Abstract
Background
Our previous study demonstrated a close relationship between NOTCH signaling pathway and salivary adenoid cystic carcinoma (SACC). HES1 is a well-known target gene of NOTCH signaling pathway. The purpose of the present study was to further explore the molecular mechanism of HES1 in SACC.
Methods
Comparative transcriptome analyses by RNA-Sequencing (RNA-Seq) were employed to reveal NOTCH1 downstream gene in SACC cells. Immunohistochemical staining was used to detect the expression of HES1 in clinical samples. After HES1-siRNA transfected into SACC LM cells, the cell proliferation and cell apoptosis were tested by suitable methods; animal model was established to detect the change of growth ability of tumor. Transwell and wound healing assays were used to evaluate cell metastasis and invasion.
Results
We found that HES1 was strongly linked to NOTCH signaling pathway in SACC cells. The immunohistochemical results implied the high expression of HES1 in cancerous tissues. The growth of SACC LM cells transfected with HES1-siRNAs was significantly suppressed in vitro and tumorigenicity in vivo by inducing cell apoptosis. After HES1 expression was silenced, the SACC LM cell metastasis and invasion ability was suppressed.
Conclusions
The results of this study demonstrate that HES1 is a specific downstream gene of NOTCH1 and that it contributes to SACC proliferation, apoptosis and metastasis. Our findings serve as evidence indicating that HES1 may be useful as a clinical target in the treatment of SACC.
Electronic supplementary material
The online version of this article (10.1186/s12885-018-4350-5) contains supplementary material, which is available to authorized users.
Keywords: SACC, RNA-Seq, HES1, Proliferation, Apoptosis, Metastasis
Background
SACC is the most common tumor in the minor salivary glands and the second most common tumor in the major salivary glands [1, 2]. Dockerty and Mayo determined that SACC exhibited aggressive features [3]. The disease is characterized by distant metastasis, a high-risk of relapse and a propensity for invading peripheral nerves. Most patients with SACC die within 5 to 20 years of diagnosis [4]. Regional lymph node metastasis was deemed clinically undetectable in affected patients, while hematogenous metastasis to the lungs, bone and liver was frequently reported in patients [5].
NOTCH signaling pathway is a traditional and complex signal pathway, which relates to tissue differentiation and proliferation. There are four NOTCH receptors and five ligands to interactive with each other in mammal. The relationship between disordered NOTCH1 and tumor development has been hotly debated. A volume of reports have demonstrated the role of NOTCH1 acting as oncogene but also tumor suppressor genetically in different cancers. Yuan and colleagues [6] reported that breast cancer patients with NOTCH1 overexpression suffered a low recurrence free survival rate; Arcaroli et al. [7] found that a NOTCH1 gene copy number gain was a worse prognostic in colorectal cancer. On the other hand, several studies reported that high NOTCH1 and NOTCH2 expression with early tumor stages might indicate a tumor-suppressive role of NOTCH signaling in gastric cancer [8]. Intriguingly, hepatocellular carcinoma [9] and medulloblastoma [10] have been observed with both functions even in the same tumor type.
NOTCH is a classical pathway that could activate HES1. Wang et al. [11] found that the expression of NOTCH1 and HES1 was up-regulated consistently in rectal neuroendocrine tumors and pancreatic neuroendocrine tumors. It means that there was a close relationship between NOTCH signaling pathway and HES gene family. HES1 is one of seven members of the HES gene family (HES1–7) [12–14]. HES1 expression is induced by the NICD and encodes a nuclear protein belonging to the hairy and enhancer of related (HESR) family of basic helix-loop-helix (bHLH)-type transcriptional repressors [15–20]. HES1 participates in cellular differentiation, cell apoptosis and cell self-renewal, and the expression level of HES1 is frequently abnormal in cancer cells. Mounting evidences support that HES1 is an oncogene in kinds of tumors. The absence of HES1 has been shown to weaken the tumorigenic capacity of oral squamous cell carcinoma cells [21], as well as colon cancer [22] and pancreatic cancer cells [23].
Our previous study has convincingly verified that NOTCH1 contributed to the cell growth, anti-apoptosis and metastasis of SACC [24]. However, as a complicated signaling pathway, we know little about the influence of this pathway’s upstream or downstream genes in human SACC. Aster’s animal study [25] uncovered harmful side effects by systemic inhibition of NOTCH signaling. So it’s quite important to find the target genes of the pathway and provide a specific means to treat cancer. The objectives of the present study were to illuminate the effects of target gene of NOTCH signaling pathway in human SACC. We analyzed the changes of transcriptome in SACC cells exhibiting NOTCH1 up-regulation by RNA-Seq and verified that HES1 was a specific downstream target of NOTCH1 signaling. HES1 was employed for further study and we compared HES1 expression levels between clinical SACC samples and normal samples using immunohistochemical staining. We also silenced HES1 expression in SACC cell line to determine the effects of HES1 on SACC cell proliferation, migration and invasion and to elucidate the mechanism by which these cells undergo apoptosis.
Methods
Cell culture and clinical samples
The SACC cell lines SACC-LM and SACC-83 were obtained from the Peking University Health Science Center. The cells were maintained in RPMI-1640 (Gibco BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Gibco). The tissue samples were obtained from Fuzhou General Hospital of Nanjing Military Command and Fujian Medical University Union Hospital. Fifty normal salivary tissue samples and 60 SACC samples were used in the study, which were approved by the Institutional Review Board of Fujian Medical University (IRB No. 35000401–11-054). Written informed consent was obtained from each participant.
RNAi transfection
A negative control (NC) siRNA and two siRNAs against HES1 were synthesized (GenePharma, Shanghai, China). The sequences of these siRNAs are listed in Table 1. The SACC LM cells were transfected with siRNAs using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, California, USA), according to the manufacturer’s instructions.
Table 1.
The sequences of the siRNAs used in the transfection experiments
| Name | Sense | Antisense |
|---|---|---|
| siRNA-HES1–425 | 5’-GGAUGCUCUGAAGAAAGAUTT-3’ | 5’-AUCUUUCUUCAGAGCAUCCTT-3’ |
| siRNA-HES1–670 | 5’-CCAACUGCAUGACCCAGAUTT-3’ | 5’-AUCUGGGUCAUGCAGUUGGTT-3’ |
| NC | 5’-UUCUCCGAACGUGUCACGUTT-3’ | 5’-ACGUGACACGUUCGGAGAATT-3’ |
Quantitative real-time PCR analysis
Total RNA was extracted from the SACC LM cells and was reverse transcribed into cDNA. The cDNA was used to detect the expression of the genes of interest by qRT-PCR, which was performed with SYBR Premix Ex Taq (Takara). The primers used in this study are listed in Table 2. The data were analyzed according to the 2-△△Ct method.
Table 2.
The primers for real-time PCR and semi-quantitative RT-PCR used in this study
| Gene | Accession no. | Forward | Reverse |
|---|---|---|---|
| ACTB | NM 001101 | CCTGGCACCCAGCACAAT | GGGCCGGACTCGTCATACT |
| HES1 | NM 005524.3 | AGGCGGACATTCTGGAAATG | CGGTACTTCCCCAGCACACTT |
| NOTCH1 | NM 017617.3 | GGAAGTTGAACGAGCATAGTCC | GCATGATGCCTACATTTCAAGA |
| CCND1 | NM 053056.2 | CCCCGCACGATTTCATTGAACA | CATGGAGGGCGGATTGGAAATG |
| Ki67 | NM 002417.4 | GCTCCCCACCTCAGAGAGTTTT | CTCTTAAGGGAGGGCTTGCAGA |
| KRT14 | NM 000526.4 | GAGCCGCATTCTGAACGAGATG | ACTGCAGCTCAATCTCCAGGTT |
| IGFBP7 | NM 001553.2 | GATGCTGGAGAATATGAGTGCC | CCATGACTACTTTTAACCATGCA |
| PSCA | NM 005672.4 | CCAGGTGAGCAACGAGGACT | TAGTCCTGTGAGTCATCCACGC |
| S100A2 | NM 005978.3 | ATAAATCCTCACCCTGGGAGCC | CCCTCTTGGCAGGAGTACTTGT |
| C9ORF3 | NM 032823.5 | TCTGCGGAAGTGGTGACCC | AGGGTCCAGCTGTATGTCCATG |
| MMP9 | NM 004994.2 | GAGGCGCTCATGTACCCTATGT | GGTGTGGTGGTGGTTGGAGG |
RNA isolation, library construction and sequencing
Total RNA was extracted using a mirVana miRNA Isolation Kit (Ambion), according to the manufacturer’s protocol, and RNA integrity was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Samples with an RNA Integrity Number (RIN) ≥ 7 were subjected to subsequent analyses. The libraries were constructed using a TruSeq Stranded mRNA LTSample Prep Kit (Illumina, San Diego, California, USA), according to the manufacturer’s instructions. These libraries were then sequenced on an Illumina sequencing platform (HiSeqTM 2500) and 150 bp paired-end reads were generated. Raw data (raw reads) were processed using custom scripts, and ploy-N-containing reads, PCR duplications and low-quality reads were removed to obtain clean reads, which were then mapped to the hg19 genome, which served as a reference, using Tophat (http://ccb.jhu.edu/software/tophat/index.shtml).
Immunohistochemistry
For the immunohistochemical assays, 5-μm-thick tissue sections were incubated with a primary antibody against HES1 (1:6400, CST, Boston, Massachusetts, USA). All the slides were reviewed independently by two pathologists who were blinded to the other’s readings. The immunohistochemical analysis results were graded with the indicated four-tier scoring system (negative, weakly positive, positive, and strongly positive).
Western blot assay
Total protein was separated by 8% SDS-PAGE and then transferred onto PVDF membranes (Amersham, Piscataway, NJ, USA), which were immunoblotted with primary antibodies against HES1 (1:1000 dilution, CST) and GAPDH (1:1000 dilution, CST) overnight and incubated with the appropriate secondary antibodies (1:2000 dilution, Abcam, London, UK). The immunoreactive protein bands were visualized using CDP STAR reagent (Roche, Basel, Switzerland).
Cell viability assay
Cell proliferation was measured by counting viable cells with Cell Counting Kit-8 (CCK-8) (Dojindo, Kumamoto, Japan). The cells were first transfected with siRNAs for 24 h and then plated in a 96-well plate. At the same time during each of the following 5 days, the absorbance of each well was measured at 450 nm with a microplate reader (BioTek, Vermont, USA).
Colony formation assay
Twenty-four hours after siRNA transfection, the cells were plated in 6-cm plates (600 cells per plate) and cultured for 2 weeks. The colonies were stained with 1% crystal violet.
Wound healing assay
SACC LM cells were transfected with siRNAs for 24 h after being seeded in a 6-well plate. A 20-μl pipette tip was used to establish a scratch-wound model until the cells amplificated and formed a monolayer covering the bottom of the plate. Then the medium was replaced with 1640 supplemented with 0.1% FBS. The width of scratch-wound was visualized to evaluate the cell invasion ability under a light microscope at the time points of 0 and 72 h and the images were captured.
Cell invasion and migration assay
Cell invasion was assessed using 24-well Matrigel-coated transwell chambers (8-μm pore size, BD Science, Franklin Lakes, New Jersey, USA). 24 h after siRNA transfection, the cells were serum starved for 24 h and then suspended in 1640 containing 1% FBS. The cells were subsequently plated in the upper transwell chamber at a density of 1.0 × 105 cells/well, and 800 μl of 1640 containing 10% FBS was added to the lower transwell chamber. After incubating for 48 h at 37 °C, the cells in the lower chamber were stained and counted. Cell migration assays were performed with transwell not coated with Matrigel.
Cell apoptosis and cell cycle assay
Cellular apoptosis was analyzed using an FITC/Annexin V Apoptosis Detection Kit (BD Pharmingen). Cell cycle activity was analyzed using a Cycletest Plus DNA Reagent Kit (BD Pharmingen). The percentage of apoptotic cells and the distributions of the cells in each cell cycle phase were determined using a BD FACS Verse Flow Cytometer.
Xenograft cancer model
The experimental animal protocol was approved by the Animal Care and Use Committee of Fujian Medical University. Female BALB/c nude mice aged 6~ 8 weeks were purchased from the Center for Animal Experiments of Fujian Medical University. Nude mice were randomly assigned to three groups, each of which comprised 5 mice. The cells (2 × 106) were suspended in 0.2 ml of serum-free 1640 and then injected into the right axillary fossa of each mouse. Tumor size was measured three times a week and was calculated using the formula V = width2 × length/2. At the end of the experiment, the tumors were harvested, washed once in PBS, and then weighed.
Statistical analysis
Statistical analysis of HES1 immunoreactivity was performed using the rank-sum test, and statistical analysis of the PCR and in vitro cell migration/invasion assay results was performed by Student’s t-test. P < 0.05 was considered statistically significant, and n.s. was indicated in the figures when P > 0.05, * when P < 0.05, **when P < 0.01 and *** when P < 0.001.
Results
HES1 expression is regulated by NOTCH1 in SACC cells
In our previous study, we proved that NOTCH1 acted as an oncogene in SACC and it promoted proliferation and migration of SACC cells [24]. To further explore the effects of downstream gene of NOTCH1 signaling pathway in SACC cells, we performed RNA-Seq to measure the changes of transcriptome after NOTCH1 intracellular domain was overexpressed in SACC-LM cells. Quality control showed a satisfactory quality of RNA with an RIN greater than 8 (Additional file 1: Figure S1 A) and the reads for the duplicate samples were relatively consistent (R = 0.989, Additional file 1: Figure S1 B). The RNA-Seq data from this study have been deposited in the NCBI Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra) under accession no. SRR5572289. A total of 1323 coding genes displayed differential expression in NOTCH1 overexpressed cells, including 806 upregulated coding genes and 517 downregulated coding genes (Additional file 1: Figure S1C, D). Gene Ontology (GO) analysis provided a measure of critical function, which showed that the upregulated genes were associated mainly with cell mobility, cell differentiation, proliferation and signal transduction, while the downregulated genes were associated with metabolism, transport and differentiation (Fig. 1a, b).
Fig. 1.
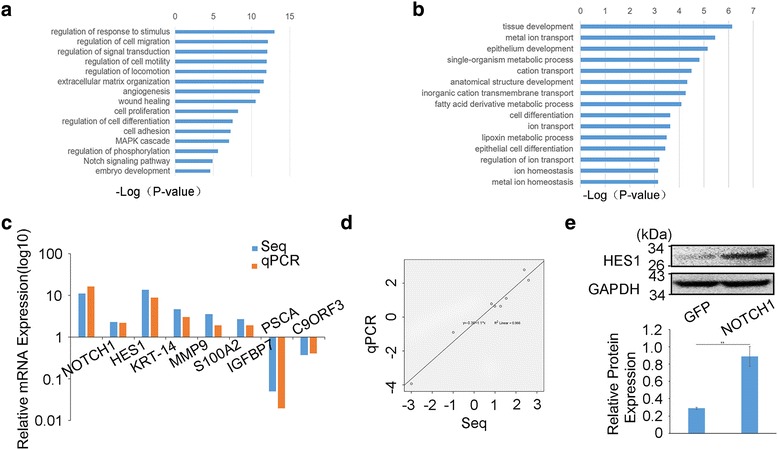
HES1 expression is regulated by NOTCH1 in SACC cells. a, b, The upregulated genes were associated mainly with cell mobility, cell differentiation and proliferation and signal transduction (a), and the downregulated genes were associated with metabolism, transport and differentiation (b). c, d, The expression of NOTCH1, HES1, KRT-14, MMP9, S100A2, IGFBP7, PSCA and C9ORF3 genes, which were selected from the RNA-Seq reports, was measured by qRT-PCR (c), and the linear regression analysis was used to compare the expression levels of the abovementioned selected genes between the RNA-Seq results and qRT-PCR results (d). e Western blot analysis and quantification of HES1 expression in SACC cells after NOTCH1 up-regulation by adenovirus infection
Based on the RNA-Seq results, we selected seven deregulated genes randomly along with NOTCH1 to validate the expression by qRT-PCR. The expression level of HES1, KRT-14, MMP9, S100A2, IGFBP7, PSCA and C9ORF3 by qPCR was consistent with the results of RNA-Seq (Fig. 1c, P < 0.001), which proved that HES1 was one specific downstream gene of the NOTCH1 signaling pathway. The linear regression analysis towards to the relationship between RNA-Seq and qPCR displayed a reliable result of RNA-seq (Fig. 1d, R2 = 0.966). Furthermore, we tested the expression of HES1 by western blot analysis (Fig. 1e), and the SACC cells transfected with NOTCH1 overexpressed plasmid displayed a higher expression of HES1 compared with control group. Additionally, the expression of HES1 has been verified decreased while NOTCH1 was restrained in SACC cells in our previous study [24]. In conclusion, we confirmed the reliability of our RNA-Seq results and focused on the potential downstream gene of NOTCH1 signaling pathway, HES1, for a series of further study.
HES1 is up-regulated in metastatic and recurrent adenoid cystic carcinomas
Basing on the results of RNA-Seq and our previous data, we explored the expression of HES1 by immunohistochemistry to validate its role in SACC. A total of 50 normal tissue samples and 60 SACC tissue samples were included in the study. We classified HES1 expression on a four-tier scale, according to the staining results. As shown in Fig. 2 and Table 3, HES1 expression was absent or very low in normal salivary gland tissues; however, HES1 expression was high in adenoid cystic carcinoma tissues (P < 0.001). The clinical stages of SACC patients were divided into early and late stages by TNM classification. Further analysis of clinical information revealed that HES1 expression was higher in late stages (tumor size>4 cm, or with lymph node or distant metastasis) than that in early stages (tumor size ≤4 cm, or without lymph node or distant metastasis) (P < 0.001, Table 4), and is especially higher in that with lymph node or distant metastasis (P < 0.05) than without. Additionally, we searched the expression level of HES1 in the publicly available Oncomine database and found that HES1 was upregulated in SACC as showed by Frierson HF, who assessed the expression of 8603 genes in 22 samples by microarray (Fig. 2b) (http://www.oncomine.org) [26]. These results indicated that HES1 might participate in the initiation and progress of SACC.
Fig. 2.
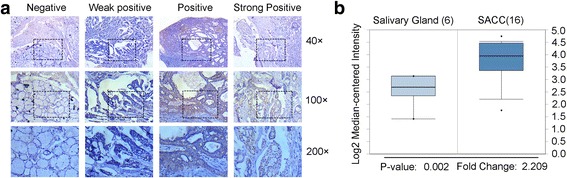
HES1 is up-regulated in metastatic and recurrent adenoid cystic carcinomas. a Representative images of HES1 expression, as determined by immunohistochemistry and the following four-tier scales: negative, weakly positive, positive and strongly positive. b The mRNA expression of HES1 was upregulated in the SACC samples compared with salivary glands, according to the Frierson dataset from the Oncomine database, the fold-change was 2.209 (P = 0.002)
Table 3.
HES1 expression in normal salivary and SACC tissues
| Samples | Cases | Negative | Weakly positive | Positive | Strongly positive | P-value |
|---|---|---|---|---|---|---|
| Normal Salivary | 50 | 43 | 7 | 0 | 0 | |
| SACC | 60 | 0 | 48 | 11 | 1 | < 0.001 |
Rank-sum test Z = −5.699
Table 4.
The expression of HES1 in clinical and pathological characteristics of SACC
| Characteristics | Total | Low HES1a | High HES1a | P value |
|---|---|---|---|---|
| Gender | ||||
| Female | 23 | 19 | 4 | 0.693 |
| Male | 37 | 29 | 8 | |
| Age | ||||
| ≤55 | 29 | 25 | 4 | 0.249 |
| > 55 | 31 | 23 | 8 | |
| Stage | ||||
| Early | 19 | 32 | 1 | 0.001*** |
| Late | 14 | 16 | 11 | |
| Invasion | ||||
| No | 28 | 25 | 3 | 0.095 |
| Yes | 32 | 23 | 9 | |
| Metastasis (Lymph node and distant) | ||||
| No | 52 | 44 | 8 | 0.024** |
| Yes | 8 | 4 | 4 | |
HES1 regulates cellular apoptosis in vitro
To investigate the effects of HES1 on cellular apoptosis, we knocked down HES1 via siRNA in SACC cells. The qRT-PCR (Fig. 3a) and western blotting (Fig. 3b) results showed that the siRNAs targeting HES1 (siRNA-425 and siRNA-670) efficiently reduced HES1 expression in SACC cells compared with negative control (NC) cells. After the cells were transfected with these siRNAs, their growth was significantly inhibited, as demonstrated by CCK8 assay (Fig. 3c, P < 0.001 on day 3, 4 and 5). Similar results were noted in the colony formation assays (Fig. 3d, P < 0.01, n = 3). To explore the effects of HES1 on cancer further, we knocked down HES1 via siRNA transfection for 48 h and then quantified the numbers of apoptotic cells via Annexin V and PI staining and flow cytometric analysis. After 48 h of transfection, the percentages of cells undergoing (Fig. 3e) early (Annexin V-positive and PI-negative) and late apoptosis (Annexin V-positive and PI-positive) were higher among HES1-silenced cells than among control cells. We performed western blotting to detect CASP3 and CASP9 expression in HES1-knockdown cells and full-length and cleaved bands were observed. Through quantification of the active bands, we concluded that the cleaved CASP3 and CASP9 protein levels (Fig. 3f) were elevated in the indicated group of cells compared with NC cells. At the same time, we also applied the PI staining flow cytometry cycle tests to explore whether HES1 knockdown affected the cell cycle phases. The results didn’t show consistent trend and there was not significant difference between NC and HES1 siRNAs (Additional file 1: Figure S2). Collectively, these results confirmed that knocking down HES1 promoted cell apoptosis in vitro, which indicated that HES1 played an oncogenic role in SACC.
Fig. 3.
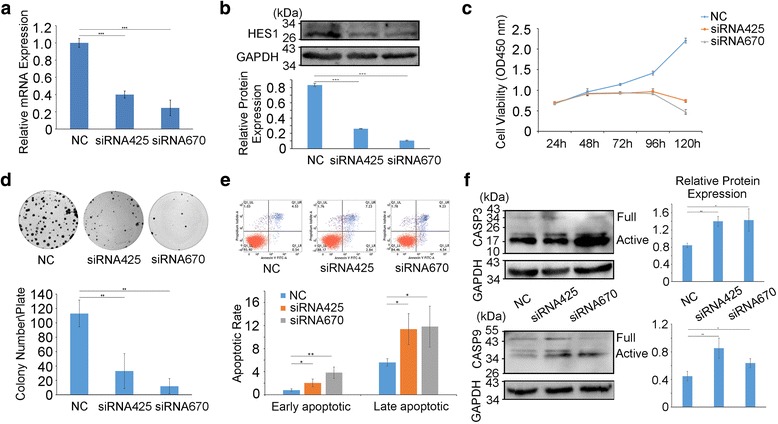
HES1 promotes cell proliferation and regulates cellular apoptosis in vitro. a, b Forty-eight hours after siRNA transfection, HES1 expression in SACC cells was measured by real-time PCR (a) and western blotting (b). c, d After siRNA transfection, SACC cell proliferation was detected by CCK-8 (C, P < 0.001 on days 3, 4 and 5) and colony formation assay (d). e The percentages of early (Annexin V-positive and PI-negative) and late-apoptosis cells (Annexin V- and PI-positive) were analyzed by flow cytometry. F, The expression of the apoptosis-related genes CASP3 and CASP9 was measured by western blotting in HES1-knockdown cells
HES1 knockdown inhibits tumorigenicity in vivo
To explore the effects of HES1 on tumorigenicity in vivo, we transfected SACC LM cells with HES1-siRNAs to silence endogenous HES1 and then subcutaneously inoculated the cells into the flanks of athymic mice. HES1 knockdown inhibited tumor growth, as determined by our results pertaining to xenograft tumor size (Fig. 4a, b) and tumor wet weight (Fig. 4c), in xenografts comprising cells transfected with siRNA425 and siRNA670 compared with xenografts comprising control cells. Moreover, we detected and analysethe expression of HES1, Ki67, CASP3 and CASP9 in the xenograft tumors tissues using immunohistochemistry (Fig. 4d, e). The immunohistochemistry expression of HES1 in the siRNA425 and siRNA670 transfected xenograft tumors groups was lower than in the control group, which supported the effective transfection with siRNA after inoculation for 18 days. The statistical proliferative index of tumor tissues stained for Ki67 came to the conclusion that HES1 knockdown also affected cell proliferation in vivo. We considered that HES1 downregulation in SACC cells led to decreases in cell proliferation and increases in cell apoptosis and apoptosis-related protein expression among the transfected groups compared with the control group, indicating that reductions in HES1 expression in SACC cells could induce cellular apoptosis to inhibit cell growth.
Fig. 4.
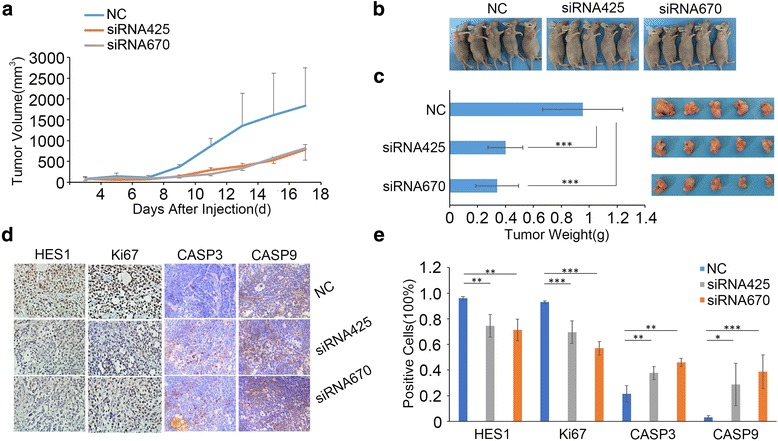
HES1 knockdown inhibits tumorigenicity in vivo. a After being transfected with siRNA, the SACC cells were subcutaneously injected into the flanks of nude mice (n = 5), and the sizes of the resultant tumors were measured three times per week using a digital caliper. b The mice were placed and photographed; c The tumors were excised, photographed and weighed. D-E, The expression of HES1, Ki67, CASP3 and CASP9 was detected in the xenograft tumors by immunohistochemistry (d) (DAB, 400×) and the positive cells were counted and compared among the groups (e)
HES1 increases cell migration and invasion in vitro
We subsequently assessed the effects of knocking down HES1 on SACC migration and invasion. Transfecting HES1-specific siRNAs into SACC cells significantly inhibited SACC cell motility and invasiveness, as demonstrated by wound healing (Fig. 5a, b) and transwell assays (Fig. 5c, d and e P< 0.05, n = 3). These results supported the idea that HES1 acted as an oncogene in SACC and contributed to adenoid cystic carcinoma cell migration and invasion.
Fig. 5.
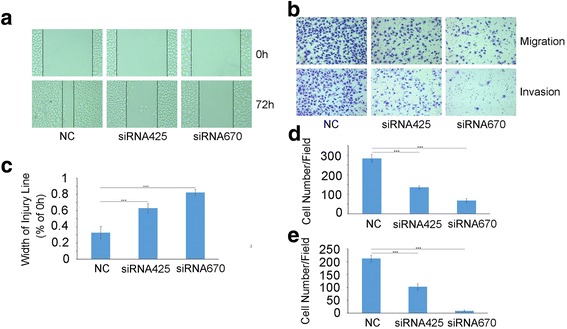
HES1 knockdown inhibits SACC cell migration and invasion ability. a, b A photomicrograph of the scratch wounds made in the siRNA-transfected SACC cell layer showing that cellular motility was inhibited in HES1-silenced cells compared with control cells (a) and the width of injury line after transfection for 72 h compared with 0 h was quantificated (b). c Representative images of transwell chambers coated without (upper panel) or with (lower panel) Matrigel after siRNA transfection. d The number of cells that migrated through the uncoated filters (i.e., no Matrigel), which represents the migratory ability of the indicated SACC cells. e The number of cells that were able to pass through the filters precoated with Matrigel, which represents the invasive ability of the indicated SACC cells. The cell counts are presented as the mean number of cells per field from at least five randomly selected low-powered fields (200×) from three independent experiments (error bars, means ± SD)
Discussion
There are emerging studies regarding SACC molecular biology. The genetic markers differentially expressed between cancerous cells and normal cells are molecules related to cell proliferation [27], growth factor receptors and ligands [28, 29], cell cycle oncogenes [30, 31], cell adhesion proteins [32], and transcription factors [33, 34]. However, HES1 has never been reported to play a role in SACC. HES1 encodes nuclear proteins to activate transcription repression in two ways [13, 35]. One is to form a non-DNA binding complex by joining with other bHLH factors via the bHLH domain, and another is to cooperate with co-repressor transducin-like enhancer of split (TLE) to prevent itself from binding to the N box through its WRPW motif and forming complexes [36, 37]. HES1 induction may depend on several disease-specific and cell-dependent signaling pathways, such as the NOTCH pathway [38], the hedgehog pathway [39, 40], the c-Jun N-terminal kinase (JNK) signaling pathway [41, 42] and the MAP kinase ERK pathway [43]. These pathways appear to be involved in cross-talk with one another at the molecular level. NOTCH is a canonical pathway in the abovementioned pathways, and HES1 plays a prominent role in the NOTCH-HES1 axis. HES1 expression was activated upon the induction of the transcriptional complex CSL-NICD in the nucleus [44]. The NOTCH signaling pathway has been reported to link various microenvironmental factors to the occurrence and development of malignant tumors. Following our previous study regarding the role of the NOTCH1 signaling pathway in SACC, we devoted our attention to systemically studying NOTCH1 [24]. In this study, we revalidate HES1 as the definite downstream gene of NOTCH1 in SACC cells via RNA-Seq analysis. Immunochemistry showed that HES1 expression levels in SACC cancerous tissues were much higher than in those in para-cancerous tissues, too. All of these give us clues that HES1 is a valuable target gene of NOTCH1 signaling pathway and captivate us to carry out more research in SACC.
Cell proliferation is a recognized key contributor to the rapid development of cancer. And stem cells are the source of cancer cells endless proliferation. Many studies indicate that HES1 has the potential to induce cancer stem cells with self-transforming ability and to trigger apoptosis resistance and oncogenesis progression. In addition, the NOTCH-HES1 pathway was verified to affect stem cell maintenance in breast cancer [45]. Our study found that the cells growth ability of SACC cells was affected by HES1. As for illuminating the relationship between HES1 and the stem cells of SACC, we will need an in-depth study in the future. It has also been reached to a consensus that HES1 is associated with apoptosis [22]. Cancer cell differentiation and apoptosis can be stimulated via NOTCH and hedgehog pathway inhibition. It is suspected that differentiation is suppressed by HES1-mediated histone deacetylase (HDAC) inhibition. HDACs have been shown to induce differentiation or apoptosis in tumors and may thus be useful as anti-tumor therapeutic agents [46]. HES1 downregulation induces growth arrest and apoptosis in acute myeloid leukemia (AML) cells; thus, HES1 may be a novel target for the treatment of AML [47]. The execution-phase of cell apoptosis can be sequentially activated by CASP. CASP3 and CASP9 are common members of the CASP family and interact with each otherCASP9 can process and activate CASP3 [48]. To explore the relationship between HES1 downregulation and apoptosis inducement in SACC cells, flow cytometry was involved in our research. It provided new insight that HES1 shared a close association with apoptosis by attracting our attention on apoptosis-related genes, such as CASP3 and CASP9, which might induce apoptosis in SACC cells.
The fate of invasion and metastasis is associated with epithelial-to-mesenchymal transition (EMT), as previous studies have shown that EMT is triggered in metastatic prostate cancer PC3 cells [49]. Otherwise, Fei Gao [22] demonstrated that EMT was enhanced by HES1, which facilitates colon cancer cell aggressiveness and inhibited by HES1 silencing. Weng MT found that HES1 controls invasiveness via the STAT3-MMP14 pathway in colorectal cancer (CRC) cells [50]. Our research also implied that HES1 promoted SACC cells migration and invasion ability, and the clinical samples analysis indicated that the high expression of HES1 in SACC might relate to more metastasis.
It is noteworthy that, the cell or tissue samples and data mining we conducted in the study were insufficiency, so larger sample size would be attempted in the future. In the meanwhile, further studies are needed to clarify specific molecule mechanism of the proliferation and metastasis of HES1 in SACC.
Conclusions
We have determined that HES1 played a vital role in maintaining metastasis, proliferation and apoptosis in SACC cells; thus, targeting HES1 may represent a promising means by which SACC can be treated. In addition to playing a role in the NOTCH signaling pathway, treatments targeting HES1 may cause fewer side effects than those targeting whole NOTCH signaling pathway totally. Therefore, clinicians and researchers should devote significant attention to developing clinical therapies designed to target HES1.
Additional file
Figure S1. RNA-sequencing for Notch1 over-expressing cells. A, The electrophoresis results of RNA extracted from the SACC cell samples implied a satisfactory RNA quality.1 N1 was the cells sample with NOTCH1 overexpressed vector while 1PS was the cells sample with negative vector, 3 N1 and 3PS were the duplicate samples of 1 N1 and 1PS. B, The reads of RNA-seq showed highly consistent in duplicate samples. C, Volcano plot for RNA-sequencing. Differentially expressed genes in each conditions were identified (in red or green), which is statistically significant. D, Heatmap for differentially expressed genes. Figure S2. The effect of HES1 knockdown on cell cycle. A, Representative images of Flow Cytometry analysis of cell cycle after PI staining. B, The percentage of cells in each phase of the cell cycle. Data from 2 independent experiments. (PDF 213 kb)
Acknowledgements
We thank Dr. Guo Chuanbin for the kind gift of SACC-LM and SACC-83.
Funding
This work was supported by the National Natural Sciences Foundation of China [grant numbers 81172583]; the Natural Sciences Foundation of Fujian [grant numbers 2010 J01157, 2011 J01167]; the Key Project of Science and Technology Foundation of Fujian Province of China [grant numbers 2011Y0025]; the Scientific Funding for Leading Research at FMUSS [grant numbers 2015-KQYY-LJ]. The funding body had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- AML
Acute myeloid leukemia
- bHLH
Basic helix-loop-helix
- CASP3
Caspase 3
- CASP9
Caspase 9
- CCK8
Cell Counting Kit-8
- CRC
Colorectal cancer
- EMT
Epithelial-to-mesenchymal transition
- GO
Gene Ontology
- HDAC
HES1-mediated histone deacetylase
- HES
Hairy and enhancer of split
- JNK
c-Jun N-terminal kinase
- NC
Negative control
- NICD
NOTCH intraCellular domain
- qRT-PCR
Quantitative real-time PCR analysis
- RIN
RNA Integrity Number
- RNA-Seq
RNA-Sequencing
- SACC
Salivary adenoid cystic carcinoma
- SRA
Sequence Read Archive
- TLE
Transducin-like enhancer of split
Authors’ contributions
XYH participated in the experimental design, experiments performing, data acquisition, statistical analysis and manuscript draft. RHG performed RNA-Seq analysis and animal assay. RHG and JX participated in the quality control of data and manuscript edit. LS and YZ participated in the clinical sample collection and immunohistochemistry assay. LCD and BHS participated the data analysis and interpretation. YGL and DLZ conceived the study, and also devoted to its design, coordination and manuscript drafting. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was performed in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Fujian Medical University. Written informed consent for the use of tissues for research was taken from patients at the time of procurement of tumor specimens. The experimental animal protocol was approved by the Animal Care and Use Committee of Fujian Medical University (IRB No. 35000401–11-054) and complied with the Basel Declaration outlines fundamental ethical principles.
Consent for publication
Written informed consent was obtained from all patients.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12885-018-4350-5) contains supplementary material, which is available to authorized users.
Contributor Information
Xiao-Yu Huang, Email: 770483176@qq.com.
Rui-Huan Gan, Email: 595332465@qq.com.
Jian Xie, Email: ajian1987213@163.com.
Lin She, Email: mykim2002@163.com.
Yong Zhao, Email: 297392511@qq.com.
Lin-Can Ding, Email: dentding@163.com.
Bo-Hua Su, Email: sudoctor2005@126.com.
Da-Li Zheng, Phone: +86-159-5900-6434, Email: dalizheng@fjmu.edu.cn.
You-Guang Lu, Phone: +86-591-83736429, Email: fjlyg63@fjmu.edu.cn.
References
- 1.Coca-Pelaz A, Rodrigo JP, Bradley PJ, Vander Poorten V, Triantafyllou A, Hunt JL, Strojan P, Rinaldo A, Haigentz M, Jr, Takes RP, et al. Adenoid cystic carcinoma of the head and neck--an update. Oral Oncol. 2015;51(7):652–661. doi: 10.1016/j.oraloncology.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Shaw RJ, Brown J, Ettl T, Risk JM. Prognostic factors in oral, Oropharyngeal, and salivary gland. Cancer. 2017;
- 3.Dockerty MB, Mayo CW. Primary tumors of the submaxillary gland with special reference to mixed tumors. Surg Gynecol Obstet. 1942;74:1033–1045. [Google Scholar]
- 4.Lloyd S, James BY, Wilson LD, Decker RH. Determinants and patterns of survival in adenoid cystic carcinoma of the head and neck, including an analysis of adjuvant radiation therapy. Am J Clin Oncol. 2011;34(1):76–81. doi: 10.1097/COC.0b013e3181d26d45. [DOI] [PubMed] [Google Scholar]
- 5.Kokemueller H, Eckardt A, Brachvogel P, Hausamen J-E. Adenoid cystic carcinoma of the head and neck—a 20 years experience. Int J Oral Maxillofac Surg. 2004;33(1):25–31. doi: 10.1054/ijom.2003.0448. [DOI] [PubMed] [Google Scholar]
- 6.Yuan X, Zhang M, Wu H, Xu H, Han N, Chu Q, Yu S, Chen Y, Wu K. Expression of Notch1 correlates with breast Cancer progression and prognosis. PLoS One. 2015;10(6):e0131689. doi: 10.1371/journal.pone.0131689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arcaroli JJ, Tai W, McWilliams R, Bagby S, Blatchford PJ, Varella-Garcia M, Purkey A, Quackenbush KS, Song EK, Pitts TM. A NOTCH1 gene copy number gain is a prognostic indicator of worse survival and a predictive biomarker to a Notch1 targeting antibody in colorectal cancer. Int J Cancer. 2016;138(1):195–205. doi: 10.1002/ijc.29676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer L, Takacs A, Slottahuspenina J, Langer R, Becker K, Novotny A, Ott K, Walch A, Hapfelmeier A, Keller G. Clinical significance of NOTCH1 and NOTCH2 expression in gastric carcinomas: an Immunohistochemical study. Front Oncol. 2015;5:94. doi: 10.3389/fonc.2015.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giovannini C, Bolondi L, Gramantieri L. Targeting Notch3 in hepatocellular carcinoma: molecular mechanisms and therapeutic perspectives. Int J Mol Sci. 2017;18(1):56. doi: 10.3390/ijms18010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ntziachristos P, Lim JS, Sage J, Aifantis I. From fly wings to targeted Cancer therapies: a centennial for notch signaling. Cancer Cell. 2014;25(3):318–334. doi: 10.1016/j.ccr.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Chen Y, Castillo FD, Yilmaz O, Deshpande V. Heterogeneity in signaling pathways of gastroenteropancreatic neuroendocrine tumors: a critical look at notch signaling pathway. Mod Pathol. 2013;26(1):139–147. doi: 10.1038/modpathol.2012.143. [DOI] [PubMed] [Google Scholar]
- 12.Taelman V, Van WR, Sölter M, Pichon B, Pieler T, Christophe D, Bellefroid EJ. Sequences downstream of the bHLH domain of the Xenopus hairy-related transcription factor-1 act as an extended dimerization domain that contributes to the selection of the partners. Dev Biol. 2004;276(1):47–63. doi: 10.1016/j.ydbio.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Kang SA, Seol JH, Kim J. The conserved WRPW motif of Hes6 mediates proteasomal degradation. Biochem Biophys Res Commun. 2005;332(1):33–36. doi: 10.1016/j.bbrc.2005.04.089. [DOI] [PubMed] [Google Scholar]
- 14.Kageyama R, Ohtsuka T, Kobayashi T. Roles of Hes genes in neural development. Dev Growth Differ. 2010;50(s1):S97–S103. doi: 10.1111/j.1440-169X.2008.00993.x. [DOI] [PubMed] [Google Scholar]
- 15.Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306(2):343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 16.Ishibashi M, Moriyoshi K, Sasai Y, Shiota K, Nakanishi S, Kageyama R. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J. 1994;13(8):1799–1805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sang L, Coller HA, Roberts JM. Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science. 2008;321(5892):1095–1100. doi: 10.1126/science.1155998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24(1):36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 19.Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435(7044):964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 20.Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134(7):1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- 21.Lee SH, Hong HS, Liu ZX, Kim RH, Kang MK, Park NH, Shin KH. TNFalpha enhances cancer stem cell-like phenotype via notch-Hes1 activation in oral squamous cell carcinoma cells. Biochem Biophys Res Commun. 2012;424(1):58–64. doi: 10.1016/j.bbrc.2012.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao F, Zhang Y, Wang S, Liu Y, Zheng L, Yang J, Huang W, Ye Y, Luo W, Xiao D. Hes1 is involved in the self-renewal and tumourigenicity of stem-like cancer cells in colon cancer. Sci Rep. 2014;4:3963. doi: 10.1038/srep03963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maniati E, Bossard M, Cook N, Candido JB, Emami-Shahri N, Nedospasov SA, Balkwill FR, Tuveson DA, Hagemann T. Crosstalk between the canonical NF-κB and notch signaling pathways inhibits Pparγ expression and promotes pancreatic cancer progression in mice. J Clin Invest. 2011;121(12):4685. doi: 10.1172/JCI45797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su BH, Qu J, Song M, Huang XY, Hu XM, Xie J, Zhao Y, Ding LC, She L, Chen J, et al. NOTCH1 signaling contributes to cell growth, anti-apoptosis and metastasis in salivary adenoid cystic carcinoma. Oncotarget. 2014;5(16):6885–6895. doi: 10.18632/oncotarget.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aster JC, Blacklow SC. Targeting the notch pathway: twists and turns on the road to rational therapeutics. J Clin Oncol. 2012;30(19):2418–2420. doi: 10.1200/JCO.2012.42.0992. [DOI] [PubMed] [Google Scholar]
- 26.Jr FH, El-Naggar AK, Welsh JB, Sapinoso LM, Su AI, Cheng J, Saku T, Moskaluk CA, Hampton GM. Large scale molecular analysis identifies genes with altered expression in salivary adenoid cystic carcinoma. Am J Pathol. 2002;161(4):1315–1323. doi: 10.1016/S0002-9440(10)64408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsen SR, Bjorndal K, Godballe C, Krogdahl A. Prognostic significance of Ki-67 in salivary gland carcinomas. J Oral Pathol Med. 2012;41(8):598–602. doi: 10.1111/j.1600-0714.2012.01148.x. [DOI] [PubMed] [Google Scholar]
- 28.Shao Z, Zhu F, Song K, Zhang H, Liu K, Shang Z. EphA2/ephrinA1 mRNA expression and protein production in adenoid cystic carcinoma of salivary gland. J Oral Maxillofac Surg. 2013;71(5):869–878. doi: 10.1016/j.joms.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 29.Ivanov SV, Panaccione A, Brown B, Guo Y, Moskaluk CA, Wick MJ, Brown JL, Ivanova AV, Issaeva N, El-Naggar AK. TrkC signaling is activated in adenoid cystic carcinoma and requires NT-3 to stimulate invasive behavior. Oncogene. 2013;32(32):3698–3710. doi: 10.1038/onc.2012.377. [DOI] [PubMed] [Google Scholar]
- 30.Ivanov SV, Panaccione A, Nonaka D, Prasad ML, Boyd KL, Brown B, Guo Y, Sewell A, Yarbrough WG. Diagnostic SOX10 gene signatures in salivary adenoid cystic and breast basal-like carcinomas. Br J Cancer. 2013;109(2):444–451. doi: 10.1038/bjc.2013.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piha-Paul SA, Cohen PR, Kurzrock R. Salivary duct carcinoma: targeting the phosphatidylinositol 3-kinase pathway by blocking mammalian target of rapamycin with temsirolimus. J Clin Oncol. 2011;29(26):e727–e730. doi: 10.1200/JCO.2011.36.2095. [DOI] [PubMed] [Google Scholar]
- 32.Zhao D, Yang K, Tang XF, Lin NN, Liu JY. Expression of integrin-linked kinase in adenoid cystic carcinoma of salivary glands correlates with epithelial-mesenchymal transition markers and tumor progression. Med Oncol. 2013;30(3):619. doi: 10.1007/s12032-013-0619-3. [DOI] [PubMed] [Google Scholar]
- 33.Tsuneki M, Maruyama S, Yamazaki M, Essa A, Abe T, Babkair HA, Ahsan MS, Cheng J, Saku T. Podoplanin is a novel myoepithelial cell marker in pleomorphic adenoma and other salivary gland tumors with myoepithelial differentiation. Virchows Arch. 2013;462(3):297–305. doi: 10.1007/s00428-012-1359-z. [DOI] [PubMed] [Google Scholar]
- 34.Stenman G, Persson F, Andersson MK. Diagnostic and therapeutic implications of new molecular biomarkers in salivary gland cancers. Oral Oncol. 2014;50(8):683–690. doi: 10.1016/j.oraloncology.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Dhanesh SB, Subashini C, James J. Hes1: the maestro in neurogenesis. Cellular & Molecular Life Sciences. 2016;73(21):1–24. [DOI] [PMC free article] [PubMed]
- 36.Paroush ZE, Jr RLF, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79(5):805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 37.Fisher AL, Ohsako S, Caudy M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol Cell Biol. 1996;16(6):2670–2677. doi: 10.1128/MCB.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuno K, Ito M, Hori K, Miyashita F, Suzuki S, Kishi N, Artavanis-Tsakonas S, Okano H. Involvement of a proline-rich motif and RING-H2 finger of Deltex in the regulation of notch signaling. Development. 2002;129(4):1049–1059. doi: 10.1242/dev.129.4.1049. [DOI] [PubMed] [Google Scholar]
- 39.Ingram WJ, Mccue KI, Tran TH, Hallahan AR, Wainwright BJ. Sonic hedgehog regulates Hes1 through a novel mechanism that is independent of canonical notch pathway signalling. Oncogene. 2008;27(10):1489–1500. doi: 10.1038/sj.onc.1210767. [DOI] [PubMed] [Google Scholar]
- 40.Wall DS, Mears AJ, Mcneill B, Mazerolle C, Thurig S, Wang Y, Kageyama R, Wallace VA. Progenitor cell proliferation in the retina is dependent on notch-independent sonic hedgehog/Hes1 activity. J Cell Biol. 2009;184(1):101–112. doi: 10.1083/jcb.200805155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curry CL, Reed LL, Nickoloff BJ, Miele L, Foreman KE. Notch-independent regulation of Hes-1 expression by c-Jun N-terminal kinase signaling in human endothelial cells. Lab Investig. 2006;86(8):842–852. doi: 10.1038/labinvest.3700442. [DOI] [PubMed] [Google Scholar]
- 42.Jin WK, Kim MJ, Kim KJ, Yun HJ, Ji SC, Sang GH, Chang TS, Park HS, Lee KW, Han PL. Notch interferes with the scaffold function of JNK-interacting protein 1 to inhibit the JNK signaling pathway. Proc Natl Acad Sci U S A. 2005;102(40):14308–14313. doi: 10.1073/pnas.0501600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stockhausen MT, Sjolund J, Axelson H. Regulation of the notch target gene Hes-1 by TGFalpha induced Ras/MAPK signaling in human neuroblastoma cells. Exp Cell Res. 2005;310(1):218–228. doi: 10.1016/j.yexcr.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 44.Rani A, Greenlaw R, Smith RA, Galustian C. HES1 in immunity and cancer. Cytokine Growth Factor Rev. 2016;30:113–117. doi: 10.1016/j.cytogfr.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 45.So JY, Wahler J, Das Gupta S, Salerno DM, Maehr H, Uskokovic M, Suh N. HES1-mediated inhibition of Notch1 signaling by a Gemini vitamin D analog leads to decreased CD44(+)/CD24(−/low) tumor-initiating subpopulation in basal-like breast cancer. J Steroid Biochem Mol Biol. 2015;148:111–121. doi: 10.1016/j.jsbmb.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sang L, Roberts JM, Coller HA. Hijacking HES1: how tumors co-opt the anti-differentiation strategies of quiescent cells. Trends Mol Med. 2010;16(1):17–26. doi: 10.1016/j.molmed.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian C, Yu Y, Jia Y, Zhu L, Zhang Y. HES1 activation suppresses proliferation of leukemia cells in acute myeloid leukemia. Ann Hematol. 2015;94(9):1477–1483. doi: 10.1007/s00277-015-2413-0. [DOI] [PubMed] [Google Scholar]
- 48.Chen WL, Pan L, Kinghorn AD, Swanson SM, Burdette JE. Silvestrol induces early autophagy and apoptosis in human melanoma cells. BMC Cancer. 2016;16(1):17. doi: 10.1186/s12885-015-1988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kong D, Li Y, Wang Z, Sarkar FH. Cancer stem cells and epithelial-to-Mesenchymal transition (EMT)-phenotypic cells: are they cousins or twins? Cancers (Basel) 2011;3(1):716–729. doi: 10.3390/cancers30100716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weng MT, Tsao PN, Lin HL, Tung CC, Change MC, Chang YT, Wong JM, Wei SC. Hes1 increases the invasion ability of colorectal Cancer cells via the STAT3-MMP14 pathway. PLoS One. 2015;10(12):e0144322. doi: 10.1371/journal.pone.0144322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. RNA-sequencing for Notch1 over-expressing cells. A, The electrophoresis results of RNA extracted from the SACC cell samples implied a satisfactory RNA quality.1 N1 was the cells sample with NOTCH1 overexpressed vector while 1PS was the cells sample with negative vector, 3 N1 and 3PS were the duplicate samples of 1 N1 and 1PS. B, The reads of RNA-seq showed highly consistent in duplicate samples. C, Volcano plot for RNA-sequencing. Differentially expressed genes in each conditions were identified (in red or green), which is statistically significant. D, Heatmap for differentially expressed genes. Figure S2. The effect of HES1 knockdown on cell cycle. A, Representative images of Flow Cytometry analysis of cell cycle after PI staining. B, The percentage of cells in each phase of the cell cycle. Data from 2 independent experiments. (PDF 213 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


