Abstract
Background: Insufficient specificity and invasiveness of currently used diagnostic methods raises the need for new markers of urological tumors. The aim of this study was to find a link between the urinary excretion of amino acids and the presence of urological tumors. Materials and Methods: Using ion-exchange chromatography, we tested urine samples of patients with prostate cancer (n=30), urinary bladder cancer (n=28), renal cell carcinoma (n=16) and healthy volunteers (control group; n=21). Results: In each category, we found a group of amino acids which differed in concentration compared to the control group. These differences were most significant in sarcosine in patients with prostate cancer; leucine, phenylalanine and arginine in those with bladder cancer; and sarcosine, glutamic acid, glycine, tyrosine and arginine in the those with renal cell carcinoma. Conclusion: Results of our research imply a possible connection between the occurrence of specific types of amino acids in the urine and the presence of urological tumors.
Keywords: Amino acids, prostate cancer, renal cell carcinoma, urinary bladder cancer
The incidence of urological malignancies including cancer of the prostate, bladder and kidney is relatively high. Prostate cancer is the second most common malignancy in the United States among men aside from non-melanoma skin cancer, with estimated 172,000 diagnoses and 28,000 deaths in 2014 (1). Bladder cancer is the 11th most commonly diagnosed cancer in the world and 14th leading cause of cancer death (2), while renal cell carcinoma represents 2-3% of all cancer and in most European countries, its incidence is still rising (3).
Despite the frequency of these types of cancer, reliable biomarkers still need to be discovered. Many possible biomarkers of prostate cancer have been tested, including prostate cancer antigen 3 (4), alpha-methyl coenzyme A racemase (5), transmembrane protease, serine 2 (TMPRSS2)– ERG, ETS transcription factor (ERG) translocation (6), urinary prostate-specific antigen (PSA) (7) or urinary thiosulfate (8). However, the only routinely used marker is serum PSA, which still causes many controversies due to the non-specificity of its elevation (9).
For patients with renal tumors, early diagnosis is crucial, nevertheless the majority of renal masses are detected incidentally during the investigation of unrelated problems. Numerous molecular markers have been tested such as carbonic anhydrase IX (CaIX) (10), vascular endothelial growth factor (VEGF) (11), hypoxia-inducible factor (HIF) (12), Ki67, p53, phosphatase and tensin homolog (PTEN) and others (13), however none of these has been recommended for routine practice.
Bladder cancer is associated with the need for lifelong surveillance by cystoscopy and urine cytology (14) and the highest cost from diagnosis to death among all cancer types (15). Currently tested markers include DNA, RNA, miRNA and protein markers. However, none are routinely used in daily clinical practice.
Urine represents an attractive biological material for testing due to the non-invasiveness of its collection, its relative stability during storage and, most of all, a natural association with the biology of tumors in the urinary tract. As shown in numerous studies, urine metabolomic profiling is suitable for urological malignancies. Changes in xenobiotic metabolism are believed to be causal in the genesis of bladder cancer (16). A metabolomic approach was used in a study of kidney cancer which demonstrated higher expression of metabolites associated with energetic metabolism, high tumor protein breakdown and utilization, and the Warburg effect (17). Sreekumar et al. used metabolomic analysis to discover markers usable for noninvasive diagnosis and prognosis of prostate cancer (18).
Our research focused on the possibility that urinary excretion of amino acids or specific groups of amino acids may be associated with the presence of prostate cancer (PCa), urinary bladder cancer (UBC) and renal cell carcinoma (RCC).
Materials and Methods
Patient selection. Institutional Review Board approval (reference: EK-377/13) was obtained for the analysis of this patient population and the population of healthy volunteers, all samples were collected and analyzed with written informed consent of the participant.
The control group we used consisted of 30 healthy volunteers older than 50 years without oncological history and no signs of current cancer according to physical, ultrasound and laboratory examinations. Spontaneously voided urine samples were obtained and immediately frozen at –75˚C until analysis. Samples of 30 patients with histologicaly confirmed PCa, 28 patients diagnosed with UBC, and 16 patients with RCC were handled in the same way. Pathological characteristics are described in Table I. For technical and organizational reasons, it was not possible to provide a standardized diet for all tested participants. Since we are aware of the impact of diet on the excretion of amino acids, we decided to use the first morning urine after fasting overnight.
Table I. Histological characteristics of tumors.
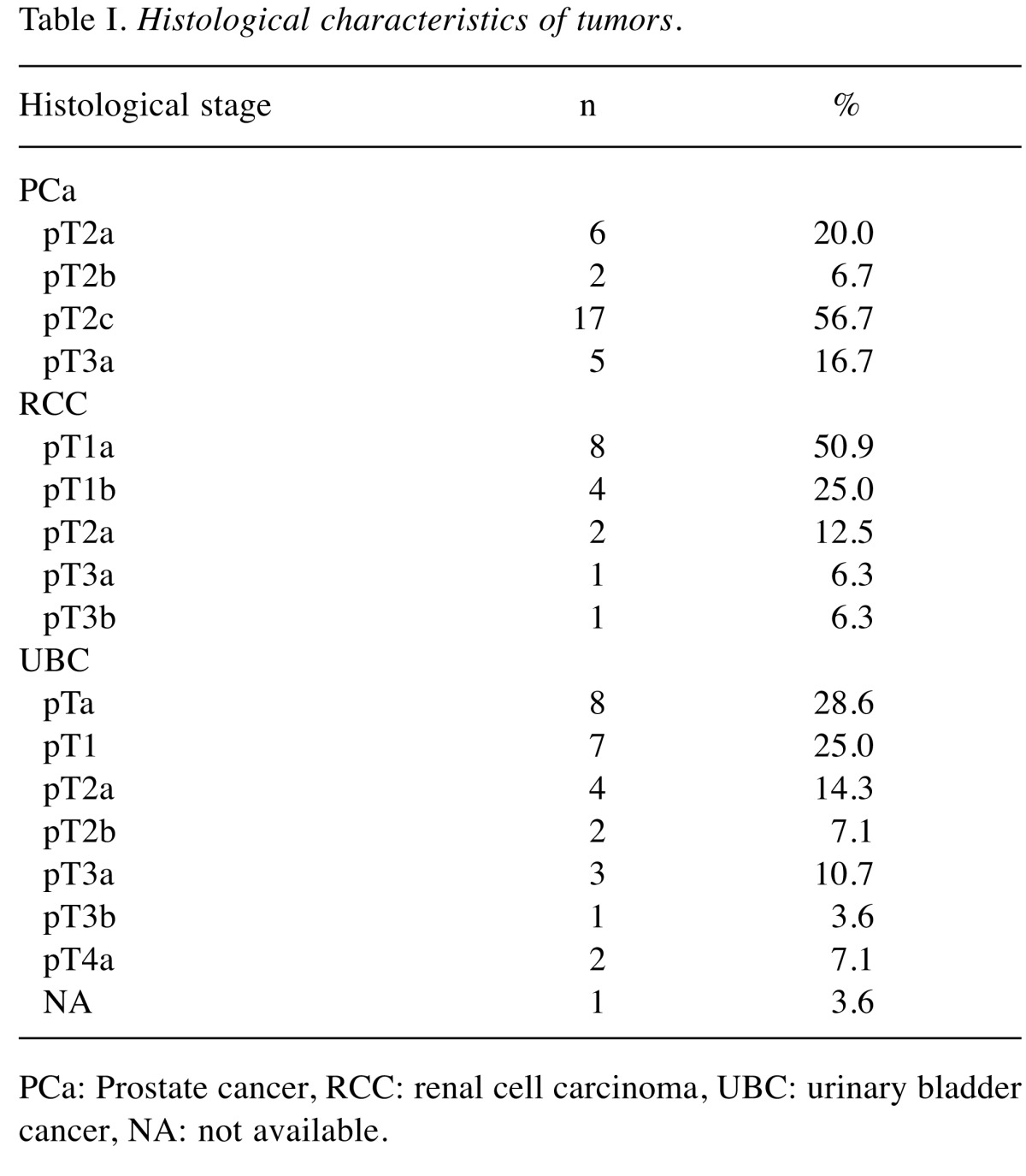
PCa: Prostate cancer, RCC: renal cell carcinoma, UBC: urinary bladder cancer, NA: not available
Sample preparation for determination of urinary amino acid patterns. For analysis of amino acid patterns, excluding sarcosine, 500 μl of urine sample was mixed with 500 μl of 35% HCl and mineralized using microwave equipment MW 3000 (Anton Paar, Graz, Austria). One hundred microliters of mineralized sample were diluted with 900 μl of dilution buffer and centrifuged at 25,000 × g for 20 min using a 5417R centrifuge (Eppendorf, Hamburg, Germany) for 20 min. Subsequently, 500 μl of sample was diluted in 500 μl of 0.6 M NaOH prior to being analyzed by ion-exchange chromatography with a visual spectrum detector (IEC-Vis). Sarcosine, as a non-proteinogenic amino acid was determined by a simple evaporation of 500 μl of urine using an Ultravap 96 nitrogen blow-down evaporator with spiral needles (Porvair Sciences limited, Leatherhead, Surrey, UK). After this procedure, the sample was diluted with 500 μl of dilution buffer and analyzed using IEC-Vis.
Ion-exchange liquid chromatography. For determination of sarcosine an IEC-Vis (Model AAA-400; Ingos, Prague, Czech Republic) with post column derivatization by ninhydrin was used. A glass column with inner diameter of 3.7 mm and 350 mm length was filled manually with strong cation exchanger (Ostion LG ANB; Ingos) in sodium cycle with particles of approximately 12 μm diameter and 8% porosity. The column temperature was maintained at 60˚C. Double channel VIS detector with inner cell of 5 μl was set to two wavelengths: 440 and 570 nm. A prepared solution of ninhydrin was stored under nitrogen in the dark at 4˚C. Elution of amino acids was achieved by buffer containing 10.0 g of citric acid, 5.6 g of sodium citrate, and 8.36 g of sodium chloride per liter of solution and pH was 3.0. The flow rate was 0.25 ml per min. The reactor temperature was set at 120˚C. Analytical parameters of the analyzed amino acids are shown in Table II.
Table II. Analytical parameters for amino acids analyzed by IEC-Vis.
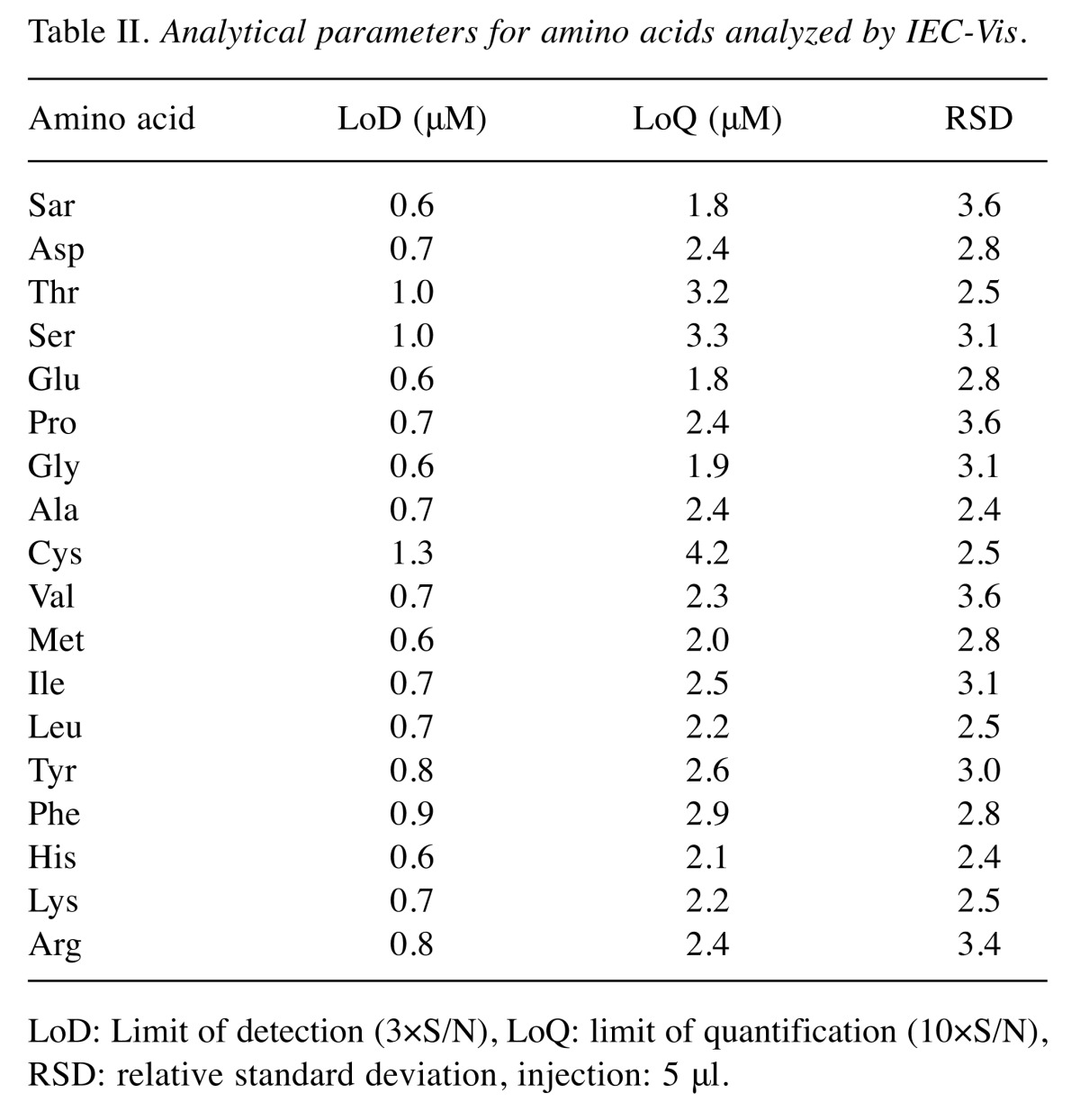
LoD: Limit of detection (3×S/N), LoQ: limit of quantification (10×S/N), RSD: relative standard deviation, injection: 5 μl.
Normalization of amino acid values obtained by IEC-Vis analysis. All obtained results were normalized using urinary creatinine. Creatinine was determined using a creatinine kit (Greiner, Stuttgart, Germany) on a BS-400 automated spectrophotometer (Mindray, Shenzhen, China), according to the manufacturer´s instructions.
Statistical analysis. Due to the non-normality of the recorded data, non-parametric statistical methods were used. Differences between groups of patients and controls were subject to batteries of Mann–Whitney two-sample tests. p-Values adjusted for multiple comparisons by a Holm’s method of less than 0.05 were considered statistically significant. Analyses were conducted using R statistical package, version 3.1.2, R Core Team (2014).
Results
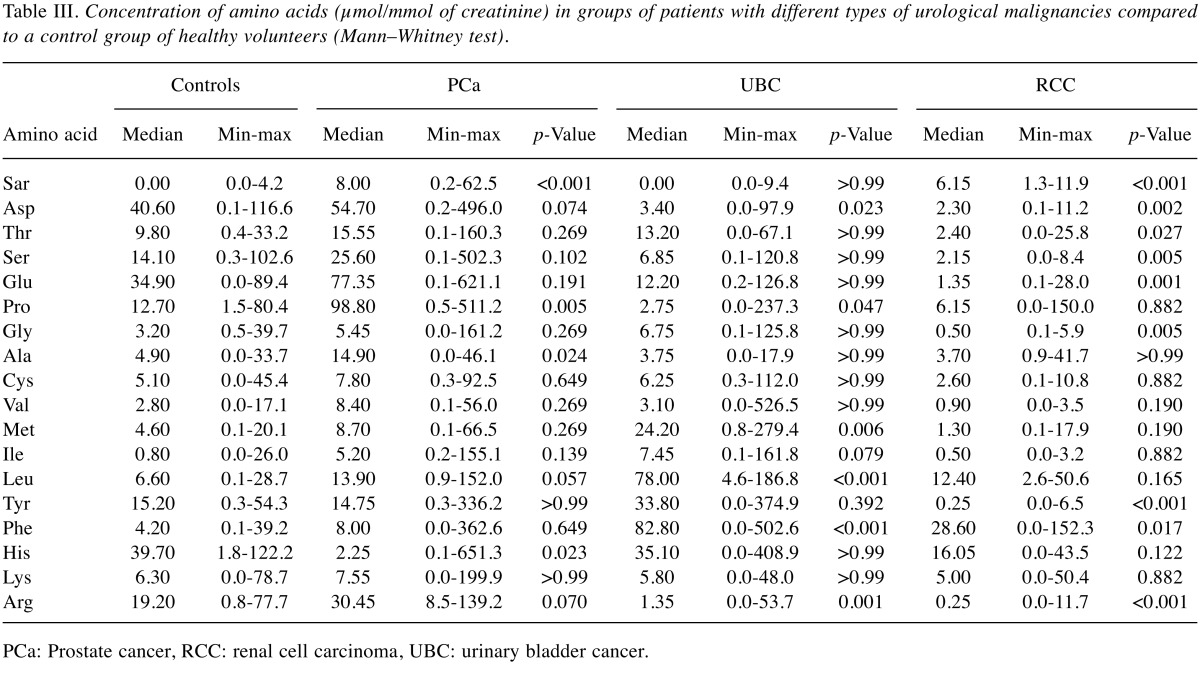
All differences in amino acid concentrations of patients with various types of urological malignancies compared with a control group of healthy volunteers are detailed in Table III. In the prostate cancer group, the analysis showed significantly (p<0.05) higher concentrations of sarcosine, proline and alanine and a lower concentration of histidine. In the group of patients with UBC, we found increased levels of methionine, leucine and phenylalanine and reduced levels of asparagine, proline and arginine. In comparison with the control group, patients with RCC showed increased concentrations of sarcosine and phenylalanine and reduced levels of asparagine, threonine, serine, glutamic acid, glycine, tyrosine and arginine.
Table III. Concentration of amino acids (μmol/mmol of creatinine) in groups of patients with different types of urological malignancies compared to a control group of healthy volunteers (Mann–Whitney test).
PCa: Prostate cancer, RCC: renal cell carcinoma, UBC: urinary bladder cancer.
There was no difference in the gender distribution between the control group and groups of patients with urological malignancies. The percentage of females in the control group, PCa, RCC and UBC groups was 28.6%, 0.0%, 31.3% and 28.6%, respectively. The age distribution also showed only negligible variation, with median ages of 59.0, 64.0, 62.5 and 66.5 years, respectively.
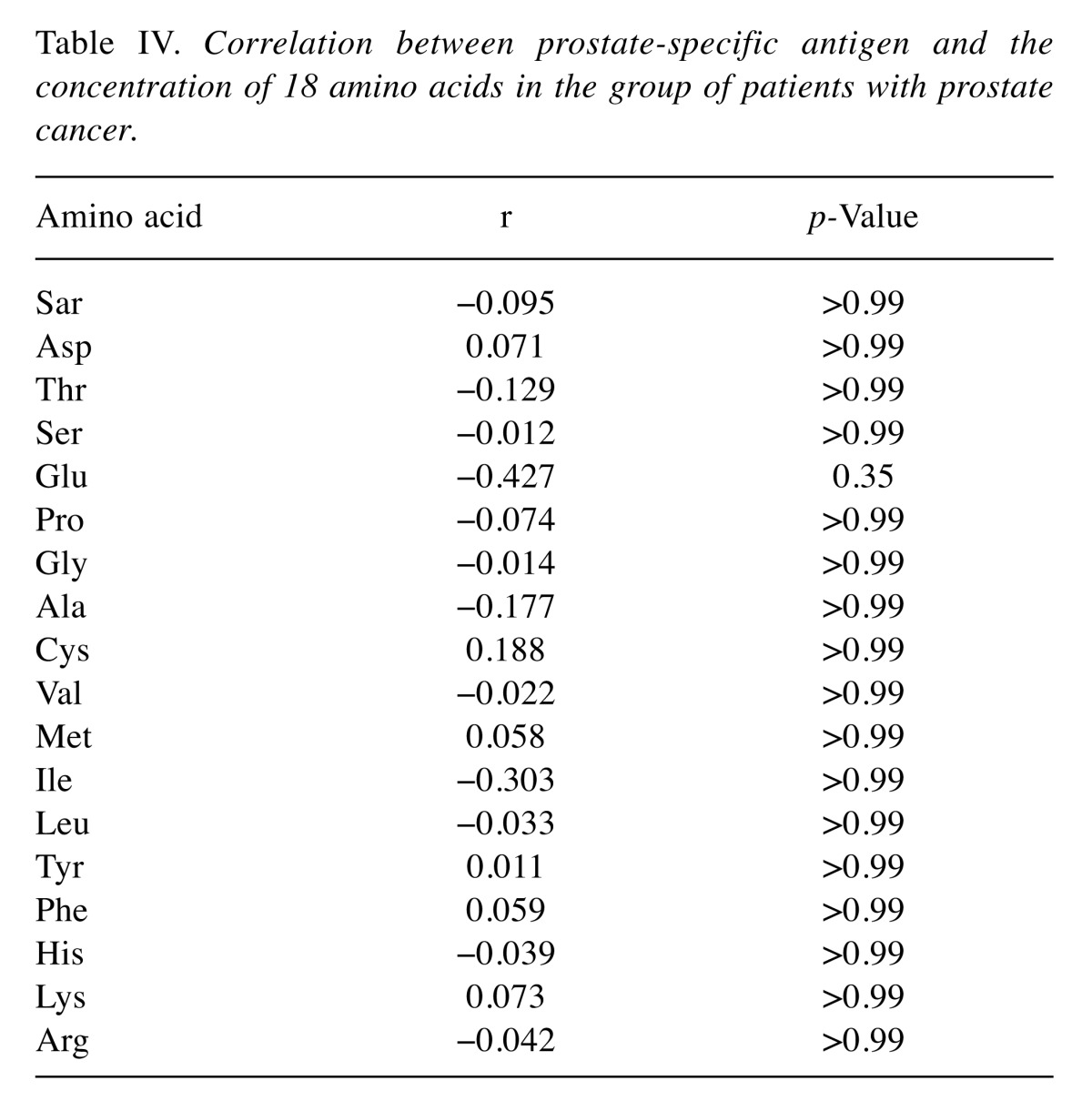
Out of 18 tested amino acids, there was a statistical difference only in serine (p<0.001) and lysine (p=0.012) between male and female controls. No correlation was found between PSA and any of the analyzed amino acids in the group of patients with PCa (Table IV).
Table IV. Correlation between prostate-specific antigen and the concentration of 18 amino acids in the group of patients with prostate cancer.
Discussion
Since Otto Warburg’s discovery of high consumption of glucose and secretion of lactate (even in excess of oxygen) by cancer cells, called the Warburg effect (19,20), highly proliferative cancer cells have been shown to exhibit alterations in various metabolic pathways (21). These changes can also be observed at the proteomic and the genomic levels (22).
A new branch of molecular biology called Metabolomics describes the complete set of small-molecule chemicals that can be found within a biological sample as the ‘metabolome’ (23). Since these molecules are products of numerous interactions and reflect actual processes occurring in the body (24), they could possibly lead to the discovery of potential biomarkers.
The easy accessibility of urine allows detailed analysis of ions and metabolites and shows that human urine contains a significant amount of hydrophilic molecules (amino acids and their derivatives, carbohydrates and carbohydrate conjugates, amongst others) (25). Interindividual variability of the human population, environmental and lifestyle influences such as diet and toxins, represent a great complication of clinical research in metabolomics. Lenz et al. observed that in urine samples of people whose diet was not standardized the metabolites which varied the most were hippurate and creatinine (26).
One of the first urine biomarkers, used for centuries, is the metabolite glucose in patients with diabetes. Regarding the kidneys, published studies showed urinary metabolic signatures not only of kidney injury (27) but also RCC. Kyoungmi et al. tested 1766 metabolites and demonstrated that urine metabolomic profiling can differentiate patients with RCC from healthy controls (28). One of the largest studies of patients with bladder cancer analyzed over 9,000 unique features and identified a few up-regulated metabolites in the patient group (29). The role of amino acids in urine as a potential oncological marker of PCa was discovered by Sreekumar et al. (30). Their research identified sarcosine as a metabolite which is markedly increased in the urine after digital rectal examination. However, the presumption that urinary sarcosine can discriminate between patients with and without prostate cancer better than PSA was disproved by Jentzmik et al. (31).
According to the Human Urine Metabolome Project (32), only few amino acids have been identified as possible oncological markers – alanine (33) and valine (34), associated with lung cancer. The only investigated and commercially used method of using amino acid concentration in urine is represented by Prostarix (Metabolon Inc.). This method based on study of 120 men is able to stratify pre-biopsy patients to risk groups according to alanine, sarcosine, glycine and glutamate level in urinary sediment obtained after digital rectal examination.
Despite controversies in previous studies, our results confirmed a strong link between the presence of sarcosine in urine and PCa. In the group of patients with RCC, we found elevated concentrations of serine and glycine. This finding is in line with a study of anabolic pathways for proteins, nucleic acids and lipids which are crucial to cancer cell growth (35).
Considering the natural proximity between urinary tract tumors and urine, our research implies a possible connection between the occurrence of specific types of amino acids in urine (or groups of amino acids) and the presence of these tumors. Monitoring the concentration of these amino acids as tumor makers in the urine can be considered a promising non-invasive method. Future use of this method should be confirmed by further research.
Acknowledgements
This study was supported by the Ministry of Health of the Czech Republic grant no. 15-33910A and a research grant AZV CZ 15-33.
References
- 1.U.S. Cancer Statistics Working Group Cancer Statistics: 1999–2014 Incidence and Mortality Web-based Report. Atlanta (GA): Department of Health and Human Services, Centers for Disease Control and Prevention, and National Cancer Institute. 2017 [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet] Lyon, France: International Agency for Research on Cancer. 2013 [Google Scholar]
- 3.Lindblad P. Epidemiology of renal cell carcinoma. Scand J Surg. 2004;93:88–96. doi: 10.1177/145749690409300202. [DOI] [PubMed] [Google Scholar]
- 4.Thapar R, Titus MA. Recent advances in metabolic profiling and imaging of prostate cancer. Curr Metabolomics. 2014;2:53–69. doi: 10.2174/2213235X02666140301002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sroka WD, Adamowski M, Słupski P, Siódmiak J, Jarzemski P, Odrowąż-Sypniewska G, Marszałł MP. Alpha-methylacyl-CoA racemase and hepsin as urinary prostate cancer markers. Int J Biol Markers. 2015;30:401–406. doi: 10.5301/jbm.5000146. [DOI] [PubMed] [Google Scholar]
- 6.Hessels D, Smit FP, Verhaegh GW, Witjes JA, Cornel EB, Schalken JA. Detection of TMPRSS2-ERG fusion transcripts and prostate cancer antigen 3 in urinary sediments may improve diagnosis of prostate cancer. Clin Cancer Res. 2007;13:5103–5108. doi: 10.1158/1078-0432.CCR-07-0700. [DOI] [PubMed] [Google Scholar]
- 7.Sağlam HS, Köse O, Ozdemir F, Adsan O. Do the values of prostate-specific antigen obtained from fresh and dried urine reflect the serum measurements. Urol Ann. 2013;5:99–102. doi: 10.4103/0974-7796.110006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chwatko G, Forma E, Wilkosz J, Głowacki R, Jóźwiak P, Różański W, Bryś M, Krześlak A. Thiosulfate in urine as a facilitator in the diagnosis of prostate cancer for patients with prostate-specific antigen less or equal 10 ng/ml. Clin Chem Lab Med. 2013;51:1825–1831. doi: 10.1515/cclm-2013-0069. [DOI] [PubMed] [Google Scholar]
- 9.Oesterling JE. Prostatespecific antigen: a critical assessment of the most useful tumor marker for adenocarcinoma of the prostate. J Urol. 1991;145:907–923. doi: 10.1016/s0022-5347(17)38491-4. [DOI] [PubMed] [Google Scholar]
- 10.Sim SH, Messenger MP, Gregory WM, Wind TC, Vasudev NS, Cartledge J, Thompson D, Selby PJ, Banks RE. Prognostic utility of pre-operative circulating osteopontin, carbonic anhydrase IX and CRP in renal cell carcinoma. Br J Cancer. 2012;107:1131–1137. doi: 10.1038/bjc.2012.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabatino M, Kim-Schulze S, Panelli MC, Stroncek D, Wang E, Taback B, Kim DW, Deraffele G, Pos Z, Marincola FM, Kaufman HL. Serum vascular endothelial growth factor and fibronectin predict clinical response to high-dose interleukin-2 therapy. J Clin Oncol. 2009;27:2645–2652. doi: 10.1200/JCO.2008.19.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivanov S, Liao SY, Ivanova A, Danilkovitch-Miagkova A, Tarasova N, Weirich G, Merrill MJ, Proescholdt MA, Oldfield EH, Lee J, Zavada J, Waheed A, Sly W, Lerman MI and Stanbridge EJ. Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am J Pathol. 2001;158:905–919. doi: 10.1016/S0002-9440(10)64038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HL, Seligson D, Liu X, Janzen N, Bui MH, Yu H, Shi T, Figlin RA, Horvath S, Belldegrun AS. Using protein expressions to predict survival in clear cell renal carcinoma. Clin Cancer Res. 2004;10:5464–5471. doi: 10.1158/1078-0432.CCR-04-0488. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths TR. Action on Bladder Cancer. Current perspectives in bladder cancer management. Int J Clin Pract. 2013;67:435–448. doi: 10.1111/ijcp.12075. [DOI] [PubMed] [Google Scholar]
- 15.Botteman MF, Pashos CL, Redaelli A, Laskin B, Hauser R. The health economics of bladder cancer: a comprehensive review of the published literature. Pharmacoeconomics. 2003;21:1315–1330. doi: 10.1007/BF03262330. [DOI] [PubMed] [Google Scholar]
- 16.Putluri N, Shojaie A, Vasu VT, Vareed SK, Nalluri S, Putluri V, Thangjam GS, Panzitt K, Tallman CT, Butler C, Sana TR, Fischer SM, Sica G, Brat DJ, Shi H, Palapattu GS, Lotan Y, Weizer AZ, Terris MK, Shariat SF, Michailidis G, Sreekumar A. Metabolomic profiling reveals potential markers and bioprocesses altered in bladder cancer progression. Cancer Res. 2011;71:7376–7386. doi: 10.1158/0008-5472.CAN-11-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim K, Taylor SL, Ganti S, Guo L, Osier MV, Weiss RH. Urine metabolomic analysis identifies potential biomarkers and pathogenic pathways in kidney cancer. OMICS. 2011;15:293–303. doi: 10.1089/omi.2010.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C and Chinnaiyan AM. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 19.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 20.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 21.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us. Cell. 2012;148:1132–1144. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliver SG, Winson MK, Kell DB, Baganz F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998;16:373–378. doi: 10.1016/s0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- 24.Dettmer K, Hammock BD. Metabolomics-a new exciting field within the "omics" sciences. Environ Health Perspect. 2004;112:A396–397. doi: 10.1289/ehp.112-1241997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C, Bjorndahl TC, Krishnamurthy R, Saleem F, Liu P, Dame ZT, Poelzer J, Huynh J, Yallou FS, Psychogios N, Dong E, Bogumil R, Roehring C, Wishart DS. The human urine metabolome. PLoS One. 2013;8(9):e73076. doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenz EM, Bright J, Wilson ID, Morgan SR, Nash AF. A 1H NMR-based metabonomic study of urine and plasma samples obtained from healthy human subjects. J Pharm Biomed Anal. 2003;33:1103–1115. doi: 10.1016/s0731-7085(03)00410-2. [DOI] [PubMed] [Google Scholar]
- 27.Han WK, Waikar SS, Johnson A, Betensky RA, Dent CL, Devarajan P, Bonventre JV. Urinary biomarkers in the early diagnosis of acute kidney injury. Kidney Int. 2008;73:863–869. doi: 10.1038/sj.ki.5002715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim K, Aronov P, Zakharkin SO, Anderson D, Perroud B, Thompson IM, Weiss RH. Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol Cell Proteomics. 2009;8:558–570. doi: 10.1074/mcp.M800165-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen C, Sun Z, Chen D, Su X, Jiang J, Li G, Lin B, Yan J. Developing urinary metabolomic signatures as early bladder cancer diagnostic markers. OMICS. 2015;19:1–11. doi: 10.1089/omi.2014.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C and Chinnaiyan AM. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 31.Jentzmik F, Stephan C, Miller K, Schrader M, Erbersdobler A, Kristiansen G, Lein M, Jung K. Sarcosine in urine after digital rectal examination fails as a marker in prostate cancer detection and identification of aggressive tumours. Eur Urol. 2010;58:12–18. doi: 10.1016/j.eururo.2010.01.035. [DOI] [PubMed] [Google Scholar]
- 32.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:801–807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:603–610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stretch C, Eastman T, Mandal R, Eisner R, Wishart DS, Mourtzakis M, Prado CM, Damaraju S, Ball RO, Greiner R, Baracos VE. Prediction of skeletal muscle and fat mass in patients with advanced cancer using a metabolomic approach. J Nutr. 2012;142:14–21. doi: 10.3945/jn.111.147751. [DOI] [PubMed] [Google Scholar]
- 35.Kalhan SC, Hanson RW. Resurgence of serine: an often neglected but indispensable amino acid. J Biol Chem. 2012;287:19786–19791. doi: 10.1074/jbc.R112.357194. [DOI] [PMC free article] [PubMed] [Google Scholar]