China | MF | This study |
| (12) | c.602delT | p.Leu201fs∗3 | 2 | POU | China | HF | Cai et al., 2016 |
| (13) | c.603_604delGG 2 | p.Val203Aspfs∗11 | 2 | POU | China | N/A | Yang et al., 2013 |
| (14) | c.662_675del14 | p.Gly221Glufs∗14 | 2 | POU | Korea | HF | Lee et al., 2010 |
| (15) | c.665C>T | p.Ser222Leu | 2 | POU | Japan | HF | Kitano et al., 2017 |
| (16) | c.668T>C | p.Leu223Pro | 2 | POU | The Netherlands | Flat, MF, and HF | Collin et al., 2008 |
| (17) | c.680delC | p.Thr227fs∗13 | 2 | POU | Japan | MF | Kitano et al., 2017 |
| (18) | c.694G>A | p.Glu232Lys | 2 | POU | Korea | HF | Baek et al., 2012 |
| (19) | c.718A>T | p.Asn240Tyr | 2 | POU | Japan | MF | Kitano et al., 2017 |
| (20) | c.841A>G | p.Ile281Val | 2 | POU Homeobox | Japan | HF | Kitano et al., 2017 |
| (21) | c.865C>T | p.Leu289Phe | 2 | POU Homeobox | The Netherlands | Flat, MF, and HF | Collin et al., 2008 |
| (22) | c.884_891del8 | Ile295Thrfs∗5 | 2 | POU Homeobox | Israel | HF | Vahava et al., 1998 |
| (23) | c.896C>T | p.Pro299Leu | 2 | POU Homeobox | Japan | MF | Kitano et al., 2017 |
| (24) | c.932T>C | p.Leu311Pro | 2 | POU Homeobox | China | HF | He et al., 2016 |
| (25) | c.976A>T | p.Arg326Ter | 2 | POU Homeobox | Japan | HF | Kitano et al., 2017 |
| (26) | c.977G>A | p.Arg326Lys | 2 | POU Homeobox | Korea | HF | Kim et al., 2013 |
| (27) | c.982A>G | p.Lys328Glu | 2 | POU Homeobox | Taiwan | HF | Lin et al., 2017 |
| (28) | c.1007delC | p.Ala336fs | 2 | POU Homeobox | Japan | N/A | Mutai et al., 2013 |
Abbreviations. HF: high-frequency hearing loss; MF: midfrequency hearing loss.
The missense mutation p.Leu201Pro is located within the POU domain and encodes a proline at position 201 instead of the highly conserved leucine, which is close to the two reported dominant mutations of POU4F3 (c.603-604delGG [p.Leu201fs∗12] and c.602delT [p.Leu201fs]) [21, 22]. We speculate that to some extent, this area of the genome is unstable and susceptible to mutation.
In this study, we identified a novel missense mutation, c.602T>C in POU4F3, in one Chinese family (6216) with ADNSHL. Younger patients in this family demonstrated midfrequency hearing loss with no additional clinical symptoms. Our results strongly suggest that this missense mutation is related to hearing loss in this family, which has a putative autosomal dominant inheritance pattern.
5. Conclusions
In summary, we describe the clinical and genetic characteristics of a Chinese family (number 6126) with postlingual ADNSHL caused by POU4F3 c. 602 T>C (p. Leu201Pro) through multiple deafness gene capture and next-generation sequencing. Notably, the hearing impairment of affected individuals in this family is mainly midfrequency. This specific audiogram should be considered in clinical genetic diagnosis and counselling. Therefore, screening for POU4F3 in ADNSHL patients with a postlingual, progressive, and U-shape audiogram is necessary for efficient genetic diagnosis and intervention.
Acknowledgments
The investigations of Pu Dai were supported by the National Natural Science Foundation of China (81230020 and 81371096) and the National Key Research and Development Project (2016YFC1000700 and 2016YFC1000704). Grants from the National Natural Science Foundation of China (81570929) were provided to Xue Gao. Grants from the National Key Research and Development Project (2016YFC1000706) and the National Natural Science Foundation of China (81371098) were provided to Yong-Yi Yuan. Grants from the National Natural Science Foundation of China (81360159) and the National Basic Research Program of China (2014CB541706) were provided to Guo-Jian Wang. Grants from the Military Health Care Special Project (15BJZ23) were given to Jin-Cao Xu. Grants from National Natural Science Foundation of China (81200751) were presented to Sha-Sha Huang. Grants from the National Natural Science Foundation of China (81400471) were given to Yu Su. The authors sincerely thank all the family members in the study for their participation and cooperation in this study.
Contributor Information
Xi Lin, Email: xlin2@emory.edu.
Pu Dai, Email: daipu301@vip.sina.com.
Disclosure
Xue Gao, Jin-Cao Xu, and Wei-Qian Wang are listed as co-first authors. The English in this document has been checked by at least two professional editors, both native speakers of English. For a certificate, please see http://www.textcheck.com/certificate/dgjliP. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Authors' Contributions
Xue Gao, Pu Dai, and Xi Lin conceived the study and participated in its design and the draft of the manuscript. Yong-Yi Yuan participated in the next-generation sequencing. Jin-Cao Xu, Wei-Qian Wang, and Dan Bai participated in the data analysis. Guo-Jian Wang, Sha-Sha Huang, Yu Su, Jia Li, and Dong-Yang Kang participated in the collection of clinical data and blood samples. All authors read and approved the final manuscript.
Supplementary Materials
Supplemental Table 1: gene panel containing 129 deafness genes.
References
- 1.Ma Z., Xia W., Liu F., et al. SLC44A4 mutation causes autosomal dominant hereditary postlingual non-syndromic mid-frequency hearing loss. Human Molecular Genetics. 2017;26(16):p. 3234. doi: 10.1093/hmg/ddx232.ddx232 [DOI] [PubMed] [Google Scholar]
- 2.Pfister M., Thiele H., Van Camp G., et al. A genotype-phenotype correlation with gender-effect for hearing impairment caused by TECTA mutations. Cellular Physiology and Biochemistry. 2004;14(4-6):369–376. doi: 10.1159/000080347. [DOI] [PubMed] [Google Scholar]
- 3.Wayne S., Robertson N. G., DeClau F., et al. Mutations in the transcriptional activator EYA4 cause late-onset deafness at the DFNA10 locus. Human Molecular Genetics. 2001;10(3):195–200. doi: 10.1093/hmg/10.3.195. [DOI] [PubMed] [Google Scholar]
- 4.Modamio-Høybjør S., Mencía Á., Goodyear R., et al. A mutation in CCDC50, a gene encoding an effector of epidermal growth factor-mediated cell signaling, causes progressive hearing loss. American Journal of Human Genetics. 2007;80(6):1076–1089. doi: 10.1086/518311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGuirt W. T., Prasad S. D., Griffith A. J., et al. Mutations in COL11A2 cause non-syndromic hearing loss (DFNA13) Nature Genetics. 1999;23(4):413–419. doi: 10.1038/70516. [DOI] [PubMed] [Google Scholar]
- 6.Kitano T., Miyagawa M., Nishio S.-Y., et al. POU4F3 mutation screening in Japanese hearing loss patients: Massively parallel DNA sequencing-based analysis identified novel variants associated with autosomal dominant hearing loss. PLoS ONE. 2017;12(5) doi: 10.1371/journal.pone.0177636.e0177636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao X., Huang S.-S., Yuan Y.-Y., et al. Targeted gene capture and massively parallel sequencing identify TMC1 as the causative gene in a six-generation Chinese family with autosomal dominant hearing loss. American Journal of Medical Genetics. 2015;167(10):2357–2365. doi: 10.1002/ajmg.a.37206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao X., Wang G.-J., Yuan Y.-Y., et al. Novel compound heterozygous mutations in MYO7A associated with usher syndrome 1 in a Chinese family. PLoS ONE. 2014;9(7) doi: 10.1371/journal.pone.0103415.e103415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erkman L., McEvilly R. J., Luo L., et al. Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature. 1996;381(6583):603–606. doi: 10.1038/381603a0. [DOI] [PubMed] [Google Scholar]
- 10.Wei Q., Zhu H., Qian X., et al. Targeted genomic capture and massively parallel sequencing to identify novel variants causing Chinese hereditary hearing loss. Journal of Translational Medicine. 2014;12, article 311 doi: 10.1186/s12967-014-0311-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vahava O., Morell R., Lynch E. D., et al. Mutation in transcription factor POU4F3 associated with inherited progressive hearing loss in humans. Science. 1998;279(5358):1950–1954. doi: 10.1126/science.279.5358.1950. [DOI] [PubMed] [Google Scholar]
- 12.Freitas É. L., Oiticica J., Silva A. G., Bittar R. S. M., Rosenberg C., Mingroni-Netto R. C. Deletion of the entire POU4F3 gene in a familial case of autosomal dominant non-syndromic hearing loss. European Journal of Medical Genetics. 2014;57(4):125–128. doi: 10.1016/j.ejmg.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Zhang C., Wang M., Xiao Y., et al. A Novel Nonsense Mutation of POU4F3 Gene Causes Autosomal Dominant Hearing Loss. Neural Plasticity. 2016;2016 doi: 10.1155/2016/1512831.1512831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang T., Wei X., Chai Y., Li L., Wu H. Genetic etiology study of the non-syndromic deafness in Chinese Hans by targeted next-generation sequencing. Orphanet Journal of Rare Diseases. 2013;8(1, article 85) doi: 10.1186/1750-1172-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H. K., Park H.-J., Lee K.-Y., Park R., Kim U.-K. A novel frameshift mutation of POU4F3 gene associated with autosomal dominant non-syndromic hearing loss. Biochemical and Biophysical Research Communications. 2010;398(3):626–630. doi: 10.1016/j.bbrc.2010.06.061. [DOI] [PubMed] [Google Scholar]
- 16.Collin R. W. J., Chellappa R., Pauw R.-J., et al. Missense mutations in POU4F3 cause autosomal dominant hearing impairment DFNA15 and affect subcellular localization and DNA binding. Human Mutation. 2008;29(4):545–554. doi: 10.1002/humu.20693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baek J.-I., Oh S.-K., Kim D.-B., et al. Targeted massive parallel sequencing: the effective detection of novel causative mutations associated with hearing loss in small families. Orphanet Journal of Rare Diseases. 2012;7(1, article 60) doi: 10.1186/1750-1172-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H.-J., Won H.-H., Park K.-J., et al. SNP linkage analysis and whole exome sequencing identify a novel POU4F3 mutation in autosomal dominant late-onset nonsyndromic hearing loss (DFNA15) PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0079063.e79063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mutai H., Suzuki N., Shimizu A., et al. Diverse spectrum of rare deafness genes underlies early-childhood hearing loss in Japanese patients: a cross-sectional, multi-center next-generation sequencing study. Orphanet Journal of Rare Diseases. 2013;8(1, article 172) doi: 10.1186/1750-1172-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Y.-H., Lin Y.-H., Lu Y.-C., et al. A novel missense variant in the nuclear localization signal of POU4F3 causes autosomal dominant non-syndromic hearing loss. Scientific Reports. 2017;7(1, article no. 7551) doi: 10.1038/s41598-017-08236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He L., Pang X., Chen P., Wu H., Yang T. Mutation in the Hair Cell Specific Gene POU4F3 Is a Common Cause for Autosomal Dominant Nonsyndromic Hearing Loss in Chinese Hans. Neural Plasticity. 2016;2016 doi: 10.1155/2016/9890827.9890827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai X. Z., Li Y., Xia L., et al. Exome sequencing identifies POU4F3 as the causative gene for a large Chinese family with non-syndromic hearing loss. Journal of Human Genetics. 2016 doi: 10.1038/jhg.2016.102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: gene panel containing 129 deafness genes.