Transient nuclear envelope (NE) ruptures in the Caenorhabditis elegans zygote are caused by a weakened nuclear lamina during nuclear positioning. Dynein-pulling forces enhance the severity of ruptures, while lamin restricts nucleocytoplasmic mixing and allows stable NE repair. This work is the first mechanistic analysis of NE rupture and repair in an organism.
Abstract
Recent work done exclusively in tissue culture cells revealed that the nuclear envelope (NE) ruptures and repairs in interphase. The duration of NE ruptures depends on lamins; however, the underlying mechanisms and relevance to in vivo events are not known. Here, we use the Caenorhabditis elegans zygote to analyze lamin’s role in NE rupture and repair in vivo. Transient NE ruptures and subsequent NE collapse are induced by weaknesses in the nuclear lamina caused by expression of an engineered hypomorphic C. elegans lamin allele. Dynein-generated forces that position nuclei enhance the severity of transient NE ruptures and cause NE collapse. Reduction of dynein forces allows the weakened lamin network to restrict nucleo–cytoplasmic mixing and support stable NE recovery. Surprisingly, the high incidence of transient NE ruptures does not contribute to embryonic lethality, which is instead correlated with stochastic chromosome scattering resulting from premature NE collapse, suggesting that C. elegans tolerates transient losses of NE compartmentalization during early embryogenesis. In sum, we demonstrate that lamin counteracts dynein forces to promote stable NE repair and prevent catastrophic NE collapse, and thus provide the first mechanistic analysis of NE rupture and repair in an organismal context.
INTRODUCTION
The nuclear envelope (NE) is a large polarized double-membrane sheet that serves to protect the genome in eukaryotic cells. The NE is continuous with the endoplasmic reticulum (ER) but has a unique protein composition and structure. In mitosis, the NE and associated proteins dramatically reorganize to promote NE breakdown (NEBD) and spindle microtubule capture of chromosomes. Following chromosome segregation, nuclear membranes emerge from the ER to reform around decondensing chromosomes (Hetzer, 2010; Ungricht and Kutay, 2017). Recent studies have shown that NE dynamics are not restricted to NEBD and reformation (De Vos et al., 2011; Vargas et al., 2012; Denais et al., 2016; Hatch and Hetzer, 2016; Raab et al., 2016; Webster et al., 2016). In migrating cells in culture, the NE undergoes reversible ruptures, which result in transient mixing of nuclear and cytoplasmic components, to promote passage of the nucleus through tight constrictions (Denais et al., 2016; Raab et al., 2016). In budding yeast, which undergoes a closed mitosis, transient NE holes form at sites of defective nuclear pore complexes (NPCs) (Webster et al., 2016), selective gatekeepers between the nucleus and cytoplasm (Hetzer and Wente, 2009). The ESCRT-III machinery seals NE holes—ESCRT-III subunits form spiral structures that constrict opposing membranes to facilitate membrane fission (McCullough et al., 2013; Shen et al., 2014; Webster et al., 2014; Chiaruttini et al., 2015; Olmos et al., 2015; Olmos and Carlton, 2016; Vietri et al., 2015; Denais et al., 2016; Raab et al., 2016). Although NE ruptures are repaired and transient, they can adversely affect genome integrity (Denais et al., 2016; Raab et al., 2016). This highlights the need to investigate mechanisms that induce NE ruptures and promote recovery from ruptures, particularly at the organismal level, which can reveal consequences of this process in a physiologically relevant context.
Transient NE ruptures occur at NE sites that are disrupted or deficient in nuclear lamins (Denais et al., 2016; Hatch and Hetzer, 2016; Raab et al., 2016; Robijns et al., 2016). The nuclear lamina is composed of the metazoan-specific intermediate filament proteins lamin A/C, B1, and B2 and associated integral membrane proteins that include the conserved Lap2-Emerin-Man1 (LEM) family (Dechat et al., 2008, 2010; Wilson and Foisner, 2010). Lamins mechanically stabilize the NE by forming separate interdigitating meshworks that associate with the inner nuclear membrane facing chromatin (Dechat et al., 2008; Davidson and Lammerding, 2014; Shimi et al., 2015; Xie et al., 2016; Turgay et al., 2017). In addition to mechanically supporting the nucleus, lamins and lamin-binding proteins regulate gene expression and organize chromatin (Reddy et al., 2008; Wilson and Foisner, 2010; Zullo et al., 2012; Mattout et al., 2015). Genetic disruption of lamin A/C and lamin-associated proteins leads to human disorders called laminopathies, which mostly affect tissues under high mechanical strain (Burke and Stewart, 2006, 2013). Disease mutations in lamin A/C lead to nuclear deformation and loss of nuclear integrity in cells from diseased tissues (Burke and Stewart, 2006, 2013). Cultured fibroblasts from patients with lamin A/C mutations undergo transient NE ruptures, suggesting that this process may be relevant to lamin-associated diseases (De Vos et al., 2011). An important next step is to understand if transient NE ruptures give rise to more severe NE integrity defects on diseased cells. However, the low frequency and stochastic nature of NE ruptures make it challenging to investigate these events in vivo.
The reduction of each lamin subtype increases the frequency of transient NE ruptures in cancer cells in culture (Vargas et al., 2012; Denais et al., 2016). Several lines of evidence suggest a critical role for lamin B1 in this process: reduction of lamin B1 results in NE regions devoid of nuclear lamina that correspond to rupture sites (Denais et al., 2016; Hatch and Hetzer, 2016), depleting lamin B1 results in the same frequency of NE ruptures as depleting all three lamins (Vargas et al., 2012), and local lamin B1 disruption at the NE has been implicated in NE sites of viral entry (de Noronha et al., 2001; Cohen et al., 2006). While these data support a specific role for lamin B1 in preventing NE ruptures, the reported structural interdependencies of lamins make it difficult to identify individual roles for lamins in NE ruptures (Shimi et al., 2008). Also, it is challenging to separate lamin’s functions in transcriptional regulation and chromatin organization from its direct structural roles in NE integrity. The generation of double and triple lamin knockouts in mouse embryonic stem cells provides an instrumental tool for understanding essential roles for individual lamin subtypes (Kim et al., 2011, 2013); however, the need to dissect mechanistic roles for lamins in NE rupture and repair in vivo remains.
Our understanding of mechanisms that induce transient NE ruptures comes from studies in adherent cancer cells in culture that showed that, in addition to weaknesses in the nuclear lamina, actin-derived or mechanical compression forces imposed on the NE induce NE ruptures (Broers et al., 2004; Le Berre et al., 2012; Hatch and Hetzer, 2016; Takaki et al., 2017). Consistent with a role for compression forces in inducing NE ruptures, constriction size and myosin activity influence the severity of migration-induced NE ruptures (Denais et al., 2016; Raab et al., 2016). Thus, the current model includes the role of a weakened lamina and actin-, but not microtubule-generated forces in inducing NE ruptures (Lammerding and Wolf, 2016).
In addition to inducing NE ruptures when weak, lamins control the time between NE rupture and reestablishment of the nuclear permeability barrier (Vargas et al., 2012; Denais et al., 2016; Raab et al., 2016). Lamin A/C accumulates at rupture sites directly following migration-induced NE rupture, and its levels of accumulation correlate with rupture severity, suggesting that it may play a role in repair of NE holes.
The finding that repair of NE holes in fungi and sealing of the reforming NE during open mitosis both involve recruitment of ESCRT-III repair machinery by the inner nuclear membrane (INM) protein LEM2 suggests that similar mechanisms contribute to NE dynamics in interphase and mitosis (Webster et al., 2016; Gu et al., 2017). Upon entry into prophase, removal of NPCs permeabilizes the NE, which coincides with INM protein diffusion into retracted ER membranes (Terasaki et al., 2001; Burke and Ellenberg, 2002; Kutay and Hetzer, 2008). Dynein-dependent tearing of nuclear membranes and disassembly of lamins clear NE remnants from condensed chromatin (Beaudouin et al., 2002; Salina et al., 2002). At the end of mitosis, ER membranes and associated INM proteins reform around segregated chromosomes to form the nascent NE (Mattaj, 2004; Anderson and Hetzer, 2007; Anderson et al., 2009). De novo NPC assembly and ESCRT-III–mediated membrane sealing reestablishes the nuclear permeability barrier (Wandke and Kutay, 2013; Olmos and Carlton, 2016; Ungricht and Kutay, 2017). Whether analogous mechanisms are involved in the more stochastic and localized process of interphase NE rupture and repair is not known.
Here, we investigate the functions of the highly conserved single Caenorhabditis elegans lamin B isoform (Riemer et al., 1993; Stuurman et al., 1998; Liu et al., 2000) in NE rupture and repair during dynein-dependent pronuclear positioning in the C. elegans zygote. Both lamin and dynein contribute to the extent of loss of the nuclear permeability barrier upon NE damage. Lamin counteracts dynein-generated forces on damaged nuclei to allow NE repair. The rapid dynamics of NE repair in the C. elegans zygote allowed us to delineate NE reorganization at repair sites and to relate these events to reestablishment of the NE permeability barrier. Lamin also protects against premature NE collapse, induced by increasing dynein forces that facilitate pronuclear migration, to prevent chromosome scattering and mitotic defects. Thus, as dynein-generated tension imposed on the NE increases in preparation for the first embryonic division, lamin stabilizes nuclei against persistent ruptures and prevents catastrophic NE collapse to protect genome integrity.
RESULTS
A timeline of pronuclear positioning events in the C. elegans embryo to dissect structural roles for lamin
To investigate whether C. elegans lamin protects against NE ruptures in vivo, we utilized the well-defined pronuclear positioning events in the transcriptionally silent C. elegans zygote that contains a single lamin B gene (lmn-1). We monitored pronuclear position in embryos expressing GFP::Histone2B and GFP::γ-tubulin to mark nuclei and centrosomes, respectively, directly following oocyte fertilization through NEBD. The times of pronuclear positioning events are relative to pseudocleavage (PC) regression, an event that occurs from asymmetric cortical acto-myosin contraction forces that establish polarity at the one-cell stage (0 s, Figure 1A; Cowan and Hyman, 2004). In our timeline, we incorporated information from other studies on the timing and location of active cytoskeletal forces and cell cycle transitions (Edgar and McGhee, 1988; O’Connell, 2000; Cowan and Hyman, 2004; Kimura and Onami, 2005; Oegema and Hyman, 2006; Portier et al., 2007; Dammermann et al., 2008; Bahmanyar et al., 2014). Our timeline highlights the time course of dynein-dependent nuclear positioning events relative to mitotic-regulated NEBD that facilitate mixing of the oocyte- and sperm-derived haploid genomes. The C. elegans zygote offers the opportunity to investigate lamin’s function in maintaining nuclear integrity in the absence of transcription and in response to increasing dynein-generated forces relevant in vivo and in an intact organism.
FIGURE 1:
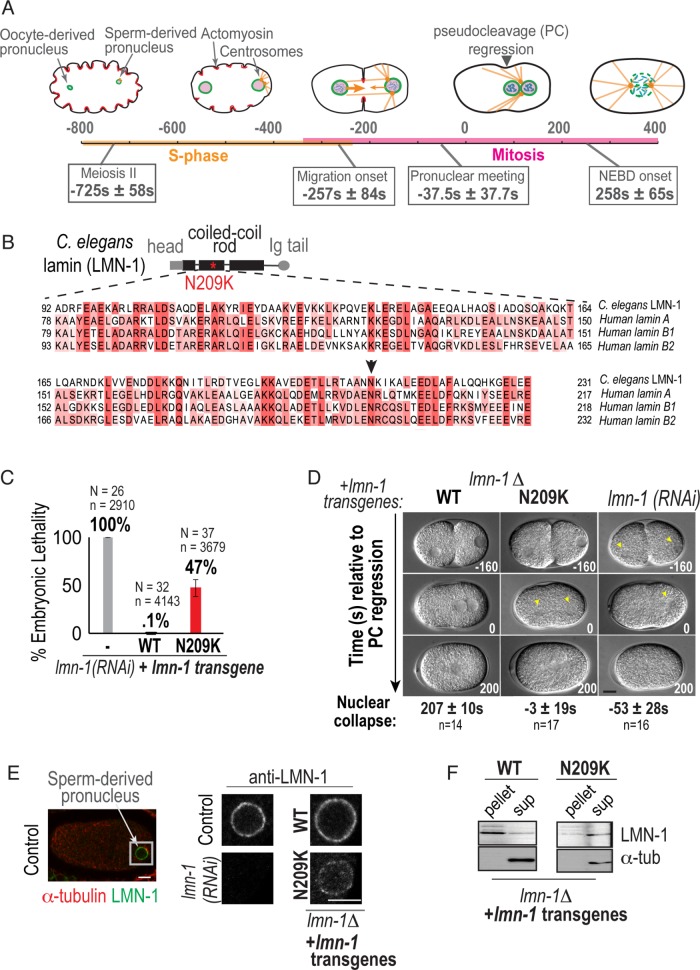
A disease-causing mutation in C. elegans lamin (lamin-N209K) is hypomorphic for lamin function. (A) Schematic illustrating the timeline of pronuclear positioning events in the one-cell stage C. elegans embryo relative to pseudocleavage (PC) regression. Times of key events are the means of n = 8 embryos ± SD. (B) Schematic of C. elegans lamin (LMN-1) protein domain structure, relative position of N209K disease mutation (red asterisk), and multiple alignment of indicated coiled-coil domain in C. elegans and human lamins. Arrowhead marks conserved asparagine residue. (C) Percentage of embryonic lethality from the indicated conditions. N = number of worms, n = number of embryos. Data are represented as mean ± SD. (D) DIC images from time-lapse series of C. elegans one-cell embryos for the indicated conditions. Arrowheads mark cleared regions that correspond to nuclear contents. Scale bar, 10 μm. Times below are the mean times in seconds ± SEM of nuclear collapse from DIC images. Time points are in seconds relative to PC regression. (E) Representative example of immunofluorescence image of a wild-type one-cell embryo stained for lamin (LMN-1) (red) and α-tubulin (green). Magnified image of sperm-derived pronucleus immunostained with antibody against endogenous LMN-1 from indicated conditions. Scale bars, 5 μm. (F) Immunoblot probed for LMN-1 and α-tubulin after preparation of lysates into soluble (sup) and insoluble (pellet) fractions of worms from indicated genetic backgrounds (N = 3). See also Supplemental Figure S1.
A disease-causing mutation in a conserved residue in C. elegans lamin (lamin-N209K) is hypomorphic for lamin function
We reasoned that a hypomorphic allele in lamin would serve as a useful tool for investigation of lamin’s structural roles in the early C. elegans embryo without potentially confounding effects of penetrant lamin depletion. C. elegans lamin is a B-type lamin with 30% amino acid sequence identity with human lamins B1, B2, and A/C (Figure 1B). Prior work aimed at using C. elegans to model laminopathies demonstrated the utility of using lamin A mutations as a guide to designing mutant alleles in C. elegans lamin for analysis of lamin function on an organismal level (Wiesel et al., 2008; Bank et al., 2011, 2012; Mattout et al., 2011; Zuela et al., 2017). To optimize for expression in the C. elegans germline and to establish an effective method of comparing wild-type and lamin mutant alleles, we used the MosI-mediated single copy insertion system (Frøkjær-Jensen et al., 2008) to engineer a RNA interference (RNAi)-resistant C. elegans lamin transgene expressed under its own cis regulatory elements. We validated this transgenic system by demonstrating that the untagged, RNAi-resistant wild-type (WT) lamin transgene (Supplemental Figure S1A) rescues the 100% embryonic lethality caused by selective depletion of endogenous lamin (Figure 1C). To generate a partially functional lamin allele, we introduced a mutation in a conserved dilated cardiomyopathy linked residue in lamin A/C (N195K in human lamin A/C, N209K in C. elegans lamin; Figure 1B; Fatkin et al., 1999; Wiesel et al., 2008). We chose this mutation because it is in a highly conserved region (Figure 1B), and the same mutation in human lamin A reduces its incorporation into the nuclear lamina and decreases the ability of the NE to withstand mechanical forces (Zwerger et al., 2013).
To more rigorously analyze the ability of engineered transgenes to support lamin’s functions, we used a lamin deletion strain (Supplemental Figure S1, B and C, referred to as lmn-1Δ), which is sterile but can be maintained as a heterozygote with a balancer (Supplemental Figure S1D; Haithcock et al., 2005). The engineered wildtype lamin (WT-lamin) transgene supports embryo production and viability in the homozygous lmn-1Δ background (Supplemental Figure S1, B and D). The lamin-N209K mutant transgene partially supports embryo production in the lmn-1Δ strain, and 75% of laid embryos do not hatch (Supplemental Figure S1, B and D). Because RNAi depletion of lamin is 100% lethal (Figure 1C), the partial viability of embryos expressing lamin-N209K upon RNAi depletion of endogenous lamin (Figure 1C) or in the lmn-1Δ strain (Supplemental Figure S1B) indicates that the lamin-N209K protein is partially functional.
Next, we performed time-lapse imaging of the first embryonic division using differential interference contrast (DIC) microscopy to determine if the lamin-N209K mutant is able to support lamin’s functions in maintaining nuclear structure. WT-lamin pronuclei maintain their shape until NEBD, whereas pronuclei in lamin RNAi–depleted embryos appear misshapen and barely visible prior to PC regression (Figure 1D). The lamin-N209K transgene supports nuclear morphology until PC regression at which time pronuclei lose their structure and become undetectable by DIC (Figure 1D). Thus, similar to its ability to partially support embryonic viability (Supplemental Figure S1B), the lamin-N209K mutation partially supports lamin’s functions in maintenance of pronuclear integrity in the lmn-1Δ strain.
Prior work in C. elegans showed abnormal localization of GFP-tagged lamin-N209K mutant protein expressed in the presence of endogenous lamin (Wiesel et al., 2008). Our attempt to generate a functional fluorescent fusion WT lamin transgene was unsuccessful (unpublished data). Using an antibody we generated against C. elegans lamin, we localized WT and N209K-lamin proteins in the lmn-1Δ background. Localization of lamin-N209K to the nuclear rim was significantly reduced in the absence of endogenous lamin (Figure 1E and Supplemental Figure S1E). Consistent with this, the lamin-N209K protein is more soluble under extraction conditions that do not normally solubilize lamin protein incorporated into the nuclear lamina (Figure 1F; Zwerger et al., 2013). Total lamin-N209K protein levels are reduced in comparison with WT lamin protein levels; however, the two transgenes express similar levels of lamin mRNA (Supplemental Figure S1, F and G). Endogenous lamin expressed from a single locus in the het deletion strain has reduced mRNA and protein levels, yet is normally localized to the nuclear rim and can support viable embryo production (“Het” in Supplemental Figure S1, D–G). Thus, the lamin-N209K mutant protein is less able to incorporate into the nuclear lamina, which results in reduced localization to the nuclear rim and an unstable soluble pool of mutant protein.
The functional and biochemical data support the conclusion that the lamin-N209K mutant protein is hypomorphic for lamin function, providing a useful tool for investigation of subtler structural functions for lamin during pronuclear positioning in vivo.
Lamin prevents transient and persistent loss of the nuclear permeability barrier during distinct stages of pronuclear positioning
To dissect the underlying cause of nuclear phenotypes in lamin RNAi–depleted and lamin-N209K mutant embryos, we used time-lapse imaging of embryos expressing fluorescently tagged markers. To generate strains that carry the lamin-N209K transgene and fluorescent markers, we took advantage of the engineered RNAi-resistant region in lamin transgenes (Supplemental Figure S1A), which allows selective depletion of endogenous lamin. Because the lamin-N209K transgene supports 100% viability in the presence of endogenous lamin (unpublished data), this strategy circumvents brood size and lethality issues that would otherwise hinder successful production of cross progeny.
Prior work in C. elegans embryos showed that lamin RNAi depletion inhibited the formation of an intact nuclear permeability barrier (Hayashi et al., 2012). To investigate the ability of the lamin-N209K mutant protein to support the NE permeability function for lamin, we monitored exclusion of soluble GFP::α-tubulin from the nuclear interior prior to pronuclear migration. GFP::α-tubulin was excluded from the sperm-derived pronucleus in embryos expressing the WT lamin transgene RNAi–depleted of endogenous lamin (hereafter referred to as WT-lamin) (Figure 2, A–D). In embryos expressing lamin-N209K RNAi depleted of endogenous lamin (hereafter referred to as lamin-N209K), nuclear GFP::α-tubulin abruptly entered the nucleus, followed by a gradual decrease in nuclear GFP::α-tubulin signal intensity (Figure 2, A and B; Supplemental Figure S2, A and B; Supplemental Movie 1). To quantitatively determine the percentage of nuclei that reestablish the nuclear permeability barrier, we measured the average fluorescence intensity of nuclear GFP::α-tubulin normalized to the initial fluorescence intensity prior to nuclear entry (Figure 2B; Supplemental Figure S2C). We defined full recovery as the time point when the normalized fluorescence intensity value of GFP::α-tubulin in the nucleus reached or fell below the initial value (Figure 2B). This criterion revealed that 45% of lamin-N209K nuclei and 44% of lamin-depleted nuclei undergo entry and full recovery of nuclear GFP::α-tubulin prior to pronuclear migration (Figure 2C). In contrast, upon pronuclear migration and rotation, none of the nuclei recovered from nuclear localization of GFP::α-tubulin (Figure 2D; Supplemental Figure S2, D and E). Instead, soluble GFP::α-tubulin persisted in the region of deformed and collapsed nuclei (Supplemental Figure S2D) in 100% of lamin RNAi–depleted and 65% of lamin-N209K embryos (magenta and black bars in Figure 2D). These data indicate that expression of the lamin-N209K transgene and RNAi depletion of lamin lead to transient and persistent disruption of the barrier function of the NE during distinct pronuclear positioning events known to result from increasing dynein forces imposed on nuclei.
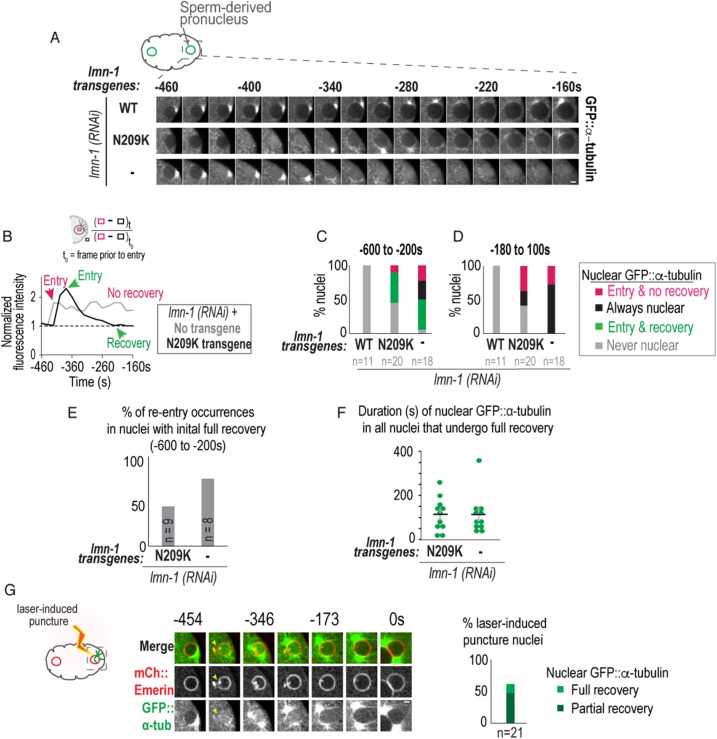
FIGURE 2:
Lamin prevents transient and persistent nuclear ruptures during pronuclear positioning. (A) Representative confocal images of a sperm-derived pronucleus from time-lapse series of indicated conditions. Scale bar, 2.5 μm. (B) (Top) Schematic of method used to measure nuclear GFP::α-tubulin fluorescence intensity levels. (Bottom) Plot of normalized nuclear GFP::α-tubulin fluorescence intensity measurements of indicated nuclei in A. (C, D) Plots of percentage of nuclei during indicated time periods relative to PC regression. (E) Percentage of GFP::α-tubulin nuclear reentry occurrences in nuclei that fully recovered from initial nuclear entry event (green bar in part C). Also see Supplemental Figure S2, A–C. (F) Plot of duration from initial entry to full recovery of nuclear GFP::α-tubulin under indicated conditions. Individual times (green) and mean time ± SEM (black) in seconds are shown. Mann–Whitney test, p = 0.71 n.s. (G) Select images from time series of laser-induced puncture and recovery of sperm-derived pronucleus. The dimensions of this laser-induced puncture are 4 × 2 × 0.5 μm. Arrow points to site of nuclear envelope puncture. Scale bar, 2.5 μm. Time in seconds relative to PC regression. Plot (Right) of percentage of sperm-derived nuclei that fully recover from nuclear GFP::α-tubulin levels (light green) or reach 50% of the nuclear GFP::α-tubulin levels at t0 (partial recovery, dark green). Plot combines data sets of nuclei cut with two different laser punctures sizes (dimensions: 4 × 2 × 0.5 μm [large cut] and 4 × 1 × 0.5 μm [small cut]). n = number of embryos. See also Supplemental Figure S2 and Supplemental Movies 1–3.
Movie S1.
Transient nuclear entry and recovery of GFP::α tubulin in lamin-N209K mutant embryos selectively depleted of endogenous lamin, related to Figure 2 and Figure S2. Confocal fluorescence images of embryos expressing GFP::α-tubulin (green) and mCherry::Histone2B (red). Embryos expressing the lamin-WT transgene (left)and lamin-N209K transgene (right) are shown. Sequences start during sperm-derived pronuclear positioning at the posterior end of the embryo and end at completion of pronuclear rotation. WT-lamin embryos exclude soluble GFP::α-tubulin from the nuclear interior until pronuclear rotation is complete. (Left panel, arrow marks entry ofGFP::α-tubulin into the nucleus). GFP::α-tubulin transiently enters the sperm-derived pronucleus in lamin-N209K mutant embryos prior to pronuclear migration(Right panel, arrows mark GFP::α-tubulin entry into pronucleus). The oocyte-derived pronucleus goes in and out of the focal plane in this embryo. The oocyte-derived pronucleusand sperm-derived pronucleus deform and lose their integrity upon pronuclear meeting and upon pronuclear rotation respectively at which time GFP::α-tubulin enters the pronucleus. Images were acquired every 20s. Playback rate is 80X real time.
At time points prior to pronuclear migration, we noticed reentry of nuclear GFP::α-tubulin following recovery in some nuclei, suggesting that these nuclei remained unstable (Supplemental Figure S2, A–C; Supplemental Movie 2). Consistent with this idea, we found that 75% of lamin-depleted nuclei that fully recovered from nuclear entry of GFP::α-tubulin (green bars in Figure 2C) had subsequent events of nuclear entry of GFP::α-tubulin prior to pronuclear migration (Figure 2E); reentry occurred in only 44% of lamin-N209K embryos (Figure 2E). The duration of nuclear GFP::α-tubulin was similar for nuclei that fully recovered under lamin depletion and lamin-N209K conditions (113 ± 27 s for lamin RNAi and 115 ± 23 s for lamin-N209K, mean ± SEM; Figure 2F); however, nuclear GFP::α-tubulin was more likely to persist in lamin-depleted nuclei during these early time points (50% in lamin RNAi–depleted vs. 10% in lamin-N209K; magenta and black bars in Figure 2C). In addition, irreversible entry of GFP::α-tubulin occurred significantly earlier in lamin-depleted embryos than in lamin-N209K expressing embryos (Figure S2E). We conclude that lamin prevents transient loss of the nuclear permeability barrier and promotes stable reestablishment of nuclear compartmentalization prior to pronuclear migration. Although lamin-N209K expression leads to disruption of the nuclear permeability barrier, its mechanical properties (Zwerger et al., 2013) and/or partial assembly at the nuclear rim are sufficient to support stable reestablishment of nuclear compartmentalization during early nuclear positioning events. When dynein forces imposed on nuclei are sufficiently strong to facilitate pronuclear meeting, lamin protects against persistent loss of the nuclear permeability barrier.
Movie S2.
Cycles of nuclear entry and reentry of GFP::α-tubulin in embryo expressing lamin-N209K transgene and depleted of endogenous lamin, related to Figure 2 and Supplemental Figure S2. Confocal fluorescence images of embryo expressing GFP::α-tubulin (green) and mCherry::Histone2B (red) depleted of endogenous lamin and expressing lamin-N209K transgene. Sequence starts during sperm-derived pronuclear positioning at the posterior end of the embryo and ends after completion of pronuclear migration. Arrow marks entry and re-entry of GFP::α-tubulin into the nucleus. The sperm-derived pronucleus deforms and loses it integrity during pronuclear migration. Images were acquired every 20s. Playback rate is 80X real time.
NE recovery following laser-induced puncture in wild-type embryos
To recapitulate recovery following nuclear entry of GFP::α-tubulin in wild-type nuclei, we monitored the normalized fluorescence intensity of nuclear GFP::α-tubulin after laser-induced puncture of the NE at time points prior to pronuclear migration (Figure 2G; Supplemental Movie 3). Laser-induced puncture caused rapid equilibration of nuclear and cytoplasmic soluble GFP::α-tubulin followed by full or partial recovery (partial recovery defined as reaching 50% of full recovery) (plot in Figure 2G; Supplemental Movie 3). These data demonstrate that wild-type nuclei reestablish the nuclear permeability barrier following a breach in the NE in the C. elegans zygote. Neither recovery percentage nor recovery kinetics was affected by the size of laser punctures (Figure 2G). However, the low percentage of full recovery following laser-induced puncture and the long duration of nuclear GFP::α-tubulin (504 ± 44 s from entry to full recovery and 254 ± 23 s to partial recovery, mean ± SEM) suggest that micrometer-scale holes generated by laser-induced puncture are larger, and thus less favorable to recovery, than holes caused by expression of the lamin-N209K mutant.
Movie S3.
The NE recovers from laser-induced puncture of the NE, related to Figure 2. Confocal fluorescence images of wildtype embryo expressing GFP::α tubulin (green) and mCherry::Emerin (red) are shown. Sequence selected starts during sperm-derived pronuclear centering and end at pronuclear rotation. Laser-induced puncture of the sperm-derived NE (Arrow appears in frame corresponding to time of puncture) leads to nuclear entry of soluble GFP::α-tubulin and deformation of the NE at the site of puncture. As sequence progresses, the sperm-derived pronucleus recovers from laser-induced puncture: GFP::α-tubulin exits the nuclear interior upon pronuclear meeting and the NE reestablishes its shape. Images were acquired every 3.6 s and shown here in 21.6 s intervals. Playback rate is 86 X real time.
NE ruptures coincide with transient reorganization of nuclear pore complexes and chromatin at rupture sites
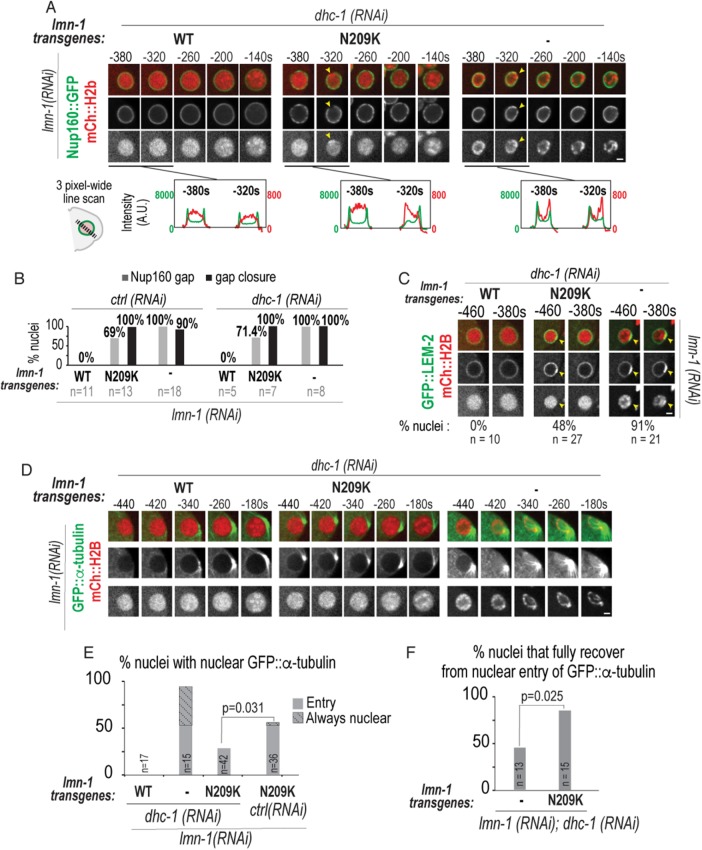
In mammalian cells, transient ruptures occur in regions that are devoid of NPCs and lamins (De Vos et al., 2011; Denais et al., 2016; Raab et al., 2016). To determine whether pronuclei in lamin RNAi–depleted or mutant embryos displayed NE regions devoid of NPCs, we imaged Nup160::GFP, a member of the NPC scaffold, and mCherry:Histone2B to mark chromatin (Hattersley et al., 2016). In S-phase, WT-lamin pronuclei contain decondensed chromatin surrounded by a smooth continuous ring of Nup160::GFP (–420 s in Figure 3, A and B). In 69% of lamin-N209K and 93% of lamin RNAi–depleted embryos, a distinct gap in the Nup160::GFP signal at the nuclear rim was present at time points that correspond to the time range of transient loss of the permeability barrier (Figure 3, A–D; ∼–600 to ∼–380 s in Figure 3C; Supplemental Movie 4). Thus, lamin inhibition causes a gap in Nup160::GFP at the nuclear rim that likely marks the site of NE rupture.
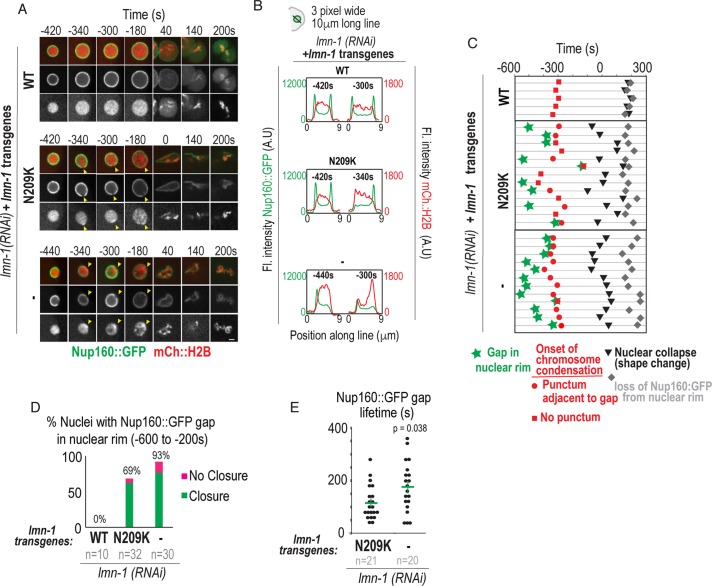
FIGURE 3:
Analysis of Nup160::GFP gap formation and closure and chromatin reorganization at sites of nuclear envelope rupture. (A) Select representative confocal images of sperm-derived pronuclei from time-lapse series of embryos expressing Nup160::GFP and mCherry::Histone2B from indicated conditions. Scale bar, 2.5 μm. Arrowheads in middle and bottom panels mark site of gap in Nup160::GFP at nuclear rim and focus of condensed chromatin. (B) Plots represent fluorescence intensities of Nup160::GFP (green) and mCherry::Histone2B (red) along three-pixel-wide lines drawn across nucleus shown in A at indicated times relative to PC regression. (C) Timeline of events scored from confocal time-lapse images of individual sperm-derived pronuclei (horizontal lines) from indicated conditions. All times are in seconds and relative to PC regression. (D) Plot of percent of nuclei with visible gaps that undergo closure (green) or no closure (magenta) under the indicated conditions. n = number of embryos. (E) Plot of duration from appearance of Nup160::GFP gap to closure at the nuclear rim. Time-lapse series in which gap was present in first frame were excluded from analysis. Mean ± SEM in seconds is shown. n = number of Nup160::GFP gaps. p value calculated by Mann–Whitney test. See also Supplemental Figure S3 and Supplemental Movies 4, 5, and 7.
Movie S4.
Nup160::GFP gaps transiently appear at the nuclear rim and mCherry:Histone2B condenses precisely at the position of gaps prior to pronuclear collapse in lamin-N209K mutant embryos selectively depleted of endogenous lamin, related to Figure 3, Figure 4, and Figure S3. Confocal fluorescence images of embryos expressing Nup160::GFP (green) and mCherry::Histone2B (red). (Left) Embryos expressing the WT-lamin transgene and (Right) lamin-N209K mutant embryos are shown. Sequences start during sperm-derived pronuclear positioning at the posterior end of the embryo and end at anaphase. Nup160::GFP localizes to the nuclear rim in a continuous pattern in control embryos throughout the sequence until NEBD at which point Nup160::GFP dissociates from the nuclear rim and simultaneously accumulates to kinetochores (Left). Gaps in the Nup160::GFP signal appear and resolve in lamin-N209K mutant embryos until pronuclear migration when the pronucleus deforms towards the anterior (Right panel, arrows follow Nup160::GFP gap and deformation at the nuclear rim in the sperm-derived pronucleus). Nup160::GFP dissociates from remnants of the NE and accumulates to kinetochores with similar timing to WT-lamin embryos. The oocyte-derived pronucleus goes in and out of the focal plane in these examples. Images were acquired every 20s. Playback rate is 80X real time.
Subsequent to the appearance of a Nup160::GFP gap in the nuclear rim, and coincident with entry into mitotic prophase, a highly condensed spot of mCherry:Histone2B appeared precisely in the region devoid of Nup160::GFP in lamin-N209K and lamin RNAi–depleted conditions (–340 and –300 s in Figure 3, A and B; ∼–380 to –300 s in Figure 3C; Supplemental Movie 4). Following its initial appearance at this site in lamin RNAi–depleted embryos, chromosomes became highly compacted and localized adjacent to the nuclear periphery (–180 s in Figure 3A). This phenotype was less severe in lamin-N209K embryos: discrete condensed chromosome puncta distribute throughout the nuclear interior at time points at which wild-type pronuclei do not contain distinct chromosomes (–180 s in Figure 3A). Thus, a condensed chromatin focus appears at NE rupture sites in early prophase, which is directly followed by abnormal distribution and acceleration of chromosome condensation.
We predicted that if NE regions devoid of Nup160::GFP are sites of NE ruptures, then they should be transient and appear and resolve with kinetics similar to loss and reestablishment of the NE permeability barrier (see Figure 2). In support of this idea, Nup160::GFP gap formation and closure correlated with deformation and restoration of nuclear shape (Supplemental Figure S3A), and in most embryos gaps closed prior to pronuclear migration (Supplemental Figure S3B). To directly visualize Nup160::GFP gaps and the nuclear permeability barrier, we imaged lamin-N209K embryos simultaneously expressing Nup160::GFP and mCherry::α-tubulin. Entry and full recovery of nuclear mCherry::α-tubulin occurred concomitantly with the appearance and closure of a Nup160::GFP gap in the majority of embryos (Supplemental Figure S3, C and D; Supplemental Movie 5), indicating that these events are associated. We also observed Nup160::GFP gaps that precede nuclear entry of mCherry::α-tubulin, but never the opposite, suggesting that in some cases damaged membranes can form prior to disruption of the nuclear permeability barrier (Supplemental Figure S3D).
Movie S5.
Nuclear entry of mCherry::α-tubulin corresponds with Nup160::GFP gaps in lamin-N209K mutant embryos selectively depleted of endogenous lamin, related to Figure S3. Confocal fluorescence images of embryos expressing Nup160::GFP (green) and mCherry::α-tubulin (red). Embryos expressing the lamin-WT transgene (left)and lamin-N209K transgene (right) are shown. Sequences start during sperm-derived pronuclear positioning at the posterior end of the embryo and end at the beginning of pronuclear migration. WT-lamin embryos exclude soluble mCherry::α-tubulin from the nuclear interior and lack Nup160::GFP gaps at the nuclear rim (Left panel). mCherry::α-tubulin transiently enters the sperm-derived pronucleus at the same time as the appearance of Nup160::GFP gap in lamin-N209K mutant embryos (Right panel, arrows follow Nup160::GFP gap and nuclear mCherry::α -tubulin in sperm-derived pronucleus). Images were acquired every 20s. Playback rate is 80X real time.
The percentage of nuclei that resolve Nup160::GFP gaps is similar to the percentage that recover from entry of GFP::α-tubulin in lamin-N209K embryos (at time points prior to PC regression, 91% of all Nup160::GFP gaps, green and magenta bars in Figure 3D, resolve and 82% of all nuclei containing GFP::α-tubulin, green and magenta bars in Figure 2C, fully recover). In contrast, a significantly lower percentage of lamin RNAi–depleted embryos restore the nuclear permeability barrier than of those that undergo Nup160 gap closure (closure occurs in 82% of all Nup160 gaps, green and magenta bars for lamin RNAi in Figure 3D; only 47% of all nuclei containing GFP::α-tubulin, green, magenta, and black bars for lamin RNAi in Figure 2C, fully recover; chi-squared test, p = 0.02). Moreover, the average duration of Nup160::GFP gaps was significantly longer in lamin RNAi–depleted than lamin-N209K embryos (175 ± 21 s in lamin RNAi depletion vs. 116 ± 14 s in lamin-N209K, mean ± SEM; Figure 3E), indicating that lamin promotes Nup160 gap closure. The average duration of Nup160::GFP gaps in lamin RNAi–depleted embryos does not correspond to the average time to reestablishment of the nuclear permeability barrier (Figures 2F and 3E), suggesting that some lamin-deficient nuclei that undergo Nup160::GFP gap closure do not restrict the nuclear permeability barrier. Thus, lamin-deficient nuclei with a longer Nup160::GFP gap duration may be more unstable, leading to persistent loss of the nuclear permeability barrier.
Upon pronuclear migration, lamin-N209K and lamin RNAi–depleted NEs marked by Nup160::GFP deformed and collapsed onto chromatin (Figure 3A, 0–140 s; Figure 3C, –100–100 s; Supplemental Movie 4). Dissociation of Nup160::GFP from NE remnants in lamin-N209K and lamin RNAi–depleted nuclei occurred approximately 200 s after PC regression, which corresponds to the timing of cell cycle–regulated NEBD in control embryos (Figure 3A, 200 s; Figure 3C, ∼200 s). Nup160::GFP accumulated onto kinetochores, the attachment sites for spindle microtubules, with similar timing under control and lamin-inhibited conditions (Figure 3A, 140 s). These data show that defects in nuclear structure and chromatin condensation caused by lamin inhibition in early embryos are not a consequence of abnormal cell cycle progression.
We conclude that Nup160 gap appearance and closure indicate sites of NE rupture and repair. Initial ruptures at the NE occur in S-phase and are followed by compacted chromatin puncta adjacent to rupture sites upon entry into prophase, which precede global defects in chromatin condensation and organization. Our analysis further revealed that lamin restricts the duration of Nup160 gaps and allows stable reestablishment of the nuclear permeability barrier.
LEM-2 and ESCRT-III transiently localize to NE ruptures
To further support the idea that Nup160::GFP gaps form at NE rupture and repair sites, we determined the localization of VPS-32 (Snf7/CHMP4), the main component of the ESCRT-III spiral filament (McCullough et al., 2013; Shen et al., 2014; König et al., 2017), by immunofluorescence in lamin RNAi–depleted embryos. The majority of nuclei with a detectable Nup160::GFP gap at the nuclear rim also accumulated VPS-32 (Figure 4, A and B). These data indicate that, similarly to observations made in ruptured nuclei in tissue-culture cells (Denais et al., 2016; Raab et al., 2016), ESCRT-III repair machinery is recruited to rupture sites that we directly show are devoid of functional nuclear pores.
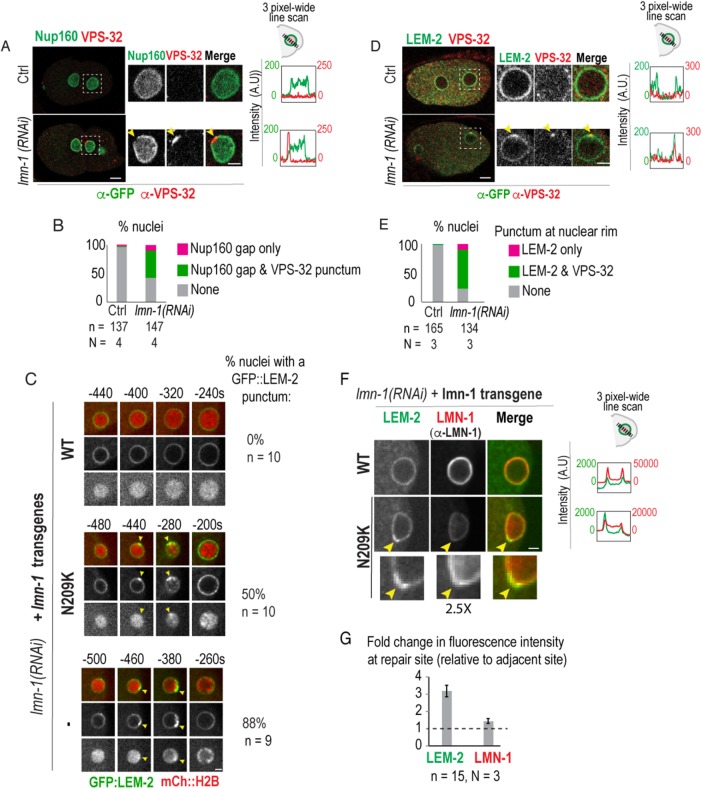
FIGURE 4:
Repair machinery is recruited to NE rupture sites marked by Nup160 gaps and LEM-2 foci at the nuclear rim. (A) Representative examples of fixed wild-type (Ctrl) and lamin RNAi–depleted two-cell stage embryos expressing Nup160::GFP immunostained against GFP (green) and VPS-32 (red). Magnified images are of posterior (P1) nucleus. Scale bars, 5 μm. Arrowheads mark Nup160::GFP gap and VPS-32 punctum. Plots (right) of fluorescence intensities of Nup160::GFP (green) and VPS-32 (red) along three-pixel-wide lines drawn across nuclei from images shown (left). (B) Plot of percent nuclei with a visible gap at nuclear rim in GFP channel in fixed early embryos (one-cell to eight-cell) under indicated conditions. N = number of experimental repeats. (C) Select images of sperm-derived pronucleus from time-lapse series. Arrowheads mark GFP::LEM-2 and mCherry::H2B foci. Time is in seconds relative to PC regression. Percentage of nuclei containing a focus of GFP::LEM-2 at the nuclear rim is shown. Scale bar, 2.5 μm. (D) Representative examples of fixed one-cell-stage embryos expressing GFP::LEM-2 immunostained against GFP (green) and VPS-32 (red). Magnified images are of sperm-derived pronucleus. Scale bars, 5 μm. Plots (right) represent fluorescence intensities of GFP::LEM-2 (green) and VPS-32 (red) along three-pixel-wide lines drawn across nuclei from images shown (left). (E) Plot of percent nuclei with visible foci at nuclear rim. (F) Select magnified images of sperm-derived pronuclei in control and lamin-N209K–expressing one-cell stage embryos expressing GFP::LEM-2 immunostained against lamin (LMN-1, red). Scale bar, 2.5 μm. Plots (right) represent fluorescence intensities of GFP::LEM-2 (green) and lamin (red) along three-pixel-wide lines drawn across nuclei from images shown. Note different scales for LMN-1 under different conditions. (G) Plot of fold change in mean fluorescence intensity values of GFP::LEM-2 and LMN-1 at NE repair site relative to an adjacent site. n = number of nuclei. N = experimental repeats.
In mammalian cells, the inner nuclear envelope protein LEM2 is the nuclear adapter that recruits ESCRT-III repair machinery to seal NE openings at the end of mitosis (Gu et al., 2017). We found that in lamin RNAi–depleted and lamin-N209K nuclei, GFP::LEM-2 transiently accumulated at the nuclear rim directly adjacent to compacted mCherry::Histone2B foci (Figure 4C). Transient GFP::LEM-2 accumulation occurred ∼–500–400 s prior to PC regression (Figure 4C), which coincides with the timing of ruptures marked by transient loss of Nup160::GFP (Figure 3, A–C) or nuclear entry of α-tubulin (Figure 2, A–C; Supplemental Figure S3, C and D). GFP::LEM-2 foci colocalized with VPS-32 at the nuclear rim in fixed lamin RNAi–depleted embryos indicating that these are sites of rupture and repair (Figure 4, D and E). Although lamin is not needed to recruit LEM-2 or ESCRT-III machinery (Figure 4, A–E), immunostaining against lamin in lamin-N209K embryos revealed that lamin localizes to regions at the nuclear rim enriched with LEM-2 (Figure 4, F and G). These data support a role for lamin in stabilizing rupture sites to allow timely sealing of NE holes.
Together, these data show that there are transient, local changes in nuclear pores and LEM-2 at NE rupture sites that coincide with recruitment of ESCRT-III fission machinery and colocalize with, but do not require, lamin.
Dynein-generated forces exacerbate NE ruptures resulting from expression of lamin-N209K
Next, we analyzed the role cytoskeletal forces play in lamin-dependent NE rupture and recovery. Prior work in cancer cells showed that actin-, and not microtubule-derived forces induce transient ruptures at NE sites devoid of lamins in interphase (Hatch and Hetzer, 2016; Lammerding and Wolf, 2016). In C. elegans embryos, actin filaments are enriched on the anterior cortex of the embryo, away from the sperm-derived pronucleus, at time points corresponding to NE ruptures (Supplemental Figure S4A; Cowan and Hyman, 2004; Munro et al., 2004; Oegema and Hyman, 2006, Davies et al., 2014), suggesting that they likely do not directly impose forces on pronuclei to cause ruptures. On the other hand, the microtubule motor dynein is directly linked to the NE through the outer nuclear membrane LINC-complex protein, ZYG-12, and functions in separating NE associated centrosomes and positioning the sperm-derived pronucleus at time points that correspond to transient NE ruptures (Gönczy et al., 1999; Malone et al., 2003; Oegema and Hyman, 2006; Luxton and Starr, 2014). However, we eliminated centrosomes as the direct cause of NE ruptures because the oocyte-derived pronucleus, which is not directly associated with centrosomes (Cowan and Hyman, 2004), also undergoes transient ruptures during this time period (Supplemental Figure S4, B–D).
To determine the role of dynein forces in NE rupture and repair, we inhibited dynein functions through RNAi of the dynein heavy chain (dhc-1) subunit of dynein and monitored the extent of depletion by observing associated phenotypes of failure of centrosome separation and pronuclear migration (Gönczy et al., 1999). We found that dynein was not responsible for the formation of Nup160::GFP gaps under lamin-N209K– or RNAi–depleted conditions (Figure 5, A and B; Supplemental Movies 6 and 7). We observed similar results with RNAi depletion of zyg-12 (Supplemental Figure S4E). We found that the transient recruitment of LEM-2 to NE rupture sites was unaffected in dynein RNAi–depleted embryos (Figure 5C) and some dynein-inhibited pronuclei also contained a condensed focus of chromatin adjacent to the Nup160::GFP gap (Figure 5A). Thus, NE rupture and repair sites in lamin-perturbed embryos are not induced by dynein forces imposed on nuclei.
FIGURE 5:
Inhibition of dynein rescues nuclear entry of GFP::α-tubulin in lamin-N209K expressing embryos. (A) (Top) Select confocal images of sperm-derived pronuclei expressing Nup160::GFP and mCherry::Histone2B from time-lapse series of lamin and dynein codepleted embryos in indicated lamin transgenic strains. Arrowheads mark sites of Nup160 gaps in each channel. (Bottom) Graphs plotting intensities of Nup160::GFP (green) and mCherry::Histone2B (red) along three-pixel-wide line scan drawn across nucleus before and after gap formation for indicated conditions above. (B) Percent of sperm-derived pronuclei with a Nup160::GFP gap and closure of the gap for the indicated conditions. (C) Select confocal images of GFP::LEM-2 (green) and mCherry::Histone2B (red) labeled nulcei for the indicated conditions. Arrowhead marks region in nuclear rim with GFP::LEM-2 accumulation. Percentage of nuclei with visible GFP::LEM-2 focus at the nuclear rim is shown. (D) Select confocal images of GFP::α-tubulin– and mCherry::Histone2B–labeled nuclei from time-lapse series of lamin and dynein codepleted embryos in indicated lamin transgenic strains. (E) Plot of percentage of nuclei from time series that undergo nuclear entry of GFP::α-tubulin (solid) or persistently contain nuclear GFP::α-tubulin (striped) from –600 to –200 s relative to PC regression. ctrl(RNAi) is RNAi against HYLS-1. n = number of embryos. (F) Percentage of nuclei with full recovery from nuclear entry of GFP::α-tubulin. n = number of GFP::α-tubulin nuclear entry occurrences. A chi-squared test was performed to test for statistical significance. Times are relative to PC regression. Scale bars, 2.5 μm. See also Supplemental Figure S4 and Supplemental Movies 6 and 7.
Movie S6.
Transient Nup160::GFP gap and mCherry::Histone2B accumulationat a NE site in a lamin-N209K mutant sperm-derived pronucleus depleted of dynein and endogenous lamin, related to Figure 5. Confocal fluorescence images of the sperm-derived pronucleus in embryos expressing Nup160::GFP (green) and mCherry::Histone2B (red). Sperm-derived pronuclei expressing the WT-lamin transgene (left) or the lamin-N209K transgene (right) in embryos co-depleted of endogenous lamin and dynein are shown. A gap in the Nup160::GFP signal appears (Left panel, arrowhead follows gap) and a condensed focus of mCherry:Histone2B appear and resolve with similar timing. Images were acquired every 20s. Playback rate is 80X real time.
Movie S7.
Nup160::GFP gaps form at the nuclear rim and chromosome condensation is abnormal in lamin RNAi-depleted embryos that are depleted of dynein, related to Figures 3 and 5. Confocal fluorescence images of embryos expressing Nup160::GFP and mCherry::Histone2B. (Left)Lamin (lmn-1) RNAi-depleted and (Right)Lamin(lmn-1) and dynein (dhc-1) co-depleted embryos are shown. Sequences from sperm-derived pronuclear positioning to pseudocleavage regression are shown. Gaps in the Nup160::GFP signal appear (Arrows follow gaps in both panels) and mCherry:Histone2B appears highly condensed and localized to the nuclear rim in both conditions. The sperm-derived pronucleus migrates towards the embryo center in lamin RNAi-depleted embryos (left) but not in lamin and dynein co-depleted embryos (right). Images were acquired every 20s. Playback rate is 80X real time.
We next determined the extent of nucleo–cytoplasmic mixing in the absence of dynein by quantifying nuclear entry and recovery of GFP::α-tubulin in dynein-depleted embryos codepleted of lamin alone or in the presence of the lamin transgenes. In lamin RNAi–depleted embryos, GFP::α-tubulin accessed the nuclear interior even in the absence of dynein (Figure 5, D and E). However, dynein inhibition significantly reduced the percentage of embryos with nuclear entry of GFP::α-tubulin in lamin-N209K–expressing embryos (53% of ctrl;lmn-1(RNAi) vs. 29% of dhc-1;lmn-1(RNAi), chi-squared test, p = 0.031; solid bars in Figure 5E). The fact that nuclear entry of GFP::α-tubulin was partially rescued by reduction of dynein forces in nuclei containing lamin-N209K, but not in lamin-deficient nuclei, indicates a requirement for lamin in restricting nucleo–cytoplasmic mixing that is counteracted by dynein forces. Consistent with this, in the absence of dynein forces, a significantly greater percentage of the GFP::α-tubulin nuclear entry occurrences fully recovered in lamin-N209K–expressing embryos than in those deficient for lamin (87% in dhc-1(RNAi); lmn-1(RNAi) lamin-N209K vs. 46% in dhc-1(RNAi); lmn-1(RNAi) nuclei, chi-squared test, p = 0.025, Figure 5F). These data, together with the fact that we observe similar percentages of GFP::LEM-2 foci and Nup160::GFP gaps, indicate that in the absence of dynein forces a weakened lamin network maintains the permeability barrier of damaged nuclei.
We conclude that dynein does not induce ruptures when lamins are weak. Instead, dynein-generated tension counteracts the ability of the weakened lamin-N209K network to stabilize damaged nuclei, which in turn exacerbates the extent of loss of the nuclear permeability barrier. Lamin-deficient nuclei are unstable even in the absence of dynein forces, indicating that other functions for lamin-mediated stability are also required to restrict the nuclear permeability barrier in damaged nuclei.
Lamin resists dynein forces during pronuclear migration to prevent chromosome scattering prior to metaphase
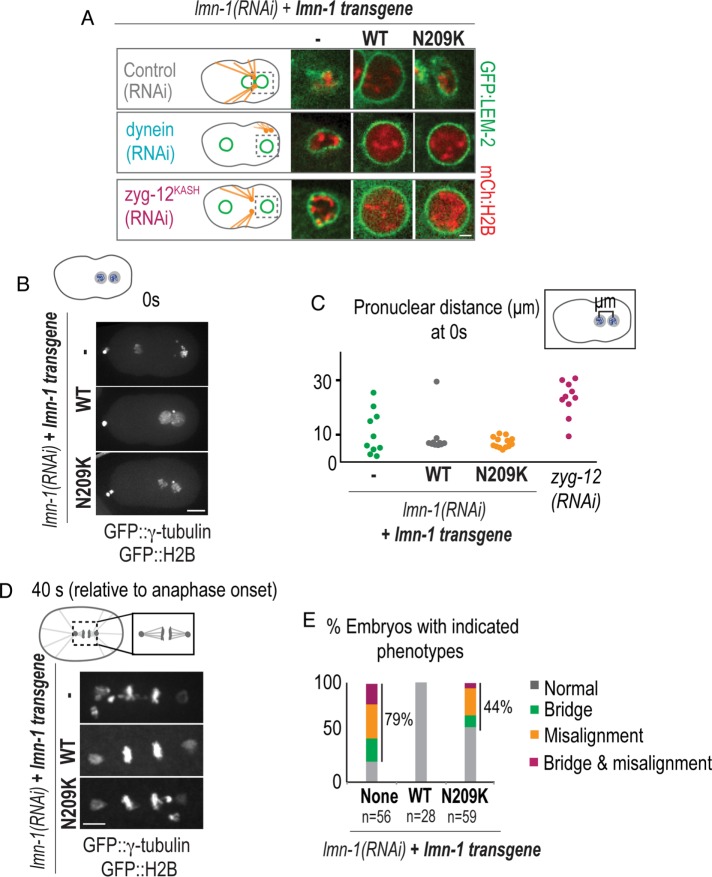
We predicted that the absence of pronuclear migration due to dynein depletion would allow us to track damaged NEs with longer recovery periods in embryos codepleted of lamin. In lamin-N209K embryos, RNAi depletion of dynein or zyg-12 inhibits pronuclear migration and rescues pronuclear collapse, which occurs at the onset of PC regression (Figure 6A). To confirm that rescue of pronuclear collapse was not because of a lack of penetrant depletion of endogenous lamin, we RNAi-depleted zyg-12 or dynein in the lmn-1Δ genetic background expressing the lamin-N209K transgene (Supplemental Figure S5A). Under these conditions, inhibition of dynein or zyg-12 rescued premature pronuclear collapse and lamin-N209K nuclei collapsed at a time similar to NEBD in WT-lamin embryos (Supplemental Figure S5A; see Figure 1D), indicating that dynein-generated forces are the cause of pronuclear collapse that occurs upon pronuclear migration in lamin-N209K embryos. In contrast, inhibition of dynein did not rescue nuclear deformation in lamin RNAi–depleted embryos, and these nuclei retained nuclear GFP::α-tubulin (Figure 6A; Figure 5, D and E). Thus, upon pronuclear migration, lamin prevents irreversible loss of the nuclear permeability barrier and NE collapse.
FIGURE 6:
Nuclear collapse leads to chromosome scattering and mitotic defects. (A) Select images of GFP::LEM-2– and mCherry::Histone2B–labeled sperm-derived pronuclei at the time of PC regression for the indicated conditions. Scale bar, 2.5 μm. (B) Maximum projection of confocal image stacks of GFP::γ-tubulin and GFP::Histone2B from lamin RNAi–depleted C. elegans embryos expressing indicated transgenes. Scale bar, 10 μm. (C) Plot of distance in micrometers between oocyte- and sperm-derived histone masses at PC regression from indicated conditions. Time in B and C is relative to PC regression. (D) Maximum projection of confocal image stacks of spindle region labeled with GFP::γ-tubulin and GFP::Histone2B 40 s after anaphase onset (defined as frame before separation of chromosome masses) from lamin RNAi–depleted embryos expressing indicated transgenes. Scale bar, 5 μm. (E) Plot of percentage of embryos at anaphase with indicated phenotypes for the indicated conditions. n = number of embryos scored. See also Supplemental Figure S5.
We noticed that despite pronuclear deformation and collapse during pronuclear migration in lamin-N209K embryos, the majority of chromosomes from each pronucleus maintain close proximity (Figure 6, B and C). The distance between chromosome masses was more severely affected in lamin RNAi–depleted embryos, which is likely an outcome of defects in nucleus–centrosome attachment (Figure 6, B and C; Meyerzon et al., 2009) as well as premature loss of NE integrity. Premature NE collapse in both conditions led to chromosomes that did not incorporate into the metaphase plate (Figure 6, D and E). In addition, some nuclei had chromosome bridges (Figure 6, D and E), a defect that has been described in prior work for later-stage embryos RNAi-depleted for lamin (Liu et al., 2000). The percentage of chromosome segregation defects was similar to the percentage of embryonic lethality in lamin-N209K–expressing embryos (44%, Figure 6E, and 47%, Figure 1C). These data suggest that in the early C. elegans embryo, an essential function of the lamin network is to structurally protect NEs during pronuclear migration.
To determine whether there is a relationship between transient NE ruptures that occur prior to pronuclear migration and chromosome missegregation in lamin-N209K, we monitored embryos through the first division cycle and scored transient nuclear GFP::α-tubulin entry events and chromosome segregation defects (Supplemental Figure S5, B and C). Surprisingly, transient nuclear entry of GFP::α-tubulin did not correlate with chromosome segregation defects (Supplemental Figure S5, B and C), and the percentage of combined phenotypes (transient NE ruptures only, chromosome segregation defects only, or both), was much greater than the percentage of embryonic lethality (77%, Figure S5C, vs. 47%, Figure 1C). We conclude that embryos survive to hatching despite transient losses of nuclear compartmentalization during early embryogenesis. These data further suggest that lethal defects in embryogenesis caused by expression of lamin-N209K are specific to the first division of the embryo, in which pronuclei must migrate across the length of the embryo to mix their haploid genomes.
DISCUSSION
This work provides the first mechanistic study of NE rupture and repair in an organismal context. By analyzing a partial-loss-of function lamin mutation in the first cell division of the C. elegans zygote, we demonstrate that lamin prevents persistent ruptures and NE collapse during dynein-dependent pronuclear positioning. Dynein-induced tension counteracts the ability of a weakened lamina to stabilize ruptured nuclei, leading to a greater extent of nucleo–cytoplasmic mixing (Figure 7). We used the relatively rapid recovery from nuclear ruptures (∼100–200 s) that occur in the C. elegans zygote to delineate the dynamics of NE reorganization at repair sites and to correlate these events to cell cycle progression. We further found that transient NE ruptures do not contribute to embryonic lethality, but rather chromosome segregation defects correlate with chromosome scattering caused by premature NE collapse. Thus, NE repair protects against catastrophic consequences that stem from loss of nuclear compartmentalization during early embryogenesis.
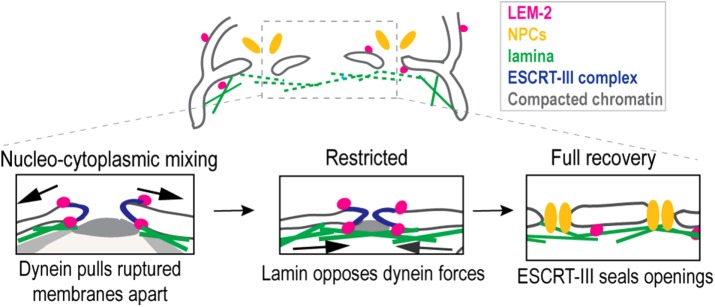
FIGURE 7:
Model for nuclear envelope repair in C. elegans. Nuclear envelope ruptures at weakened sites of the nuclear lamina are cleared of nuclear pore complexes (NPCs; orange). LEM-2 (magenta) and ESCRT-III (dark blue) are recruited to ruptured membranes. Dynein forces pull ruptured nuclear membranes apart, causing prolonged nucleo–cytoplasmic mixing. Lamin stabilizes NE rupture sites to allow narrowing of opposing membranes for fission by ESCRT-III machinery, which restricts further nucleo–cytoplasmic mixing. Full recovery of repaired nuclei reestablishes the nuclear permeability barrier and allows reinsertion of NPCs.
Dynein forces counteract lamin’s ability to limit loss of the nuclear permeability barrier upon rupture
We propose a model in which dynein forces imposed on the NE oppose lamin’s ability to stabilize ruptures at weakened sites of the NE and thus the response of the NE to damage (Figure 7). In our model, dynein-generated forces pull ruptured nuclear membranes apart, while lamin assembly at and adjacent to NE rupture sites counteracts these forces and thereby maintains a narrow opening between ruptured membranes for efficient repair by the ESCRT-III machinery.
The idea that lamin restricts the size of NE holes that are otherwise enlarged by dynein pulling forces is supported by the fact that lamin-N209K expression causes NE damage in the absence of dynein forces, as indicated by the unchanged frequency of Nup160 gaps and LEM-2 foci at the nuclear rim, yet nucleo-cytoplasmic mixing is rescued. These results suggest that damage to the NE in lamin-N209K embryos is more readily repaired without dynein forces, which in turn prevents nuclear accumulation of cytoplasmic proteins. In the absence of lamin, Nup160 gaps and nucleo–cytoplasmic mixing are more likely to persist, supporting the idea that lamin is needed to restrict NE hole size to allow efficient repair. Interestingly, ruptures that recover fully under lamin-N209K and lamin RNAi–depleted conditions do so with similar kinetics, suggesting that a hole in the NE below a threshold size can efficiently recover independently of lamin. The relatively longer recovery times of laser-induced micrometer-scale punctures in wild-type nuclei suggest that hole sizes that are sufficiently large take longer to recover. The duration of ruptures might also be affected by lamin’s role in the stability of recently sealed NE rupture sites—membranes that transiently reseal may more frequently reopen without lamin, leading to longer recovery times. Future work that directly determines the size of NE holes under these different conditions is required to distinguish between these possibilities.
In the absence of lamin, NE ruptures are unaffected by dynein forces. Thus, other functions for lamin may be necessary to allow for robust NE repair. Our data exclude the possibility that lamin is required for the recruitment of repair machinery, which is consistent with work in migrating cancer cells (Denais et al., 2016). Lamin may function to recruit other unknown factors for nuclear repair or may stabilize nuclei against other cytoskeletal forces that were not tested in this study. We also show that lamin-deficient nuclei are more likely to rerupture, suggesting that lamin functions to stabilize nuclei against multiple ruptures, which may occur at recently sealed sites, since we rarely saw multiple rupture sites in the same nucleus (unpublished data). Interestingly, ESCRT-III depletion in mammalian cells prolongs but does not prevent recovery from nucleo–cytoplasmic mixing (Denais et al., 2016; Raab et al., 2016; Robijns et al., 2016), suggesting that other ESCRT-III independent mechanisms may exist to restrict the nuclear permeability barrier upon rupture.
We show that lamin-N209K localizes to repair sites that are also marked by LEM-2 accumulation, suggesting that lamin may function in stabilizing rupture sites directly. Consistent with a role for lamin at rupture sties, lamin A, but not lamin B1, accumulates at NE rupture sites directly following ruptures caused by constricted cell migration (Denais et al., 2016). The single C. elegans lamin B protein may have conserved functions at nuclear repair sites to restrict loss of the nuclear permeability barrier upon damage.
The ability of dynein to tear nuclear membranes has been reported during NEBD when dynein forces pull on permeabilized NEs to facilitate the removal of nuclear membranes from chromatin (Beaudouin et al., 2002; Salina et al., 2002; Kutay and Hetzer, 2008). We demonstrate that the tug of war between dynein-generated tension and lamin-mediated stability is relevant in ruptured nuclei in interphase. Dynein tension imposed on interphase nuclei that facilitate centrosome attachment and nuclear positioning are relatively weak (Kimura and Onami, 2005; Oegema and Hyman, 2006), allowing the weakened lamin-N209K network to resist catastrophic tearing of damaged nuclear membranes and allow NE repair. However, when dynein forces increase upon pronuclear migration, lamin-N209K is unable to prevent catastrophic collapse. In addition to weaker dynein forces imposed on nuclei in interphase, lamin assembly is likely favored over disassembly. The function for lamin-mediated nuclear stability in preventing persistent ruptures by resisting dynein forces is likely important across different cell types that utilize dynein-dependent mechanisms to position nuclei (Zhang et al., 2009; Coffinier et al., 2011; Bone and Starr, 2016).
Mechanisms that induce NE rupture in the early C. elegans embryo
In cells in culture, constriction during cell migration or actin-derived compression forces cause nuclear herniations at regions devoid of lamins, which increases the surface tension causing transient ruptures at these sites (Harada et al., 2014; Denais et al., 2016; Hatch and Hetzer, 2016; Raab et al., 2016; Thiam et al., 2016; Irianto et al., 2017). Not all transient rupture sites correlate with chromatin herniations, suggesting that herniations are not a prerequisite for rupture (Hatch and Hetzer, 2016). We did not detect chromatin herniations upon NE rupture in C. elegans pronuclei. Instead, we found that condensed chromatin puncta appear at rupture sites—an observation also made in ruptured nuclei and nuclei under micropipette aspiration in mammalian cells (Robijns et al., 2016; Irianto et al., 2016). The delayed appearance of condensed chromatin relative to the appearance of other indicators of rupture suggests that chromatin reorganization is a consequence of rupture in C. elegans embryos.
Our data show that dynein forces during pronuclear centering do not induce NE ruptures, suggesting that another type of force is involved. Actomyosin contractile forces are not directly coupled to the NE in early C. elegans embryos (Cowan and Hyman, 2004; Munro et al., 2004; Oegema and Hyman, 2006). However, we cannot exclude the possibility that indirect actin-generated forces, such as cytoplasmic flow generated by asymmetric actomyosin contractility (Hird and White, 1993), induce NE ruptures in this system. Interestingly, micronuclei with reduced lamin undergo ruptures independent of compression forces, supporting the idea that in specific contexts disorganization of the nuclear lamina is sufficient to induce NE ruptures (Hatch et al., 2013). Further studies are needed to determine whether lamin inhibition on its own or in combination with internal or external pressure imposed on the NE causes transient ruptures.
Local NE and chromatin reorganization during NE rupture and recovery
We show that Nup160 gaps and accumulation of LEM-2 at NE ruptures sites colocalize with ESCRT-III repair machinery. We suggest that the region devoid of Nup160::GFP, which is ∼800 nm to 3 μm in the xy direction when the sperm-derived pronucleus is ∼4–7 μm in diameter, indicates the site but not the size of NE ruptures. These regions are likely cleared of nucleoporins to support restoration of NEs at and adjacent to damaged regions. The accumulation of LEM-2 at rupture sites also suggests local membrane remodeling. In addition to the reorganization of NE proteins upon rupture, it is compelling that compacted chromatin foci form directly adjacent to peripheral rupture sites upon entry into prophase. Chromatin compaction may be accelerated in ruptured nuclei due to equilibration of mitotic-activated factors. Local reorganization of NE proteins induced by rupture may also alter the distribution of chromatin. Interestingly, depletion of nucleoporins Nup205/93, which are needed to set the exclusion limit of the nuclear permeability barrier, also causes peripheral chromatin in C. elegans embryos (Galy et al., 2003). Thus, reorganization of chromatin may generally result from loss of the nuclear permeability barrier, independent of ruptures per se, in early embryos. Determining the role of chromatin reorganization and the proteins that function in local membrane remodeling at NE rupture sites is important in understanding the mechanism of NE repair.
The relationship between transient NE ruptures and the cell cycle
It has been suggested that NE rupture and repair mechanisms are restricted to interphase (Hatch and Hetzer, 2014). We found that repair mechanisms active during early prophase in the C. elegans zygote ensure maintenance of NE integrity during pronuclear positioning. Tissue culture cells couple mitotic entry to NEBD, whereas the C. elegans zygote undergoes an elongated prophase (∼450 s) upon entry into mitosis, which coincides with dynein-dependent pronuclear positioning (Lee et al., 2000; Oegema and Hyman, 2006). Nuclear envelope repair mechanisms may be inactivated by factors that trigger NEBD-mediated NE permeabilization, which coincides with diffusion of INM proteins into the ER. One possibility is that diffusion of LEM-2 into the ER upon global NE permeabilization (Ungricht and Kutay, 2017) prevents NE repair during this time by removing the nuclear specific adapter for the ESCRT-III complex. A key future direction will be determining the factors that signal repair upon NE rupture prior to mitotic entry.
MATERIALS AND METHODS
C. elegans strains
The C. elegans strains used in this paper are listed in Supplemental Table S1. Strains were maintained at 20°C on nematode growth medium (NGM) plates seeded with OP50 Escherichia coli. The engineered lmn-1 gene containing a reencoded region in exon 4 was cloned into pCFJ151 for integration into the Mos1 site on Chromosome II (MosSCI; Frøkjær-Jensen et al., 2008). Young adult hermaphrodites were injected with the transgene-containing plasmid (10 ng/µl), a plasmid encoding the Mos1 transposase (Pglh-2::transposase, pJL43.1, 10 ng/µl), and three plasmids encoding fluorescent markers: Pmyo-2::mCherry (pCFJ90, 1.4 ng/µl), Pmyo-3::mCherry (pCFJ104, 2.9 ng/µl), and Prab-3::mCherry (pGH8, 5.7 ng/µl). The fluorescent markers were used to determine integration. After injection, worms were singled out onto OP50 seeded plates and moving progeny lacking fluorescent markers were screened for integration by amplification of regions neighboring the integration site with PCR. Transgenic strains were crossed into marker strains or the lmn-1Δ strain using standard genetic procedures.
RNAi
Double-stranded RNAs (dsRNAs) were made with the oligonucleotides listed in Supplemental Table S2 to amplify regions from N2 genomic DNA by PCR. T3 and T7 transcription reactions (MEGAscript, Life Technologies) were used with purified PCRs as templates. T3 and T7 reactions were mixed and purified using a phenol-chloroform purification for dsRNA against lmn-1 or an RNAeasy Mini Kit (Qiagen) for dsRNA against zyg-12, dhc-1, or hyls-1. Purified RNA products from T3 and T7 transcription reactions were resuspended to a final concentration of 1X soaking buffer (32.7 mM Na2HPO4, 16.5 mM KH2PO4, 6.3 mM NaCl, 14.2 mM NH4Cl) and the mixture was annealed by incubation at 68°C for 10 min followed by 37°C for 30 min. dsRNA was stored in –80°C in 2-μl aliquots. A fresh aliquot was used for each new experiment.
For single depletion of endogenous lmn-1, L4 stage hermaphrodites were injected with dsRNA (1 mg/ml) and incubated at 20°C for 24–28 h postinjection prior to imaging, brood size assays, immunofluorescence, or worm preparation for immunoblotting. For codepletion of endogenous lmn-1 and an additional target, dsRNA conditions were optimized after each generation of dsRNA-targeting lmn-1. The dsRNA-targeting lmn-1 and dsRNA against the additional target were mixed in a ratio containing a total of 1.1–1.5 mg/ml in a ratio with a two- to eightfold increase in dsRNA targeting lmn-1. As a control for double depletion, dsRNAs were mixed with hyls-1 dsRNA at the same ratio and only compared with depletions under the same conditions.
Brood size and lethality assays
For lethality assays, L4 worms were injected with dsRNA, singled 24 h postinjection, and removed 48 h postinjection. Hatched and unhatched embryos were counted on each plate 24 h after removal of adult hermaphrodites. Brood sizes for lmn-1 transgenic worms in the lmn-1Δ allele, lmn-1(tm1502), were performed similarly but without dsRNA injection.
Immunoblotting
Affinity-purified antibodies against LMN-1 amino acids 464–562 were generated by amplifying the region from an N2 cDNA library, cloning into pGEX6P-1, and purifying GST tagged protein from bacteria. The antigen was outsourced for injection into rabbits (Covance) and affinity-purified using a MBP fusion of the same antigen using standard methods. For immunoblotting, 70 gravid adult worms were washed with M9 (Na2HPO4, 4.2 mM; KH2PO4, 2.2 mM; MgSO4, 1 mM; and NaCL, 8.6 mM in ddH2O) and 0.1% Triton and brought to a volume of 60 μl. A quantity of 20 μl of 4× sample buffer was added to the sample. Worms were then sonicated for 10 min at 70°C, incubated at 95°C for 5 min, and sonicated for 10 min more at 70°C. The samples were subsequently frozen at –20°C. A quantity of 20 μl (∼17.5 worms) of indicated samples was added to each lane of the protein gel. To load an equal amount of protein from lmn-1Δ worms that are sterile and lack gonads and embryos, 1.5× more adult worm extract was loaded. The samples were separated by SDS–PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane for immunoblotting. Primary antibodies were diluted to 1 μg/ml for rabbit-anti-LMN-1 and mouse-anti-α-tubulin (EMD Millipore). Secondary antibodies were diluted 1:5000 for horseradish peroxidase (HRP)-conjugated goat-anti-rabbit and 1:7000 for HRP-conjugated goat-anti-mouse.
Fractionation assay
Worms enriched for gravid adults were removed from plates using M9 + 0.1% Triton. Worms were then pelleted by centrifugation at 1000 × g for 3 min, washed twice with M9, and pelleted, and the pellet was flash frozen in liquid nitrogen. Frozen samples were crushed with a pestle and resuspended in 100 μl of lysis buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid, 1% NP40, 1 mM dithiothreitol, and ½ tablet of protease inhibitor per 5 ml of lysis buffer). A 20-μl sample was saved, and then the lysate was spun at 1000 rpm on a tabletop microfuge for 2 min at 4°C to pellet the debris. Samples from the supernatant and pellet were saved, and the remaining supernatant was spun at 14,000 rpm on a tabletop microfuge for 10 min at 4°C to fractionate soluble and insoluble extract. Pellets were prepared for subsequent analysis by resuspension in 100 μl of lysis buffer. Equal volumes of samples were loaded on a SDS–PAGE gel and immunoblotted as above.
Live microscopy
Early C. elegans embryos were dissected from adult hermaphrodites in M9, mounted on a 2% agarose slide, and imaged at room temperature (∼21°C). Imaging was performed on an inverted Nikon (Melville, NY) Ti microscope with a 60 × 1.4 NA plan Apo objective lens on an Andor Revolution XD Confocal system (Andor Technology) equipped with a confocal scanner unit (CSU-XI, Yokogawa) with solid state 150-mW 488-nm and 100-mW 560-nm lasers and a high-resolution ORCA R-3 Digital CCD Camera (Hamamatsu). Early embryos were imaged at a temporal resolution of 20 s and five z-slices were acquired with 2-μm steps.
For laser ablation experiments, images were collected on an inverted Nikon TE2000 equipped with 488- and 561-nm lasers, a Hamamatsu EM CCD camera, and an 80-MHz Ti:sapphire femtosecond pulsed laser at center wavelength 800 nm (Mai-Tai, Spectra-Physics). For ablation, a 16-kHz femtosecond pulse train with pulse energy ∼6 nJ was used, which was produced by selecting pulses from the above Ti:sapphire pulsed laser using a pulse picker (Eclipse Pulse Picker; KMLabs). Ablation of the pronuclear envelope was performed by moving the sample on a piezo stage (P-545 PInano XYZ; Physik Instrumente) in three dimensions controlled by a home-developed LabVIEW program (LabVIEW; National Instruments) at a speed of 200 µm/s. The cut volumes are 4 × 1 × 0.5 µm and 4 × 2 × 0.5 µm and were combined in an analysis shown in Figure 2G. Images were acquired every 3.6 s, and one z section was acquired at each time point.
Immunostaining
Adult worms (15–20 per slide) were picked into ddH2O (4 µl) on 0.1% poly-l-lysine–coated slides. Worms were squashed with a coverslip (18 × 18 mm) to remove embryos, and slides were freeze-cracked on a dry ice bath. After the freeze crack, slides were fixed for 20 min in 100% methanol at –20°C. Slides were next washed twice for 10 min in 1× PBS + 0.2% Tween-20 (PBST) at room temperature, after which they were blocked in 1% bovine serum albumin in PBST (50 µl per slide) for 1 h at room temperature in a humid chamber. The slides were then incubated in primary antibody in PBST overnight at 4°C in a humid chamber (45 µl per slide; rabbit α-LMN1, 1 µg/ml; mouse α-tubulin [clone DM1A; EMD Millipore], 0.5 µg/ml; 2.5 µg/ml; goat α-GFP [Hyman lab], 1 µg/ml; rabbit α-VPS-32 [Audhya lab], 0.5 µg/ml). The GFP::LEM-2 fusion protein retained signal after fixation precluding a need for antibody staining (Figure 4, F and G). Following primary antibody incubation, slides were washed twice for 10 min in PBST (50 µl per slide) and incubated for 1 h in the dark with secondary antibody in PBST (45 µl per slide) at room temperature (anti-rabbit Cy3/Rhodamine, 1:200; anti-mouse Cy5 1:200; anti-goat FITC 1:200; anti-rabbit Cy5 1:200 [Jackson Immunoresearch]). Slides were then washed with PBST twice for 10 min (50 µl/slide) in the dark and mounted with Molecular Probes ProLong Gold Antifade Reagent with DAPI. After mounting, slides were left to dry at room temperature and then kept at –20°C for storage. Images shown in Figures 1F and 4, A and D, were acquired on a Zeiss LSM 510 meta confocal microscope. Immunofluorescence images shown in Figure 4F were acquired on a Nikon (Melville, NY) Ti microscope with a 60 × 1.4 NA plan Apo objective lens on an Andor Revolution XD Confocal system (Andor Technology) equipped with a confocal scanner unit (CSU-XI, Yokogawa) with solid state 150-mW 488-nm and 100-mW 560-nm lasers and a high-resolution ORCA R-3 Digital CCD Camera (Hamamatsu). Image analyses shown in Supplemental Figure S1E and Figure 4, B and E, were measured from epifluorescence images acquired on an inverted Nikon microscope with a 60 × 1.4 NA plan Apo objective equipped with a high- resolution ORCA R-3 Digital CCD camera (Hamamatsu).
Image analysis
All fluorescence intensity measurements were quantified using ImageJ (Fiji) software (National Institutes of Health). Images were analyzed in a systematic manner to avoid bias. Representative images are shown to reflect data sets. All early embryos were included in image analysis for immunofluorescence and all live-cell one-stage embryos that were included in the time frame specified in the analysis were included.
For fluorescence intensity measurements of nuclear α-tubulin, the average value of a 2.3 × 2.3 μm region of interest placed centrally in the nucleus was determined and subtracted from a background region outside the embryo. For initial exclusion and nuclear entry, a threshold value was determined by the average intensity value in control nuclei under the same condition. Fluorescence intensities following nuclear entry were normalized to value in frame prior to nuclear entry (t0). Full recovery was defined as the time point when values reached levels at t0.
For line scans, a three-pixel-wide, 10-μm-long line was centered on the nucleus and each value was plotted along the line after subtraction of cytoplasmic background (the average of a three-pixel-wide, 10-μm-long line in the cytoplasm in the same embryo). For linescans across nuclei immunostained for lamin (Supplemental Figure S1E), gray values were measured along two 5 pixel × 25 pixel (5 µm) lines, drawn from the inside to the outside of the nucleus and centered on the nuclear rim, and the cytosolic background (the average of the outer three values of each line) was subtracted from each value along the line. The corrected values along each line were averaged for an individual pronucleus to gain an average line of gray values for that pronucleus. Fold change in lamin levels at nuclear repair sites (Figure 4F) was determined by calculating the average maximum values of a three-pixel-wide line scan drawn across the nuclear rim at the repair site (defined by LEM-2 accumulation) and dividing by maximum values of a three-pixel-wide line scan drawn across the nuclear rim at an adjacent site.
Nup160::GFP gaps, GFP::LEM-2 foci, and mCherry::H2B puncta adjacent to Nup160 gaps were scored in each nucleus in early stage embryos (for fixed analysis up to the four- to eight-cell stage; only the one-cell stage for live analysis). Localization of VPS-32 was scored subsequent to identification of a Nup160 gap or LEM-2 foci.
RNA isolation and qPCR
Worm collection and RNA isolation.
Unsynchronized adult worms (5–10 plates per strain) were washed off plates in 1 ml of 1× M9 Buffer into a 15-ml conical. Worms were washed three times in 5 ml of M9 Buffer, and the buffer was subsequently removed from the pellet. Worm pellets were then frozen at –80°C. To isolate RNA, 50–100 µl of worm pellet was ground with a micro pestle and added to 1 ml of Trizol. Total RNA was then precipitated by Trizol–chloroform precipitation. To start the precipitation, the Trizol–worm solution was vortexed for 15–30 min at room temperature, and 200 µl of chloroform was added. The solution was next vortexed for 15 s and incubated for 5 min at room temperature. After incubation, the solution was spun for 15 min at 4°C (12,000 rpm). The upper aqueous layer was transferred to a clean tube, 500 µl of isopropanol was added, and the tube was inverted six times. The solution was incubated at room temperature for 10 min to precipitate the RNA and next spun for 10 min at 4°C (12,000 rpm). The supernatant was removed and washed in 1 ml of 75% ethanol by inverting once and vortexing for 10 s. The ethanol-soaked pellet was spun for 5 min at 4°C (7500 rpm), the supernatant was removed, and the was pellet air-dried at room temperature for 5–10 min. The pellet was resuspended in 87 µl of RNase-free water. The RNA was incubated at 37°C for 15 min and either kept at –80°C or used immediately for RNA cleanup and reverse transcription.
RNA cleanup and reverse transcription.
After isolation, the RNA solution was treated with Ambion Turbo DNase to remove DNA. Briefly, the isolated RNA was mixed with 10 µl NEB Buffer 4, 1 µl of 50 µM CaCl2, and 2 µl Ambion Turbo DNase and incubated at room temperature for 15 min. The RNA/DNase reaction was cleaned with a Qiagen RNeasy Kit, eluted into 20 µl of RNase free water, and kept at –80°C or used immediately for reverse transcription. Total RNA (500 ng) was then reverse transcribed using an Invitrogen SuperScript II Reverse Transcriptase kit. The cDNA was stored at –20°C or used immediately for quantitative PCR (qPCR).
qPCR and analysis.
For qPCR, cDNA was diluted fivefold and prepared for amplification using a BioRad SYBR Green Supermix Kit with the primers for lmn-1 listed in Supplemental Table S2. Reactions were also prepared for hxk-2 with primers from Kudron et al. (2013) and listed in Supplemental Table S2. For each biological sample, two technical replicates of both the lmn-1 and hxk-2 reactions were loaded into a 384-well plate for amplification. The reactions were amplified and Ct values measured using a BioRad CFX 384 qPCR machine, using an annealing temperature of 55°C. After amplification, Ct values were normalized using primers specific to hxk-2, and the fold change relative to the control (N2) was calculated using the 2–(ΔΔCt) method (BioRad).
Statistical analysis
All measurements with statistical analysis were taken from multiple samples and independent biological repeats. The full data set is represented. Means and error bars are indicated in the figure legends and are measured from biological replicates. Groups were selected based on experimental condition. Distributions that reach a normal distribution (using the Shapiro–Wilk test) were considered sufficiently large. For those data sets that do not have Gaussian distributions, appropriate statistical tests were applied. A nonparametric two-tailed Mann–Whitney test or a chi-squared test was applied and is specified in figure legends. Statistical significance was defined as when p values were less than 0.05.
Supplementary Material
Acknowledgments
We thank Patrick Lusk, Megan King, Topher Carroll, Joshua Gendron, and Julie Canman for insightful discussions, experimental suggestions, and helpful critiques and Arshad Desai, Karen Oegema, Julie Canman, and Anjon Audhya for strains or antibodies used in experiments. We thank our sources of support from Yale University and the Ludwig Institute for Cancer Research (R.B.). We thank the National Institutes of Health for support of L.P. (T32 GM007499) and M.M. (T32GM007223-S1). D.N. was supported in part by the National Institutes of Health (1R21HD080057-01A1) and the National Science Foundation (DMR-0820484).
Abbreviations used:
- INM
inner nuclear membrane
- NE
nuclear envelope
- NEBD
nuclear envelope breakdown
- NPC
nuclear pore complex
- RNAi
RNA interference
- PC
pseudocleavage
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-06-0374) on January 31, 2018.
REFERENCES
- Anderson DJ, Vargas JD, Hsiao JP, Hetzer MW. (2009). Recruitment of functionally distinct membrane proteins to chromatin mediates nuclear envelope formation in vivo. J Cell Biol , 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW. (2007). Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat Cell Biol , 1160–1166. [DOI] [PubMed] [Google Scholar]
- Bahmanyar S, Biggs R, Schuh AL, Desai A, Müller-Reichert T, Audhya A, Dixon JE, Oegema K. (2014). Spatial control of phospholipid flux restricts endoplasmic reticulum sheet formation to allow nuclear envelope breakdown. Genes Dev , 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank EM, Ben-Harush K, Wiesel-Motiuk N, Barkan R, Feinstein N, Lotan O, Medalia O, Gruenbaum Y. (2011). A laminopathic mutation disrupting lamin filament assembly causes disease-like phenotypes in Caenorhabditis elegans. Mol Biol Cell , 2716–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank EM, Ben-Harush K, Feinstein N, Medalia O, Gruenbaum Y. (2012). Structural and physiological phenotypes of disease-linked lamin mutations in C. elegans. J Struct Biol , 106–112. [DOI] [PubMed] [Google Scholar]
- Beaudouin J, Gerlich D, Daigle N, Elis R, Ellenberg J. (2002). Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell , 83–96. [DOI] [PubMed] [Google Scholar]
- Bone CR, Starr DA. (2016). Nuclear migration events throughout development. J Cell Sci , 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broers JL, Peeters EA, Kuipers HJ, Endert J, Bouten CV, Oomens CW, Baaijens FP, Ramaekers RC. (2004). Decreased mechanical stiffness in LMNA-/- cells is caused by defective nucleo–cytoskeletal integrity: implications for the development of laminopathies. Hum Mol Genet , 2567–2580. [DOI] [PubMed] [Google Scholar]
- Burke B, Ellenberg J. (2002). Remodelling the walls of the nucleus. Nat Rev Mol Cell Biol , 487–497. [DOI] [PubMed] [Google Scholar]
- Burke B, Stewart CL. (2006). The laminopathies: the functional architecture of the nucleus and its contribution to disease. Annu Rev Genomics Hum Genet , 369–405. [DOI] [PubMed] [Google Scholar]
- Burke B, Stewart CL. (2013). The nuclear lamins: flexibility in function. Nat Rev Mol Cell Biol , 13–24. [DOI] [PubMed] [Google Scholar]
- Chiaruttini N, Redondo-Morata L, Colom A, Humbert F, Lenz M, Scheuring S, Roux A. (2015). Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell , 866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffinier C, Jung HJ, Nobumori C, Chang S, Tu Y, Barnes RH, 2nd, Yoshinaga Y, de Jong PJ, Vergnes L, Reue K, et al. (2011). Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol Biol Cell , 4683–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Behzad AR, Carroll JB, Panté N. (2006). Parvoviral nuclear import: bypassing the host nuclear-transport machinery. J Gen Virol , 3209–3213. [DOI] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA. (2004). Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu Rev Cell Dev Biol , 427–453. [DOI] [PubMed] [Google Scholar]
- Dammermann A, Maddox PS, Desai A, Oegema K. (2008). SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the gamma-tubulin-mediated addition of centriolar microtubules. J Cell Biol , 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson PM, Lammerding J. (2014). Broken nuclei—lamins, nuclear mechanics, and disease. Trends Cell Biol , 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies T, Jordan SN, Chand V, Sees JA, Laband K, Carvalho AX, Shirasu-Hiza M, Kovar DR, Dumont J, Canman JC. (2014). High-resolution temporal analysis reveals a functional timeline for the molecular regulation of cytokinesis. Dev Cell , 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. (2010). Nuclear lamins. Cold Spring Harb Perspect Biol , a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. (2008). Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev , 832–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J. (2016). Nuclear envelope rupture and repair during cancer cell migration. Science , 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Noronha CM, Sherman MP, Lin HW, Cavrois MV, Moir RD, Goldman RD, Greene WC. (2001). Dynamic disruptions in nuclear envelope architecture and integrity induced by HIV-1 Vpr. Science , 1105–1108. [DOI] [PubMed] [Google Scholar]
- De Vos WH, Houben F, Kamps M, Malhas A, Verheyen F, Cox J, Manders EM, Verstraeten VL, van Steensel MA, Marcelis CL, et al. (2011). Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum Mol Genet , 4175–4186. [DOI] [PubMed] [Google Scholar]
- Edgar LG, McGhee JD. (1988). DNA synthesis and the control of embryonic gene expression in C. elegans. Cell , 589–599. [DOI] [PubMed] [Google Scholar]
- Fatkin D, MacRae C, Sasaki T, Wolff MR, Porcu M, Frenneaux M, Atherton J, Vidaillet HJ, Jr, Spudich S, De Girolamin U, et al. (1999). Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med , 1715–1724. [DOI] [PubMed] [Google Scholar]
- Frøkjær-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. (2008). Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet , 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Mattaj IW, Askjaer P. (2003). Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell , 5104–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P, Pichler S, Kirkham M, Hyman AA. (1999). Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol , 135–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, LaJoie D, Chen OS, von Appen A, Ladinsky MS, Redd MJ, Nikolova L, Bjorkman PJ, Sundquist WI, Ullman KS, et al. (2017). LEM2 recruits CHMP7 for ESCRT-mediated nuclear envelope closure in fission yeast and human cells. Proc Natl Acad Sci USA E2166–E2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haithcock E, Dayani Y, Neufeld E, Zahand AJ, Feinstein N, Mattout A, Gruenbaum Y, Liu J. (2005). Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc Natl Acad Sci USA , 16690–16695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Swift J, Irianto J, Shin JW, Spinler KR, Athirasala A, Diegmiller R, Dingal PC, Ivanoska IL, Discher DE. (2014). Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J Cell Biol , 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. (2013). Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell , 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Hetzer MW. (2014). Breaching the nuclear envelope in development and disease. J Cell Biol , 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Hetzer MW. (2016). Nuclear envelope rupture is induced by actin-based nucleus confinement. J Cell Biol , 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattersley N, Cheerambathur D, Moyle M, Stefanutti M, Richardson A, Lee KY, Dumont J, Oegema K, Desai A. (2016). A nucleoporin docks protein phosphatase 1 to direct meiotic chromosome segregation and nuclear assembly. Dev Cell , 463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Kimura K, Kimura A. (2012). Localized accumulation of tubulin during semi-open mitosis in the Caenorhabditis elegans embryo. Mol Biol Cell , 1688–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer MW. (2010). The nuclear envelope. Cold Spring Harb Perspect Biol , a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer MW, Wente SR. (2009). Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev Cell , 606–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hird SN, White JG. (1993). Cortical and cytoplasmic flow polarity in early embryonic cells of Caenorhabditis elegans. J Cell Biol , 1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J, Pfeifer CR, Bennett RR, Xia Y, Ivanoska IL, Liu AJ, Greenberg AJ, Discher DE. (2016). Nuclear constriction segregates mobile nuclear proteins away from chromatin. Mol Biol Cell , 4011–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J, Xia Y, Pfeifer CR, Athirasala A, Ji J, Alvey C, Tewari M, Bennett RR, Harding SM, Liu AJ, et al. (2017). DNA damage follows repair factor depletion and portends genome variation in cancer cells. Curr Biol , 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan CM, Gaiano N, Ko MS, Zheng Y. (2011). Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science , 1706–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Zheng X, Zheng Y. (2013). Proliferation and differentiation of mouse embryonic stem cells lacking all lamins. Cell Res , 1420–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Onami S. (2005). Computer simulations and image processing reveal length-dependent pulling force as the primary mechanism for C. elegans male pronuclear migration. Dev Cell , 765–775. [DOI] [PubMed] [Google Scholar]
- König J, Frankel EB, Audhya A, Müller-Reichert T. (2017). Membrane remodeling during embryonic abscission in Caenorhabditis elegans. J Cell Biol , 1277–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudron M, Niu W, Lu Z, Wang G, Gerstein M, Snyder M, Reinke V. (2013). Tissue-specific direct targets of Caenorhabditis elegans Rb/E2F dictate distinct somatic and germline programs. Genome Biol , R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Hetzer MW. (2008). Reorganization of the nuclear envelope during open mitosis. Curr Opin Cell Biol , 669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J, Wolf K. (2016). Nuclear envelope rupture: actin fibers are putting the squeeze on the nucleus. J Cell Biol , 5–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Berre M, Aubertin J, Piel M. (2012). Fine control of nuclear confinement identifies a threshold deformation leading to lamina rupture and induction of specific genes. Integr Biol (Camb) , 1406–1414. [DOI] [PubMed] [Google Scholar]
- Lee KK, Gruenbaum Y, Spann P, Liu J, Wilson KL. (2000). C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol Biol Cell , 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ben-Shahar TR, Riemer D, Treinin M, Spann P, Weber K, Fire A, Gruenbaum Y. (2000). Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell , 3937–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GWG, Starr DA. (2014). KASHing up with the nucleus: novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Curr Opin Cell Biol , 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. (2003). The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell , 825–836. [DOI] [PubMed] [Google Scholar]
- Mattaj IW. (2004). Sorting out the nuclear envelope from the endoplasmic reticulum. Nat Rev Mol Cell Biol , 65–69. [DOI] [PubMed] [Google Scholar]
- Mattout A, Pike BL, Towbin BD, Bank EM, Gonzalez-Sandoval A, Stadler MB, Meister P, Gruenbaum Y, Gasser SM. (2011). An EDMD mutation in C. elegans lamin blocks muscle-specific gene relocation and compromises muscle integrity. Curr Biol , 1603–1614. [DOI] [PubMed] [Google Scholar]
- Mattout A., Cabianca DS, Gasser SM. (2015). Chromatin states and nuclear organization in development—a view from the nuclear lamina. Genome Biol , 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Colf LA, Sundquist WI. (2013). Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem , 663–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerzon M, Gao Z, Liu J, Wu JC, Malone CJ, Starr DA. (2009). Centrosome attachment to the C. elegans male pronucleus is dependent on the surface area of the nuclear envelope. Dev Biol , 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E, Nance J, Priess JR. (2004). Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell , 413–424. [DOI] [PubMed] [Google Scholar]
- O’Connell K. (2000). The centrosome of the early C. elegans embryo: inheritance, assembly, replication, and developmental roles. Curr Top Dev Biol , 365–384. [DOI] [PubMed] [Google Scholar]
- Oegema K, Hyman AA. (2006). Cell division. WormBook 1–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y, Carlton JG. (2016). The ESCRT machinery: new roles at new holes. Curr Opin Cell Biol , 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y, Hodgson L, Mantell J, Verkade P, Carlton JG. (2015). ESCRT-III controls nuclear envelope reformation. Nature , 236–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier N, Audhya A, Maddox PS, Green RA, Dammermann A, Desai A, Oegema K. (2007). A microtubule-independent role for centrosomes and aurora a in nuclear envelope breakdown. Dev Cell , 515–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M, Gentili M, de Belly H, Thiam HR, Vargas P, Jimenez AJ, Lautenschlaeger F, Voituriez R, Leoon-Duménil AM, Manel N, et al. (2016). ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science , 359–362. [DOI] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. (2008). Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature , 243–247. [DOI] [PubMed] [Google Scholar]
- Riemer D, Dodemont H, Weber K. (1993). A nuclear lamin of the nematode Caenorhabditis elegans with unusual structural features; cDNA cloning and gene organization. Eur J Cell Biol , 214–223. [PubMed] [Google Scholar]
- Robijns J, Molenberghs F, Sieprath T, Corne TDJ, Verschuuren M, De Vos WH. (2016). In silico synchronization reveals regulators of nuclear ruptures in lamin A/C deficient model cells. Sci Rep , 30325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B. (2002). Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell , 97–107. [DOI] [PubMed] [Google Scholar]
- Shen Q-T, Schuh AL, Zheng Y, Quinney K, Wang L, Hanna M, Mitchell JC, Otegui MS, Ahlquist P, Cui Q, Audhya A. (2014). Structural analysis and modeling reveals new mechanisms governing ESCRT-III spiral filament assembly. J Cell Biol , 763–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, Jaqaman K, Goldman RD. (2015). Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol Biol Cell , 4075–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Pfleghaar K, Kojima SI, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, et al. (2008). The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev , 3409–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman N, Heins S, Aebi U. (1998). Nuclear lamins: their structure, assembly, and interactions. J Struct Biol , 42–66. [DOI] [PubMed] [Google Scholar]
- Takaki T, Montagner M, Serres MP, Le Berre M Russell M, Collinson L, Szuhai K, Howell M, Boulton SJ, Sahai E, et al. (2017). Actomyosin drives cancer cell nuclear dysmorphia and threatens genome stability. Nat Commun , 16013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M, Campagnola P, Rolls MM, Stein PA, Ellenberg J, Hinkle B, Slepchenko B. (2001). A new model for nuclear envelope breakdown. Mol Biol Cell , 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam HR, Vargas P, Carpi N, Crespo CL, Raab M, Terriac E, King MC, Jacobelli J, Alberts AS, Stradal T, et al. (2016). Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat Commun , 10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay Y, Eibauer M, Goldman AE, Shimi T, Khayat M, Ben-Harush K, Dubrovsky-Gaupp A, Sapra KT, Goldman RD, Medalia O. (2017). The molecular architecture of lamins in somatic cells. Nature , 261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungricht R, Kutay U. (2017). Mechanisms and functions of nuclear envelope remodelling. Nat Rev Mol Cell Biol , 229–245. [DOI] [PubMed] [Google Scholar]
- Vargas JD, Hatch EM, Anderson DJ, Hetzer MW. (2012). Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus , 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri M, Schink KO, Campsteijn C, Wegner CS, Schultz SW, Christ L, Thoresen SB, Brech A, Raiborg C, Stenmark H. (2015). Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature , 231–235. [DOI] [PubMed] [Google Scholar]
- Wandke C, Kutay U. (2013). Enclosing chromatin: reassembly of the nucleus after open mitosis. Cell , 1222–1225. [DOI] [PubMed] [Google Scholar]
- Webster BM, Colombi P, Jäger J, Lusk CP. (2014). Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell , 388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BM, Thaller DJ, Jäger J, Ochmann SE, Borah S, Lusk CP. (2016). Chm 7 and Heh 1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing. EMBO J , 2447–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesel N, Mattout A, Melcer S, Melamed-Book N, Herrmann H, Medalia O, Aebi U, Gruenbaum Y. (2008). Laminopathic mutations interfere with the assembly, localization, and dynamics of nuclear lamins. Proc Natl Acad Sci USA , 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KL, Foisner R. (2010). Lamin-binding proteins. Cold Spring Harb Perspect Biol , a000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Chojnowski A, Boudier T, Lim JSY, Ahmed S, Ser Z, Stewart C, Burke B. (2016). A-type lamins form distinct filamentous networks with differential nuclear pore complex associations. Curr Biol , 2651–2658. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. (2009). SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron , 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuela N, Dorfman J, Gruenbaum Y. (2017). Global transcriptional changes caused by an EDMD mutation correlate to tissue specific disease phenotypes in C. elegans. Nucleus , 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo JM, Demarco IA, Piqué-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Berstein BE, Pritchard JK, Reddy KL, Singh H. (2012). DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell , 1474–1487. [DOI] [PubMed] [Google Scholar]
- Zwerger M, Jaalouk DE, Lombardi ML, Isermann P, Mauermann M, Dialynas G, Herrmann H, Wallrath LL, Lammerding J. (2013). Myopathic lamin mutations impair nuclear stability in cells and tissue and disrupt nucleo–cytoskeletal coupling. Hum Mol Genet , 2335–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.