Abstract
Background/Aim: To explore the relationship between p53, p63, c-kit, Ki67, cMet, claudin7, CK5/6, CK17, AR, PTEN, EGFR, ALK, PDL-1 and c-MYC expression with the clinicopathological features of triple- negative breast cancer. Materials and Methods: Immunohistochemistry was performed in 84 triple-negative breast cancer samples. Results: A statistically significant relationship between tumour grade and claudin-7 (p=0.004) and between protein p53 and positive lymph nodes (p=0.015) was found. High expression of claudin-7 (OR=65.8, 95%CI=4.35-995.19, p-value=0.003) and low expression of c-kit (OR=0.14, 95%CI=0.025-0.793, p-value=0.026) and protein p63 (OR=0.18 95%CI=0.035-0.978, p-value=0.047) was associated with higher tumour grade. Higher AR expression (OR=13.44, 95%CI=1.28-141.56, p-value=0.031) and lower expression of CK5/6 cytokeratins was found in patients with positive lymphovascular invasion (LVI) (OR=0.072, 95%CI=0.007-0.732, p-value=0.026). Only the cell proliferation index (Ki67) has been proven to be statistically significant for disease-free survival (p-value=0.0378), and overall survival (p-value=0.0186). Conclusion: High expression of claudin-7 and low expression of c-kit and protein p63 are associated with higher tumour grade. AR and CK5/6 expression seem to be important in LVI.
Keywords: Triple-negative breast cancer, p53, p63, c-kit, Ki67, cMet, claudin 7, CK 5/6, CK17, AR, PTEN, EGFR, ALK, PDL-1 and c-MYC
With over 1 million women affected worldwide, breast cancer (BC) is currently the commonest malignancy in females and the second leading cause of cancer related death (1). Triple negative breast cancer (TNBC) immunohistochemically characterized by the lack of expression of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor type 2 (HER2), is responsible for a disproportionate share of mortality owing to its aggressive clinical behavior, poor prognosis and lack of targeted therapies (2).
Recent studies have classified TNBC into 4 major subtypes: basal-like (BLBCs), mesenchymal, luminal androgen receptor and immune enriched. Of the total cases of TNBC, 70-80% are Basal like Breast Cancers sharing numerous clinical and pathological features and these terms are often used interchangeably by clinicians although they are not synonymous. New therapeutic approaches have been proposed based on different factors, however they are not currently used to stratify patients for decisions about clinical therapy. In addition, there are no predictive markers that have been widely accepted and proven to be significant across all different studies.
The aim of the study was to explore the relationship between p53, p63, c-kit, Ki67, cMet, claudin 7, CK 5/6, CK17, AR, PTEN, EGFR, ALK, PDL-1 and c-MYC with tumor grade, number of positive lymph nodes, lymphovascular invasion (LVI), local recurrence, distant recurrence, disease-free survival and overall survival in TNBC.
Materials and Methods
The study was performed in Hygeia General Hospital, Athens Greece from 2003-2011. A total of 84 TNBC cases were included from a prospectively collected database. Samples with an absent or incomplete immunohistochemistry report for the respective pathology were excluded. Tissue samples were fixed in 10% buffered formalin and embedded in paraffin wax. The tumors were classified and graded according to the suggested criteria of World Health Organisation (WHO) 2014 and Tumor Node Metastasis status/American Joint Committee on Cancer (TNM/AJCC) 2009 system. Suitable selected paraffin blocks containing representative tumor areas were identified on corresponding hematoxylin-eosin –stained sections. Areas of interest were identified and marked on the source block. The source block was cored and a 1.5 mm core was transferred to the recipient “master block” using the Tissue Microarrayer (MTA-1, Beecher Instruments, Sun Prairie, WI, USA). Two, representative tumor cores were arrayed per specimen as cores of normal tissue from breast, endometrium, colon and spleen and used as reliability indicators (control). Immunohistochemical staining was performed for 14 antigens: p53, p63, c-kit, Ki67, cMet, claudin 7, CK 5/6, CK17, AR, PTEN, EGFR, ALK, PDL-1 and c-MYC.
Statistical analysis. Univariate analysis was performed to explore the association of biomarkers’ expression with all the dependent variables of interest: tumor grade, number of positive lymph nodes, LVI, local recurrence (LR) and distant metastasis. Categorical variables were reported as counts and percentages and compared with Fisher’s Exact test.
Multivariate logistic regression models were used for modelling the effect of biomarkers to the dependent variables tumor grade, lymph nodes and LVI. The backward elimination method with removal criterion p=5% has been used, resulting to models with statistically significant effects. Odds ratios (OR), 95%CI and Likelihood ratio test values were reported for each analysis.
Cox proportional-hazards regression models, with backward elimination with p=10%, are provided for the disease-free survival (DFS) and overall survival (OS). We also performed the Log-rank test to test the equality of survival functions.
All comparisons were two tailed and a p-value of <0.05 was considered statistically significant. Data analysis was performed using STATA statistical software version 12.0.
Results
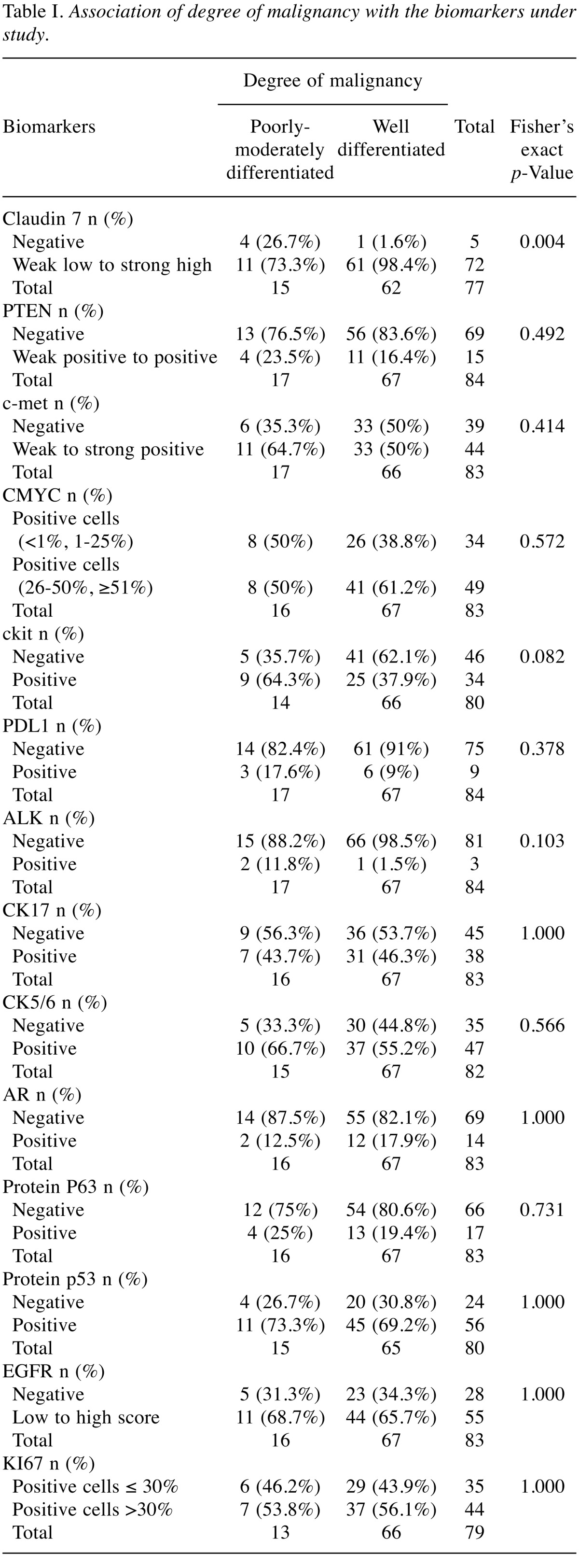
A total of 84 patients were included in the analysis. All patients were females with median age 49 years (range=25-79 years). Seventeen (20.2%) were treated with mastectomy and 67 (79.8%) with lumpectomy and radiotherapy. All patients received adjuvant chemotherapy and all histological types were invasive ductal carcinoma. The association of biomarkers with tumor grade is summarized in Table I. The analysis of the tumor grade was conducted using the degree of malignancy as a dependent variable. Fisher’s exact test resulted in a statistically significant relationship between the degree of malignancy and binding of protein claudin 7 for the two different values of the biomarker, negative and weak low to strong high. Independent predictors of the grade of cancer were determined in a multivariate logistic regression model with stepwise backward elimination of variables with a p-value=0.05. The expression of claudin 7 appeared to be much higher in patients with higher tumor grade (OR=65.8, 95%CI=4.35-995.19, p-value=0.003). According to the logistic regression model for the degree of malignancy with 71 observations, there is also a statistically significant relationship with c-kit (p-value=0.026), and protein p63 (p-value=0.047). Their expression appeared to be lower in patients with higher tumor grade with values of odds ratio OR=0.14, 95%CI=0.025-0.793, p-value=0.026 and OR=0.18 95%CI=0.035-0.978, p-value=0.047, respectively.
Table I. Association of degree of malignancy with the biomarkers under study.
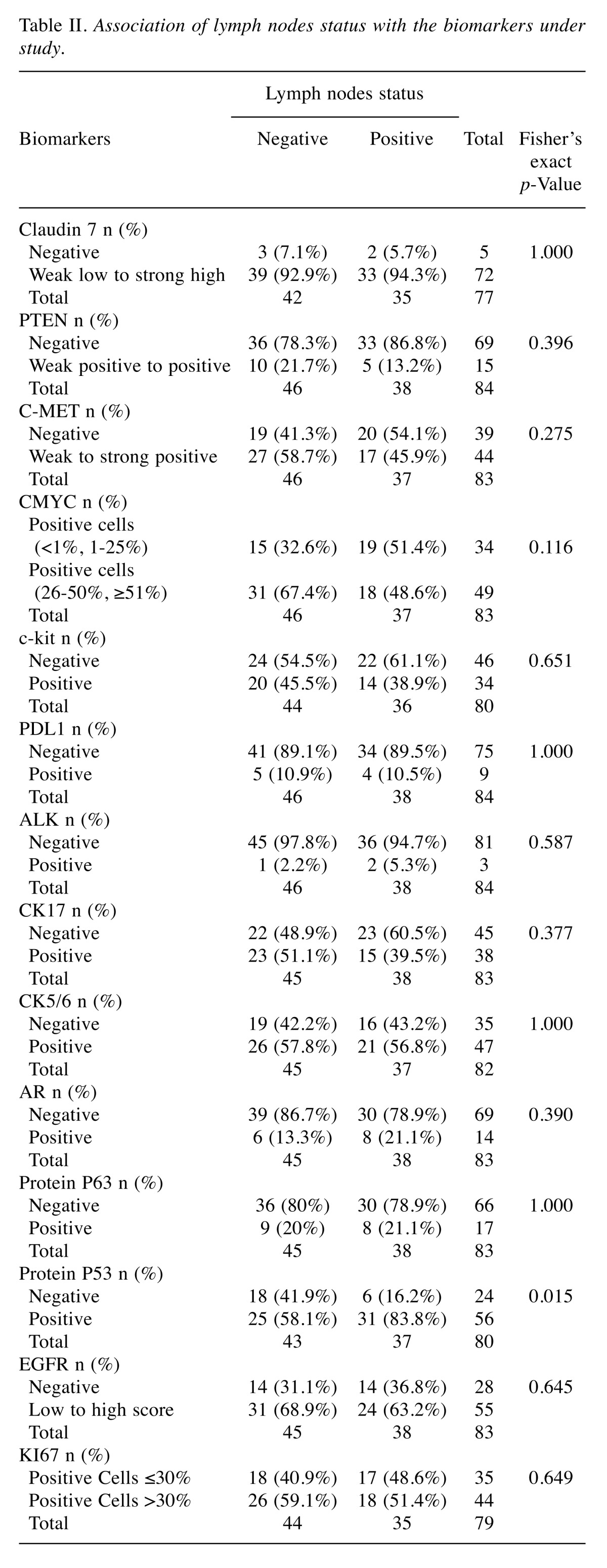
The association of lymph nodes status with the biomarkers under study is summarized in Table II. There is a statistically significant relationship between the protein p53 biomarker and the lymph nodes status according to Fisher’s exact test and the multivariate logistic regression model with 71 observations, where we examined the stepwise backward elimination of variables with a p-value=0.05. The expression of protein p53 was higher in patients with positive lymph nodes (OR=3.84, 95%CI=1.22-12.07, p-value=0.021).
Table II. Association of lymph nodes status with the biomarkers under study.
Independent predictors for LVI were determined in a multivariate logistic regression model with 69 observations and stepwise backward elimination of variables with a p-value=0.05. The expression of androgen receptor (AR) appeared to be much higher in those patients with positive LVI process (OR=13.44, 95%CI=1.28-141.56, p-value=0.031). Also, the expression of ck5/6 cytokeratins appeared to be lower in patients with positive LVI (OR=0.072, 95%CI=0.007-0.732, p-value=0.026).
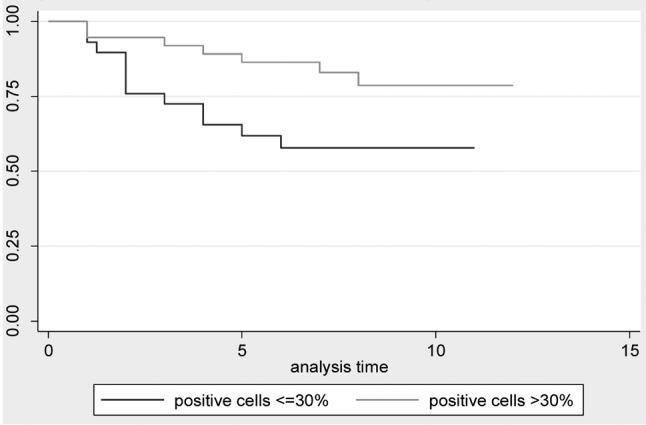
Disease free survival and overall survival. DFS and OS status between groups of patients with different levels of each biomarker have been compared using log-rank test with a 5% level of statistical significance. Only the cell proliferation index (Ki67) has been proven to be statistically significant for DFS (p-value=0.0378), and for OS (p-value=0.0186), which means that the survival function is differentiated for the two levels of the biomarker. The significance of this biomarker has also been confirmed by the Cox regression model for DFS with backward elimination (HR=0.39, 95%CI=0.153-0.994, p-value=0.048). We run the same Cox regression model with backward elimination for OS and it concluded that biomarker Ki67 is an important predictor for the overall survival of patients (HR=0.277, 95%CI=0.087-0.886, p-value=0.03). Table III presents the results for the Log-rank test for equality of survival functions in 2 different groups of each biomarker. Kaplan-Meier graphs are provided for both OS (Figure 1), and DFS status (Figure 2).
Table III. Log-rank test for equality of survival functions in 2 different groups of each biomarker.
Figure 1. Kaplan-Meir overall survival estimates for cell proliferation index ki67.
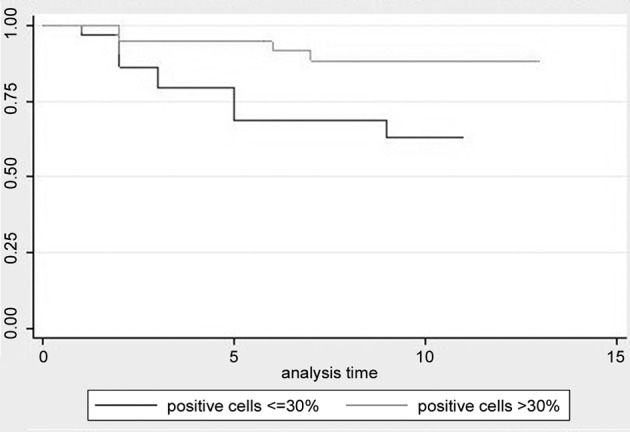
Figure 2. Kaplan-Meir disease free survival estimates for cell proliferation index ki67.
Discussion
TNBC has unfavourable prognosis characterized by larger size and features of aggressive behavior. Many published studies have attempted to identify new biomarkers to sub-classify TNBC into different prognostic groups and to select patients who are candidates for more aggressive targeted therapy regimens.
Claudins are transmembrane proteins which have a major role on the regulation of cell adhesion, proliferation and differentiation, in controlling paracellular permeability and the maintenance of epithelial cell polarity (3-6). TNBC is often characterized by low claudin expression, especially claudin-3, -4, -7 (7). However, in our study, higher expression of claudin 7 was significantly correlated with higher tumor grade (OR=65.8, 95%CI=4.35-995.19, p-value=0.003). Although this observation seems to contradict the hypothesis that claudin low cells are associated with poor outcome, it is consistent with other recent studies, as the Bernardi et al. study, who did not find any association between claudin-7 and different subtypes and supported that claudin-7 expression in invasive ductal carcinoma was associated with a shorter time of recurrence, suggesting a contribution of this marker to the aggressiveness of breast cancer (8).
C-Met is a transmembrane tyrosine kinase receptor that plays a vital role in cell-cell detachment, invasiveness, tumour angiogenesis, proliferation, metastasis and survival (9). The evidence of the influence of c-Met expression on survival outcomes is inconclusive. In the present study, there was no significant correlation of c-Met expression with tumour grade, LVI and number of positive lymph nodes. However, Fisher’s exact test resulted in a statistically significant relationship between AR (p-value=0.041) and was also significantly correlated (p-value=0.002) with cell proliferation index Ki67 behaviour. In a recent meta-analysis by Yan et al. (10), it was shown that c-Met overexpression was associated with 1.41-fold increased risk of recurrence in the hormone positive group compared to 2.31-fold in TNBC, suggesting that c-Met could be a therapeutic target for TNBC. Additional high-quality data is needed in order to draw more reliable conclusions.
PTEN is a tumour suppressor 200 kb gene, located on chromosome 10q23 that regulates many cellular functions including cell growth, proliferation and migration. It is lost or mutated in many types of cancer including breast, prostate, and lung cancer (11). PTEN losses have been observed in up to 37-74% of TNBCs (12-14), suggesting a rationale to evaluate mTOR inhibitors in patients with TNBC as there is no alternative targeted treatment. Inanc et al. detected 44.3% PTEN loss among 99 TNBC tumors, which was also associated with shorter DFS, increased recurrence and mortality risk compared to patients without a PTEN loss. No relationship was found between PTEN loss and other clinical and pathological parameters, only that it was higher among patients with LVI (15). In the present study, no statistically significant relationship was found between PTEN loss and tumor grade, LVI, and the number of positive lymph nodes.
C-MYC is a basic helix-loop-helix zipper motif transcription factor. Its amplification is one of the most frequent aberrations in BC that has been detected in 1-94% of patients in different studies and found to be associated with the basal subtype. Bouchalova et al. in a study of 187 patients with TNBC, c-MYC protein expression was found in the majority of the TNBCs (164 out of 187 patients) and was associated with worse survival (16). Horiuchi et al. investigated the biology of TNBC and identified that MYC signalling is elevated in these tumours and the expression of multiple MAX (another basic helix-loop-helix zipper protein) binding partners, which regulate MYC activity, was altered in TNBC and may therefore contribute to increased MYC pathway activity and worse patient outcome (17). In the present study, no statistically significant relationship was found between c-MYC protein expression and a role in regulation of cell proliferation, tumor grade, LVI, and the number of positive lymph node.
C-kit is a transmembrane tyrosine kinase receptor present on the surface of hematopoietic stem cells and also of other cells. It induces apoptosis and also increases the invasiveness of cancer cells (12). In this study the expression of c-kit appeared to be lower in patients with higher tumor grade (OR=0.14, 95%CI=0.025-0.793, p-value=0.026). Fisher’s exact test resulted in a statistically significant relationship between c-kit (p-value=0.022) and cytokeratins ck5/6 (p-value=0.022). Furthermore, there was a statistically significant association between c-kit, and EGFR (p-value=0.045).
Similarly Tsuitsui et al. found that loss of c-kit expression was associated with lymph node metastases, and worse prognosis as it was associated with an advanced stage of breast cancer (18). Opposite results were presented by Diallo et al. who underlined that c-kit expression represents an independent negative prognostic marker in high-risk breast cancer (19). Zhu et al. evaluated the expression of c-kit protein and the mutations of the c-kit gene in triple negative breast cancers; 41.7% of the TNBCs positive for c-kit might benefit from tyrosine kinase inhibitors (20). Kanapathy Pillai et al. reported c-kit expression in 89% of TNBCs and this was also associated with EGFR, CK5/6 and high Ki67 proliferation index (21). The significance of c-kit expression is controversial and the rate of positive c-kit varies from 1 to 82%, which is likely attributable to the different methods of determination of c-kit expression.
PD-L1, programmed cell death 1 ligand 1, is a 40kDa transmembrane protein, expressed on the surface of activated cytotoxic T cells, causing inhibition of IL-2 production and T cell migration and proliferation (22). Few studies have investigated PD-L1 expression in BC, however the results varied regarding its expression rate and prognostic value. The present study demonstrated PD-L1 expression in 10.4% of patients while 69 out of 77 (89.6%) patients did not express PD-L1 however, no statistically significant relationship was found between PD-L1 and tumor grade, LIV, and the number of the positive lymph nodes. Similarly, Beckers et al. in a study of 161 primary TNBCs, PD-L1 expression was very common in TNBC but was not an independent prognostic marker as there were differences in the outcome depending on which cellular compartment PDL-1 was expressed (tumour cell membrane, cytoplasm, and stromal cellular compartment) (23). Qin et al. has shown high PD-L1 expression associated with significantly decreased survival, higher tumor grade (24).
ALK is a tyrosine receptor kinase whose pathway, has been identified by Lehmann et al. as an important signalling pathway that is common in TNBCs (25). In the present study only two women with TNBC expressed this receptor and 75 out of 77 women did not.
Cytokeratins 5/6 and 17 are important markers for the identification of the basaloid group and are correlated with poor patient outcome in TNBC (26-29). In our study 57.3% of women were found positive for CK5/6 while 35 out of 82 were negative. The expression of CK5/6 cytokeratins appeared to be lower in patients with LVI (OR=0.072, 95%CI=0.007-0.732, p-value=0.026). Also, a significant association was found between CK5/6 and EGFR (p-value=0.00417) and CK17 and Ki67 (p-value=0.041). These results are contradictory to many results in the literature where high expression of CK5/6 is significantly associated with worse clinicopathological features in TNBC (30)-(31). CK5/6 positive expression rates vary from 24 to 72 % in the literature as, there are different scoring systems used in IHC studies for CK5/6 and no precise cutoff value exists (32).
Androgen receptor is a member of the steroid hormone receptor family, which functions as a classic ligand-activated intracellular transcription factor. AR expression in TNBC is reported lower than estrogen receptor positive breast cancers, and the prognostic value of this is still unclear. In the present study, 82.9% of women were found positive for AR while 14 out of 82 were negative. The expression of AR appeared to be much higher in patients with positive LVI (OR=13.44, 95%CI=1.28-141.56, p-value=0.031). Hu et al. who analyzed AR expression in 211 TNBCs, found that patients with AR positive tumours had an 83% increase in overall mortality compared with the AR-negative group (33). McGhan et al. found AR positive TNBCs to correlate with higher grade and LN metastases (34). Millis et al. in the largest to date cohort study were 6341 breast cancers were evaluated (2,111 TNBC and 4,230 non-TNBC) showed that higher AR expression in TNBC was associated with lower ki-67 levels suggesting that androgens might have an antiproliferative effect (35). These differences in the prognostic role of AR expression are due to the variability in antibodies, scoring systems and cut-offs used to define AR positivity but also on the complexity of this pathway.
TP53 is a tumour suppressor gene that encodes a nuclear phosphoprotein. Mutation in TP53 results in loss of the usual wt-p53 tumour suppressor functions and it often exerts opposing effects. 20-35% of all breast cancers have a TP53 mutation; in TNBC TP53 mutation is present in almost all of them (25,36). Previous studies have shown that p53 expression is higher in TNBC and that may play a role in the worse prognosis of TNBC (37). It has also been shown that breast tumours with mutant p53 were generally TNBC and were associated with decreased survival (38,39). Similarly, in the present study, the expression of p53 was higher in patients with positive lymph nodes (OR=3.84, 95%=CI=1.22-12.07,p-value=0.021) suggesting worse prognosis of these patients.
Epidermal growth factor receptor (EGFR), is one of the most notable cancer molecular targets. In breast cancer, EGFR expression level or gene mutation status is increasingly being used to select patients for selective treatments. In our study, no statistical significant correlation was found between EGFR tumor grade, lymphovascular invasion, and the number of the positive LNs. A statistically significant association according to a Fisher’s exact test was found between EGFR and c-kit(p-value=0.045) and CK5/6 and EGFR (p-value= 0.004). The uncertainty in establishing EGFR as a prognostic and predictive factor in breast cancer is mostly due to the different methods used for the detection of EGFR dysregulation.
Ki67 immune expression is closely associated with the cell cycle and can be used as a prognostic and predictive marker (40,41). In TNBC patients, high expression of Ki67 has a direct correlation with tumour size, grade and higher levels (>35% staining) have been linked with an increased risk of death. Ki67 levels were significantly increased in ductal TNBC compared to other histologic types (80% in TNBC vs. 10-30% in other types) (42). Li et al. investigated the expression of Ki67 in TNBC and found that it was significantly correlated with tumour size and lymph node metastases, no correlation was observed with age and clinical stage; suggesting that Ki67 may be an indicator of poor prognosis in TNBC patients (43). Niikura et al. on the other hand showed that Ki67 was not associated with survival in the hormone receptor (HR)-negative group of 716 patients (44).
In contrast to the established predictive and prognostic value of Ki67 expression in patients with HR-positive tumours (45-47), there is only little evidence to support Ki67 as a predictive marker for chemotherapeutic efficacy and defining good prognosis in HR-negative breast cancers after neoadjuvant chemotherapy. Sueta et al. reported that Ki67 had no predictive value for pathologic Complete Response (pCR) in HER2 and triple-negative subtypes with neoadjuvant chemotherapy (NAC) (45). Similarly, Joneset al. also reported that Ki67 had no predictive value for pCR in triple-negative subtypes, as a greater chemotherapy sensitivity was generally observed in these tumours (48).
However, one clinical trial with 552 breast cancer patients showed that Ki67 independently improved the prediction of treatment response in a group of luminal tumours as well as triple-negative tumours post neoadjuvant chemotherapy (49). Tan et al. examined Ki67 expression as a predictor of pCR after anthracycline and/or taxane-containing neoadjuvant chemotherapy in a total 183 HR-negative patients, 61 of which were TNBC. Ki67 labelling index was a predictive marker for pathologic complete response and higher Ki67 expression was associated with HER2 status, tumor size, lymph node status, LVI and tumor grade. Also, high Ki67 expression in residual tumours was strongly correlated with poor disease-free, but not overall survival (50). How et al. has concluded that lower Ki67 has poor prognosis relevance in TNBC patients diagnosed at ≤50 years-old in a study of 571 patients (51).
In our study, Fisher’s exact test resulted in a statistically significant relationship (p-value=0.041) between cell proliferation index Ki67, which was positive in 44 out of the 79 patients, and cytokeratin 17 which was positive in 38 cases out of the 79.
DFS and OS between groups of patients with different levels of each biomarker have been compared using log-rank test with a 5% level of statistical significance. According to the Log-rank test, only the cell proliferation index (Ki67) has been proven to be statistically significant for the DFS(p-value=0.0378), and for the OS (p-value=0.0186), which means that the survival function is differentiated for the two levels of the biomarker. The significance of this biomarker has also been confirmed by the Cox regression model for DFS with backward elimination (HR=0.39, 95%CI=0.153-0.994, p-value=0.048). We run the same Cox regression model with backward elimination for OS and it concluded that biomarker Ki67 is an important predictor for the overall survival of patients (HR=0.277, 95%CI=0.087-0.886, p-value=0.03).
Further studies are required to assess the benefit of Ki67 assessment in TNBC.
P63 is a transcription factor member of the p53 gene family. Recent data suggest a complex role for p63 in breast cancer with certain studies suggesting an oncogenic role for ΔNp63, and others a tumor suppressor role (52,53). In our study, the expression of protein p63 appeared to be lower in patients with higher tumor grade (OR=0.18, 95%CI=0.035-0.978, p-value=0.047).
Conclusion
High expression of claudin-7 and low expression of c-kit and protein p63 are associated with higher tumour grade. AR and CK5/6 expression seem to be important in LVI. These findings suggest that these biomarkers may be useful as prognostic or predictive indicators, as well as possible markers for novel therapies.
References
- 1.Glass AG, Lacey JV Jr., Carreon JD, Hoover RN. Breast cancer incidence, 1980-2006: Combined roles of menopausal hormone therapy, screening mammography, and estrogen receptor status. J Natl Cancer Inst. 2007;99(15):1152–1161. doi: 10.1093/jnci/djm059. [DOI] [PubMed] [Google Scholar]
- 2.Quinn JE, Kennedy RD, Mullan PB, Gilmore PM, Carty M, Johnston PG, Harkin DP. Brca1 functions as a differential modulator of chemotherapy-induced apoptosis. Cancer Res. 2003;63(19):6221–6228. [PubMed] [Google Scholar]
- 3.Lioni M, Brafford P, Andl C, Rustgi A, El-Deiry W, Herlyn M, Smalley KS. Dysregulation of claudin-7 leads to loss of e-cadherin expression and the increased invasion of esophageal squamous cell carcinoma cells. Am J Pathol. 2007;170(2):709–721. doi: 10.2353/ajpath.2007.060343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh AB, Sharma A, Dhawan P. Claudin family of proteins and cancer: An overview. J Oncol. 2010;2010:541957. doi: 10.1155/2010/541957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myal Y, Leygue E, Blanchard AA. Claudin 1 in breast tumorigenesis: Revelation of a possible novel "Claudin high" Subset of breast cancers. J Biomed Biotechnol. 2010;2010:956897. doi: 10.1155/2010/956897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Janne PA. Met amplification leads to gefitinib resistance in lung cancer by activating erbb3 signaling. Science. 2007;316(5827):1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 7.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, Backlund MG, Yin Y, Khramtsov AI, Bastein R, Quackenbush J, Glazer RI, Brown PH, Green JE, Kopelovich L, Furth PA, Palazzo JP, Olopade OI, Bernard PS, Churchill GA, Van Dyke T, Perou CM. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8(5):R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernardi MA, Logullo AF, Pasini FS, Nonogaki S, Blumke C, Soares FA, Brentani MM. Prognostic significance of cd24 and claudin-7 immunoexpression in ductal invasive breast cancer. Oncol Rep. 2012;27(1):28–38. doi: 10.3892/or.2011.1477. [DOI] [PubMed] [Google Scholar]
- 9.Sierra JR, Tsao MS. C-met as a potential therapeutic target and biomarker in cancer. Ther Adv Med Oncol 3(1. 2011;Suppl):S21–35. doi: 10.1177/1758834011422557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan S, Jiao X, Zou H, Li K. Prognostic significance of c-met in breast cancer: A meta-analysis of 6010 cases. Diagn Pathol. 2015;10:62. doi: 10.1186/s13000-015-0296-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salmena L, Carracedo A, Pandolfi PP. Tenets of pten tumor suppression. Cell. 2008;133(3):403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 12.Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, Richon C, Yan K, Wang B, Vassal G, Delaloge S, Hortobagyi GN, Symmans WF, Lazar V, Pusztai L. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res. 2009;15(2):441–451. doi: 10.1158/1078-0432.CCR-08-1791. [DOI] [PubMed] [Google Scholar]
- 13.Martin V, Botta F, Zanellato E, Molinari F, Crippa S, Mazzucchelli L, Frattini M. Molecular characterization of egfr and egfr-downstream pathways in triple negative breast carcinomas with basal like features. Histol Histopathol. 2012;27(6):785–792. doi: 10.14670/HH-27.785. [DOI] [PubMed] [Google Scholar]
- 14.Stefansson OA, Jonasson JG, Olafsdottir K, Hilmarsdottir H, Olafsdottir G, Esteller M, Johannsson OT, Eyfjord JE. Cpg island hypermethylation of brca1 and loss of prb as co-occurring events in basal/triple-negative breast cancer. Epigenetics. 2011;6(5):638–649. doi: 10.4161/epi.6.5.15667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inanc M, Ozkan M, Karaca H, Berk V, Bozkurt O, Duran AO, Ozaslan E, Akgun H, Tekelioglu F, Elmali F. Cytokeratin 5/6, c-met expressions, and pten loss prognostic indicators in triple-negative breast cancer. Med Oncol. 2014;31(1):801. doi: 10.1007/s12032-013-0801-7. [DOI] [PubMed] [Google Scholar]
- 16.Bouchalova K, Svoboda M, Kharaishvili G, Vrbkova J, Bouchal J, Trojanec R, Koudelakova V, Radova L, Cwiertka K, Hajduch M, Kolar Z. Bcl2 is an independent predictor of outcome in basal-like triple-negative breast cancers treated with adjuvant anthracycline-based chemotherapy. Tumour Biol. 2015;36(6):4243–4252. doi: 10.1007/s13277-015-3061-7. [DOI] [PubMed] [Google Scholar]
- 17.Horiuchi D, Kusdra L, Huskey NE, Chandriani S, Lenburg ME, Gonzalez-Angulo AM, Creasman KJ, Bazarov AV, Smyth JW, Davis SE, Yaswen P, Mills GB, Esserman LJ, Goga A. Myc pathway activation in triple-negative breast cancer is synthetic lethal with cdk inhibition. J Exp Med. 2012;209(4):679–696. doi: 10.1084/jem.20111512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsutsui S, Yasuda K, Suzuki K, Takeuchi H, Nishizaki T, Higashi H, Era S. A loss of c-kit expression is associated with an advanced stage and poor prognosis in breast cancer. Br J Cancer. 2006;94(12):1874–1878. doi: 10.1038/sj.bjc.6603183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diallo R, Ting E, Gluz O, Herr A, Schutt G, Geddert H, Mohrmann S, Gabbert HE, Nitz U, Poremba C. C-kit expression in high-risk breast cancer subgroup treated with high-dose or conventional dose-dense chemotherapy. Verh Dtsch Ges Pathol. 2006;90:177–185. [PubMed] [Google Scholar]
- 20.Zhu Y, Wang Y, Guan B, Rao Q, Wang J, Ma H, Zhang Z, Zhou X. C-kit and pdgfra gene mutations in triple negative breast cancer. Int J Clin Exp Pathol. 2014;7(7):4280–4285. [PMC free article] [PubMed] [Google Scholar]
- 21.Kanapathy Pillai SK, Tay A, Nair S, Leong CO. Triple-negative breast cancer is associated with egfr, ck5/6 and c-kit expression in malaysian women. BMC Clin Pathol. 2012;12:18. doi: 10.1186/1472-6890-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheppard KA, Fitz LJ, Lee JM, Benander C, George JA, Wooters J, Qiu Y, Jussif JM, Carter LL, Wood CR, Chaudhary D. Pd-1 inhibits t-cell receptor induced phosphorylation of the zap70/cd3zeta signalosome and downstream signaling to pkctheta. FEBS Lett. 2004;574(1-3):37–41. doi: 10.1016/j.febslet.2004.07.083. [DOI] [PubMed] [Google Scholar]
- 23.Beckers RK, Selinger CI, Vilain R, Madore J, Wilmott JS, Harvey K, Holliday A, Cooper CL, Robbins E, Gillett D, Kennedy CW, Gluch L, Carmalt H, Mak C, Warrier S, Gee HE, Chan C, McLean A, Walker E, McNeil CM, Beith JM, Swarbrick A, Scolyer RA, O’Toole SA. Programmed death ligand 1 expression in triple-negative breast cancer is associated with tumour-infiltrating lymphocytes and improved outcome. Histopathology. 2016;69(1):25–34. doi: 10.1111/his.12904. [DOI] [PubMed] [Google Scholar]
- 24.Qin T, Zeng YD, Qin G, Xu F, Lu JB, Fang WF, Xue C, Zhan JH, Zhang XK, Zheng QF, Peng RJ, Yuan ZY, Zhang L, Wang SS. High pd-l1 expression was associated with poor prognosis in 870 chinese patients with breast cancer. Oncotarget. 2015;6(32):33972–33981. doi: 10.18632/oncotarget.5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 27.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning PE, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van de Rijn M, Perou CM, Tibshirani R, Haas P, Kallioniemi O, Kononen J, Torhorst J, Sauter G, Zuber M, Kochli OR, Mross F, Dieterich H, Seitz R, Ross D, Botstein D, Brown P. Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. Am J Pathol. 2002;161(6):1991–1996. doi: 10.1016/S0002-9440(10)64476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutton LM, Han JS, Molberg KH, Sarode VR, Cao D, Rakheja D, Sailors J, Peng Y. Intratumoral expression level of epidermal growth factor receptor and cytokeratin 5/6 is significantly associated with nodal and distant metastases in patients with basal-like triple-negative breast carcinoma. Am J Clin Pathol. 2010;134(5):782–787. doi: 10.1309/AJCPRMD3ARUO5WPN. [DOI] [PubMed] [Google Scholar]
- 31.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363(20):1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 32.Ryu DW, Jung MJ, Choi WS, Lee CH. Clinical significance of morphologic characteristics in triple negative breast cancer. J Korean Surg Soc. 2011;80(5):301–306. doi: 10.4174/jkss.2011.80.5.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu R, Dawood S, Holmes MD, Collins LC, Schnitt SJ, Cole K, Marotti JD, Hankinson SE, Colditz GA, Tamimi RM. Androgen receptor expression and breast cancer survival in postmenopausal women. Clin Cancer Res. 2011;17(7):1867–1874. doi: 10.1158/1078-0432.CCR-10-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGhan LJ, McCullough AE, Protheroe CA, Dueck AC, Lee JJ, Nunez-Nateras R, Castle EP, Gray RJ, Wasif N, Goetz MP, Hawse JR, Henry TJ, Barrett MT, Cunliffe HE, Pockaj BA. Androgen receptor-positive triple negative breast cancer: A unique breast cancer subtype. Ann Surg Oncol. 2014;21(2):361–367. doi: 10.1245/s10434-013-3260-7. [DOI] [PubMed] [Google Scholar]
- 35.Millis SZ, Gatalica Z, Winkler J, Vranic S, Kimbrough J, Reddy S, O’Shaughnessy JA. Predictive biomarker profiling of >6000 breast cancer patients shows heterogeneity in tnbc, with treatment implications. Clin Breast Cancer. 2015;15(6):473–481. doi: 10.1016/j.clbc.2015.04.008. e473. [DOI] [PubMed] [Google Scholar]
- 36.Bertheau P, Espie M, Turpin E, Lehmann J, Plassa LF, Varna M, Janin A, de The H. Tp53 status and response to chemotherapy in breast cancer. Pathobiology. 2008;75(2):132–139. doi: 10.1159/000123851. [DOI] [PubMed] [Google Scholar]
- 37.Han JS, Cao D, Molberg KH, Sarode VR, Rao R, Sutton LM, Peng Y. Hormone receptor status rather than her2 status is significantly associated with increased ki-67 and p53 expression in triple-negative breast carcinomas, and high expression of ki-67 but not p53 is significantly associated with axillary nodal metastasis in triple-negative and high-grade non-triple-negative breast carcinomas. Am J Clin Pathol. 2015;135(2):230–237. doi: 10.1309/AJCP9DV3EVZUATFV. [DOI] [PubMed] [Google Scholar]
- 38.Petit T, Wilt M, Velten M, Millon R, Rodier JF, Borel C, Mors R, Haegele P, Eber M, Ghnassia JP. Comparative value of tumour grade, hormonal receptors, ki-67, her-2 and topoisomerase ii alpha status as predictive markers in breast cancer patients treated with neoadjuvant anthracycline-based chemotherapy. Eur J Cancer. 2004;40(2):205–211. doi: 10.1016/s0959-8049(03)00675-0. [DOI] [PubMed] [Google Scholar]
- 39.Tewari M, Krishnamurthy A, Shukla HS. Predictive markers of response to neoadjuvant chemotherapy in breast cancer. Surg Oncol. 2008;17(4):301–311. doi: 10.1016/j.suronc.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 40.de Azambuja E, Cardoso F, de Castro G Jr., Colozza M, Mano MS, Durbecq V, Sotiriou C, Larsimont D, Piccart-Gebhart MJ, Paesmans M. Ki-67 as prognostic marker in early breast cancer: A meta-analysis of published studies involving 12,155 patients. Br J Cancer. 2007;96(10):1504–1513. doi: 10.1038/sj.bjc.6603756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA. Ki67 in breast cancer: Prognostic and predictive potential. Lancet Oncol. 2010;11(2):174–183. doi: 10.1016/S1470-2045(09)70262-1. [DOI] [PubMed] [Google Scholar]
- 42.Munzone E, Botteri E, Sciandivasci A, Curigliano G, Nole F, Mastropasqua M, Rotmensz N, Colleoni M, Esposito A, Adamoli L, Luini A, Goldhirsch A, Viale G. Prognostic value of ki-67 labeling index in patients with node-negative, triple-negative breast cancer. Breast Cancer Res Treat. 2012;134(1):277–282. doi: 10.1007/s10549-012-2040-6. [DOI] [PubMed] [Google Scholar]
- 43.Li H, Han X, Liu Y, Liu G, Dong G. Ki67 as a predictor of poor prognosis in patients with triple-negative breast cancer. Oncol Lett. 2015;9(1):149–152. doi: 10.3892/ol.2014.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niikura N, Masuda S, Kumaki N, Xiaoyan T, Terada M, Terao M, Iwamoto T, Oshitanai R, Morioka T, Tuda B, Okamura T, Saito Y, Suzuki Y, Tokuda Y. Prognostic significance of the ki67 scoring categories in breast cancer subgroups. Clin Breast Cancer. 2014;14(5):323–329. doi: 10.1016/j.clbc.2013.12.013. e323. [DOI] [PubMed] [Google Scholar]
- 45.Sueta A, Yamamoto Y, Hayashi M, Yamamoto S, Inao T, Ibusuki M, Murakami K, Iwase H. Clinical significance of pretherapeutic ki67 as a predictive parameter for response to neoadjuvant chemotherapy in breast cancer: Is it equally useful across tumor subtypes. Surgery. 2014;155(5):927–935. doi: 10.1016/j.surg.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 46.Viale G, Giobbie-Hurder A, Regan MM, Coates AS, Mastropasqua MG, Dell’Orto P, Maiorano E, MacGrogan G, Braye SG, Ohlschlegel C, Neven P, Orosz Z, Olszewski WP, Knox F, Thurlimann B, Price KN, Castiglione-Gertsch M, Gelber RD, Gusterson BA, Goldhirsch A. Prognostic and predictive value of centrally reviewed ki-67 labeling index in postmenopausal women with endocrine-responsive breast cancer: Results from breast international group trial 1-98 comparing adjuvant tamoxifen with letrozole. J Clin Oncol. 2008;26(34):5569–5575. doi: 10.1200/JCO.2008.17.0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshioka T, Hosoda M, Yamamoto M, Taguchi K, Hatanaka KC, Takakuwa E, Hatanaka Y, Matsuno Y, Yamashita H. Prognostic significance of pathologic complete response and ki67 expression after neoadjuvant chemotherapy in breast cancer. Breast Cancer. 2015;22(2):185–191. doi: 10.1007/s12282-013-0474-2. [DOI] [PubMed] [Google Scholar]
- 48.Jones RL, Salter J, A’Hern R, Nerurkar A, Parton M, Reis-Filho JS, Smith IE, Dowsett M. Relationship between oestrogen receptor status and proliferation in predicting response and long-term outcome to neoadjuvant chemotherapy for breast cancer. Breast Cancer Res Treat. 2010;119(2):315–323. doi: 10.1007/s10549-009-0329-x. [DOI] [PubMed] [Google Scholar]
- 49.Fasching PA, Heusinger K, Haeberle L, Niklos M, Hein A, Bayer CM, Rauh C, Schulz-Wendtland R, Bani MR, Schrauder M, Kahmann L, Lux MP, Strehl JD, Hartmann A, Dimmler A, Beckmann MW, Wachter DL. Ki67, chemotherapy response, and prognosis in breast cancer patients receiving neoadjuvant treatment. BMC Cancer. 2011;11:486. doi: 10.1186/1471-2407-11-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan QX, Qin QH, Yang WP, Mo QG, Wei CY. Prognostic value of ki67 expression in hr-negative breast cancer before and after neoadjuvant chemotherapy. Int J Clin Exp Pathol. 2014;7(10):6862–6870. [PMC free article] [PubMed] [Google Scholar]
- 51.Hao S, He ZX, Yu KD, Yang WT, Shao ZM. New insights into the prognostic value of ki-67 labeling index in patients with triple-negative breast cancer. Oncotarget. 2016;7(17):24824–24831. doi: 10.18632/oncotarget.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Memmi EM, Sanarico AG, Giacobbe A, Peschiaroli A, Frezza V, Cicalese A, Pisati F, Tosoni D, Zhou H, Tonon G, Antonov A, Melino G, Pelicci PG, Bernassola F. P63 sustains self-renewal of mammary cancer stem cells through regulation of sonic hedgehog signaling. Proc Natl Acad Sci USA. 2015;112(11):3499–3504. doi: 10.1073/pnas.1500762112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buckley NE, Conlon SJ, Jirstrom K, Kay EW, Crawford NT, O’Grady A, Sheehan K, Mc Dade SS, Wang CW, McCance DJ, Johnston PG, Kennedy RD, Harkin DP, Mullan PB. The deltanp63 proteins are key allies of brca1 in the prevention of basal-like breast cancer. Cancer Res. 2011;71(5):1933–1944. doi: 10.1158/0008-5472.CAN-10-2717. [DOI] [PubMed] [Google Scholar]