Connexin43 mRNA can be internally translated to generate N-terminally truncated isoforms including GJA1-20k. GJA1-20k expression is suppressed during TGF-β–induced epithelial–mesenchymal transition, stabilizing full-length Connexin43 in the Golgi, reducing channel oligomerization, cell surface expression, and connexin43 gap junction formation.
Abstract
Epithelial–mesenchymal transition (EMT) is activated during development, wound healing, and pathologies including fibrosis and cancer metastasis. Hallmarks of EMT are remodeling of intercellular junctions and adhesion proteins, including gap junctions. The GJA1 mRNA transcript encoding the gap junction protein connexin43 (Cx43) has been demonstrated to undergo internal translation initiation, yielding truncated isoforms that modulate gap junctions. The PI3K/Akt/mTOR pathway is central to translation regulation and is activated during EMT, leading us to hypothesize that altered translation initiation would contribute to gap junction loss. Using TGF-β–induced EMT as a model, we find reductions in Cx43 gap junctions despite increased transcription and stabilization of Cx43 protein. Biochemical experiments reveal suppression of the internally translated Cx43 isoform, GJA1-20k in a Smad3 and ERK-dependent manner. Ectopic expression of GJA1-20k does not halt EMT, but is sufficient to rescue gap junction formation. GJA1-20k localizes to the Golgi apparatus, and using superresolution localization microscopy we find retention of GJA1-43k at the Golgi in mesenchymal cells lacking GJA1-20k. NativePAGE demonstrates that levels of GJA1-20k regulate GJA1-43k hexamer oligomerization, a limiting step in Cx43 trafficking. These findings reveal alterations in translation initiation as an unexplored mechanism by which the cell regulates Cx43 gap junction formation during EMT.
INTRODUCTION
A variety of growth factors, including transforming growth factor β (TGF-β), can induce epithelial cells to undergo the transdifferentiation process of epithelial–mesenchymal transition (EMT). Although EMT occurs normally in development and wound healing, it can also contribute to pathological processes such as fibrosis and cancer progression (Lamouille et al., 2014). During EMT, cellular junctions are disassembled and remodeled in concert with cytoskeletal alterations and acquisition of a mesenchymal phenotype (Kalluri and Weinberg, 2009). Gap junctions, composed of connexin proteins, provide direct intercellular communication between polarized epithelial cells and are subject to down-regulation with loss of polarity during EMT (Goodenough and Paul, 2009; Bax et al., 2011). Of the 21 human connexins, connexin43 (Cx43; gene name GJA1) is the most ubiquitously expressed gap junction protein (Beyer et al., 1987). A rate-limiting step in Cx43 intracellular transport occurs at the trans-Golgi network (TGN) where six Cx43 monomers must oligomerize to form Cx43 hemichannels, or connexons, to permit cytoskeleton-based trafficking to the plasma membrane (Vanslyke et al., 2009). Connexons on the surface of apposing cells dock to form continuous channels which coalesce into dense arrays, termed gap junction plaques, to effect direct intercellular flow of ions and small molecules (Kumar and Gilula, 1996). Suppression of gap junctional intercellular coupling (GJIC) was thought to be a necessary step for EMT progression as cells become migratory; however, involvement of Cx43 in critical cellular processes including cell migration and cell cycle regulation implicate a more complex role for this protein during EMT (Langlois et al., 2010; Machtaler et al., 2014; Hino et al., 2015; Qiu et al., 2015).
Investigation of the molecular mechanisms governing EMT has focused on induction of transcriptional programs and posttranslational modifications. Translation, however, is gaining increasing appreciation as a dynamic regulator of the proteome during cellular processes including EMT (Tumbarello et al., 2005; Lindsay et al., 2011). Translation of Cx43 can be regulated at the RNA level during development, demonstrated in mice through alternative splicing and alternative promoter usage within its 5′ untranslated region (5′ UTR; Pfeifer et al., 2004). This process is further complicated by the existence of an internal ribosome entry site (IRES) within the GJA1 5′ UTR, and the recent finding that the coding sequence of GJA1 is also subject to internal translation initiation events to yield N-terminally truncated Cx43 isoforms (Schiavi et al., 1999; Smyth and Shaw, 2013; Salat-Canela et al., 2014; Ul-Hussain et al., 2014). Alternative modes of translation initiation such as internal translation are historically recognized as a mechanism used by viruses to increase their protein coding capacity (May et al., 2017). More recently, alternative translation has been recognized to occur in eukaryotes, playing a role in the biological programs of apoptosis, differentiation, and metastasis (Steder et al., 2013; Marques-Ramos et al., 2017). High-throughput experiments in mammalian cells have suggested a prominent role for alternative translation in modulating protein diversity, and genome-wide analysis of translation initiation sites using ribosome profiling identified a substantial degree of novel and “noncanonical” translation initiation start sites (Ingolia et al., 2011; Popa et al., 2016; Weingarten-Gabbay et al., 2016). These sites can generate N-terminal truncations of proteins as is found with GJA1, upstream open reading frames, or alternate reading frames (Ingolia et al., 2011).
The most predominantly expressed N-terminally truncated Cx43 isoform is termed GJA1-20k, with full-length Cx43 termed GJA1-43k (Smyth and Shaw, 2013; Salat-Canela et al., 2014; Ul-Hussain et al., 2014). Importantly, GJA1-20k is understood to reside at intracellular structures including the endoplasmic reticulum (ER) and Golgi apparatus and has been demonstrated to regulate Cx43 gap junction formation. Limiting the cell’s ability to express GJA1-20k results in distribution of Cx43 throughout the cytosol and loss of gap junctions at cell borders (Smyth and Shaw, 2013). In cardiac cells, ectopic expression of GJA1-20k induces actin stabilization and promotes gap junction formation at intercalated discs, revealing therapeutic potential in restoring gap junction coupling during ischemia (Basheer et al., 2017). More recently, studies have also localized GJA1-20k to mitochondria where a protective role in mitochondrial dynamics and integrity was identified (Fu et al., 2017). Levels of GJA1-20k are responsive to cellular stress such as hypoxia and simulated ischemia, and alteration of GJA1 mRNA internal translation can be regulated by signal transduction pathways including PI3K/Akt/mTOR and Mnk1/2 (Smyth and Shaw, 2013; Ul-Hussain et al., 2014). TGF-β induces EMT through both Smad and nonSmad signaling (Gal et al., 2008). One way that TGF-β signaling contributes to EMT is through activation of the PI3K/Akt/mTOR pathway, an important regulator of protein translation and GJA1-20k synthesis (Chen et al., 2012; Smyth and Shaw, 2013; Lamouille et al., 2014). We therefore hypothesized that alterations in translation initiation could contribute to changes in gap junction formation during EMT, independent of posttranslational or transcriptional regulation.
RESULTS
Internal translation of Gja1 is suppressed during TGF-β–induced EMT
To investigate the contribution of internal translation initiation in EMT-associated alterations in Cx43 gap junctions, we utilized normal mouse mammary gland epithelial cells (NMuMG). NMuMG cells are an established cell model which readily undergoes EMT upon stimulation with TGF-β (Piek et al., 1999). Expression of mesenchymal markers including fibronectin and N-cadherin with concomitant suppression of the epithelial marker E-cadherin were detected by Western blot, reverse transcription quantitative PCR (RT-qPCR), and immunofluorescence, confirming induction of the EMT process within 24 h of TGF-β addition (Supplemental Figure 1). Phase contrast microscopy reveals morphological changes from an epithelial “cobblestone” phenotype to larger spindle-shaped mesenchymal cells by 48 h after TGF-β stimulation (Figure 1A). Given the role of PI3K/AKT/mTOR in TGF-β–induced EMT, and in regulation of GJA1-20k translation (Smyth and Shaw, 2013), we investigated the status of Gja1 translation by Western blot. Cx43 was detected with an antibody directed against the C-terminus, capable of detecting GJA1-20k and its full-length counterpart GJA1-43k. We find significantly increased levels of GJA1-43k in mesenchymal cells in comparison to controls treated only with vehicle (Figure 1B). GJA1-20k levels, however, are significantly reduced relative to GJA1-43k (Figure 1, B, quantified in C). To ask whether increased GJA1-20k protein degradation is responsible, we used replication incompetent lentiviruses to generate stable cell lines encoding LacZ, the entire GJA1 coding sequence, or GJA1-20k alone. Again, levels of GJA1-43k were increased and relative levels of GJA1-20k reduced following stimulation with TGF-β in LacZ and GJA1 cell lines. The GJA1-20k stable cells, however, maintained GJA1-20k expression following stimulation with TGF-β, where the protein is constitutively synthesized from an independent mRNA and therefore not subject to suppression of internal translation (Figure 1, D, quantified in E). These data demonstrate that altered GJA1-43k and GJA1-20k levels in mesenchymal cells are likely not due to increased degradation of GJA1-20k, but rather inhibition of translation initiation within GJA1 mRNA with concomitant enhanced GJA1-43k translation from the initial start codon. Specificity to TGF-β signaling is confirmed with the TGF-β receptor inhibitor SB431542, which inhibits EMT induction and rescues expression of GJA1-20k. Together, these data suggest a shift in translation initiation may occur during EMT to regulate Cx43 localization and function at the posttranscriptional level.
FIGURE 1:
TGF-β treatment induces EMT and suppresses internal translation of Gja1 mRNA. NMuMG cells were treated with 2 ng/ml TGF-β for 48 h to induce EMT. (A) Phase contrast microscopy of NMuMG cells ×20. (B) Western blot of cell lysates probed with Cx43 C-terminal antibody to detect full-length GJA1-43k and internally translated GJA1-20k. α-Tubulin serves as loading control. (C) Quantification of GJA1-20k band intensity relative to GJA1-43k from B. (D) Stable pools of NMuMG cells transduced with pLenti6.3-LacZ, pLenti6.3-GJA1, or pLenti6.3-GJA1-20k were treated with vehicle, 2 ng/ml TGF-β, 5 μM SB431542, or TGF-β + SB431542 for 48 h. Western blot of cell lysates probed with Cx43 C-terminal antibody to detect full-length GJA1-43k and GJA1-20k. Arrows indicate TGF-β treated samples; GAPDH serves as loading control. (E) Quantification of GJA1-20k band intensity relative to GJA1-43k from D. Graphs represent mean ± SEM. Statistical analysis performed using the Student’s t test (C, n = 3) and one-way analysis of variance (ANOVA) with Tukey’s multiple comparison posttest (E, n = 3). *p ≤ 0.05.
GJA1-43k protein localizes to nonjunctional soluble pools during TGF-β–induced EMT
Dissolution of intercellular junctions is an established hallmark of EMT, yet conflicting reports exist regarding the fate of Cx43 as cells acquire the mesenchymal phenotype (van der Heyden et al., 2000; de Boer et al., 2007; Tacheau et al., 2008; Bax et al., 2011; Hills et al., 2012). Our finding that GJA1-20k, which is known to regulate gap junction formation, occurs at lower levels in mesenchymal cells (Figure 1), would suggest alterations in GJA1-43k intracellular trafficking. Immunofluorescence confocal microscopy reveals diffuse redistribution of Cx43 to the cytosol during EMT (Figure 2A). Cx43 gap junctions still occur at cell–cell borders in mesenchymal cells, but are fewer and more punctate than those of epithelial cells. To complement immunofluorescence studies, we employed the Triton X-100 solubility assay to fractionate and quantify junctional GJA1-43k in mesenchymal cells and their unstimulated epithelial counterparts at 48 h after induction. Consistent with our immunofluorescence data, significantly lower levels of junctional Cx43 were detected following TGF-β stimulation relative to total Cx43 (Figure 2, B, quantified in C). Together, these data demonstrate limited gap junction formation in mesenchymal cells despite increased Cx43 protein levels.
FIGURE 2:
Cytosolic enrichment of Cx43 occurs during TGF-β–induced EMT. NMuMG cells were treated with 2 ng/ml TGF-β for 48 h to induce EMT before fixation for immunofluorescence analysis or protein harvesting for solubility assay and Western blot. (A) Fixed cell confocal immunofluorescence (×100) of cells labeled with an antibody directed against the Cx43 C-terminus (green) with borders detected using E-cadherin in vehicle-treated cells (red) and N-cadherin in TGF-β–treated cells (red). Cx43 depicted in white in monochrome images with zoom of cell border shown below. Nuclei counterstained with DAPI (blue). Scale bar: 20 μm. (B) Western blots of total, soluble (nonjunctional), and insoluble (junctional) GJA1-43k following fractionation using the 1% Triton X-100 solubility assay. (C) Quantification of insoluble (junctional) GJA1-43k band intensity relative to soluble (nonjunctional) GJA1-43k from B (n = 3). Graphs represent mean ± SEM. Statistical analysis performed using the Student’s t test. *p ≤ 0.05.
Levels of GJA1-43k protein and Gja1 transcript increase during TGF-β–induced EMT and GJA1-43k protein is stabilized
In Figures 1 and 2 we report that TGF-β stimulation increases GJA1-43k levels in concert with reduced GJA1-20k expression and redistributes GJA1-43k to nonjunctional compartments. Interestingly, total levels of GJA1-43k protein increase significantly over the 72 h time course of EMT induction (Figure 3, A, quantified in B). We also find by RT-qPCR that Gja1 mRNA increased continuously throughout the 72 h time course (Figure 3C). These findings led us to investigate changes in Cx43 protein regulation, where altered GJA1-43k stability could explain the lack of correlation between protein and mRNA levels, and gap junction formation. Click chemistry was utilized to perform nonradioactive pulse-chase experiments to determine GJA1-43k stability in the presence of TGF-β stimulation (Breinbauer and Köhn, 2003). We find that despite reduced gap junction formation, GJA1-43k half-life increased from 1.3 h in unstimulated cells to 2 h following 48 h TGF-β stimulation (Figure 3, D, quantified in E).
FIGURE 3:
Levels of GJA1-43k protein and Gja1 transcript increase during TGF-β–induced EMT and GJA1-43k is stabilized. NMuMG cells were treated with vehicle, 2 ng/ml TGF-β, 5 μM SB431542 (SB), or TGF-β + SB431542 and sampled at 24, 48, and 72 h after stimulation for Western blot and RT-qPCR. (A) Western blot of GJA1-43k in NMuMG cells after treatment (left panels) with α-tubulin serving as loading control (right panels). (B) Quantification of GJA1-43k band intensity relative to α-tubulin from A. (C) qRT-PCR of Gja1 mRNA levels after treatment relative to vehicle at 24, 48, and 72 h after treatment (n = 6). (D) Western blot of total GJA1-43k and TAMRA pulsed GJA1-43k in NMuMG cells following 48 h treatment with vehicle or TGF-β. Time points indicate hours after pulse with TAMRA alkyne. (E) GJA1-43k exponential decay curves of blots described in E; vehicle t1/2 = 1.3 h, TGF-β t1/2 = 2.0 h (n = 3). Graphs represent mean ± SEM. Statistical analysis performed using one-way ANOVA with Tukey’s multiple comparison posttest (B, n = 3; C, n = 5). *p ≤ 0.05; **p ≤ 0.01.
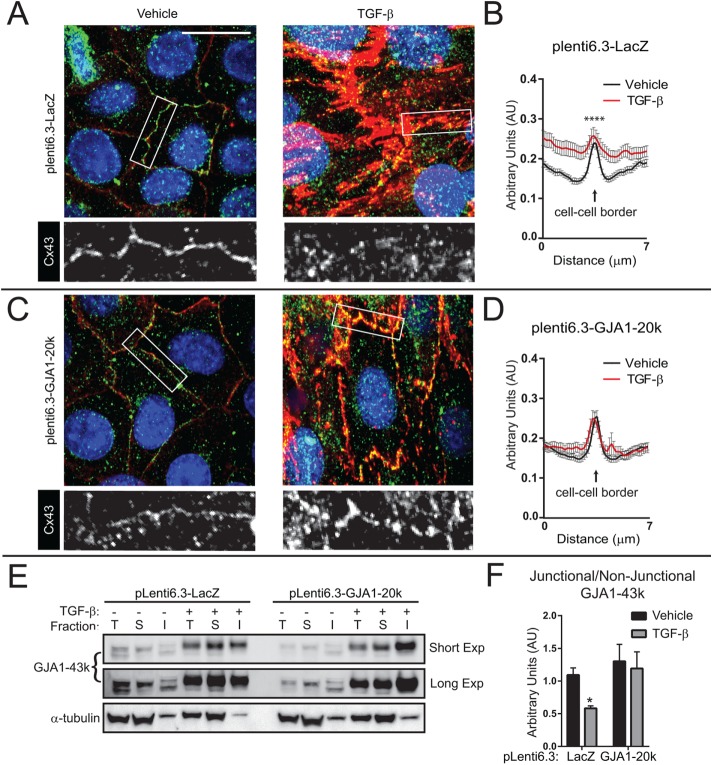
Ectopic stable expression of GJA1-20k rescues gap junction formation and does not alter EMT marker induction in NMuMG cells
Having demonstrated that internal translation of GJA1-20k is suppressed during EMT, we next removed cellular control of GJA1-20k expression in order to test the necessity of GJA1-20k suppression for gap junction down-regulation. Utilizing lentivirally transduced clonal NMuMG cell lines expressing LacZ as control or the GJA1-20k coding sequence, we find that both LacZ and GJA1-20k cell lines underwent EMT following TGF-β stimulation. Both cell lines displayed comparable morphological changes by phase contrast microscopy (Figure 4A) and immunofluorescence confocal imaging reveals comparable fibronectin (green) induction with actin (red) stress-fiber formation in mesenchymal cells (Figure 4B). Induction of EMT markers fibronectin and N-cadherin was confirmed biochemically by Western blot (Figure 4, C, quantified in D). To determine effects on GJA1-43k subcellular localization and gap junction formation, confocal immunofluorescence microscopy was employed (Figure 5, A–D). We detected GJA1-43k using an antibody targeting the cytoplasmic loop of Cx43, in order to exclude a confounding signal from truncated Cx43 isoforms. To measure changes in GJA1-43k gap junction formation and subcellular distribution, quantification was performed by fluorescence intensity line scans across cell borders (Figure 5, B and D; Smyth et al., 2012). Cell borders are identified with a pan-cadherin antibody (red). Upon TGF-β stimulation, GJA1-20k cells are protected against cytosolic redistribution of Cx43 and gap junctions are maintained with practically no change in intensity profiles detected (Figure 5, C and D), despite comparable increases in GJA1-43k to LacZ cells (Figure 1, D and E). To complement immunofluorescence studies, we again employed the Triton X-100 solubility assay to fractionate and quantify junctional GJA1-43k relative to nonjunctional protein 48 h after EMT induction in cell lines expressing LacZ or GJA1-20k. Consistent with our immunofluorescence data, significantly lower levels of junctional Cx43 occur following TGF-β stimulation in cells expressing LacZ, while cells expressing GJA1-20k maintained junctional Cx43 during EMT (Figure 5, E, quantified in F). These data demonstrate that suppression of GJA1-20k translation has no apparent effect on EMT induction, but is necessary for limiting gap junction formation in mesenchymal cells.
FIGURE 4:
NMuMG cells transduced with pLenti6.3-LacZ or pLenti6.3-GJA1-20k undergo EMT. Stable NMuMG cells transduced with pLenti6.3-LacZ or pLenti6.3-GJA1-20k were treated with vehicle, 2 ng/ml TGF-β, 5 μM SB431542 (SB), or TGF-β + SB431542 for 48 h. (A) Phase contrast microscopy at 48 h after stimulation, ×20. Scale bar: 100 μm. (B) Fixed cell confocal immunofluorescence (×100) of cells labeled with antibody directed against EMT markers actin (red) and fibronectin (green). Nuclei counterstained with DAPI (blue). Scale bar: 20 μm. (C) Western blot probed for EMT markers fibronectin and N-cadherin with α-tubulin serving as loading control. (D) Quantification of Western blot band intensity in C. Graphs represent mean ± SEM. Statistical analysis performed using one-way ANOVA with Tukey’s multiple comparison posttest (n = 3). ns: p value not significant.
FIGURE 5:
Ectopic stable expression of GJA1-20k rescues gap junction loss during TGF-β–induced EMT. Stable NMuMG cells were treated with vehicle or TGF-β for 48 h and subsequently fixed for confocal immunofluorescence imaging. (A) Cells stably expressing pLenti6.3-LacZ labeled for GJA1-43k with antibody directed against the Cx43 cytoplasmic loop (green) to prevent labeling for GJA1-20k. Cell borders are identified with pan-cadherin (red) and nuclei counterstained with DAPI (blue). (B) Quantification of average fluorescence intensity profiles of 7-μm lines bisecting, and perpendicular to, cell–cell borders from confocal microscopy maximum intensity projections. Three cell–cell borders per image, n = 11 images. (C) Cells stably expressing pLenti6.3-GJA1-20k labeled for GJA1-43k with antibody directed against the Cx43 cytoplasmic loop (green). Cell borders are identified with pan-cadherin (red) and nuclei counterstained with DAPI. (D) Quantification of average fluorescence intensity profiles as in B. (E) Western blots of total (T), soluble (S; nonjunctional), and insoluble (I; junctional) GJA1-43k following fractionation using the 1% Triton X-100 solubility assay in NMuMG cells transduced with pLenti6.3-LacZ or pLenti6.3-GJA1-20k. Cells were treated with vehicle or TGF-β for 48 h. (F) Quantification of insoluble (junctional) GJA1-43k band intensity relative to soluble (nonjunctional) GJA1-43k from E (n = 5). Graphs represent mean ± SEM. Statistical analysis performed using the Student’s t test. *p ≤ 0.05; ****p ≤ 0.0001.
GJA1-20k localizes to the Golgi apparatus and regulates Cx43 hemichannel oligomerization
Given that GJA1-20k is known to play a role in GJA1-43k trafficking, we investigated localization of GJA1-20k/GJA1-43k within subcellular compartments of the vesicular transport pathway. Confocal immunofluorescence microscopy revealed colocalization of V5-epitope tagged GJA1-20k with the Golgi apparatus marker GM130 (Figure 6A). Direct stochastic optical reconstruction microscopy (STORM) was employed to localize the Golgi apparatus (GM130) at subdiffraction limit resolution with GJA1-43k in cells stably expressing LacZ or GJA1-20k (Figure 6, B–E). Cross-pair correlation generates a probability distribution of finding a given molecule as a function of a given distance from another distinct molecule, with a pair correlation amplitude of 1 indicating no colocalization between two proteins and higher values indicating a correlation. Utilizing an antibody directed against the Cx43 cytoplasmic loop, pair correlation analysis reveals an increase in GJA1-43k colocalization with GM130 following TGF-β stimulation in cells expressing LacZ, suggesting retention of GJA1-43k in the Golgi apparatus (Figure 6C). In cells expressing GJA1-20k, however, GJA1-43k colocalization with GM130 remains comparable to that in vehicle-treated cells (Figure 6E). GJA1-43k oligomerization into hexamers at the TGN is a rate-limiting step in Cx43 trafficking (Musil and Goodenough, 1993). Given our GJA1-43k and GJA1-20k localization data, we hypothesized a role for GJA1-20k in promoting hemichannel formation, thereby enhancing Golgi exit, transport, and Cx43 gap junction formation during TGF-β stimulation. We turned to nondenaturing blue NativePAGE gel electrophoresis and quantified the ratio of hexameric (∼258 kDa) versus monomeric (43 kDa) Cx43 in cell lines stably expressing LacZ or GJA1-20k, with and without TGF-β stimulation (Figure 6, F, quantified in G). Quantification of band intensity by densitometry reveals a modest increase in hexamer formation in LacZ cells following EMT induction. GJA1-20k cells, however, display significantly larger increases in hexamer formation than that observed in LacZ control cells (Figure 6G).
FIGURE 6:
GJA1-20k promotes GJA1-43k oligomerization and release from the Golgi apparatus. NMuMG cells were transfected with C-terminally V5 tagged GJA1-20k and subsequently fixed for confocal immunofluorescence imaging. (A) Fixed cell confocal immunofluorescence (×100) of NMuMG cells 24 h posttransfection. The Golgi apparatus is labeled with an antibody directed against GM130 (green) and GJA1-20k detected with antibody directed against the V5 epitope tag (red). (B, D) Stable NMuMG cells transduced with pLenti6.3-LacZ or pLenti6.3-GJA1-20k were treated with vehicle or TGF-β for 48 h and subsequently fixed for superresolution immunofluorescence imaging. 3D STORM imaging detecting GJA1-43k (green) and GM130 (pink) with localizations depicted as 50-nm point clouds in an x-y cross-section. Scale bars: 2 µm. (C, E) Cross-pair correlation functions for GJA1-43k and GM130 from stable NMuMG cells treated with vehicle (black) or TGF-β (red). Graphs represent mean ± SEM, n = 10 cells. (F) Western blot of blue NativePAGE probed for Cx43 hexamers (∼258 kDa) and monomers (∼43 kDa). (G) Quantification of Western blot band intensity in F (n = 5). Graphs represent mean ± SEM. Statistical analysis performed using the Student’s t test. *p ≤ 0.05; **p ≤ 0.01.
Smad3 and ERK1/2 activity are necessary to suppress internal translation of GJA1 mRNA during TGF-β stimulation
To determine which pathway(s) activated by TGF-β regulate the balance between GJA1-20k and GJA1-43k, we employed chemical inhibitors targeting protein kinases downstream from TGF-β signaling. We then measured the ratio of GJA1-20k/GJA1-43k by Western blot after 24 h of inhibitor treatment with or without the addition of TGF-β. Canonical TGF-β signaling involves activation of Smad transcription factors in order to modulate gene expression. We used the chemical inhibitor SIS3 to inhibit phosphorylation of the receptor Smad, Smad3 (Jinnin et al., 2006), to ask whether Smad signaling was necessary to suppress internal translation of GJA1-20k. Incubation with SIS3 combined with TGF-β treatment resulted in a significantly larger ratio of GJA1-20k/GJA1-43k compared with TGF-β treatment alone. This confirms a role for this arm of TGF-β signaling in altered translation initiation. TGF-β signaling also activates ERK1/2 independent of Smad-induced transcriptional modulation. We inhibited the map kinases ERK1/2 with the chemical inhibitor SCH 772984 and found a greater than ninefold increase in the ratio of GJA1-20k/GJA1-43k compared with vehicle. When ERK1/2 inhibition is combined with TGF-β treatment there is a significantly larger ratio of GJA1-20k/GJA1-43k compared with TGF-β treatment alone. In addition, both p38 MAPK and JNK are activated by TGF-β in a Smad-independent manner. We employed the chemical inhibitors SB 202190 and SP 600125 to inhibit p38 and JNK, respectively. Neither p38 nor JNK inhibition combined with TGF-β has a significant effect on the ratio of GJA1-20k/GJA1-43k when compared with TGF-β treatment alone (Figure 7, A, quantified in B). Together, these findings confirm a role for internal translation of GJA1 mRNA in regulation of full-length Cx43 (GJA1-43k) isoform transport and oligomerization, therefore controlling trafficking and gap junction formation at the posttranscriptional stage (Figure 7C).
FIGURE 7:
TGF-β activates Smad and ERK signaling to alter translation initiation and limit gap junction formation during EMT. NMuMG cells were treated with vehicle (veh; DMSO), 10 µM SIS3, 20 µM SB 202190 (SB2), 20 µM SP 600125 (SP6), or 0.1 µM SCH 772984 (SCH), for 15 min followed by the addition of 2 ng/ml TGF-β for 24 h. (A) Western blot of cell lysates probed with Cx43 C-terminal antibody to detect full-length GJA1-43k and internally translated GJA1-20k. (B) Quantification of GJA1-20k band intensity relative to GJA1-43k from A. (C) Schematic illustrating suppression of GJA1-20k translation during TGF-β–induced EMT to limit GJA1-43k oligomerization and gap junction formation. Graph represents mean ± SEM. Statistical analysis performed using one-way ANOVA with Dunnett’s multiple comparison posttest (B, n = 5–7). ***p ≤ 0.0005; ****p ≤ 0.0001.
DISCUSSION
Research on Cx43 and gap junction regulation has historically focused upon transcription, trafficking, and posttranslational modification of the full-length GJA1-43k protein. The existence of an IRES within the 5′ UTR of GJA1 mRNA, together with the recent finding by several groups that GJA1 mRNA can undergo internal translation, has highlighted translation initiation as a novel and key regulatory step in Cx43 gap junction regulation (Schiavi et al., 1999; Smyth and Shaw, 2013). Internal translation of GJA1 mRNA yields N-terminally truncated isoforms, including GJA1-20k, and their expression influences GJA1-43k localization and gap junction formation. However, implementation of this dynamic translational regulation by the cell in gap junction formation and maintenance has yet to be demonstrated. We utilized EMT as a physiologically relevant model system with which to investigate the role of internal translation and GJA1-20k on GJA1-43k gap junction formation and maintenance. The process of EMT involves reorganization of intercellular junctional structures as cells acquire mesenchymal traits (Lamouille et al., 2014). As the primary means of direct intercellular communication, gap junctions undergo significant remodeling during this process (McLachlan et al., 2006; Bax et al., 2011). In this study, we report that suppression of Gja1 mRNA internal translation in mesenchymal cells is sufficient to limit gap junction formation.
Although the loss of adherens and tight junctions during EMT has been well documented, data regarding the effects of TGF-β on GJA1 transcription and Cx43 protein levels are conflicting (Larson et al., 1997; van der Heyden et al., 2000; de Boer et al., 2007; Tacheau et al., 2008; Hills et al., 2012; Chen et al., 2015b). Electrical conduction between epicardial cells is decreased significantly during and after EMT, coincident with decreased expression of Cx43 as well as other gap junction proteins (Bax et al., 2011). EMT induction in human kidney cells also results in decreased GJIC, and again results in decreased Cx43 expression as cells become mesenchymal (Hills et al., 2012). Overexpression of Cx43 in glioma and breast cancer cells results in an epithelial morphology, supporting a role of Cx43 and gap junctions in maintaining an epithelial phenotype (Huang et al., 1998; McLachlan et al., 2006). Tumor-suppressive properties of Cx43 have been documented in a number of contexts beyond EMT, and Cx43 expression is down-regulated in many human cancers (Tsai et al., 1996; Laird et al., 1999; Sirnes et al., 2012). Ectopic expression of Cx43 in colon cancer epithelial cells does not appear to form gap junctions but limits cancer cell growth by antagonizing apoptotic pathways (Sirnes et al., 2012). In contrast, other studies have reported induction of Cx43 expression during EMT and associated high levels of Cx43 with more invasive and migratory cells (Larson et al., 1997; Bates et al., 2007; Tacheau et al., 2008; Zhang et al., 2015). Finally, overexpression of connexins has been associated with inhibition of EMT in hepatocellular and lung carcinoma cell populations resistant to chemotherapeutic agents (Yu et al., 2014, 2017). Together with our work revealing lack of correlation between Cx43 protein, RNA levels, and gap junction formation, this highlights the complexity of connexin biology during EMT progression and the contribution of translational regulation.
Based on our data, the question remains as to why mesenchymal cells must harbor such high levels of intracellular GJA1-43k relative to gap junctional GJA1-43k. Interestingly, we did not observe complete gap junction dissolution in mesenchymal cells, although gap junctions were less dense and more punctate than in epithelial cells. Nonchannel/junctional functions for Cx43 have been reported where intracellular localization is sufficient to negatively regulate cell cycle progression and participate in cytoskeleton nucleation (Olbina and Eckhart, 2003; Chen et al., 2015a). Indeed, cell proliferation is suppressed during TGF-β–induced EMT, and intracellular GJA1-43k may play a role here (Roberts et al., 1985). The existence of internally translated Cx43 isoforms which encompass the C-terminus further complicates interrogation of this biology, as many nonjunctional functions of Cx43 have been attributed its C-terminus. The Cx43 C-terminus alone is sufficient to restore actin-mediated B-cell spreading and adhesion, and the cytoskeleton plays an essential role in migration of mesenchymal cells (Yilmaz and Christofori, 2009; Machtaler et al., 2011). In addition to intercellular communication, gap junction maintenance may assist in cell migration where the adhesive properties of gap junction channels have previously been shown as necessary for cell motility (Elias et al., 2007; Machtaler et al., 2014). Importantly, such high levels of intracellular GJA1-43k demonstrate that transport is not constitutive and that a posttranscriptional mechanism is therefore in place to limit gap junction formation irrespective of GJA1-43k protein levels.
Activation of the Smad-independent mTOR pathway is known to suppress internal translation of the GJA1 transcript (Smyth and Shaw, 2013). In this study, we investigated pathways upstream and independent of mTOR. The TGF-β receptor rapidly activates ERK1/2 signaling through recruitment and phosphorylation of ShcA (Lee et al., 2007). TGF-β also activates TAK1, independent of its receptor kinase activity, leading to activation of the MAP kinases p38 and JNK (Yamaguchi et al., 1995; Watkins et al., 2006; Sorrentino et al., 2008). Following chemical inhibition of ERK1/2 signaling, but not that of p38 or JNK, we report a significant increase in the ratio of GJA1-20k to GJA1-43k. ERK1/2 signaling is necessary for TGF-β–induced EMT, further suggesting a role for internal translation of GJA1 in EMT (Xie et al., 2004). Of relevance, ERK pathway activation promotes canonical translation through ribosomal protein S6 phosphorylation (Roux et al., 2007). Our findings are also consistent with previous reports that Mnk1/2 suppresses GJA1-20k translation as ERK activation is upstream of Mnk (Salat-Canela et al., 2014). We find inhibition of Smad3 activity with the inhibitor SIS3 also rescues GJA1-20k expression, implying transcriptional effects may also regulate GJA1 translation, such as alternate UTR usage. However, our data in Figure 1D, in which we overexpress the coding sequence of GJA1 alone, support a dominant shift in cellular processes regulating translation initiation independent of changes to the mRNA. Moreover, internal translation of an independent mRNA transcript, paxillin (Pxn), has also been reported in EMT, suggesting a global shift in translation initiation during TGF-β–induced EMT (Tumbarello et al., 2005). Given the role of paxillin in EMT through focal adhesion formation, it is tempting to speculate that an entire “nonjunctional” mesenchymal and motile phenotype may be affected through suppression of internal translation relevant mRNA families (Turner, 2000).
Direct posttranslational modification through phosphorylation of the GJA1-43k C-terminus is known to promote gap junction internalization and degradation of GJA1-43k (Hesketh et al., 2010; Smyth et al., 2014; Solan and Lampe, 2014). A phosphorylation cascade encompassing Akt, PKC, and MAPK converges at the GJA1-43k C-terminus to effect internalization from the plasma membrane (Smyth et al., 2014). Indeed, alterations in GJA1-43k phosphoisoforms are observed in our biochemical solubility studies where we find reduced junctional GJA1-43k in mesenchymal cells. Together with our localization data and the fact that we report GJA1-43k is stabilized during TGF-β–induced EMT, however, it seems probable that GJA1-43k predominantly remains within intracellular compartments in these cells, the majority most likely not having reached the cell surface where such phosphorylation events are understood to occur. Moreover, the internalization of gap junctions occurs rapidly after activation of growth factor signaling cascades and so may play a role in dissolution of gap junctions earlier in the EMT process than the 48–72 h time points examined in this study (Dunn and Lampe, 2014; Fong et al., 2014; Smyth et al., 2014).
Oligomerization of GJA1-43k into hemichannels occurs at the TGN, later in the vesicular transport pathway than most ion channels (Musil and Goodenough, 1993). Only two proteins are known to interact with GJA1-43k in the ER/Golgi and regulate its transport. ERP29 interacts with GJA1-43k within the ER preventing premature oligomerization, and the GTPase Rab20 can also regulate Cx43 transport from the ER (Das Sarma et al., 2008; Das et al., 2009). Previous findings demonstrated that GJA1-20k resides in the ER/Golgi, interacts with GJA1-43k, and regulates gap junction formation (Smyth and Shaw, 2013). Recently, GJA1-20k has been localized to the mitochondria, where immunogold electron microscopy revealed GJA1-20k at the microtubule/mitochondria interface in addition to ER/Golgi. Here, we have utilized confocal microscopy to confirm localization GJA1-20k to the Golgi apparatus. Therefore, GJA1-20k may complement ERP29/Rab20 activity by chaperoning GJA1-43k through the Golgi apparatus transport pathway, spatially and temporally regulating Cx43 hemichannel oligomerization and vesicular loading at the TGN. The stoichiometry of GJA1-43k/GJA1-20k is difficult to measure in the biochemical assays herein, but it does seem that relatively low levels of GJA1-20k are sufficient to promote GJA1-43k transport. Given that GJA1-20k has not been detected on the cell surface and resides primarily within the Golgi apparatus, it is likely that one GJA1-20k molecule may interact transiently with several GJA1-43k monomers/hemichannel intermediates as they pass through the Golgi apparatus.
As a membrane channel, Cx43 hemichannels can exchange molecules such as ATP and glutamate with the extracellular milieu in addition to its more established function as a gap junction (Stout et al., 2002; Orellana et al., 2011). Although our biochemical Triton X-100 solubility findings demonstrate a relative reduction in Cx43 gap junction formation during EMT, they do not exclude the possibility of increased surface hemichannel expression. Accrual of surface Cx43 hemichannels into gap junction structures is limited by binding of the C-terminus to ZO-1, a scaffolding protein down-regulated during TGF-β–induced EMT (Rhett et al., 2011). Therefore, in mesenchymal cells we would expect the absence of negative regulation of gap junction accretion by ZO-1, but did not observe increases in gap junction size with TGF-β stimulation. Furthermore, data presented in Figures 3 and 6 show that TGF-β stimulation alters Cx43 protein stability and oligomerization, which are key limiting steps in the trafficking of hemichannels to the cell surface. These findings, together with previous studies indicating GJA1-20k expression is restricted to intercellular compartments, lead us to conclude that the observed increases in soluble GJA1-43k is primarily intracellular. The role of Cx43 internally translated isoforms in regulation of surface hemichannel activity and/or accrual into gap junctions, however, is an interesting topic of future research.
Several internally translated isoforms of Cx43 have been detected in addition to GJA1-20k (Smyth and Shaw, 2013). In this study, we focus on GJA1-20k as the most predominantly expressed isoform but given that all three preceding methionine codons (M100, 125, and 147) must also be mutated to limit GJA1-43k trafficking, some redundancy in function is likely to exist between these molecules (Smyth and Shaw, 2013). The Cx43 C-terminal truncation mutant does not display perturbed trafficking (Fishman et al., 1991; Morley et al., 1996; Maass et al., 2004), but would encode all four isoforms mentioned above also lacking their C-termini. Intriguingly, this may point toward the fourth transmembrane domain of Cx43, which is intact in this mutant and contained in GJA1-20k as most important in regulation of GJA1-43k transport. Together, our data indicate that levels of GJA1-20k provide a means for the cell to fine-tune Cx43 hemichannel oligomerization and thus gap junctions rapidly at the posttranscriptional level. Indeed, pathological Cx43 remodeling in the heart during ischemia can be prevented through actin stabilization by ectopic GJA1-20k expression, suggesting internal translation is a common pathway for gap junction regulation across diverse cell types and tissues (Basheer et al., 2017).
At least 600 human genes are now understood to undergo alternate mechanisms of translation initiation and this study supports the importance of this biological process in health and disease (Ingolia et al., 2011; Weingarten-Gabbay et al., 2016;, Karginov et al., 2017). Regulation of translation initiation can be thought of globally, whereby signal transduction cascades can shunt the entire translational landscape of the cell, or gene-specifically, where alternate promoter usage/splicing can lead to changes in specific mRNAs and render them subject to such translation initiation. Our data reveal that a shift in the translational landscape of the cell occurs during EMT affecting the proteome and contributing to alterations in gap junction formation. This suggests a potent mechanism whereby the cell can rapidly regulate Cx43 transport and gap junction formation at the point of GJA1-20k translation, without inducing transcription of mRNA or synthesis of full-length GJA1-43k.
MATERIALS AND METHODS
Molecular biology
Human GJA1 cDNA was obtained from GE Dharmacon and cloned into pDONR221 using Gateway BP cloning (forward primer GGGGACAAGTTTGTACAAAAAAGCAGGCTTAGTGGTGCCCAGGCAAC; reverse primer GGGGACCACTTTGTACAAGAAAGCTGGGTTCTAGATCTCCAGGTCATCAGGCC) to create an entry clone (BP clonase II; Thermo Scientific). GJA1 was then inserted into pcDNA3.2-V5/DEST from pDONR221/hGJA1 entry clone using Gateway LR cloning to generate pcDNA3.2/hGJA1. pcDNA3.2/GJA1-20k-V5 was generated using forward primer GGGGACAAG TTTGTACAAAAAAGCAGGCTTAATGCTGGTGGTGTCCTTGGT and reverse primer GGGGACCACTTTGTACAAGAAAGCTGGGTTGATCTCCAGGTCATCAGGCCG to insert the GJA1-20k coding sequence into pDONR221 using Gateway BP cloning (BP clonase II; Thermo Scientific). GJA1-20k was then inserted into pcDNA3.2/V5-DEST from pDONR221/hGJA1-20k entry clone using Gateway LR cloning (LR clonase; Thermo Scientific). Lentiviral expression vectors were created using TOPO cloning to insert the GJA1 coding sequence (forward primer GTGGTGCCCAGGCAACATG; reverse primer CTAGATCTCCAGGTCATCAGGCC) or GJA1-20k (forward primer ATGCTGGTGGTGTCCTTGGTG; reverse primer CTAGATCTCCAGGTCATCAGGCC) into pLenti6.3/V5-TOPO (Thermo Scientific). pLenti6.3/V5-GW/lacZ and pcDNA3.2/GW-CAT served as control expression vectors (Thermo Scientific). All recombinant DNA constructs were verified by Sanger sequencing.
Cell culture
NMuMG cells were a kind gift from Rik Derynck (University of California, San Francisco) and maintained in DMEM+Glutamax supplemented with 10% fetal bovine serum (FBS), sodium pyruvate, nonessential amino acids (Thermo Scientific), bovine insulin solution (5 μg/ml; Sigma) and MycoZAP (Lonza). MycoZAP is included to prevent Mycoplasma contamination. Cells were maintained in a humidified atmosphere of 5% CO2 at 37°C. For experiments, cells were plated at 1.6 × 104 cells/cm2 in 100-mm plates or six-well dishes for protein harvesting and eight-well chamber slides for imaging. Cells were incubated for 24 h before addition of media containing one of the following treatments: TGF-β1 (2 ng/ml; Humanzyme), the TβR1 blocker SB 431542 (SB; 5 µM; Sigma), Smad3 inhibitor SIS3 (10 µM; Selleckchem), p38 inhibitor SB 202190 (20 µM; Cayman), JNK inhibitor SP 600125 (20 µM; Cayman), ERK1/2 inhibitor SCH 772984 (0.1 µM; Cayman), or vehicle (veh; dimethyl sulfoxide [DMSO]). 293FT cells were obtained from Thermo Scientific and maintained in DMEM+GlutaMAX supplemented with 10% FBS, sodium pyruvate, nonessential amino acids (Thermo Scientific), and MycoZAP (Lonza).
Lentivirus production, cell transduction, and stable cell line generation
Lentivirus was created from pLenti6.3-hGJA1, pLenti6.3/GJA1-20k, and pLenti6.3-LacZ according to the manufacturer’s instructions (ViraPower Lentiviral Expression System; Thermo Scientific). Titered and normalized viruses were used to infect NMuMG cells plated in six-well dishes in the presence of hexadimethrine bromide (4 µg/ml; Sigma). After 48 h, cells were split 1:40 into 100-mm dishes and 10 µg/ml blasticidin added to medium for selection. Medium was changed every 2 d and healthy colonies picked using cloning rings (Scienceware). Clones were expanded and screened for stable expression by Western blotting and immunofluorescence.
Western blotting
Cells were lysed in RIPA buffer (0.1% SDS, 50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 2 mM NaF, 200 μM Na3VO4) supplemented with HALT Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific) and Western blotting conducted as previously described (Smyth et al., 2010, 2012). Briefly, cells were scraped into RIPA buffer and sonicated before centrifugation at 10,000 × g for 20 min at 4°C. Protein concentration was quantified using the Bio-Rad DC Protein Assay and lysates normalized to the same concentration. 4X Bolt LDS sample buffer (Thermo Scientific) supplemented with dithiothreitol (DTT; 400 mM) was added and samples heated for 10 min at 70°C before SDS–PAGE and Western blotting with the following primary antibodies: mouse anti–N-cadherin and mouse anti–E-cadherin (1:1000; BD Biosciences), rabbit anti-Cx43 (1:3000; Sigma), rabbit anti-fibronectin (1:3000; Sigma), and mouse anti–α-tubulin (1:3000; Sigma). Goat secondary antibodies conjugated to Alexa Fluor 555 and 647 (Thermo Scientific) were used at 1:1000 and membranes imaged on a ChemiDoc MP (Bio-Rad).
Immunofluorescence
Cells were fixed in 37°C 4% paraformaldehyde for 20 min and stored in PBS at 4°C until immunostaining was conducted as previously described (Smyth et al., 2012) with the following primary antibodies: rabbit anti-fibronectin (Sigma; 1:500), mouse anti–N-cadherin (BD Biosciences; 1:200), mouse anti–E-cadherin (BD Biosciences; 1:1000), rabbit anti-Cx43 (Sigma; 1:3000), and mouse anti–α-tubulin (Sigma; 1:3000). Secondary antibodies used were goat anti-rabbit Alexa Fluor 488 and anti-mouse Alexa Fluor 647 (Thermo Scientific; 1:500) with Alexa Fluor 647 phalloidin employed to detect actin (Thermo Scientific; 1:20). For immunofluorescence using antibody directed against the Cx43 cytoplasmic loop, cells grown on 35-mm glass-bottomed dishes (Mat-Tek) or eight-well chambered slides were fixed in methanol for 5 min on ice. Antibodies used were rabbit anti-Cx43 cytoplasmic loop (Abcam; 1:500), mouse anti-pan cadherin (BD Biosciences; 1:500), and anti-rabbit Alexa Fluor 488 and anti-mouse Alexa Fluor 647 (ThermoScientific; 1:500). To localize GJA1-20k, NMuMG cells were transfected in 35-mm glass-bottomed dishes with 2 µg pcDNA3.2/hGJA1-20k-V5, or pcDNA3.2/GW-CAT using Lipofectamine 3000 (Thermo Scientific). Posttransfection (24 h) cells were fixed in methanol for 5 min on ice. Immunostaining was performed as described above using the following primary antibodies: rabbit anti-V5 (Sigma; 1:1000) and mouse anti-GM130 (BD Biosciences; 1:250).
Superresolution microscopy
Cells plated in 35-mm Mat-Tek dishes were fixed in methanol for 5 min on ice. Cells were washed three times in PBS before blocking for 1 h at room temperature in 5% normal donkey serum (Jackson ImmunoResearch), 0.5% Triton X-100, in PBS (blocking buffer). Cells were incubated with the following primary antibodies diluted in blocking buffer: mouse anti-GM130 (BD Biosciences; 1:1000) and rabbit anti-Cx43 cytoplasmic loop (Abcam,; 1:500). Following PBS washes, cells were incubated with donkey secondary antibodies conjugated to Alexa Fluor 647 (Thermo Scientific) or CF568 (Biotium) diluted in blocking buffer. After PBS washes, two-color direct STORM imaging was conducted using a Vutara 350 (Bruker). Cells were imaged in 50 mM Tris-HCl, 10 mM NaCl, 10% (wt/vol) glucose buffer containing 20 mM mercaptoethylamine, 1% (vol/vol) 2-mercaptoethanol, 168 active units/ml glucose oxidase, and 1404 active units/ml catalase. Videos of 5000 frames were acquired for each probe and three-dimensional (3D) images reconstructed in Vutara SRX software. Intracellular regions of interest were selected and coordinates of localized molecules used to calculate pair correlation functions in the Vutara SRX software (Sengupta et al., 2011, 2013).
Image analysis
For quantification of Cx43 expression at cell–cell borders, pan-cadherin images were used to identify cell–cell borders for vehicle and TGF-β–treated cells, respectively. Lines (10 µm) were drawn perpendicular to cell–cell borders every 10 pixels for the length of each border. The plot profile function in ImageJ was used to quantify Cx43 fluorescence intensity from each line and data averaged. Images were acquired and analyzed by separate individuals, with image identity blinded to the analyst.
Separation of soluble (nonjunctional) and insoluble (junctional) Cx43
Cells were harvested in 1% Triton X-100 buffer (50 mM Tris, pH 7.4, 1% Triton X-100, 2 mM EDTA, 2 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid [EGTA], 250 mM NaCl, 1 mM NaF, 0.1 mM Na3VO4) supplemented with HALT Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific). Samples were lysed in 1% Triton X-100 buffer for 1 h nutating at 4°C. After lysing, 25 µl of lysate was removed and snap-frozen to serve as total protein fraction. The remaining lysate was centrifuged at 15,000 × g for 30 min, and supernatant removed to serve as the soluble fraction. Pellets containing insoluble protein were resuspended in 4X Bolt LDS sample buffer (Thermo Scientific). All samples were sonicated and centrifuged for 20 min at 10,000 × g following the addition of 4X Bolt LDS sample buffer (Thermo Scientific) supplemented with DTT (400 mM) and SDS–PAGE Western blotting performed as described above.
Real-time quantitative PCR
RNA was extracted from NMuMG cells using the Purelink RNA Mini Kit (Thermo Scientific) and homogenized by passage 10 times through an 18-gauge needle. DNA was digested on column using PureLink DNase (Thermo Scientific). RNA (1 µg) was reverse transcribed with iScript cDNA Synthesis Kit (Bio-Rad). RT-qPCR was performed using SYBR Select Master Mix for CFX (Thermo Scientific) on a CFX Connect Real-Time System (Bio-Rad). Validated PrimeTime qPCR primers were purchased from Integrated DNA Technologies. Cycling parameters consisted of 50°C, 2 min; 95°C, 2 min; 39 cycles of 95°C, 15 s; 55°C, 15 s; 72°C, 1 min followed by a melt curve.
Click chemistry pulse-chase assay
NMuMG cells were plated in six-well dishes and fully supplemented DMEM and treated with TGF-β or vehicle as described above. After 48 h TGF-β stimulation, cells were rinsed and starved for 1 h in methionine/cysteine-free DMEM (Thermo Scientific) supplemented with 10% dialyzed FBS (GE Healthcare). Cells were then pulsed for 1 h with Click-iT L-Azidohomoalanine (ThermoScientific; 50 µM final concentration) added directly to starvation media. Samples were harvested over an 8 h period at 2 h intervals into 200 μl lysis buffer (1% SDS, 50 mM Tris-HCl) supplemented with HALT Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific). Lysates were incubated on ice for 15 min and sonicated before centrifugation at 10,000 × g for 20 min at 4°C. Protein concentration was quantified using the Bio-Rad DC Protein Assay and all samples normalized to 400 µg/100 µl. The click reaction was performed on 400 µg protein using the Click-iT Protein Reaction Buffer Kit and tetramethylrhodamine (TAMRA) alkyne (ThermoScientific; 40 µM final concentration) as per manufacturer’s instructions. Immunoprecipitation was performed as previously described (Smyth et al., 2014). Briefly, Cx43 was immunoprecipitated from precleared lysates using 1 µg rabbit anti-Cx43 (Sigma) and protein G Dynabeads (Thermo Scientific). Western blotting was performed as described above and pulsed Cx43 detected using antibody directed against TAMRA (ThermoScientific; 1:1000). Blots were stripped using Re-Blot Plus Strong Solution (Millipore) and reprobed for total Cx43 with rabbit anti-Cx43 (Sigma; 1:3000).
Blue NativePAGE
NMuMG cells were washed twice with PBS and harvested by scraping into 1X NativePAGE sample buffer (Thermo Scientific) containing 1% Digitonin and HALT Protease Inhibitor Cocktail (Thermo Scientific). Lysates were centrifuged at 100,000 × g for 15 min at 4°C using a Beckman Coulter Optima TLX ultracentrifuge. G-250 sample additive was added to supernatants to 0.25%. Samples were loaded on a 4–16% Bis-Tris Novex NativePAGE Gel (Thermo Scientific) and electrophoresed at 4°C according to the manufacturer’s instructions. Proteins were transferred to PVDF membranes in Bolt transfer buffer (Thermo Scientific) for 1 h at 25 V. GJA1-43k was detected with rabbit anti-Cx43 (Sigma; 1:3000) followed by goat anti-rabbit horseradish peroxidase (HRP) (Abcam; 1:5000). Membranes were imaged following the addition of Clarity Western ECL Substrate on a ChemiDoc MP (Bio-Rad).
Supplementary Material
Acknowledgments
We thank Darlon Jan, Gillian E. Jones, and Daniel Purcell for technical support. This work was supported by National Institutes of Health National Heart, Lung, and Blood Institute (NIH NHLBI) R01 grant HL-132236 (to J.W.S.) and NIH NHLBI F31 grant HL-140909 (to C.C.J.).
Abbreviations used:
- Cx43
connexin 43
- EMT
epithelial–mesenchymal transition
- IRES
internal ribosome entry site
- mTOR
mammalian target of rapamycin
- ORF
open reading frame
- TGF-β
transforming growth factor beta
- TGN
trans-Golgi network
- UTR
untranslated region.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-06-0406) on February 21, 2018.
REFERENCES
- Basheer WA, Xiao S, Epifantseva I, Fu Y, Kleber AG, Hong T, Shaw RM. (2017). GJA1-20k arranges actin to guide Cx43 delivery to cardiac intercalated discs. Circ Res , 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates DC, Sin WC, Aftab Q, Naus CC. (2007). Connexin43 enhances glioma invasion by a mechanism involving the carboxy terminus. Glia , 1554–1564. [DOI] [PubMed] [Google Scholar]
- Bax NA, Pijnappels DA, Van Oorschot AA, Winter EM, De Vries AA, Van Tuyn J, Braun J, Maas S, Schalij MJ, Atsma DE, et al. (2011). Epithelial-to-mesenchymal transformation alters electrical conductivity of human epicardial cells. J Cell Mol Med , 2675–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer EC, Paul DL, Goodenough DA. (1987). Connexin43: a protein from rat heart homologous to a gap junction protein from liver. J Cell Biol , 2621–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breinbauer R, Köhn M. (2003). Azide–alkyne coupling: a powerful reaction for bioconjugate chemistry. Chem Bio Chem , 1147–1149. [DOI] [PubMed] [Google Scholar]
- Chen YC, Chang HM, Cheng JC, Tsai HD, Wu CH, Leung PC. (2015b). Transforming growth factor-β1 up-regulates connexin43 expression in human granulosa cells. Hum Reprod , 2190–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Mayo JN, Gourdie RG, Johnstone SR, Isakson BE, Bearden SE. (2015a). The connexin 43/ZO-1 complex regulates cerebral endothelial F-actin architecture and migration. Am J Physiol Cell Physiol , C600–C607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X-F, Zhang H-J, Wang H-B, Zhu J, Zhou W-Y, Zhang H, Zhao M-C, Su J-M, Gao W, Zhang L, et al. (2012). Transforming growth factor-β1 induces epithelial-to-mesenchymal transition in human lung cancer cells via PI3K/Akt and MEK/Erk1/2 signaling pathways. Mol Biol Rep , 3549–3556. [DOI] [PubMed] [Google Scholar]
- Das S, Smith TD, Sarma JD, Ritzenthaler JD, Maza J, Kaplan BE, Cunningham LA, Suaud L, Hubbard MJ, Rubenstein RC, Koval M. (2009). ERp29 restricts Connexin43 oligomerization in the endoplasmic reticulum. Mol Biol Cell , 2593–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Sarma J, Kaplan BE, Willemsen D, Koval M. (2008). Identification of rab20 as a potential regulator of connexin 43 trafficking. Cell Commun Adhes , 65–74. [DOI] [PubMed] [Google Scholar]
- De Boer TP, Van Veen TA, Bierhuizen MF, Kok B, Rook MB, Boonen KJ, Vos MA, Doevendans PA, De Bakker JM, Van Der Heyden MA. (2007). Connexin43 repression following epithelium-to-mesenchyme transition in embryonal carcinoma cells requires Snail1 transcription factor. Differentiation , 208–218. [DOI] [PubMed] [Google Scholar]
- Dunn CA, Lampe PD. (2014). Injury-triggered Akt phosphorylation of Cx43: a ZO-1-driven molecular switch that regulates gap junction size. J Cell Sci , 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias LA, Wang DD, Kriegstein AR. (2007). Gap junction adhesion is necessary for radial migration in the neocortex. Nature , 901–907. [DOI] [PubMed] [Google Scholar]
- Fishman GI, Moreno AP, Spray DC, Leinwand LA. (1991). Functional analysis of human cardiac gap junction channel mutants. Proc Natl Acad Sci USA , 3525–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong JT, Nimlamool W, Falk MM. (2014). EGF induces efficient Cx43 gap junction endocytosis in mouse embryonic stem cell colonies via phosphorylation of Ser262, Ser279/282, and Ser368. FEBS Lett , 836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Zhang S-S, Xiao S, Basheer WA, Baum R, Epifantseva I, Hong T, Shaw RM. (2017). Cx43 isoform GJA1-20k promotes microtubule dependent mitochondrial transport. Front Physiol , 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal A, Sjoblom T, Fedorova L, Imreh S, Beug H, Moustakas A. (2008). Sustained TGF β exposure suppresses Smad and non-Smad signalling in mammary epithelial cells, leading to EMT and inhibition of growth arrest and apoptosis. Oncogene , 1218–1230. [DOI] [PubMed] [Google Scholar]
- Goodenough DA, Paul DL. (2009). Gap junctions. Cold Spring Harb Perspect Biol , a002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh GG, Shah MH, Halperin VL, Cooke CA, Akar FG, Yen TE, Kass DA, Machamer CE, Van Eyk JE, Tomaselli GF. (2010). Ultrastructure and regulation of lateralized connexin43 in the failing heart. Circ Res , 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills CE, Siamantouras E, Smith SW, Cockwell P, Liu KK, Squires PE. (2012). TGFβ modulates cell-to-cell communication in early epithelial-to-mesenchymal transition. Diabetologia , 812–824. [DOI] [PubMed] [Google Scholar]
- Hino H, Dai P, Yoshida T, Hatakeyama T, Harada Y, Otsuji E, Okuda T, Takamatsu T. (2015). Interaction of Cx43 with Hsc70 regulates G1/S transition through CDK inhibitor p27. Sci Rep , 15365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RP, Fan Y, Hossain MZ, Peng A, Zeng ZL, Boynton AL. (1998). Reversion of the neoplastic phenotype of human glioblastoma cells by connexin 43 (cx43). Cancer Res , 5089–5096. [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. (2011). Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell , 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinnin M, Ihn H, Tamaki K. (2006). Characterization of SIS3, a novel specific inhibitor of Smad3, and its effect on transforming growth factor-β1-induced extracellular matrix expression. Mol Pharmacol , 597–607. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. (2009). The basics of epithelial-mesenchymal transition. J Clin Invest , 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov TA, Pastor DPH, Semler BL, Gomez CM. (2017). Mammalian polycistronic mRNAs and disease. Trends Genet , 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar NM, Gilula NB. (1996). The gap junction communication channel. Cell , 381–388. [DOI] [PubMed] [Google Scholar]
- Laird DW, Fistouris P, Batist G, Alpert L, Huynh HT, Carystinos GD, Alaoui-Jamali MA. (1999). Deficiency of connexin43 gap junctions is an independent marker for breast tumors. Cancer Res , 4104–4110. [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. (2014). Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol , 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois S, Cowan KN, Shao Q, Cowan BJ, Laird DW. (2010). The tumor-suppressive function of Connexin43 in keratinocytes is mediated in part via interaction with caveolin-1. Cancer Res , 4222–4232. [DOI] [PubMed] [Google Scholar]
- Larson DM, Wrobleski MJ, Sagar GD, Westphale EM, Beyer EC. (1997). Differential regulation of connexin43 and connexin37 in endothelial cells by cell density, growth, and TGF-β1. Am J Physiol , C405–C415. [DOI] [PubMed] [Google Scholar]
- Lee MK, Pardoux C, Hall MC, Lee PS, Warburton D, Qing J, Smith SM, Derynck R. (2007). TGF-β activates Erk MAP kinase signalling through direct phosphorylation of ShcA. Embo J , 3957–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay J, Mcdade SS, Pickard A, Mccloskey KD, Mccance DJ. (2011). Role of ΔNp63γ in epithelial to mesenchymal transition. J Biol Chem , 3915–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass K, Ghanem A, Kim J-S, Saathoff M, Urschel S, Kirfel G, Grümmer R, Kretz M, Lewalter T, Tiemann K, et al. (2004). Defective epidermal barrier in neonatal mice lacking the C-terminal region of Connexin43. Mol Biol Cell , 4597–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machtaler S, Choi K, Dang-Lawson M, Falk L, Pournia F, Naus CC, Matsuuchi L. (2014). The role of the gap junction protein connexin43 in B lymphocyte motility and migration. FEBS Lett , 1249–1258. [DOI] [PubMed] [Google Scholar]
- Machtaler S, Dang-Lawson M, Choi K, Jang C, Naus CC, Matsuuchi L. (2011). The gap junction protein Cx43 regulates B-lymphocyte spreading and adhesion. J Cell Sci , 2611–2621. [DOI] [PubMed] [Google Scholar]
- Marques-Ramos A, Candeias MM, Menezes J, Lacerda R, Willcocks M, Teixeira A, Locker N, Romão L. (2017). Cap-independent translation ensures mTOR expression and function upon protein synthesis inhibition. RNA , 1712–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May J, Johnson P, Saleem H, Simon AE. (2017). A sequence-independent, unstructured internal ribosome entry site is responsible for internal expression of the coat protein of Turnip crinkle virus. J Virol , e02421-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclachlan E, Shao Q, Wang HL, Langlois S, Laird DW. (2006). Connexins act as tumor suppressors in three-dimensional mammary cell organoids by regulating differentiation and angiogenesis. Cancer Res , 9886–9894. [DOI] [PubMed] [Google Scholar]
- Morley GE, Taffet SM, Delmar M. (1996). Intramolecular interactions mediate pH regulation of connexin43 channels. Biophys J , 1294–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musil LS, Goodenough DA. (1993). Multisubunit assembly of an integral plasma membrane channel protein, gap junction connexin43, occurs after exit from the ER. Cell , 1065–1077. [DOI] [PubMed] [Google Scholar]
- Olbina G, Eckhart W. (2003). Mutations in the second extracellular region of connexin 43 prevent localization to the plasma membrane, but do not affect its ability to suppress cell growth. Mol Cancer Res , 690–700. [PubMed] [Google Scholar]
- Orellana JA, Froger N, Ezan P, Jiang JX, Bennett MV, Naus CC, Giaume C, Saez JC. (2011). ATP and glutamate released via astroglial connexin 43 hemichannels mediate neuronal death through activation of pannexin 1 hemichannels. J Neurochem , 826–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer I, Anderson C, Werner R, Oltra E. (2004). Redefining the structure of the mouse connexin43 gene: selective promoter usage and alternative splicing mechanisms yield transcripts with different translational efficiencies. Nucleic Acids Res , 4550–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piek E, Moustakas A, Kurisaki A, Heldin CH, Ten Dijke P. (1999). TGF-β type I receptor/ALK-5 and Smad proteins mediate epithelial to mesenchymal transdifferentiation in NMuMG breast epithelial cells. J Cell Sci (Pt 24), 4557–4568. [DOI] [PubMed] [Google Scholar]
- Popa A, Lebrigand K, Barbry P, Waldmann R. (2016). Pateamine A-sensitive ribosome profiling reveals the scope of translation in mouse embryonic stem cells. BMC Genomics , 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Cheng JC, Zhao J, Chang HM, Leung PC. (2015). Transforming growth factor-β stimulates human ovarian cancer cell migration by up-regulating connexin43 expression via Smad2/3 signaling. Cell Signal , 1956–1962. [DOI] [PubMed] [Google Scholar]
- Rhett JM, Jourdan J, Gourdie RG. (2011). Connexin 43 connexon to gap junction transition is regulated by zonula occludens-1. Mol Biol Cell , 1516–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AB, Anzano MA, Wakefield LM, Roche NS, Stern DF, Sporn MB. (1985). Type β transforming growth factor: a bifunctional regulator of cellular growth. Proc Natl Acad Sci USA , 119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux PP, Shahbazian D, VU H, Holz MK, Cohen MS, Taunton J, Sonenberg N, Blenis J. (2007). RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem , 14056–14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salat-Canela C, Sese M, Peula C, Ramon Y, Cajal S, Aasen T. (2014). Internal translation of the connexin 43 transcript. Cell Commun Signal , 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavi A, Hudder A, Werner R. (1999). Connexin43 mRNA contains a functional internal ribosome entry site. FEBS Lett , 118–122. [DOI] [PubMed] [Google Scholar]
- Sengupta P, Jovanovic-Talisman T, Lippincott-Schwartz J. (2013). Quantifying spatial organization in point localization superresolution images using pair correlation analysis. Nat Protoc , 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P, Jovanovic-Talisman T, Skoko D, Renz M, Veatch SL, Lippincott-Schwartz J. (2011). Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat Methods , 969–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirnes S, Bruun J, Kolberg M, Kjenseth A, Lind GE, Svindland A, Brech A, Nesbakken A, Lothe RA, Leithe E, Rivedal E. (2012). Connexin43 acts as a colorectal cancer tumor suppressor and predicts disease outcome. Int J Cancer , 570–581. [DOI] [PubMed] [Google Scholar]
- Smyth JW, Hong TT, Gao D, Vogan JM, Jensen BC, Fong TS, Simpson PC, Stainier DY, Chi NC, Shaw RM. (2010). Limited forward trafficking of connexin 43 reduces cell-cell coupling in stressed human and mouse myocardium. J Clin Invest , 266–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth JW, Shaw RM. (2013). Autoregulation of connexin43 gap junction formation by internally translated isoforms. Cell Rep , 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth JW, Vogan JM, Buch PJ, Zhang SS, Fong TS, Hong TT, Shaw RM. (2012). Actin cytoskeleton rest stops regulate anterograde traffic of connexin 43 vesicles to the plasma membrane. Circ Res , 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth JW, Zhang SS, Sanchez JM, Lamouille S, Vogan JM, Hesketh GG, Hong T, Tomaselli GF, Shaw RM. (2014). A 14-3-3 mode-1 binding motif initiates gap junction internalization during acute cardiac ischemia. Traffic , 684–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solan JL, Lampe PD. (2014). Specific Cx43 phosphorylation events regulate gap junction turnover in vivo. FEBS Lett , 1423–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino A, Thakur N, Grimsby S, Marcusson A, von Bulow V, Schuster N, Zhang S, Heldin CH, Landstrom M. (2008). The type I TGF-β receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat Cell Biol , 1199–1207. [DOI] [PubMed] [Google Scholar]
- Steder M, Alla V, Meier C, Spitschak A, Pahnke J, Furst K, Kowtharapu BS, Engelmann D, Petigk J, Egberts F, et al. (2013). DNp73 exerts function in metastasis initiation by disconnecting the inhibitory role of EPLIN on IGF1R-AKT/STAT3 signaling. Cancer Cell , 512–527. [DOI] [PubMed] [Google Scholar]
- Stout CE, Costantin JL, Naus CC, Charles AC. (2002). Intercellular calcium signaling in astrocytes via ATP release through connexin hemichannels. J Biol Chem , 10482–10488. [DOI] [PubMed] [Google Scholar]
- Tacheau C, Fontaine J, Loy J, Mauviel A, Verrecchia F. (2008). TGF-β induces connexin43 gene expression in normal murine mammary gland epithelial cells via activation of p38 and PI3K/AKT signaling pathways. J Cell Physiol , 759–768. [DOI] [PubMed] [Google Scholar]
- Tsai H, Werber J, Davia MO, Edelman M, Tanaka KE, Melman A, Christ GJ, Geliebter J. (1996). Reduced connexin 43 expression in high grade, human prostatic adenocarcinoma cells. Biochem Biophys Res Commun , 64–69. [DOI] [PubMed] [Google Scholar]
- Tumbarello DA, Brown MC, Hetey SE, Turner CE. (2005). Regulation of paxillin family members during epithelial-mesenchymal transformation: a putative role for paxillin delta. J Cell Sci , 4849–4863. [DOI] [PubMed] [Google Scholar]
- Turner CE. (2000). Paxillin and focal adhesion signalling. Nat Cell Biol , E231–E236. [DOI] [PubMed] [Google Scholar]
- Ul-Hussain M, Olk S, Schoenebeck B, Wasielewski B, Meier C, Prochnow N, May C, Galozzi S, Marcus K, Zoidl G, Dermietzel R. (2014). Internal ribosomal entry site (IRES) activity generates endogenous carboxyl-terminal domains of Cx43 and is responsive to hypoxic conditions. J Biol Chem , 20979–20990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Heyden MA, Veltmaat JM, Hendriks JA, Destree OH, Defize LH. (2000). Dynamic connexin43 expression and gap junctional communication during endoderm differentiation of F9 embryonal carcinoma cells. Eur J Cell Biol , 272–282. [DOI] [PubMed] [Google Scholar]
- Vanslyke JK, Naus CC, Musil LS. (2009). Conformational maturation and post-ER multisubunit assembly of gap junction proteins. Mol Biol Cell , 2451–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins SJ, Jonker L, Arthur HM. (2006). A direct interaction between TGF-β activated kinase 1 and the TGF-β type II receptor: implications for TGF-β signalling and cardiac hypertrophy. Cardiovasc Res , 432–439. [DOI] [PubMed] [Google Scholar]
- Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E. (2016). Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science , 241–253. [DOI] [PubMed] [Google Scholar]
- Xie L, Law BK, Chytil AM, Brown KA, Aakre ME, Moses HL. (2004). Activation of the Erk pathway is required for TGF-β1-induced EMT in vitro. Neoplasia , 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K. (1995). Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science , 2008–2011. [DOI] [PubMed] [Google Scholar]
- Yilmaz M, Christofori G. (2009). EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev , 15–33. [DOI] [PubMed] [Google Scholar]
- Yu M, Han G, Qi B, Wu X. (2017). Cx32 reverses epithelial-mesenchymal transition in doxorubicin-resistant hepatocellular carcinoma. Oncol Rep , 2121–2128. [DOI] [PubMed] [Google Scholar]
- Yu M, Zhang C, LI L, Dong S, Zhang N, Tong X. (2014). Cx43 reverses the resistance of A549 lung adenocarcinoma cells to cisplatin by inhibiting EMT. Oncol Rep , 2751–2758. [DOI] [PubMed] [Google Scholar]
- Zhang A, Hitomi M, Bar-Shain N, Dalimov Z, Ellis L, Velpula KK, Fraizer GC, Gourdie RG, Lathia JD. (2015). Connexin 43 expression is associated with increased malignancy in prostate cancer cell lines and functions to promote migration. Oncotarget , 11640–11651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.