Abstract
Caffeoyl coenzyme A-3-O-methyltransferase (CCoAOMT) plays an important role in lignin biosynthesis and is encoded by two genes in poplar (Populus trichocarpa). Here, we describe the expression pattern conferred by the two CCoAOMT promoters when fused to the gus-coding sequence in transgenic poplar (Populus tremula × Populus alba). Both genes were expressed similarly in xylem and differentially in phloem. In xylem, expression was preferentially observed in vessels and contact rays, whereas expression was barely detectable in storage rays and fibers, suggesting different routes to monolignol biosynthesis in the different xylem types. Furthermore, after wounding, fungal infection, and bending, the expression of both genes was induced concomitantly with de novo lignin deposition. Importantly, upon bending and leaning of the stem, the cell-specific expression pattern was lost, and both genes were expressed in all cell types of the xylem. CCoAOMT promoter activity correlated well with the presence of the CCoAOMT protein, as shown by immunolocalization. These expression data may explain, at least in part, the heterogeneity in lignin composition that is observed between cell types and upon different environmental conditions.
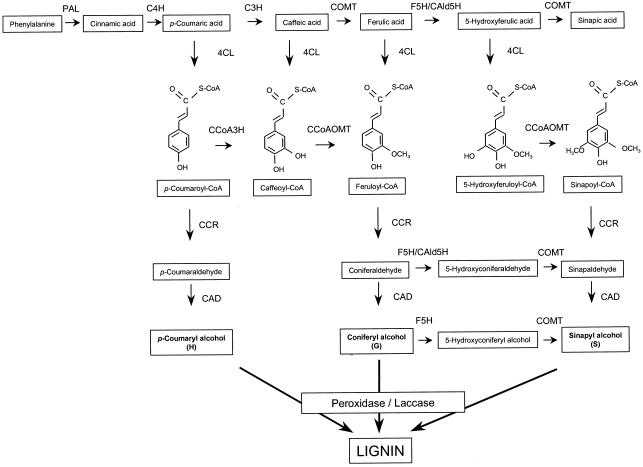
Lignin is a major structural component of plant secondary cell walls and is, after cellulose, the most abundant organic polymer on earth. In vascular plants, lignin provides rigidity to the cell walls, confers impermeability to xylem vessels, and forms a physicochemical barrier against microbial attack (Monties, 1989; Northcote, 1989; Moerschbacher et al., 1990). Lignin is mainly derived from the dehydrogenative polymerization of three different hydroxycinnamyl alcohols, p-coumaryl, coniferyl, and sinapyl alcohol, which give rise to the p-hydroxyphenyl, guaiacyl (G), and syringyl (S) units of the lignin polymer, respectively. These units differ by the degree of methoxylation at the 3 and/or 5 positions of the aromatic ring (Fig. 1). Therefore, the hydroxylation and methylation reactions are important in determining lignin composition.
Figure 1.
Phenylpropanoid and monolignol biosynthesis pathways. C3H, Coumarate 3-hydroxylase; CCoA3H, coumaroyl-CoA 3-hydroxylase; CCR, cinnamoyl-CoA reductase; F5H, ferulate 5-hydroxylase; CAld5H, coniferaldehyde 5-hydroxylase.
Although the overall biochemical route to lignin has been studied for many years, it is still largely unknown which reactions control the amount and composition of lignin (Boudet et al., 1995; Whetten and Sederoff, 1995; Dixon et al., 1996; Douglas, 1996; Baucher et al., 1998; Boudet, 1998; Whetten et al., 1998). The lignin polymer is complex and heterogenous with respect to the relative proportions of the three monolignol units and the different types of interunit linkages (for review, see Campbell and Sederoff, 1996). For example, lignin from gymnosperms consists mainly of G units, whereas lignin from angiosperms is predominantly made up of both G and S units. Lignin from grasses incorporates also considerable amounts of p-hydroxyphenyl units. The content and composition of lignin do not only differ among taxa but also between different cell types of a single tissue. Even within a single cell wall, lignin structure and/or composition can show considerable variation (Joseleau and Ruel, 1997). Furthermore, lignin heterogeneity is influenced by environmental stress. This heterogeneity is probably caused by differences in the spatiotemporal expression of certain enzymes of the lignin biosynthetic pathway and by differences in their substrate specificities (Campbell and Sederoff, 1996).
Both lignin content and composition are known to have an impact on several agro-industrial uses of plants. For instance, the amount of lignin and the S/G ratio are of critical importance in paper pulping and forage crop digestibility (for review, see Dixon et al., 1996; Baucher et al., 1998). A better understanding of the biosynthesis of lignin would provide opportunities to develop strategies that allow a more economic use of raw materials in the agro-industry. Therefore, there is considerable interest in genetic engineering of lignin levels and/or composition to improve digestibility of forage crops and pulping properties of trees (Baucher et al., 1998).
The biosynthesis pathway of the lignin precursors proceeds through the common phenylpropanoid pathway starting from Phe and leading to the synthesis of cinnamoyl-coenzyme A (CoA) esters. Subsequently, the cinnamoyl-CoA esters are channeled into the monolignol branch pathway to produce cinnamyl alcohols. Caffeic acid/5-hydroxyferulic acid O-methyl-transferase (COMT; EC 2.1.1.68) has been characterized in a number of species (Baucher et al., 1998; Whetten et al., 1998) and has long been considered as the only methylating enzyme involved in lignification. However, it has been shown that the O-methylation of the lignin precursors can also occur at the level of the hydroxycinnamoyl-CoA esters (Ye et al., 1994; Inoue et al., 1998; Martz et al., 1998; Meng and Campbell, 1998; Li et al., 1999). A specific O-methyltransferase, caffeoyl-CoA-3-O-methyltransferase (CCoAOMT; EC 2.1.1.104), which catalyzes the methylation of caffeoyl-CoA to feruloyl-CoA, was initially characterized in cell suspensions of parsley and carrot challenged with a fungal elicitor (Kneusel et al., 1989; Pakusch et al., 1989; Kühnl et al., 1989) and hence was believed to play a role in disease resistance. Later, based on the observation that CCoAOMT was markedly induced during lignification of in vitro differentiating tracheary elements of zinnia, Ye et al. (1994) suggested that this gene could also have an important function in lignification. Immunolocalization, tissue printing studies, and promoter β-glucuronidase (GUS) fusion analyses in various plant species have shown that expression of CCoAOMT is highly correlated with lignifying tissues (Ye and Varner, 1995; Ye, 1997; Inoue et al., 1998; Li et al., 1999). The possible involvement of CCoAOMT in lignification was further supported by studies in which COMT had been down-regulated in transgenic tobacco and poplar (Populus tremula × Populus alba). In these transgenic plants, thioacidolysis experiments revealed that the S to G ratio had decreased because of a drop in the number of S units, whereas the number of G units had remained the same (Atanassova et al., 1995) or had even increased (Van Doorsselaere et al., 1995; Tsai et al., 1998; Lapierre et al., 1999). Because inhibition of the activity of COMT did not reduce the production of G units, CCoAOMT was hypothesized to play a role in bypassing the COMT-mediated methylation of the lignin precursors at the cinnamoyl-CoA ester level (Van Doorsselaere et al., 1995; Lapierre et al., 1999). Finally, the involvement of CCoAOMT in lignification was recently demonstrated in transgenic tobacco, down-regulated for CCoAOMT (Zhong et al., 1998). Lignin analysis of these plants showed an increased S to G ratio and a decreased lignin content because of an overall decrease in both G and S units, indicating a role for CCoAOMT in the pathway toward G and S lignin.
We have recently isolated two genes encoding CCoAOMT from poplar (Chen et al., 1998, 1999). Here, we have focused on the spatiotemporal expression directed by the two poplar CCoAOMT promoters in transgenic poplar. Consistent with a predominant role for CCoAOMT in lignification, the expression driven by both promoters was closely associated with lignifying tissues, both during normal development and upon biotic (fungal infection) and abiotic (wounding and bending) stress, which is known to influence lignin deposition. In non-stressed xylem tissue both promoters were shown to confer expression preferentially in vessel elements and in contact rays, whereas expression was barely detectable in storage rays and fibers. However, upon bending of the stem, cell specificity in the xylem was lost and the expression of the chimeric genes was observed in all cell types of the xylem. The CCoAOMT promoter activity correlated well with the presence of CCoAOMT protein, as shown by immunolocalization, indicating that CCoAOMT production is transcriptionally regulated. This is the first report demonstrating differential gene expression between different ray cell types and cell-specific alterations in CCoAOMT expression upon mechanical stress in plants. Our data furthermore underscore differential and conditional expression of lignin biosynthesis genes as a molecular mechanism to explain lignin heterogeneity.
RESULTS
The Two CCoAOMT Promoters Confer Expression in Lignifying Cells
We have previously demonstrated that CCoAOMT is encoded by two genes in poplar (Chen et al., 1999). To analyze the spatiotemporal expression pattern conferred by the two CCoAOMT promoters, the 2.0-kb and the 1.4-kb upstream regions of gPtCCoAOMT1 and of gPtCCoAOMT2, respectively, were fused to the coding sequence of the gus gene. Poplar was transformed with a T-DNA containing either the chimeric PCCoAOMT1-GUS or the PCCoAOMT2-GUS gene. For each construct, 20 independent primary transformants were isolated. Histochemical GUS assays were carried out on 3-month-old greenhouse-grown transgenic poplars (60 cm height, 25 internodes). All transformants harboring the same construct exhibited a similar qualitative pattern of GUS localization. At the macroscopic level, GUS staining revealed that the two chimeric genes were expressed in vascular tissue (leaves, petioles, stems, and roots). Seven representative transgenic lines for each construct were chosen for further detailed analyses.
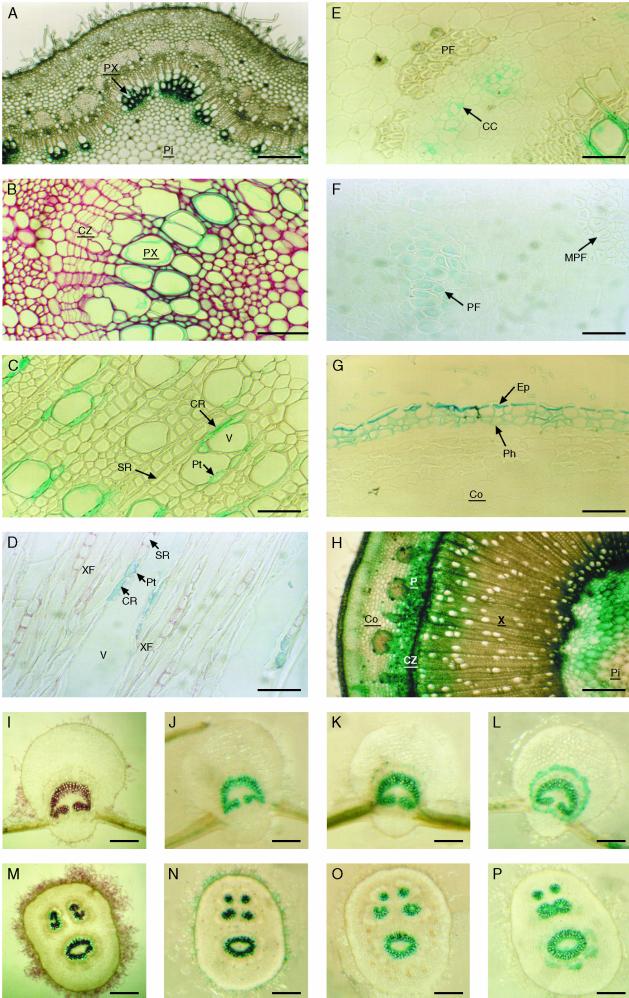
In agreement with the results from Osakabe et al. (1996), in transversal sections of top internodes (internode 2–4), only the primary xylem but not the phloem tissue was lignified as indicated by phloroglucinol-HCl (P-HCl) staining (data not shown). GUS activity driven by both promoters was detected in the primary xylem region (Fig. 2A). No staining was observed in the phloem, cambium, and pith. Observations under higher magnification revealed GUS activity in the vessels and in the primary xylem parenchyma cells between and surrounding the vessels (Fig. 2B). In internodes located at middle positions of the stem (internodes 8–15), in addition to the primary xylem, secondary xylem and phloem fibers had developed and were lignified, as revealed by P-HCl staining (data not shown). GUS staining driven by both promoters was mainly localized in young differentiating xylem and in the phloem region. Under higher magnification, we observed that both promoters directed a similar expression pattern in xylem, whereas they conferred differential expression in phloem tissue.
Figure 2.
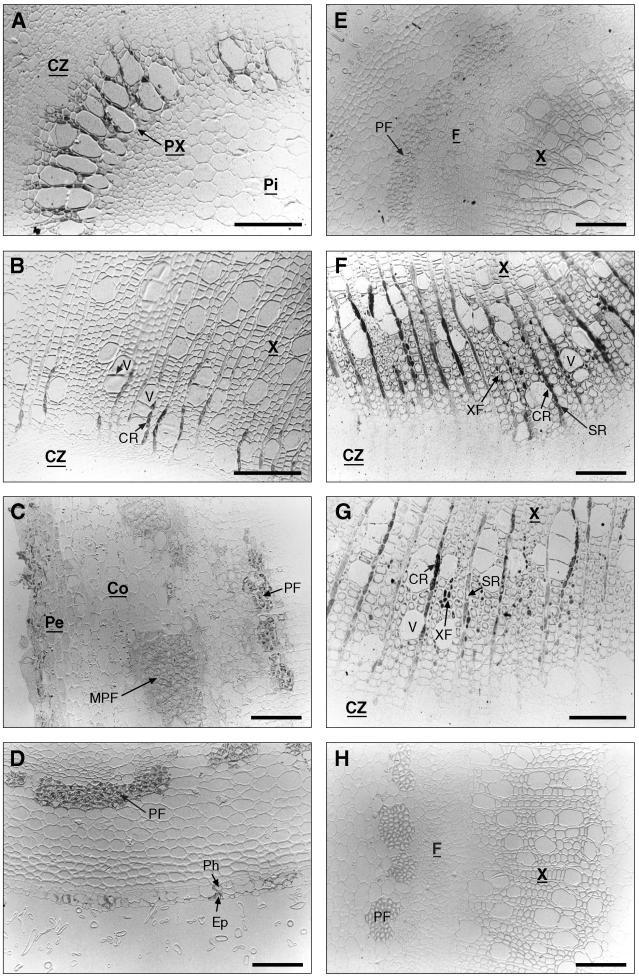
Histochemical analysis in transgenic poplar showing GUS activity or lignin deposition. A Thick transversal section of young stem of poplar transformed with PBINPOP1. B, Enlargement of A showing GUS activity in young xylem cells. C, Thin transversal section of the xylem in the middle part of the stem (PBINPOP1). D, Longitudinal section of the same stem as shown in C. E, Thin transversal section of the bark of the middle part of the stem (PBINPOP1). F, Thin section of the bark of the middle part of the stem (PBINPOP2). G, Thin transversal section of the bark showing GUS staining (PBINPOP1). H, (Legend continues on facing page.)Thick transversal section of a poplar stem transformed with a PCaMV35S-GUS construct. I and M, Thick transversal sections of young leaf and petiole stained with P-HCl. J and N, Thick transversal sections of young leaf and petiole (PBINPOP1). K and O, Thick transversal section of old leaf and petiole (PBINPOP1). L and P, Thick transversal section of old leaf and petiole (PBINPOP2) at the same developmental stage as sections in K and O. CC, Companion cell; Co, cortex; CR, contact ray cell; CZ, cambial zone; Ep, epidermis; MPF, mature phloem fibers; P, phloem; PF, phloem fibers; Ph, phellem; Pi, pith; Pt, pit; PX, primary xylem; SR, storage ray cell; V, vessel; X, xylem; XF, xylem fiber. Bars = 200 μm (in A and H–P) and 50 μm (in B–G).
In xylem, GUS activity was preferentially localized in developing vessel cells and in the ray cells adjacent to these vessels (Fig. 2C). A longitudinal section shows that only those ray cells that are connected to the adjacent vessel by pits (contact ray cells) stained blue but not the storage (isolation) rays (Fig. 2D). No GUS activity was detected in xylem fibers except for some fibers that were adjacent to the vessels that showed GUS activity. In phloem tissue of the top one-half of the plant, PCCoAOMT1-GUS was expressed in companion cells, whereas expression in phloem fibers was barely detectable (Fig. 2E). However, expression was observed in a limited number of cells surrounding the phloem fibers (data not shown). In contrast, GUS activity directed by PCCoAOMT2-GUS was located preferentially in differentiating phloem fibers and the adjacent cells but was undetectable in companion cells. No expression was detected in mature phloem fibers (Fig. 2F). In the lower part of the stem (internode 22–25) however, GUS activity conferred by PCCoAOMT1-GUS shifted from companion cells to the phloem fibers. Therefore, in the lower part of the stem, both genes were similarly expressed in young differentiating phloem fibers. The cell-specific expression in xylem remained the same throughout the whole stem. Both chimeric genes were also expressed in the epidermis and in the phellem of the periderm (Fig. 2G).
Poplars transformed with a PCaMV35S-GUS construct were used as controls (Fig. 2H). In agreement with the data of Nilsson et al. (1996), in cross sections of either young or older stems of the plants, GUS activity was localized in the cortex, the phloem tissue, the cambial zone, and the pith. Less-intense staining was observed in differentiating xylem and was not restricted to the contact ray cells. Therefore, we believe that the cell-specific expression of both chimeric genes truly reflects CCoAOMT promoter activity.
A similar temporal and cell-specific expression pattern was observed in leaves and petioles. In the leaves and petioles from the top part of the plant, expression of both chimeric genes was located in the primary xylem where lignification takes place (Fig. 2, I, J, M, and N). In the leaves and petioles from the middle part of the plant, both phloem fibers and xylem cells were highly lignified as indicated by P-HCl staining (data not shown). GUS staining revealed that both chimeric genes were highly expressed in the xylem and PCCoAOMT2-GUS was additionally expressed in phloem fibers (Fig. 2, K, L, O, and P). In the leaves and petioles from the basal part of the plant, staining in the vascular tissue was less intense, presumably because xylem and phloem fibers were fully lignified at this stage. Quantitative GUS analyses indicated a GUS activity gradient in the leaves, with 2- to 3-fold higher levels in the youngest leaves than in the oldest leaves (data not shown).
In addition to the expression in xylem, phloem, and periderm, expression conferred by both chimeric genes was often detected in hair cells of young stems, petioles, and leaves. In agreement with the function of CCoAOMT, these cells stained positively for phenolic compounds when treated with P-HCl (Fig. 2, I and M). Additional expression was observed in the meristem of apical and axillary buds (data not shown). In root tissue, both chimeric genes were expressed in phloem fibers, vessels, and contact rays, as in the stem (data not shown).
Both CCoAOMT Promoters Are Responsive to Biotic and Abiotic Stress
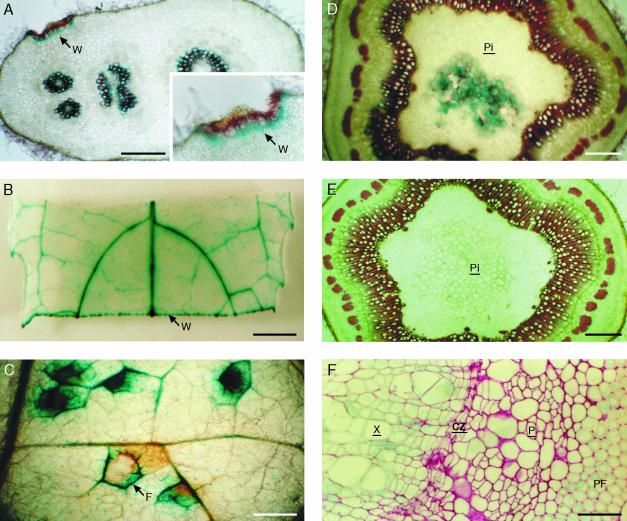
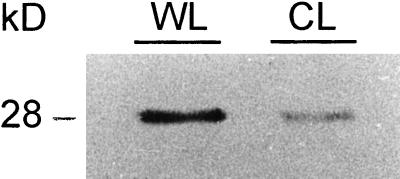
In addition to the developmentally regulated deposition of lignin, de novo formation of lignin can be induced at sites of wounding or pathogen attack. At these sites, lignin deposition plays a major role in plant defense by strengthening the cell wall to prevent the spread of the invading pathogen (Vance et al., 1980; Borg-Olivier and Monties, 1993; Hawkins and Boudet, 1996). To investigate whether the expression of the chimeric genes is associated with lignin deposition upon wounding, leaves and petioles of 2-month-old in vitro as well as 3-month-old greenhouse-grown transgenic poplar were wounded (see “Materials and Methods”). Regardless the tissue type (leaves or petioles), the developmental state (young or mature leaves), or the growth conditions (in vitro or greenhouse-grown), de novo lignin deposition occurred at the site of injury between 4 and 5 d after wounding, as visualized by P-HCl staining (Fig. 3A). GUS activity driven by both promoters was induced at the wounded site (Fig. 3B) and in the cells adjacent to those staining for lignin 4 d after wounding (Fig. 3A). GUS staining was barely detectable at the wounded sites until 3 d after wounding (data not shown). The induction of CCoAOMT production upon wounding of leaves was investigated by protein gel blotting as well. CCoAOMT was induced 4 d after wounding compared to the control (Fig. 4).
Figure 3.
CCoAOMT promoter activity in transgenic poplar upon biotic and abiotic stress conditions. A, Transversal section of a petiole stained for both lignin and GUS. B, GUS activity in a mechanically wounded leaf. C, M. pinitorca-infected leaf stained for both lignin deposition and GUS activity. D, Double staining of a thick transversal section of a bent stem for lignin and GUS activity. E, Thick transversal section of a non-bent stem stained by P-HCl as control. F, Enlargement of a thin transversal section of the bent stem, transformed with PBINPOP1. CZ, Cambial zone; F, site of fungal infection; P, phloem; PF, phloem fibers; Pi, pith; W, wound; X, xylem. Bars = 200 μm (in A, B, D, and E), 100 μm (in C), and 50 μm (in F).
Figure 4.
Protein gel-blot analysis of poplar protein extracts. Crude protein extracts from poplar control leaf (CL) and wounded leaf (WL) separated by SDS-PAGE, immobilized on a nitrocellulose membrane, and incubated with anti-CCoAOMT antibodies. The protein molecular mass is shown at the left in kD.
Detached leaves from transgenic and wild-type poplar were sprayed with spores of the fungus Melampsora pinitorca, a natural pathogen for poplars belonging to the Leuce section. Seven days after infection, the orange uredosores became visible, and necrotic lesions developed at the site of infection. At this stage, both promoters were induced in the cells surrounding the uredosores and necrotic lesions, as shown by the intense GUS staining, whereas no GUS staining was observed before the formation of the lesions. Figure 3C shows that the blue precipitate was closely associated with the lignified region at the infection site, indicating a close correlation between the induced expression of both chimeric genes and lignification at the site of necrosis.
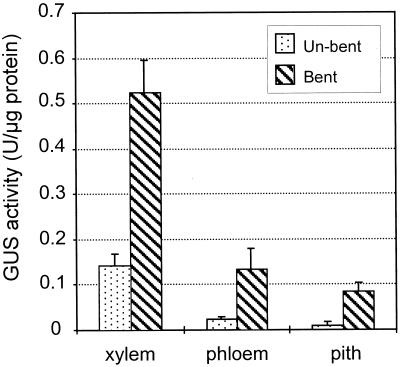
To study the expression of both chimeric genes upon mechanical stress, histochemical GUS analyses and quantitative fluorimetric assays were conducted on stems of 3-month-old greenhouse-grown poplar of which the stem had been gently fixed to an angle of 90° (see “Materials and Methods”). Quantitative GUS assays showed that after 9 d of bending, GUS activity had increased almost 4-fold in xylem and 5.5-fold in phloem tissue (Fig. 5). GUS activity was induced at similar levels in the tensed and the opposite part of the xylem and phloem. GUS activity in the pith, which was undetectable in the non-bent condition, increased to approximately 70% of the non-bent xylem value. Transversal sections were made after 9 d of bending. For both chimeric genes, a more intensive GUS staining was observed in xylem and phloem, compared with that of the non-bent condition, as well as in the pith, where no GUS activity was detected in the non-bent condition (Fig. 3, D and E). In agreement with a role for CCoAOMT in lignification, P-HCL staining was observed in the pith (Fig. 3D). Interestingly, in contrast to the non-bent condition, GUS staining was detected in all xylem cell types (Fig. 3F), although not uniformly over the entire xylem. The expression of PCCoAOMT1-GUS was also up-regulated in phloem fibers upon bending (Fig. 3F), whereas its expression was barely detectable in this tissue in the non-bent condition (Fig. 2E). However, bending did not induce GUS activity in phloem companion cells in PCCoAOMT2-GUS-transformed poplars. Thus, bending stress resulted in the loss of the cell-specific expression pattern in the vascular tissue and in a patchy expression of both genes in all lignifying cells of the xylem and phloem.
Figure 5.
Induction of GUS activity in transgenic poplar stems as a consequence of mechanical bending. GUS activity was measured in xylem, phloem, and pith tissues. The data represent the average of three independent experiments. The se is shown.
Immunolocalization of CCoAOMT in Poplar Stem
To verify whether the cell-specific and conditional expression driven by the CCoAOMT promoters coincided with the localization of the CCoAOMT protein, immunolocalization experiments were carried out on similar stem sections as those used for the GUS assays with antiserum against alfalfa CCoAOMT (Kersey et al., 1999). With these antibodies, a single 28-kD protein band was detected in the protein extracts from wild-type poplar stem by protein gel-blot analysis, whereas no signal was detected in protein extracts from transgenic poplars down-regulated for CCoAOMT (H. Meyermans and W. Boerjan, unpublished results). This observation indicates that the alfalfa CCoAOMT antibodies can specifically recognize CCoAOMT in poplar. Poplar CCoAOMT was localized by immunogold complexes with silver enhancement and examined by light microscopy. In general, labeling was present in differentiating phloem and xylem. Virtually no labeling appeared in the pith, cambium, and cortex. In sections through the top part of the stem, CCoAOMT was present in primary xylem bundles (Fig. 6A), which is in agreement with the result from promoter-GUS assays (Fig. 2B). In the middle and lower part of stem, CCoAOMT was detected in differentiating xylem (Fig. 6B), in differentiating phloem fibers but not in mature ones (Fig. 6C), as well as in the epidermis (Fig. 6D). Very faint labeling was observed in the companion cells (data not shown). In xylem tissue, as shown in Figure 6B, CCoAOMT was preferentially localized in contact ray cells associated with vessels and in the vessels themselves. This cell-specific localization of CCoAOMT in secondary xylem is similar to that of GUS directed by both CCoAOMT promoters (Fig. 2, A–G). No signal could be observed in control sections of the same stem treated with the preimmune serum (Fig. 6H) nor in the sections of the transgenic poplar down-regulated for CCoAOMT (Fig. 6E).
Figure 6.
Immunolocalization of CCoAOMT in transversal sections of poplar stem by light microscopy. A, Young stem of 3-month-old greenhouse-grown poplar. B, Xylem tissue of the middle part of a 3-month-old greenhouse-grown poplar stem. C and D, Bark tissue of old and young stem. E, Stem of transgenic poplar down-regulated for CCoAOMT. F, Bent stem. G, Leaned stem. H, Control section treated with preimmune serum. Co, Cortex; CR, contact ray cell; CZ, cambial zone; Ep, epidermis; F, phloem; MPF, mature phloem fibers; Pe, periderm; PF, phloem fibers; Ph, phellem; Pi, pith; PX, primary xylem; SR, storage ray cell; V, vessel; X, xylem; XF, xylem fiber. Bars = 100 μm.
To confirm that upon mechanical stress the cell-specific expression of CCoAOMT was altered, immunolocalization experiments were carried out on bent stems. As shown in Figure 6F, CCoAOMT was significantly induced in xylem tissue of stems that were bent for 9 d. In agreement with the induction of CCoAOMT promoter activity upon bending, immunolabeling was detected in all cell types of the xylem tissue and most intensely in the rays cells. In addition to mechanical bending, poplars were leaned at an angle of 45°, and sections were made through the curved part of the stem (see “Materials and Methods”). Also under this condition, the expression of CCoAOMT was significantly induced in all cell types of the xylem (Fig. 6G) and revealed an expression pattern similar to that after bending.
Subcellular Localization of CCoAOMT
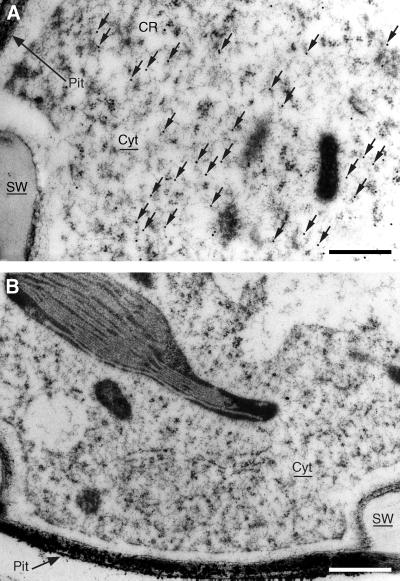
It is still largely unclear where monolignol biosynthesis occurs at the subcellular level. Several studies have suggested the existence of multienzyme complexes, other studies have shown the association of lignin biosynthesis enzymes with cellular organelles (Smith et al., 1994; Nakashima et al., 1997; Šamaj et al., 1998; Rasmussen and Dixon, 1999). To investigate whether CCoAOMT is entirely cytosolic or fractionally associated with membranes, we have localized CCoAOMT by electron microscopy on sections similar to those used for light microscopy and found a similar cellular distribution of CCoAOMT. Along the differentiating xylem, immunogold particles appeared mostly in the contact rays and in the vessels, although in the vessels only a small layer of cytoplasm remained prior to autolysis. Interestingly, at more advanced stages of xylem development, after autolysis of the vessels, no immunogold particles could be detected anymore neither in mature vessels nor in the contact rays adjacent to those mature vessels. Furthermore, labeling was observed in differentiating but not in mature phloem fibers, in the companion cells, in the epidermis, and in the phellem of the periderm, consistent with the promoter-GUS data and the immunolocalizations carried out by light microscopy. No signal was found in the cambium, pith, or cortex region (data not shown). A more detailed observation showed that the silver-enhanced particles were randomly localized in the cytoplasm and were not located on the cell wall or associated with any organelles (Fig. 7A). When similar sections were incubated with preimmune serum, no specific binding of gold particles was observed (Fig. 7B).
Figure 7.
Subcellular localization of CCoAOMT in poplar stem by electron microscopy. A, Contact ray cell from a stem section of the middle part of a 3-month-old greenhouse-grown poplar. The labeling with alfalfa CCoAOMT antibodies is concentrated in the cytosol. B, control section treated with preimmune serum. CR, Contact ray cell; Cyt, cytoplasm; SW, secondary wall. Arrows indicate gold particles. Bars = 1 μm.
DISCUSSION
Here, we have shown that the two CCoAOMT promoters direct expression in lignifying cells and in cells closely associated with lignifying cells, both during normal development and upon stress conditions known to influence lignin deposition (wounding, pathogen attack, and mechanical bending). These data are consistent with a role for CCoAOMT in lignification. Furthermore, both chimeric CCoAOMT genes were expressed in the epidermis (including hair cells) of young stems, leaves, petioles, and in the shoot apex. The gPtCCoAOMT1 promoter additionally directed expression in phloem companion cells, which are generally accepted to be not lignified or only poorly lignified. The deposition of phenolic compounds in non-lignified cells or tissues may serve important protective functions. In these cells, CCoAOMT may for example be involved in the biosynthesis of lignans, which share the same monolignol precursors as lignin. Lignans are an abundant class of phytochemicals in plants that play a role in defense, but little is known about their biosynthesis and their site of accumulation (Davin et al., 1992; Higuchi, 1997). The promoter of the eucalyptus cinnamyl alcohol dehydrogenase (CAD2) gene also conferred expression in poorly lignified cells, such as the periderm of stems, cortical cells containing crystals, companion cells, and cambial cells in transgenic poplar (Feuillet et al., 1995; Hawkins et al., 1997; Šamaj et al., 1998). CAD2 catalyzes the conversion of cinnamaldehydes to cinnamyl alcohols (Fig. 1) and may also be involved in lignan synthesis in these cells (Hawkins et al., 1997). The expression data additionally show that both promoters are multifunctional rather than diverged where each promoter has adopted separate functions. This result is in contrast to the 4-coumarate:CoA ligase (4CL) genes of poplar: Pt4CL1 is expressed in cells that participate in the developmentally regulated lignification process (mainly in xylem), whereas the Pt4CL2 confers expression in the epidermis of stem and leaf where it functions in the biosynthesis of phenylpropanoids other than lignin (Hu et al., 1998). Because immunolocalization of CCoAOMT showed the same tissue and cell-specific localization as revealed by GUS assays, we can conclude that the expression of CCoAOMT is mainly regulated at the transcriptional level.
CCoAOMT Is Expressed in Vessels and Contact Ray Cells
For several genes involved in lignin biosynthesis, the tissue-specific expression has been analyzed. For example, a bean Phe ammonia-lyase (PAL)-GUS fusion in tobacco and potato was expressed in all developing xylem cells including ray cells (Bevan et al., 1989). Hauffe et al. (1991) reported that P4CL-GUS expression was localized preferentially in differentiating xylem tissue and in xylem ray cells between highly lignified vessels and fibers in transgenic tobacco. The promoter of the CAD2 gene from eucalyptus conferred expression in phloem fibers, in the rays of differentiating xylem, and in the vascular cambium in transgenic poplar (Feuillet et al., 1995; Hawkins et al., 1997).
In contrast to the expression pattern conferred by the PAL, 4CL, and CAD2 promoters, the promoter of CCoAOMT was differentially expressed in different ray cell types. Under normal, non-stressed growth conditions, PCCoAOMT-GUS expression was preferentially observed in the contact ray cells that are connected to vessels through pits and not in the storage ray cells (isolation rays). These two types of ray cells have first been reported by Czaninski (1977), and they differ with respect to their contacts, through pits, with vessels. In dicotyledonous plants, pits are formed in the secondary xylem between a parenchyma cell and a conducting cell (vessel). Such pits are considered to play an important role during xylem cell differentiation, providing channels through which not only wall precursors and other metabolites may be transferred into the developing cell, but also signals that control their differentiation (for review, see Mezitt and Lucas, 1996). Our electron microscopy studies furthermore showed that at the stage that vessels are still alive, as judged by the presence of cytoplasm, CCoAOMT was present both in vessels and in the adjacent contact rays. However, upon autolysis of the vessel, expression of CCoAOMT was coincidentally switched off both in the vessels and in the contact rays. The similarity in timing of CCoAOMT expression in vessels and contact rays suggests that the vessels and contact rays share a common signal that triggers expression of the CCoAOMT genes in both cell types and that may be transported through the connecting pits. This hypothesis is in agreement with recent data of Murakami et al. (1999), who have shown by UV absorption of cell walls that lignification of the cell wall of contact rays and vessels in Populus maximowiczii occurs simultaneously and prior to that of the storage rays and fibers.
The observation that the CCoAOMT promoters drive expression in vessels and contact rays further raises the question whether the lignin composition of these cells differs from that of xylem fibers and storage rays. Higuchi (1997) has reported the predominant presence of G units in the lignin of vessel cell walls in angiosperm trees. The analysis of transgenic plants down-regulated for CCoAOMT and in vitro enzymatic assays support the idea that the difference in lignin composition is indeed due to differential expression of CCoAOMT. Zhong et al. (1998) have shown that lignin from transgenic tobacco plants down-regulated for CCoAOMT, apart from having a lower total Klason lignin content, is more depressed in G than in S units. Similar results were obtained in transgenic poplar, down-regulated for CCoAOMT (H. Meyermans, unpublished data).
Based on in vitro enzymatic assays, Osakabe et al. (1999) have recently proposed that the lignin precursors are predominantly synthesized via coniferaldehyde and not via ferulic acid, 5-hydroxyferulic acid, and sinapic acid. These data are based on the observation that coniferaldehyde inhibits the coniferaldehyde 5-hydroxylase/ferulic acid 5-hydroxylase reaction from ferulate to 5-hydroxyferulate in vitro. The cell-specific expression of CCoAOMT suggests that in vessels and contact rays, G and S units may be formed predominantly via caffeoyl-CoA and feruloyl-CoA, whereas in fibers and storage ray cells G and S units may be formed preferentially via caffeic acid, ferulic acid, and feruloyl-CoA. Based on the fact that COMT plays an important role in controlling S-unit biosynthesis (Atanassova et al., 1995; Van Doorsselaere et al., 1995; Tsai et al., 1998; Lapierre et al., 1999) by converting 5–hydroxyconiferaldehyde and 5–hydroxyconiferyl alcohol to sinapaldehyde and sinapyl alcohol, respectively (Humphreys et al., 1999; Osakabe et al., 1999), the conversion of caffeic acid to ferulic acid may also be less efficient in cells that make less S units. In vessels, where the S to G ratio is low compared with fibers (Saka and Goring, 1985), CCoAOMT may effectively bypass the COMT-mediated conversion of caffeic acid to ferulic acid. Together, the cell-specific expression of CCoAOMT and the different lignin composition in these cells suggest that the metabolic flux through the phenylpropanoid and lignin biosynthesis pathway is different in different cell types.
CCoAOMT Promoters Are Responsive to Wounding and Fungal Attack
The responses of plants to mechanical wounding and pathogen attack are often very similar and include the rapid accumulation of phenolic compounds, the production of phytoalexins, the synthesis of hydrolytic enzymes, and the reinforcement of cells with lignin and/or suberin at the site of injury (Blanchette and Biggs, 1992). In most of the cases examined, the appearance of such defense substances is the result of increased gene expression either within the affected tissue or throughout the plant. Most of the genes that code for enzymes of the lignin biosynthesis pathway, such as PAL, 4CL, COMT, and cinnamate 4-hydroxylase (C4H), have been reported to be induced by wounding, elicitors, and/or fungal infection (Baucher et al., 1998). CCoAOMT activity has been extensively studied in response to elicitors in cell suspension culture (Kneusel et al., 1989; Kühnl et al., 1989; Schmitt et al., 1991; Ni et al., 1996; Busam et al., 1997). Transient expression assays performed with PCCoAOMT-GUS constructs in parsley protoplasts showed a high expression level in response to elicitation (Grimmig and Matern, 1997). In tobacco, RNA gel-blot analyses showed an increased steady-state CCoAOMT mRNA level upon infection with tobacco mosaic virus or fungal elicitor (Martz et al., 1998). In our study the expression of both chimeric CCoAOMT genes was strongly induced at the wound site and at the infection site of leaves, concomitantly with the deposition of lignin or lignin-like material. That GUS activity was observed in the cell layer adjacent to the lignifying cell layer suggests that these cells provide monolignols to their neighboring cells for lignification or the production of lignin-like material. These results show that the induced formation of lignin or lignin-like material is closely correlated with the cellular localization of CCoAOMT gene expression in response to both wounding and fungal attack in plants.
CCoAOMT Promoters Are Responsive to Mechanical Stress
Mechanical stress caused by leaning stems results in compression wood in gymnosperms and tension wood in angiosperms (Timell, 1986; Castéra et al., 1994). Both compression and tension wood have been shown to have an altered lignin content and composition compared with normal non-stressed wood (Timell, 1986; Rolando et al., 1992). In contrast to the intensive study of lignin biosynthesis enzymes, such as PAL, 4CL, COMT, CAD, and cinnamoyl-CoA reductase in compression wood (Kutsuki and Higuchi, 1981; Zhang and Chiang, 1997), little information is available on gene expression in tension wood. Here, we report on a gene that is induced upon bending in a hardwood species. The expression of both chimeric CCoAOMT genes was up-regulated in all lignifying tissues of the stem 9 d after mechanical bending. There was a pronounced increase in GUS activity in xylem and phloem and in the pith cells concomitantly with the deposition of lignin or lignin-like material in the pith. It is noteworthy that in response to mechanical bending, the cell-specific expression pattern directed by both CCoAOMT promoters in the stem was lost.
Mechanical stress may also be the reason why PCCoAOMT1-GUS expression was observed in phloem fibers at the base of the stem and in the root, whereas no activity was observed in phloem fibers in the top one-half of the stem. Immunolocalization of CCoAOMT on sections of stems that were bent or leaned were in agreement with the promoter-GUS analyses and show that the induction of CCoAOMT expression upon mechanical stress is regulated at the transcriptional level. Considering the involvement of CCoAOMT in lignification, our observations suggest that the up-regulation of CCoAOMT in a non-cell-type-specific manner may alter lignin composition or content. However, Vander Mijnsbrugge et al. (2000) have recently reported that a poplar phenylcoumaran benzylic ether reductase, which is the most abundant protein in poplar xylem and involved in the biosynthesis of lignans (Gang et al., 1999), is up-regulated in xylem, phloem, and pith tissues of poplar stem in response to mechanical bending. Therefore, up-regulation of CCoAOMT in response to bending may also be involved in the synthesis of lignans apart from lignin. The analysis of lignin amount and composition as well as soluble phenolics from transgenic poplar, down-regulated for CCoAOMT, may shed further light on the fate of the produced monolignol intermediates upon mechanical stress.
CCoAOMT Is a Cytosolic Protein
The phenylpropanoid pathway is involved in the biosynthesis of a wide variety of natural products from plants. Little is known about the mechanism the cell uses to regulate the flux into the different end products of the pathway. Metabolic labeling experiments have suggested the existence of multienzyme complexes that channel intermediates of phenylpropanoid synthesis without their release into general metabolic pools (Stafford, 1981; Hrazdina, 1992; Rasmussen and Dixon, 1999). Several of the enzymes of the phenylpropanoid and lignin-specific pathways have been studied for their subcellular distribution by electron microscopy. In French bean, PAL has been localized in the cytosol whereas C4H was associated with the endoplasmic reticulum membrane and Golgi bodies (Smith et al., 1994). In poplar, CAD2 has been localized in the cytosol, on endoplasmic reticulum membranes and on Golgi-derived vesicles (Šamaj et al., 1998). In tracheary elements derived from zinnia mesophyll cells, two different types of PAL and CAD exist. For both enzymes, one isoform was shown to be cytosolic and the other associated to Golgi-derived vesicles and secondary cell walls (Nakashima et al., 1997). Recently, Rasmussen and Dixon (1999) have presented evidence for metabolic channeling, involving coupling of PAL and C4H, based on in vivo and in vitro labeling. These authors demonstrated that one form of PAL (PAL1) was associated with tobacco microsomes and was involved in channeling, and suggested that this isoform is in close physical association with C4H on microsomal membranes. Our results show that CCoAOMT is randomly distributed in the cytoplasm and is not directly involved in multienzyme complexes that are present at the membrane.
In summary, our data show that the CCoAOMT promoters are responsive to signals that control lignin deposition throughout plant development and adjust lignin quality according to environmental conditions. Taken together with the differential CCoAOMT promoter activity in the different xylem cell types, our observations may explain, at least part of, the large heterogeneity in lignin amount and composition that is observed between different cell types and within individual cell walls (Joseleau and Ruel, 1997; Baucher et al., 1998). Extensive promoter comparisons and deletion analyses are necessary to identify the cis elements that are involved in the responses to the different signals that induce CCoAOMT promoter activity.
MATERIALS AND METHODS
Plasmid Constructions
The characterization of gPtCCoAOMT1 (accession no. AJ223620) and gPtCCoAOMT2 (accession no. AJ223621) has been described previously (Chen et al., 1998, 1999). To fuse the 5′-untranslated region sequences of both CCoAOMT genes to the coding sequence of the GUS gene, an NcoI site was generated by PCR three codons downstream of the CCoAOMT start codon by using an oligonucleotide complementary to the 5′ flanking vector DNA in combination with the 21-mer oligonucleotide 5′-CTCTCCCATGGTGGCCATTAT-3′ and 5′-CTCTCCCATGGCGGCCATTAT-3′ for gPtCCoAOMT1 and gPtCCoAOMT2, respectively (a single line indicates the NcoI site and the reversed original start codon is bold and underlined. By using these oligonucleotides, the 1,994-bp and 1,363-bp promoter fragments of gPtCCoAOMT1 and gPtCCoAOMT2 were generated by PCR, respectively. Subsequently, both PCR products were digested with NcoI and SacI and cloned into the NcoI/SacI site of pGUS1 (Peleman et al., 1989), yielding the plasmids PGUSPOP1 and PGUSPOP2. Both chimeric PCCoAOMT1-GUS and PCCoAOMT2-GUS genes were subsequently isolated from PGUSPOP1 and PGUSPOP2, respectively, by an XbaI digest, and cloned into the XbaI site of the binary vector PBIN19 (Bevan, 1984), resulting in the plasmids PBINPOP1 and PBINPOP2, respectively.
Plant Material and Transformation
PBINPOP1, PBINPOP2, and PCaMV35-GUS (Nilsson et al., 1996) were mobilized to Agrobacterium tumefaciens strain C58C1Rif harboring the plasmid pMP90 by the freeze-thaw method described by Zahm et al. (1984). Poplar (Populus tremula × Populus alba Institut National de la Recherche Agronomique clone 717-1B4) was stably transformed with all three constructs according to Leplé et al. (1992). Each primary transformant was derived from a different explant. The plants were maintained and micropropagated in vitro on one-half Murashige and Skoog medium at 24°C with a 16-h light/8-h dark cycle. Five-week-old plants were transferred to soil and further grown in a greenhouse at 21°C with the same light/dark cycle.
Histochemical GUS Assays
Histochemical staining for GUS activity was performed essentially as described by Jefferson et al. (1987) and Hawkins et al. (1997). Small pieces of stems, roots, leaves, and petioles were excised and pretreated with 95% (v/v) acetone for 30 min at room temperature to prevent wound induction and rinsed three times with 100 mm potassium phosphate buffer (pH 7.0). GUS staining was carried out by incubating sections with 2 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 0.1 mm potassium ferricyanide, and 0.1 mm potassium ferrocyanide in 100 mm potassium phosphate buffer (pH 7.0). Staining was allowed to proceed at 37°C until blue stain developed in the samples (1–4 h). Subsequently, samples were fixed in 3% (v/v) glutaraldehyde in 100 mm potassium phosphate buffer (pH 7.0) overnight at 4°C, washed in the same buffer, and embedded in Histoform according to the manufacturer's protocol (Historesin Embedding Kit, Heraeus Kulzer, Wehrheim, Germany). Thin sections (8 μm) were cut on a microtome (Reichert-Jung, Nussloch, Germany). For thick slices, small pieces of stems, leaves, and petioles were pretreated with 95% (v/v) acetone, embedded in 7% (w/v) agarose, and sectioned (75–150 μm) with a vibroslicer (Laborimpex, Brussels). GUS staining was carried out as described above. All the sections were mounted on microscope slides for photography (Diaplan, Leitz, Wetzlar, Germany).
Quantitative GUS Assays
Quantitative GUS analyses were performed as described by Jefferson et al. (1987). Protein concentrations were measured with the method of Bradford (1976). GUS activity was assayed by enzymatic conversion of 4-methylumbelliferyl-β-d-glucuronide to 4-methylumbelliferone which was quantified with a fluorimeter (Labsystems Fluoreskan II, Helsinki). GUS activity was expressed in units per microgram of protein.
Lignin Staining
Lignin and/or phenolic compounds were revealed by P-HCl staining according to Speer (1987). Sections and samples were incubated for 2 min in phloroglucinol solution (1% in ethanol:water, 92:8 [v/v]) and then mounted in 25% (v/v) HCl, prior to microscopic examination.
Wounding, Bending, Leaning, and Pathogen Infection
For the wounding experiments, petioles of 3-month-old, greenhouse-grown plants were wounded by making a 0.5-cm-long slit with a scalpel. Wounding of the leaves was done on plants that were grown axenically in glass jars for 2 months. From each of six comparable plants, one-half of a leaf was removed using a sterile scalpel, and a strip of leaf tissue of approximately 2 mm bordering the cutting site was harvested after 0, 1, 2, 3, 4, and 5 d after wounding. The harvested samples were analyzed by histochemistry for GUS activity and lignin deposition as well as by protein gel-blot analysis.
For the bending experiments, stems of 3-month-old greenhouse-grown wild-type and transgenic poplars were gently fixed at a 90° angle at internode 3 to 4 counted from the top of the stem for various periods. Non-bent stems of the same, vegetatively propagated transgenic line were used as control. The bent part of the stem was collected for GUS assays and immunolocalization experiments. For quantitative GUS assays, the xylem, phloem, and pith tissues from the bent stems were collected separately and immediately frozen in liquid nitrogen. The harvested samples were stored at −70°C until analysis.
Alternatively, 3-month-old greenhouse-grown poplars were leaned at 45° for 9 d. The upper part of the stem (approximately 15–25 cm) grows vertically after leaning the plant. The samples for immunolocalization were taken from the curved part of the plant, i.e. between the oblique and the vertical part of the stem.
Spores of the fungus Melampsora pinitorca were collected from leaves of outside-grown poplar trees and determined according to Pinon (1973). For fungal infection, detached leaves were put upside down on wet filter paper in 9-cm Petri dishes. Subsequently, these leaves were sprayed for 2 s with spores of the fungus, in a concentration of 200,000 spores mL−1, using a spray machine (Aku-Sprühpistole W50, Wagner, Markdorf, Germany). The infected leaves were subsequently floated upside down on water in 9-cm Petri dishes and incubated at 22°C in a greenhouse for various periods.
Protein Gel Blotting
Proteins were extracted with 100 mm Tris (tris- [hydroxymethyl]aminomethane)-HCl (pH 7.5), 2 mm EDTA, 20% (v/v) glycerol, and 1 mm dithiothreitol. Protein (5 μg) from leaves was separated by SDS-PAGE. The protein gel was subsequently blotted on Hybond-C super membrane according to the manufacturer's instructions (Amersham, Aylesbury, UK). After incubation with antiserum raised against alfalfa CCoAOMT (Kersey et al., 1999), filters were incubated with anti-rabbit IgG alkaline phosphatase conjugate (Roche Diagnostics, Brussels) at a dilution of 1/2,500. The immunodetection was performed using an alkaline phosphate p-toluidine salt (Duchefa, Haarlem, The Netherlands) and p-nitroblue tetrazolium chloride (Duchefa) in the detection reaction.
Sample Preparation for Light and Electron Microscopy
Small pieces of tissue harvested from non-stressed stem, wounded stem, and stems that had been bent or leaned were cut into pieces of approximately 1 mm3. These pieces were immersed in fixation solution (2.5% [v/v] paraformaldehyde and 0.3% [v/v] glutaraldehyde in 0.1 m sodium-cacodylate buffer [21.4 g Na(CH3)2AsO2 · 3H2O2 in 1,000 mL of distilled water], pH 7.2) under vacuum for 4 h at room temperature and then incubated for 14 h at 4°C under rotation. Following three washes for 2 h in 0.1 m sodium-cacodylate buffer (pH 7.2) at 4°C, the samples were dehydrated through a graded ethanol series under rotation at 4°C as follows: 30% (v/v) ethanol for 2 h, 50% (v/v) ethanol for 2 h, 70% (v/v) ethanol overnight, 95% (v/v) ethanol for 2 h, and 95% (v/v) ethanol overnight. The samples were embedded by incubating them successively in a 1:1 (v/v) ratio of 95% (v/v) ethanol:LR White (London Resin Co., Basingstoke, UK) at 4°C overnight and then for three changes, at least 8 h each, in pure resin at 4°C. The samples were placed in gelatin capsules containing fresh nitrogen-fluxed resin. Polymerization was performed by UV illumination at 4°C for 24 h followed by 24 h at 65°C to ensure complete polymerization of the resin. These LR White-embedded samples were ready to be sectioned for immunocytochemical localization of proteins using light or electron microscopy.
Light Microscopy
LR White-embedded samples were cut in semithin sections (1–3 μm) using a 2050 microtome (Reichert-Jung). The sections were collected on Vectabond-coated glass slides. Rabbit polyclonal anti-CCoAOMT antibodies (Kersey et al., 1999) were used as primary antibody. The AuroProbe One and IntenSE reagents were used for immunogold silver staining according to the manufacturer's protocol (Amersham). Sections were examined with a light microscope Jenalumar (Zeiss, Jena, Germany).
Electron Microscopy
Ultrathin sections of gold interference color (60–90 nm) were made from the LR White-embedded samples using an ultracut E (Reichert-Jung), and were collected on collodion-coated Cu grids. These sections were floated with the tissue-containing side downward for 5 min on a droplet of blocking solution (0.1% [w/v] bovine serum albumin and 0.05% [w/v] NaN3 in phosphate-buffered saline) to avoid non-specific adsorption of the antibodies. Subsequently, the sections were incubated for 1 h with primary antiserum raised against alfalfa CCoAOMT (Kersey et al., 1999), diluted 1:100 in blocking solution, followed by a 30-min incubation with a protein A-gold conjugate (15 nm; Amersham) diluted 1:50 in gold buffer (1% [w/v] bovine serum albumin and 0.05% [w/v] NaN3 in phosphate-buffered saline). Sections were washed three times with gold buffer, three times with distilled water, and then allowed to air-dry. The grids were post-stained for 12 min with 2% (w/v) uranyl acetate, washed five times with distilled water, and then left to air dry. Sections were examined using transmission electron microscopy (Elmiskop 101, Siemens, Karlsruhe, Germany).
ACKNOWLEDGMENTS
The authors thank Björn Sundberg for helpful discussions, Sabrina Neyrinck for technical assistance, Jan Van Doorsselaere and Jørgen Holst Christensen for critical reading of the manuscript, Martine De Cock for help with preparing it, and Rebecca Verbanck for the figures.
Footnotes
This work was supported by the Flemish Government (grant no. IBW/3/1998) and the European Union (grant nos. FAIR CT95–0424 and INCO–DC IC18–CT97–0203). G.J.E. is a Research Engineer of the Institut National de la Recherche Agronomique (France).
LITERATURE CITED
- Atanassova R, Favet N, Martz F, Chabbert B, Tollier MT, Monties B, Fritig B, Legrand M. Altered lignin composition in transgenic tobacco expressing O-methyltransferase sequences in sense and antisense orientation. Plant J. 1995;8:465–477. [Google Scholar]
- Baucher M, Monties B, Van Montagu M, Boerjan W. Biosynthesis and genetic engineering of lignin. Crit Rev Plant Sci. 1998;17:125–197. [Google Scholar]
- Bevan M. Binary Agrobacteriumvectors for plant transformation. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M, Shufflebottom D, Edwards K, Jefferson R, Schuch W. Tissue- and cell-specific activity of a phenylalanine ammonia-lyase promoter in transgenic plants. EMBO J. 1989;8:1899–1906. doi: 10.1002/j.1460-2075.1989.tb03592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette RA, Biggs AR. Defense Mechanisms of Woody Plants against Fungi (Springer Series in Wood Science). Berlin: Springer-Verlag; 1992. [Google Scholar]
- Borg-Olivier O, Monties B. Lignin, suberin, phenolic acids and tyramine in the suberized, wound-induced potato periderm. Phytochemistry. 1993;32:601–606. [Google Scholar]
- Boudet A-M. A new view of lignification. Trends Plant Sci. 1998;3:67–71. [Google Scholar]
- Boudet AM, Lapierre C, Grima-Pettenati J. Biochemistry and molecular biology of lignification. New Phytol. 1995;129:203–236. doi: 10.1111/j.1469-8137.1995.tb04292.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Busam G, Junghanns KT, Kneusel RE, Kassemeyer H-H, Matern U. Characterization and expression of caffeoyl-coenzyme A 3-O-methyltransferase proposed for the induced resistance response of Vitis viniferaL. Plant Physiol. 1997;115:1039–1048. doi: 10.1104/pp.115.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MM, Sederoff RR. Variation in lignin content and composition: mechanisms of control and implications for the genetic improvement of plants. Plant Physiol. 1996;110:3–13. doi: 10.1104/pp.110.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castéra P, Nepveu G, Mahé F, Valentin G. A study on the growth stresses, tension wood distribution and other related wood defects in poplar (Populus euramericanacv I214): end splits, specific gravity and pulp yield. Ann Sci For. 1994;51:301–313. [Google Scholar]
- Chen C, Ardiles-Diaz W, Van Montagu M, Boerjan W. A poplar gene for caffeoyl-coenzyme A 3-O-methyltransferase (accession no. AJ223620) (PGR 99-085) Plant Physiol. 1999;120:635. [Google Scholar]
- Chen C, Meyermans H, Van Doorsselaere J, Van Montagu M, Boerjan W. A gene encoding caffeoyl coenzyme A 3-O-methyltransferase (CCoAOMT) from Populus trichocarpa (accession no. AJ223621) (PGR 98-104) Plant Physiol. 1998;117:719. [Google Scholar]
- Czaninski Y. Vessel-associated cells. Int Assoc Wood Anat Bull. 1977;3:51–55. [Google Scholar]
- Davin LB, Lewis NG, Umezawa T. Phenylpropanoid metabolism: biosynthesis of monolignols, lignans and neolignans, lignins and suberins. In: Stafford HA, Ibrahim RK, editors. Phenolic Metabolism in Plants, Recent Advances in Phytochemistry. Vol. 26. New York: Plenum Press; 1992. pp. 325–375. [Google Scholar]
- Dixon RA, Lamb CJ, Masoud S, Sewalt VJ, Paiva NL. Metabolic engineering: prospects for crop improvement through the genetic manipulation of phenylpropanoid biosynthesis and defense responses: a review. Gene. 1996;179:61–71. doi: 10.1016/s0378-1119(96)00327-7. [DOI] [PubMed] [Google Scholar]
- Douglas C. Phenylpropanoid metabolism and lignin biosynthesis: from weeds to trees. Trends Plant Sci. 1996;1:171–178. [Google Scholar]
- Feuillet C, Lauvergeat V, Deswarte C, Pilate G, Boudet A, Grima-Pettenati J. Tissue- and cell-specific expression of a cinnamyl alcohol dehydrogenase promoter in transgenic poplar plants. Plant Mol Biol. 1995;27:651–667. doi: 10.1007/BF00020220. [DOI] [PubMed] [Google Scholar]
- Gang DR, Kasahara H, Xia Z-Q, Vander Mijnsbrugge K, Bauw G, Boerjan W, Van Montagu M, Davin LB, Lewis NG. Evolution of plant defense mechanisms: relationships of phenylcoumaran benzylic ether reductases to pinoresinol-lariciresinol and isoflavone reductases. J Biol Chem. 1999;274:7516–7527. doi: 10.1074/jbc.274.11.7516. [DOI] [PubMed] [Google Scholar]
- Grimmig B, Matern U. Structure of the parsley caffeoyl-CoA O-methyltransferase gene, harbouring a novel elicitor responsive cis-acting element. Plant Mol Biol. 1997;33:323–341. doi: 10.1023/a:1005780529457. [DOI] [PubMed] [Google Scholar]
- Hauffe KD, Paszkowski U, Schulze-Lefert P, Hahlbrock K, Dangl JL, Douglas CJ. A parsley 4CL-1 promoter fragment specifies complex expression patterns in transgenic tobacco. Plant Cell. 1991;3:435–443. doi: 10.1105/tpc.3.5.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins S, Boudet A. Wound-induced lignin and suberin deposition in a woody angiosperm (Eucalyptus gunniiHook.): histochemistry of early changes in young plants. Protoplasma. 1996;191:96–104. [Google Scholar]
- Hawkins S, Samaj J, Lauvergeat V, Boudet A, Grima-Pettenati J. Cinnamyl alcohol dehydrogenase: identification of new sites of promoter activity in transgenic poplar. Plant Physiol. 1997;113:321–325. doi: 10.1104/pp.113.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi T. Biochemistry and Molecular Biology of Wood (Springer Series in Wood Science). Berlin: Springer-Verlag; 1997. [Google Scholar]
- Hrazdina G. Compartmentation in aromatic metabolism. In: Stafford HA, Ibrahim RK, editors. Phenolic Metabolism in Plants, Recent Advances in Phytochemistry. Vol. 26. New York: Plenum Press; 1992. pp. 1–23. [Google Scholar]
- Hu W-J, Kawaoka A, Tsai C-J, Lung J, Osakabe K, Ebinuma H, Chiang VL. Compartmentalized expression of two structurally and functionally distinct 4-coumarate:CoA ligase genes in aspen (Populus tremuloides) Proc Natl Acad Sci USA. 1998;95:5407–5412. doi: 10.1073/pnas.95.9.5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys JM, Hemm MR, Chapple C. New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci USA. 1999;96:10045–10050. doi: 10.1073/pnas.96.18.10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Sewalt VJH, Ballance GM, Ni W, Stürzer C, Dixon RA. Developmental expression and substrate specificities of alfalfa caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase in relation to lignification. Plant Physiol. 1998;117:761–770. doi: 10.1104/pp.117.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseleau J-P, Ruel K. Study of lignification by noninvasive techniques in growing maize internodes: an investigation by Fourier transform infrared cross-polarization-magic angle spinning 13C-nuclear magnetic resonance spectroscopy and immunocytochemical transmission electron microscopy. Plant Physiol. 1997;114:1123–1133. doi: 10.1104/pp.114.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey R, Inoue K, Schubert KR, Dixon RA. Immunolocalization of two lignin O-methyltransferases in stems of alfalfa (Medicago sativaL.) Protoplasma. 1999;209:46–57. doi: 10.1007/BF01415700. [DOI] [PubMed] [Google Scholar]
- Kneusel RE, Matern U, Nicolay K. Formation of trans-caffeoyl CoA from trans-4-coumaroyl CoA by Zn2+-dependent enzymes in cultured plant cells and its activation by an elicitor-induced pH shift. Arch Biochem Biophys. 1989;269:455–462. doi: 10.1016/0003-9861(89)90129-x. [DOI] [PubMed] [Google Scholar]
- Kühnl T, Koch U, Heller W, Wellmann E. Elicitor inducedS-adenosyl-l-methionine:caffeoyl-CoA3-O- methyltransferase from carrot cell suspension cultures. Plant Sci. 1989;60:21–25. [Google Scholar]
- Kutsuki H, Higuchi T. Activities of some enzymes of lignin formation in reaction wood of Thuja orientalis, Metasequoia glyptostroboides and Robinia pseudoacacia. Planta. 1981;152:365–368. doi: 10.1007/BF00388263. [DOI] [PubMed] [Google Scholar]
- Lapierre C, Pollet B, Petit-Conil M, Toval G, Romero J, Pilate G, Leplé J-C, Boerjan W, Ferret V, De Nadai V, Jouanin L. Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid O-methyltransferase activity have opposite impact on the efficiency of industrial kraft pulping. Plant Physiol. 1999;119:153–163. doi: 10.1104/pp.119.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplé JC, Brasileiro ACM, Michel MF, Delmotte F, Jouanin L. Transgenic poplars: expression of chimeric genes using four different constructs. Plant Cell Rep. 1992;11:137–141. doi: 10.1007/BF00232166. [DOI] [PubMed] [Google Scholar]
- Li L, Osakabe Y, Joshi CP, Chiang VL. Secondary xylem-specific expression of caffeoyl-coenzyme A 3-O-methyltransferase plays an important role in the methylation pathway associated with lignin biosynthesis in loblolly pine. Plant Mol Biol. 1999;40:555–565. doi: 10.1023/a:1006244325250. [DOI] [PubMed] [Google Scholar]
- Martz F, Maury S, Pinçon G, Legrand M. cDNA cloning, substrate specificity and expression study of tobacco caffeoyl-CoA 3-O-methyltransferase, a lignin biosynthetic enzyme. Plant Mol Biol. 1998;36:427–437. doi: 10.1023/a:1005969825070. [DOI] [PubMed] [Google Scholar]
- Meng H, Campbell WH. Substrate profiles and expression of caffeoyl coenzyme A and caffeic acid O-methyltransferases in secondary xylem of aspen during seasonal development. Plant Mol Biol. 1998;38:513–520. doi: 10.1023/a:1006071708728. [DOI] [PubMed] [Google Scholar]
- Mezitt LA, Lucas WJ. Plasmodesmal cell-to-cell transport of proteins and nucleic acids. Plant Mol Biol. 1996;32:251–273. doi: 10.1007/BF00039385. [DOI] [PubMed] [Google Scholar]
- Moerschbacher BM, Noll U, Gorrichon L, Reisener H-J. Specific inhibition of lignification breaks hypersensitive resistance of wheat to stem rust. Plant Physiol. 1990;93:465–470. doi: 10.1104/pp.93.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monties B. Lignins. In: Dey PM, Harborne JB, editors. Methods in Plant Biochemistry: Plant Phenolics. Vol. 1. New York: Academic Press; 1989. pp. 113–157. [Google Scholar]
- Murakami Y, Funada R, Sano Y, Ohtani J. The differentiation of contact cells and isolation cells in the xylem ray parenchyma of Populus maximowiczii. Ann Bot. 1999;84:429–435. [Google Scholar]
- Nakashima J, Awano T, Takabe K, Fujita M, Saiki H. Immunocytochemical localization of phenylalanine ammonia-lyase and cinnamyl alcohol dehydrogenase in differentiating tracheary elements derived from Zinniamesophyll cells. Plant Cell Physiol. 1997;38:113–123. [Google Scholar]
- Ni W, Sewalt VJH, Korth KL, Blount JW, Ballance GM, Dixon RA. Stress response in alfalfa: XXI. Activation of caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase genes does not contribute to changes in metabolite accumulation in elicitor-treated cell-suspension cultures. Plant Physiol. 1996;112:717–726. doi: 10.1104/pp.112.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson O, Little CHA, Sandberg G, Olsson O. Expression of two heterologous promoters, Agrobacterium rhizogenes rolC and cauliflower mosaic virus 35S, in the stem of transgenic hybrid aspen plants during the annual cycle of growth and dormancy. Plant Mol Biol. 1996;31:887–895. doi: 10.1007/BF00019475. [DOI] [PubMed] [Google Scholar]
- Northcote DH. Control of plant cell wall biosynthesis: an overview. In: Lewis NG, Paice MG, editors. Plant Cell Wall Polymers, ACS Symposium Series. Vol. 399. Washington, DC: American Chemical Society; 1989. pp. 1–15. [Google Scholar]
- Osakabe Y, Nanto K, Kitamura H, Kawai S, Kondo Y, Fujii T, Takabe K, Katayama Y, Morohoshi N. Immunocytochemical localization of phenylalanine ammonia-lyase in tissues of Populus kitakamiensis. Planta. 1996;200:13–19. doi: 10.1007/BF00196643. [DOI] [PubMed] [Google Scholar]
- Osakabe K, Tsao CC, Li L, Popko JL, Umezawa T, Carraway DT, Smeltzer RH, Joshi CP, Chiang VL. Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc Natl Acad Sci USA. 1999;96:8955–8960. doi: 10.1073/pnas.96.16.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakusch A-E, Kneusel RE, Matern U. S-Adenosyl-l-methionine:trans-caffeoyl-coenzyme A 3-O-methyltransferase from elicitor-treated parsley cell suspension cultures. Arch Biochem Biophys. 1989;271:488–494. doi: 10.1016/0003-9861(89)90299-3. [DOI] [PubMed] [Google Scholar]
- Peleman J, Boerjan W, Engler G, Seurinck J, Botterman J, Alliotte T, Van Montagu M, Inzé D. Strong cellular preference in the expression of a housekeeping gene of Arabidopsis thaliana encoding S-adenosylmethionine synthetase. Plant Cell. 1989;1:81–93. doi: 10.1105/tpc.1.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon J. Les rouilles du peuplier en France: systématique et répartition du stade urédien. Eur J For Pathol. 1973;3:221–228. [Google Scholar]
- Rasmussen S, Dixon RA. Transgene-mediated and elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell. 1999;11:1537–1551. doi: 10.1105/tpc.11.8.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolando C, Monties B, Lapierre C. Thioacidolysis. In: Lin SY, Dence CW, editors. Methods in Lignin Chemistry. Berlin: Springer-Verlag; 1992. pp. 334–349. [Google Scholar]
- Saka S, Goring DAI. Localization of lignins in wood cell walls. In: Higuchi T, editor. Biosynthesis and Biodegradation of Wood Components. Orlando, FL: Academic Press; 1985. pp. 51–62. [Google Scholar]
- Šamaj J, Hawkins S, Lauvergeat V, Grima-Pettenati J, Boudet A. Immunolocalization of cinnamyl alcohol dehydrogenase 2 (CAD 2) indicates a good correlation with cell-specific activity of CAD 2 promoter in transgenic poplar shoots. Planta. 1998;204:437–443. doi: 10.1007/s004250050277. [DOI] [PubMed] [Google Scholar]
- Schmitt D, Pakusch A-E, Matern U. Molecular cloning, induction, and taxonomic distribution of caffeoyl-CoA 3-O-methyltransferase, an enzyme involved in disease resistance. J Biol Chem. 1991;266:17416–17423. [PubMed] [Google Scholar]
- Smith CG, Rodgers MW, Zimmerlin A, Ferdinando D, Bolwell GP. Tissue and subcellular immunolocalisation of enzymes of lignin synthesis in differentiating and wounded hypocotyl tissue of French beans (Phaseolus vulgarisL.) Planta. 1994;192:155–164. [Google Scholar]
- Speer EO. A method of retaining phloroglucinol proof of lignin. Stain Technol. 1987;62:279–280. doi: 10.3109/10520298709108008. [DOI] [PubMed] [Google Scholar]
- Stafford HA. Phenylalanine ammonia-lyase. In: Conn EE, editor. Secondary Plant Products, The Biochemistry of Plants. Vol. 7. New York: Academic Press; 1981. pp. 117–137. [Google Scholar]
- Timell TE. Compression Wood in Gymnosperms. Heidelberg: Springer-Verlag; 1986. [Google Scholar]
- Tsai C-J, Popko JL, Mielke MR, Hu W-J, Podila GK, Chiang VL. Suppression of O-methyltransferase gene by homologous sense transgene in quaking aspen causes red-brown wood phenotypes. Plant Physiol. 1998;117:101–112. doi: 10.1104/pp.117.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance CP, Kirk TK, Sherwood RT. Lignification as a mechanism of disease resistance. Annu Rev Phytopathol. 1980;18:259–288. [Google Scholar]
- Vander Mijnsbrugge K, Beeckman H, De Rycke R, Van Montagu M, Engler G, Boerjan W (2000) Phenylcoumaran benzylic ether reductase, a prominent poplar xylem protein, is strongly associated with phenylpropanoid biosynthesis in lignifying cells. Planta (in press) [DOI] [PubMed]
- Van Doorsselaere J, Baucher M, Chognot E, Chabbert B, Tollier M-T, Petit-Conil M, Leplé J-C, Pilate G, Cornu D, Monties B, Van Montagu M, Inzé D, Boerjan W, Jouanin L. A novel lignin in poplar trees with a reduced caffeic acid/5-hydroxyferulic acid O-methyltransferase activity. Plant J. 1995;8:855–864. [Google Scholar]
- Whetten R, Sederoff R. Lignin biosynthesis. Plant Cell. 1995;7:1001–1013. doi: 10.1105/tpc.7.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetten RW, MacKay JJ, Sederoff RR. Recent advances in understanding lignin biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:585–609. doi: 10.1146/annurev.arplant.49.1.585. [DOI] [PubMed] [Google Scholar]
- Ye Z-H. Association of caffeoyl coenzyme A 3-O-methyltransferase expression with lignifying tissues in several dicot plants. Plant Physiol. 1997;115:1341–1350. doi: 10.1104/pp.115.4.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z-H, Kneusel RE, Matern U, Varner JE. An alternative methylation pathway in lignin biosynthesis in Zinnia. Plant Cell. 1994;6:1427–1439. doi: 10.1105/tpc.6.10.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z-H, Varner JE. Differential expression of two O-methyltransferases in lignin biosynthesis in Zinnia elegans. Plant Physiol. 1995;108:459–467. doi: 10.1104/pp.108.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahm P, Hohmeyer C, Geider K. Site-specific mutagenesis of the Ti plasmid by transformation of Agrobacterium tumefaciens with mutagenized T-DNA fragments cloned in E. coliplasmids. Mol Gen Genet. 1984;194:188–194. [Google Scholar]
- Zhang X-H, Chiang VL. Molecular cloning of 4-coumarate:coenzyme A ligase in loblolly pine and the roles of this enzyme in the biosynthesis of lignin in compression wood. Plant Physiol. 1997;113:65–74. doi: 10.1104/pp.113.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Morrison WH, III, Negrel J, Ye Z-H. Dual methylation pathways in lignin biosynthesis. Plant Cell. 1998;10:2033–2046. doi: 10.1105/tpc.10.12.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]