Live-cell single-molecule imaging demonstrates that long-lasting telomerase–telomere interactions correspond to telomerase ribonucleoproteins that are engaged with the chromosome end. Furthermore, processive telomere elongation by telomerase is shown to be a key contributing factor to telomere length maintenance in vivo.
Abstract
The reverse transcriptase telomerase adds telomeric repeats to chromosome ends to counteract telomere shortening and thereby assures genomic stability in dividing human cells. Key parameters in telomere homeostasis are the frequency with which telomerase engages the chromosome end and the number of telomeric repeats it adds during each association event. To study telomere elongation in vivo, we have established a live-cell imaging assay to track individual telomerase ribonucleoproteins in CRISPR-edited HeLa cells. Using this assay and the drug imetelstat, which is a competitive inhibitor of telomeric DNA binding, we demonstrate that stable association of telomerase with the single-stranded overhang of the chromosome end requires telomerase-DNA base pairing. Furthermore, we show that telomerase processivity contributes to telomere elongation in vivo. Together, these findings provide new insight into the dynamics of telomerase recruitment and the importance of processivity in maintaining telomere length in human cancer cells.
INTRODUCTION
Chromosomes in human cells are capped by telomeres, repetitive DNA tracts bound by the shelterin protein complex (de Lange, 2005). Telomeres shorten during each cell cycle due to the failure of the DNA-replication machinery to copy the very end of each chromosome (Harley et al., 1990). To counteract this shortening, continuously dividing cells, such as stem cells and most cancer cells, express telomerase (Stewart and Weinberg, 2006; Schmidt and Cech, 2015). Telomerase is an RNA-containing reverse transcriptase, which adds DNA to the 3′ single-stranded overhang of human chromosomes specified by the template region of the telomerase RNA (TR) (Cech, 2004).
The requirement of cancer cells to express telomerase is highlighted by the frequent occurrence of mutations in the promoter of the gene for telomerase reverse transcriptase (TERT) (Horn et al., 2013; Huang et al., 2013). These mutations activate the mono-allelic expression of TERT (Bell et al., 2015; Borah et al., 2015; Stern et al., 2015; Chiba et al., 2017), which is normally down-regulated when human cells differentiate. In addition to its importance in cancer formation and survival, defects in telomerase-mediated telomere maintenance are associated with a number of premature aging diseases, such as Dyskeratosis congenita (Armanios and Blackburn, 2012). Thus, telomere maintenance by telomerase plays a key role in several human pathologies, and understanding its basic biology could lead to new approaches to treat these diseases.
Human telomerase is a ribonucleoprotein (RNP), composed of the TERT protein and the telomerase RNA (TR). In addition, the telomerase holoenzyme contains accessory subunits, for example, dyskerin, NHP2, and NOP10, which associate with TR to stabilize the RNA and ensure its nuclear localization (Schmidt and Cech, 2015). In vitro, human telomerase can processively synthesize multiple telomeric repeats without dissociating from its DNA substrate (Wu et al., 2017b). In vivo, telomerase is thought to add ∼50–60 nucleotides to most chromosome ends in a single processive step (Zhao et al., 2009). In support of this hypothesis, recent results have implicated telomerase processivity as an important contributor to telomere maintenance in vivo (Wu et al., 2017a). The nucleus of a human cancer cell only contains ∼250 fully assembled telomerase RNPs (Xi and Cech, 2014), which is approximately stoichiometric with the number of chromosome ends after DNA replication has occurred.
Telomerase is recruited to telomeres during the S-phase of the cell cycle by a direct interaction between the oligonucleotide/oligosaccharide-binding (OB)-fold domain of the shelterin component TPP1 and the telomerase essential N-terminal (TEN) domain of TERT (Nandakumar and Cech, 2012; Sexton et al., 2012; Zhong et al., 2012; Schmidt et al., 2014). Using live-cell single-molecule imaging and CRISPR genome editing to produce telomerase with a 3xFLAG-HaloTag (referred to as Halo or HaloTag throughout the rest of the article), we have recently demonstrated that telomerase rapidly diffuses through the nucleus of human cells, searching for telomeres to bind (Schmidt et al., 2016). When telomerase encounters a chromosome end, it can form two types of interactions: short “probing” interactions and long “static” interactions. Importantly, the specific binding of TPP1 and TERT is required for the formation of both types of interactions. We postulated that, in addition, the long-static interactions require base pairing of TR to single-stranded telomeric DNA and therefore represent telomerase RNPs that are actively elongating the telomere, but we did not provide direct evidence for this hypothesis.
Here we demonstrate that long-static interactions indeed require base pairing of TR with the chromosome end by utilizing the cancer drug imetelstat (JNJ-63935937, also known as GRN163L), a first-in-class telomerase inhibitor currently in clinical development in hematologic malignancies. Imetelstat, a 13-mer thiophosphoramidate oligonucleotide, is complementary to the template region of TR and prevents its base pairing with telomeric DNA (Herbert et al., 2005). Furthermore, we demonstrate that Halo-telomerase, which has normal activity but reduced processivity, elongates telomeres at a lower rate than wild-type (WT) telomerase in cells; this highlights the importance of the intrinsic processivity of telomerase for telomere maintenance. Together, these observations provide new insight into telomerase recruitment to telomeres and the contribution of telomerase processivity to telomere maintenance.
RESULTS
Halo-telomerase is active but has reduced processivity
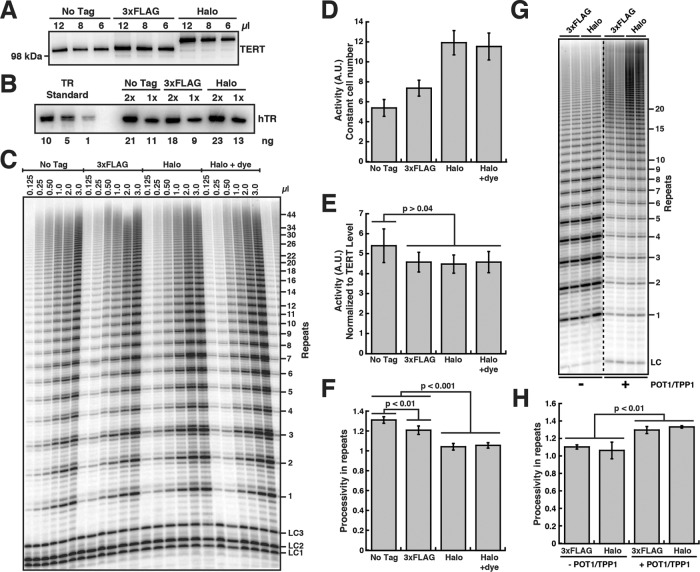
Because our live-cell single-molecule imaging utilizes Halo-TERT, we determined the enzymatic properties of telomerase modified with N-terminal tags. We overexpressed tagged TERT proteins with TR in HEK293T cells (Figure 1A and Supplemental Figure S1A). WT TERT and Halo-TERT associated with comparable amounts of TR, indicating that the HaloTag does not prevent the assembly of TERT and TR (Figure 1B and Supplemental Figure S1B). To measure the catalytic properties of Halo-telomerase, we carried out direct telomerase extension assays (Figure 1C). While telomerase activity normalized to the number of cells used as input material was increased in Halo-TERT samples relative to WT TERT (Figure 1D), the presumably more accurate normalization to the amount of TERT purified showed similar amounts of telomerase activity (Figure 1E). The presence of a 3xFLAG-tag led to a small decrease in processivity, while the HaloTag led to a larger ∼20% reduction, both decreases being statistically significant (Figure 1F). The Halo-telomerase had the same activity and processivity with or without the fluorescent dye used for live-cell imaging. Furthermore, the HaloTag did not affect the functional interaction of telomerase with the telomeric protein TPP1, demonstrated by the increase in processivity in the presence of the shelterin component POT1/TPP1 (Figure 1, G and H). We conclude that the introduction of the HaloTag reduces telomerase processivity, as previously shown for Halo- and 3xFLAG-TERT (Chiba et al., 2016; Schmidt et al., 2016). Importantly, the HaloTag does not affect telomerase activity or the interaction of TERT with its telomeric partner, TPP1.
FIGURE 1:
The HaloTag on TERT has no effect on telomerase properties except for a reduction in processivity. (A) Western blot of telomerase immunopurified from HEK293T cells overexpressing various untagged and tagged TERT proteins and TR, probed with an anti-TERT antibody. (B) Northern blot of RNA extracted from immunopurified telomerase variants, probed with three TR probes. Standards are in vitro–transcribed full-length TR and were used to quantify the amount of TR in IPs (values below the lanes). (C) Direct telomerase extension assay at 50 mM KCl of various immunopurified telomerase variants. LC1, LC2, and LC3, labeled DNA loading controls. (D) Quantification of telomerase activity normalized to the loading controls and the number of cells used as input for immunopurification (n = 6, mean ± SD). (E) Quantification of telomerase activity normalized to the loading controls and the TERT level (see panel A, n = 6, mean ± SD, t test). (F) Quantification of telomerase processivity using the decay method (n = 5, mean ± SD, t test). (G) Direct telomerase extension assay at 150 mM KCl (to limit processivity) of 3xFLAG- and 3xFLAG-HaloTag-telomerase immunopurified from HEK293T cells using anti-FLAG resin in the absence and presence of POT1/TPP1. LC, labeled DNA loading control. (H) Quantification of 3xFLAG- and 3xFLAG-HaloTag-telomerase processivity in the absence and presence of POT1/TPP1 using the decay method (n = 5, mean ± SD, t test).
Halo-telomerase elongates telomeres in vivo
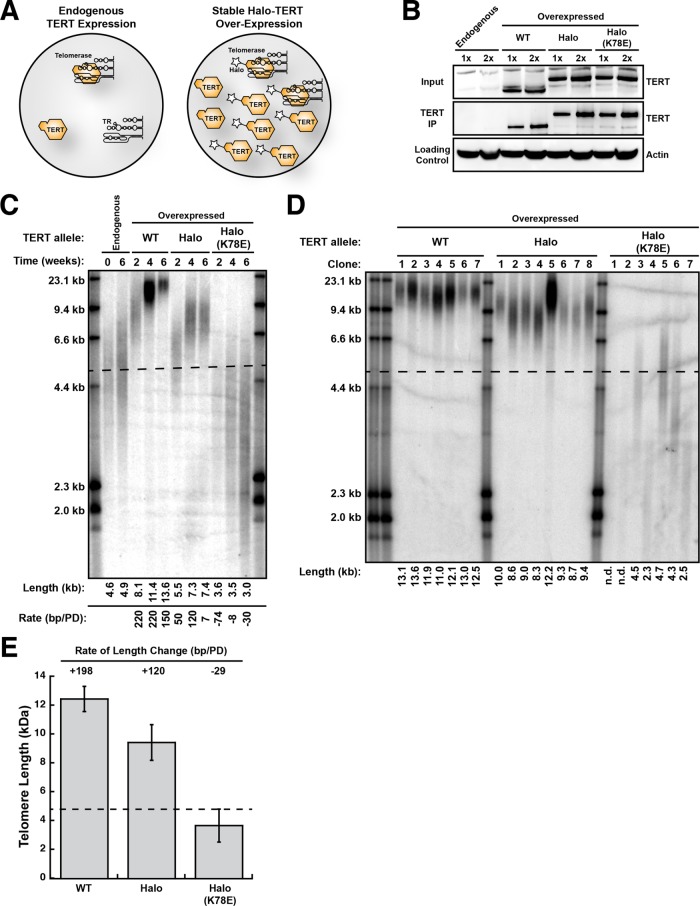
To test whether Halo-telomerase can elongate telomeres in vivo, we stably introduced WT TERT, Halo-TERT, and Halo-TERT harboring the K78E recruitment-deficient mutation into HeLa cells by retroviral transduction (Figure 2A). This approach leads to overexpression of the respective TERT allele (Figure 2B), which elicits a dominant effect by outcompeting the endogenous TERT for assembly with TR into the mature telomerase RNP (Figure 2A). TERT was overexpressed to a similar degree in all polyclonal, virally transduced cell lines (Figure 2B). To measure the telomerase activity in these cells, we immunopurified telomerase and subjected it to direct telomerase assays (Figure 2B and Supplemental Figure S1C). Similarly to telomerase overexpressed in HEK293T cells (see above), we observed comparable catalytic activity for all TERT variants (Supplemental Figure S1D) and a reduction of processivity of telomerase RNPs that were modified with the HaloTag (Supplemental Figure S1E). As previously shown (Schmidt et al., 2014), TERT overexpression increased telomerase activity per cell in all cell lines (Supplemental Figure S1D). Importantly, Halo-TERT harboring the K78E mutation displayed enzymatic properties that were indistinguishable from its WT counterpart (Figure 2B and Supplemental Figure S1, C–E).
FIGURE 2:
Halo-telomerase elongates telomeres in vivo. (A) Experimental design. Overexpression of TERT (star represents tag) will increase telomerase levels in HeLa cells by driving the assembly of free TR into telomerase RNPs. Owing to its higher levels, exogenous tagged TERT will outcompete endogenous TERT for assembly with TR. (B) Western blots of cell lysates (Input) and TERT immunopurified (TERT IP) from cell lines overexpressing various TERT alleles probed with an anti-TERT antibody. Cell lysates were probed with an anti-beta-Actin antibody as loading control. (C) Telomere length analysis of polyclonal HeLa cell lines stably overexpressing various TERT proteins by Southern blot of telomeric restriction fragments. Each rate of telomere extension was calculated relative to the previous time point recorded. (D) Telomere length analysis of single-cell clones of HeLa cells stably overexpressing various TERT proteins by Southern blot of telomeric restriction fragments. In C and D, the dashed line represents the mean length of parental cell telomeres. (E) Quantification of the rate of telomere length change by averaging the telomere length of all single-cell clones (n = 5–8, see panel D), calculating their length relative to those of the parental HeLa cells (see panel C) and dividing by the number of population doublings between introduction of the TERT transgene and analysis of telomere length.
To determine whether Halo-telomerase can elongate telomeres in cells, we measured telomere lengths in virally transduced cell lines by Southern blotting. It is important to note that although TERT is substantially overexpressed, the TR subunit becomes limiting, so telomerase activity increases only ∼1.5–2-fold, as seen previously (Supplmental Figure S1D) (Cristofari et al., 2007; Schmidt et al., 2014; Xi and Cech, 2014). Telomere length in the parental HeLa cells remained constant over the time course of the experiment (Figure 2C). Expression of WT TERT led to telomere elongation from ∼4.6 to ∼13.6 kb over the time course of 6 wk, which corresponds to a growth rate of 150–220 base pairs per population doubling (bp/PD) (Figure 2C). Telomere length in cells expressing Halo-TERT also increased (from ∼4.6 to 7.4 kb), but at a slower rate of ∼50–120 bp/PD, and telomeres reached their new length set point by 4 wk (Figure 2C). Importantly, telomere length in cells expressing Halo-TERT harboring the K78E mutation, which has full enzymatic activity (Supplemental Figure S1, A–C) but cannot localize to telomeres (Schmidt et al., 2014), shrunk from ∼4.6 to ∼3.0 kb over the 6-wk time course (Figure 2C), confirming that TERT overexpressed from the transgene is dominant over endogenous TERT.
As an additional approach to determine the impact of TERT overexpression on telomere length, we isolated single-cell clones from the polyclonal populations 1 wk after viral transduction and determined their telomere length 5 wk after introduction of the TERT transgene (Figure 2D). Telomeres in clones expressing WT TERT and Halo-TERT grew to an average of ∼12.5 and ∼9.4 kb, corresponding to growth rates of ∼200 and ∼120 bp/PD, respectively (Figure 2E). These growth rates are consistent with those observed in the polyclonal cell populations (Figure 2C). Clones expressing K78E Halo-TERT shortened to ∼3.7 kb at a rate of ∼30 bp/PD (Figure 2, D and E). In total, these observations demonstrate that Halo-telomerase elongates telomeres in vivo, but it does so at a reduced rate compared with WT telomerase.
Imetelstat prevents the association of telomerase with its ssDNA substrate
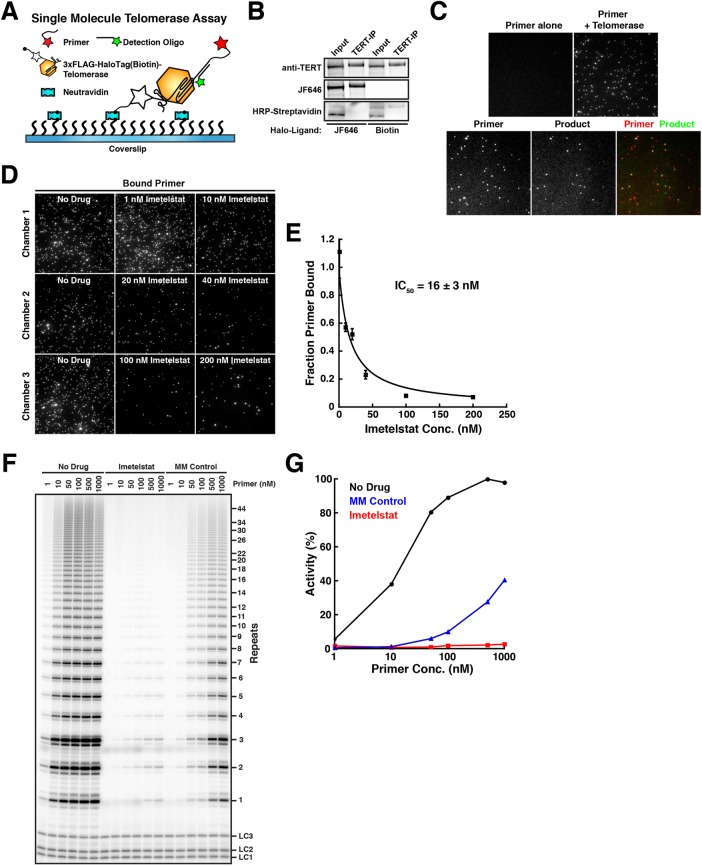
Imetelstat is complementary to the template region of TR and therefore should be a competitive inhibitor of single-stranded (ss) telomeric DNA binding to telomerase (Herbert et al., 2005). To test this hypothesis, we established a single-molecule telomerase primer-binding assay (Figure 3A). Halo-telomerase purified from HEK293T cells was modified with a HaloTag-ligand conjugated to a biotin molecule (Figure 3B) to allow immobilization on a coverslip surface derivatized with neutradivin (Figure 3A). Primer binding by telomerase was analyzed by telomerase-dependent recruitment of a fluorescently labeled telomeric oligonucleotide to the surface of the coverslip, visualized by TIRF microscopy (Figure 3, A and C). Importantly, a substantial fraction (34% ± 8%, mean ± SD, N = 6) of the telomerase RNPs immobilized by this approach were enzymatically active, as determined by a single-molecule telomerase activity assay previously established by Sua Myong and colleagues (Figure 3, A and C) (Hwang et al., 2014). It is worth noting that this method underestimates the fraction of active telomerase RNPs, because only products that are at least three telomeric repeats in length can be detected. We then carried out this single-molecule primer-binding assay in the presence of increasing concentrations of imetelstat (Figure 3, D and E). Imetelstat progressively decreased the amount of telomeric primer bound to telomerase (Figure 3, D and E), consistent with the hypothesis that imetelstat competes with ssDNA for telomerase binding. Half-maximal inhibition (IC50) of primer binding occurred at ∼16 nM imetelstat (Figure 3E), which is comparable to the concentration (20 nM) of telomeric primer used in this experiment.
FIGURE 3:
Imetelstat (GRN163L) is a competitive inhibitor of primer-substrate binding by telomerase. (A) Experimental design of single-molecule telomerase primer binding and activity assay. Halo-telomerase is modified with a biotin-HaloTag-ligand and immobilized on the coverslip surface using NeutrAvidin. Primer binding is visualized by telomerase-dependent recruitment of a fluorescent primer to the coverslip surface. The telomerase extension product is detected using a fluorescently labeled oligonucleotide anti-sense to the telomerase extension product. (B) Western blot and fluorescence imaging of Halo-telomerase modified with a fluorescent dye (JF646) or biotin, probed with an anti-TERT antibody or HRP-conjugated streptavidin. (C) Single-molecule TIRF imaging of primer molecules recruited to the coverslip surface by telomerase (top) and its colocalization with telomerase extension products after incubation with nucleotide substrate (bottom). (D) Single-molecule TIRF imaging of primer binding by telomerase in the presence of increasing concentrations of imetelstat. (E) Quantification of primer binding to telomerase as a function of imetelstat concentration (n = 5 fields of view per concentration, data points plotted as mean ± SD, error on IC50 reflects error in the corresponding fit of the data to a simple binding curve). (F) Direct telomerase assay at 150 mM KCl in the absence and presence of imetelstat (10 nM), or mismatched control oligonucleotide (MM Control, 10 nM), and increasing concentrations of primer substrate. LC1, LC2, and LC3, labeled DNA loading controls. (G) Quantification of telomerase activity as a function of primer concentration in absence and presence of imetelstat (10 nM) or mismatched control oligonucleotide (MM Control, 10 nM).
As an alternative approach to address how imetelstat affects primer binding to telomerase, we carried out direct telomerase assays in the presence of imetelstat or a control oligonucleotide that carries mismatches at four positions in the nucleotide sequence at varying concentrations of telomeric substrate primer (Figure 3F) (Asai et al., 2003). Imetelstat (10 nM) strongly inhibited telomerase at all primer concentrations used (Figure 3, F and G). The mismatched control also inhibited telomerase activity, but activity was recovered at higher primer concentrations (Figure 3, F and G), demonstrating that the control compound is a less-effective inhibitor of telomerase activity. Together, these results demonstrate that imetelstat acts as competitive inhibitor of telomerase binding to its ssDNA substrate.
Imetelstat inhibits telomerase in vivo
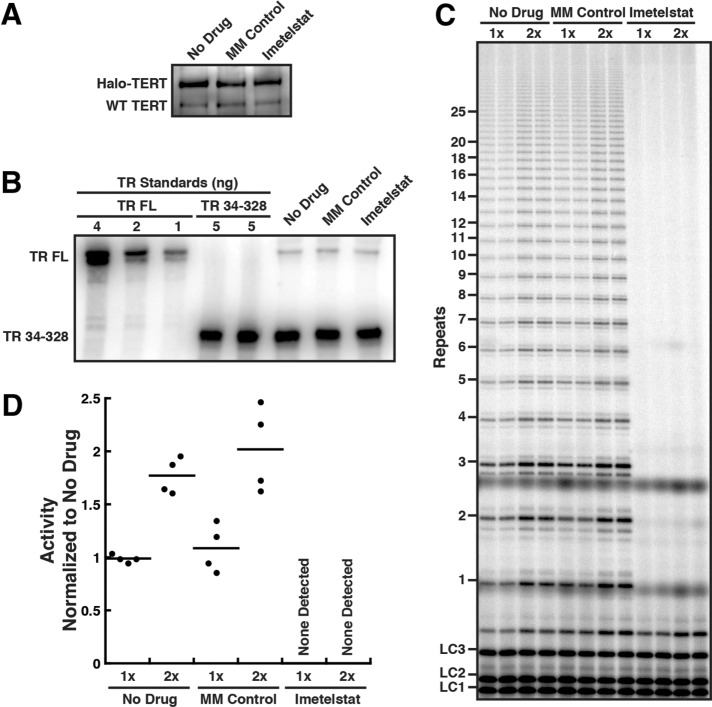
Because we planned to use imetelstat as an inhibitor of telomerase–telomere base pairing in live-cell imaging experiments, we tested how the drug affected telomerase RNP activity in vivo. We treated HeLa cells expressing Halo-TERT from the endogenous TERT locus with 2 µM of imetelstat or mismatch (MM) control oligonucleotide for 24 h and immunopurified telomerase RNPs from the treated cells. Similar amounts of TERT and TR were purified from treated and untreated cells (Figure 4, A and B). This demonstrates that a 24-h treatment with imetelstat does not affect telomerase RNP assembly in HeLa cells. While telomerase from untreated cells and cells treated with the mismatch control oligonucleotide showed similar telomerase activity, no activity was detected in telomerase preparations from cells treated with imetelstat (Figure 4, C and D). Together, these observations demonstrate that 24 h of treatment with imetelstat completely inhibits telomerase activity in HeLa cells, without affecting telomerase RNP assembly. In addition, imetelstat likely has a very slow dissociation rate from telomerase, since no telomerase activity is recovered even after a prolonged purification procedure (∼3 h). In contrast, while the MM control can inhibit telomerase weakly in an in vitro activity assay (Figure 3, F and G), it likely dissociates from telomerase throughout the purification process or does not bind telomerase in vivo at the concentrations used.
FIGURE 4:
Imetelstat inhibits telomerase in vivo but does not affect RNP assembly. (A) Western blot of telomerase immunopurified from HeLa cells expressing Halo-TERT from the endogenous TERT locus, after 24-h treatment with 2 µM imetelstat or 2 µM mismatched control oligonucleotide, probed with an anti-TERT antibody. (B) Northern blot of RNA extracted from telomerase immunopurified from HeLa cells expressing Halo-TERT after 24-h treatment with 2 µM imetelstat, 2 µM mismatched control oligonucleotide, or no drug. In vitro–transcribed full-length (FL) TR was included as size standard and truncated TR 34-328 as loading and recovery control. Blots were probed with three radiolabeled oligonucleotides antisense to TR. (C) Direct telomerase assays at 150 mM KCl of telomerase immuno-purified from HeLa cells expressing Halo-TERT from the endogenous TERT locus, after 24-h treatment with 2 µM imetelstat, 2 µM mismatched control oligonucleotide, or no drug; each concentration in duplicate. LC1, LC2, and LC3, labeled DNA loading controls. (D) Quantification of the activity of telomerase purified from HeLa cells expressing Halo-TERT from the endogenous TERT locus, after 24-h treatment with 2 µM imetelstat or 2 µM mismatched control oligonucleotide, normalized to No Drug and loading control (n = 4, mean).
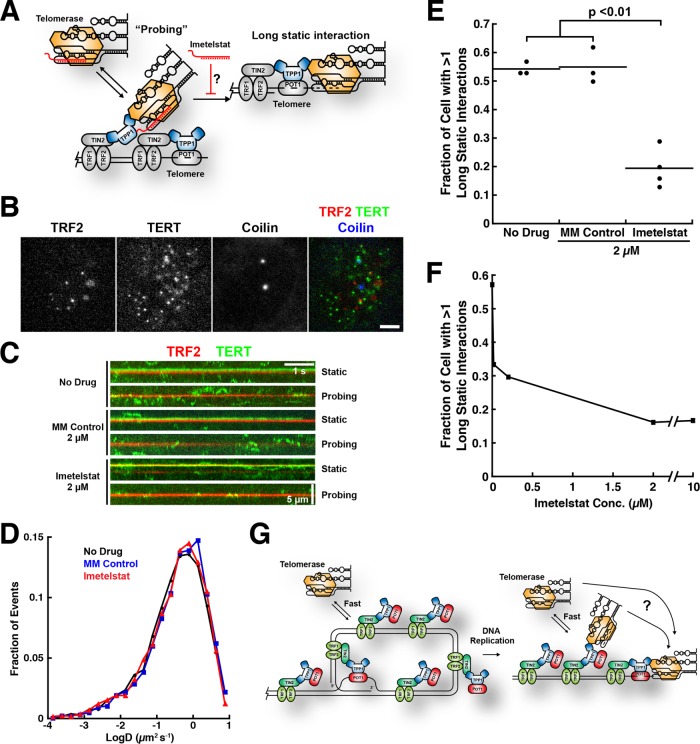
Imetelstat inhibits the formation of long-static telomerase–telomere interactions in vivo
We previously demonstrated that telomerase forms two types interactions with telomeres: short, dynamic “probing” interactions and long-lasting static interactions (Figure 5A) (Schmidt et al., 2016). We speculated that the long-lasting static interactions represent telomerase RNPs that are base paired with the single-stranded overhang of the chromosome end but were not able to provide direct evidence for this hypothesis (Schmidt et al., 2016). To test the base-pairing hypothesis, we carried out live-cell single-molecule imaging of telomerase trafficking in HeLa cells in the presence of 2 μM imetelstat or the mismatched control oligonucleotide (Figure 5B and Supplemental Movies 1–3). We observed dynamic “probing” and long-static interactions under all conditions (Figure 5C and Supplemental Movies 1–3), and the overall distribution of diffusion coefficients was unaffected by imetelstat treatment (Figure 5D), indicating no gross changes in RNP assembly or behavior. However, the number of cells in which we observed long-static interactions was reduced significantly (p < 0.01) in the presence of imetelstat in a dose-dependent manner (Figure 5, E and F) but remained unchanged when cells were treated with the mismatched control oligonucleotide (Figure 5E). These observations demonstrate that the long-static interactions depend on the ability of telomerase to base pair with the single-stranded overhang of the chromosome end.
FIGURE 5:
Imetelstat inhibits the formation of long-static telomerase–telomere interactions. (A) Experimental rationale. If long-static telomerase–telomere interactions require the base pairing of TR to the chromosome end, then imetelstat should prevent their formation. (B) Still images from three-color live-cell single-molecule imaging experiments. Telomeres were marked by mEOS3.2-TRF2, telomerase was detected using a 3xFLAG-HaloTag-TERT conjugated to JF646, and Cajal bodies were visualized using BFP-coilin (scale bar = 5 µm). (C) Kymographs of TERT and TRF2 trajectories from HeLa cells stably expressing 3xFLAG-HaloTag-TERT and mEOS3.2-TRF2 from their respective endogenous loci. Cells were treated with 2 µM imetelstat, 2 µM mismatched control oligonucleotide, or no drug. (D) Diffusion coefficient distributions of TERT trajectories from HeLa cells stably expressing 3xFLAG-HaloTag-TERT treated with 2 µM imetelstat, 2 µM mismatched control oligonucleotide, or no drug (n = 4000–5000 trajectories per condition). (E) Quantification of the fraction of cells with at least one long-static telomerase–telomere interaction after treatment with 2 µM imetelstat, 2 µM mismatched control oligonucleotide, or no drug (NNo Drug = 3, NMM Control = 3, Nimetelstat = 4, where N = number of independent experiments, n = 13–36 cells per condition, mean ± SD, t test). (F) Quantification of the fraction of cells with at least one long-static telomerase–telomere interaction as a function of imetelstat concentration (n = 28–44 cells per condition). (G) Model for telomerase recruitment to chromosome ends derived from the current study. Short probing interactions require TERT–TPP1 protein–protein binding, while the productive interaction of telomerase with the DNA 3′-end (far right) also requires base pairing with the template region of TR. Whether telomerase can hop or slide to the 3′ end from internal binding sites or, alternatively, is released back into the nucleus and must collide directly with the 3-terminus is an open question (?).
Movie S1.
Imaging of Halo-TERT in nuclei of living HeLa cells. 2D-movies of Halo-TERT (green, λex = 647 nm, λem = 670 nm) labeled with JF646 in the nuclei of a HeLa cells treated with no drug (Movie 1), 2 μM mismatched control oligonucleotide (Movie 2), or 2μM imetelstat (Movie 3). Movies were acquired with an exposure time of 20 ms for an effective frame rate of 46 frames per second. HA-mEOS3.2-TRF2 signals (red, λex = 561 nm, λem = 590 nm) are a maximum intensity projection of the first 50 frames of the movie acquired simultaneously to TERT. BFP-coilin signals (blue, λex = 405 nm, λem = 450 nm) are maximum intensity projections of the first 50 frames of a movie acquired immediately before simultaneous imaging TERT and TRF2 (Scale bar = 5 μm, Timestamp in ms).
Movie S2.
Imaging of Halo-TERT in nuclei of living HeLa cells. 2D-movies of Halo-TERT (green, λex = 647 nm, λem = 670 nm) labeled with JF646 in the nuclei of a HeLa cells treated with no drug (Movie 1), 2 μM mismatched control oligonucleotide (Movie 2), or 2μM imetelstat (Movie 3). Movies were acquired with an exposure time of 20 ms for an effective frame rate of 46 frames per second. HA-mEOS3.2-TRF2 signals (red, λex = 561 nm, λem = 590 nm) are a maximum intensity projection of the first 50 frames of the movie acquired simultaneously to TERT. BFP-coilin signals (blue, λex = 405 nm, λem = 450 nm) are maximum intensity projections of the first 50 frames of a movie acquired immediately before simultaneous imaging TERT and TRF2 (Scale bar = 5 μm, Timestamp in ms).
Movie S3.
Imaging of Halo-TERT in nuclei of living HeLa cells. 2D-movies of Halo-TERT (green, λex = 647 nm, λem = 670 nm) labeled with JF646 in the nuclei of a HeLa cells treated with no drug (Movie 1), 2 μM mismatched control oligonucleotide (Movie 2), or 2μM imetelstat (Movie 3). Movies were acquired with an exposure time of 20 ms for an effective frame rate of 46 frames per second. HA-mEOS3.2-TRF2 signals (red, λex = 561 nm, λem = 590 nm) are a maximum intensity projection of the first 50 frames of the movie acquired simultaneously to TERT. BFP-coilin signals (blue, λex = 405 nm, λem = 450 nm) are maximum intensity projections of the first 50 frames of a movie acquired immediately before simultaneous imaging TERT and TRF2 (Scale bar = 5 μm, Timestamp in ms).
Another part of our hypothesis was that the short “probing” interactions would not be affected by treating cells with imetelstat, because they are stabilized by protein–protein interactions. We therefore carried out a kinetic analysis of the residence times of TERT particles in proximity to telomeres, Cajal bodies, and other nuclear locations (Supplemental Figure S2, A–C). All residence time distributions fit well to the sum of two exponential decay functions (Supplemental Figure S2, A–C), one with a very rapid off-rate and one with a ∼10-fold slower off-rate, indicating that two distinct molecular processes underlie the behavior of the TERT RNPs at all nuclear locations. We propose that the fast component reflects TERT particles that are not actually associated with any subnuclear structure and by chance did not diffuse very far between two consecutive frames. Consistent with this interpretation, the half-lives of the fast components were similar to the sampling rate of the experiment, ∼18–27 and 22 ms, respectively (Supplemental Figure S2E). In contrast, the slow component likely represents TERT particles that are forming an interaction with a subnuclear structure. Importantly, the slower off-rate of TERT particles at telomeres, Cajal bodies, and other nuclear locations were similar under all experimental conditions (Supplemental Figure S2D), indicating that the underlying molecular interactions are unaffected by treatment with imetelstat. Furthermore, the off-rate of TERT particles at telomeres and Cajal bodies was significantly (p < 0.05) lower than at other nuclear locations (Supplemental Figure S2D), consistent with their forming specific interactions with telomeres and Cajal bodies. The fraction of TERT particles that dissociate from telomeres with a slower rate constant was reduced after treatment with imetelstat (Supplemental Figure S2F). This decrease is likely due to the contributions of long-static interactions to this analysis, because these are reduced by imetelstat treatment. In total, these observations demonstrate that the long-static interactions we observe by live-cell single-molecule imaging represent telomerase RNPs that are engaged with the telomere by base pairing of TR of the chromosome end.
DISCUSSION
Long-static telomerase–telomere interactions require base pairing of TR with the chromosome end
Telomere maintenance is essential for the proliferation of all actively dividing cells in the human body, including stem cells and cancer cells (Stewart and Weinberg, 2006). Telomerase compensates for telomere shrinkage that occurs during semiconservative DNA replication by adding telomeric repeats to the chromosome ends (Schmidt and Cech, 2015). Telomerase is a unique reverse transcriptase that synthesizes DNA using the template sequence present in its RNA subunit TR (Wu et al., 2017b). A critical step in telomere lengthening is telomerase recruitment to telomeres, which is not trivial due to the low abundance of human telomerase even in cancer cells (Xi and Cech, 2014). We have previously described two different types of interactions that telomerase can from with telomeres (Schmidt et al., 2016): short dynamic “probing” interactions and long-lasting static interactions. Both types of interactions require binding of the TEN-domain of TERT with the TEL-patch of TPP1, because both are eliminated by a point mutation in TERT (K78E).
Imetelstat, a lipid-modified thio-phosphoramidate oligonucleotide that is complementary to the template region of TR (Herbert et al., 2005), allowed us to analyze the contributions of this region of TR to the interactions that we observe in vivo. We proposed that the long-static TERT-telomere associations represent telomerase RNPs that are base paired with the chromosome end and are actively elongating the telomere. Since imetelstat is a competitive inhibitor of the association of telomerase with its DNA substrate, it should interfere with the formation of long-static interactions if they required base pairing of TR with the chromosome end. Indeed, the frequency of the formation of long-static interactions was dramatically reduced in the presence of imetelstat but remained unchanged when cells were treated with a mismatched control oligonucleotide. Therefore, since telomerase RNPs that are engaged in long-static interactions are base paired to the chromosome end, they are likely actively elongating the telomere.
Telomerase processivity contributes to telomere maintenance in vivo
To processively synthesize multiple telomeric repeats, the template region of TR must be repositioned relative to the substrate DNA in a step called translocation. In vitro human telomerase can processively synthesize multiple telomeric repeats, but how much processivity contributes to telomere maintenance in vivo remains unclear (Wu et al., 2017b). In vivo, the amount of telomere lengthening that occurs at a given telomere depends on two key parameters: the number of times that telomerase binds to the DNA at the chromosome end per cell cycle (frequency) and the number of repeats telomerase adds to the telomere per association event (processivity).
The introduction of the HaloTag on the N-terminus of TERT allowed us to visualize telomerase in living cells and inadvertently generated a TERT variant with reduced telomerase processivity without affecting activity, which is the number of nucleotides added per unit time. We could therefore test the contribution of telomerase processivity to telomere elongation in vivo by comparing Halo-telomerase with the wild-type RNP. The presence of the HaloTag led to a substantial reduction in the rate of telomere lengthening and in the plateau length when TERT was overexpressed in human cancer cells. Overexpression of telomerase likely leads to telomere growth by increasing the frequency of lengthening events that occur per telomere in a given S-phase. The intrinsic processivity of telomerase dictates how many repeats are added in each individual telomere lengthening event and is unlikely to be affected by overexpressing TERT. Under the conditions in our experiments, the amount of TERT protein vastly exceeds the amount of TR, making TR the limiting component for telomerase assembly. Therefore, the telomerase RNP concentrations and thus the frequency of telomere lengthening events are likely similar for all TERT alleles tested. The difference in telomere lengthening can therefore be attributed to the difference in intrinsic processivity of WT versus Halo-TERT. Importantly, telomere lengthening depended on the ability of Halo-TERT to localize to telomeres, demonstrating that even though it occurs at a lower overall rate, telomere elongation is carried out by Halo-telomerase. We conclude that a small decrease in telomerase processivity (∼20%) can have a substantial effect on telomere length when aggregated over multiple cell divisions. This observation could provide an explanation for why previous attempts to establish human embryonic stem cell lines expressing HaloTag-TERT were unsuccessful (Chiba et al., 2016) if embryonic stem cells were more sensitive to reduced telomerase processivity than cancer cells. In total, our results highlight the importance of telomerase processivity for telomere lengthening in vivo, consistent with recent observations made by others (Wu et al., 2017a).
Implications for telomere maintenance
We have now defined elements of the molecular basis of the short “probing” and long-static interactions observed in our live-cell single-molecule imaging experiments. Because the protein–protein interaction between TERT and TPP1 is required for both types of interactions and base pairing of TR with the chromosome end only for the formation of long-static interactions, it is tempting to speculate that “probing” interactions are an intermediate between free telomerase and telomerase that is elongating the telomere (Figure 5G). Probing the telomere could then increase the chance of telomerase finding the chromosome end, for example, by sliding, hopping, or hand-off. Furthermore, because “probing” is transient, it would not trap the small pool of telomerase RNPs at telomeres that do not have the 3′ overhang available for binding. If telomere elongation were possible only during a brief window of time after DNA replication (Zhao et al., 2009), then this “probing” mechanism would increase the probability of telomerase finding the chromosome end during this time window (Figure 5G). Whether telomerase is recruited to specific telomeres (e.g., short telomeres) during a particular time window will be the subject of future investigation. For now, the results presented in this study lay the groundwork for a comprehensive quantitative analysis of telomere elongation by telomerase in human cancer cells.
MATERIALS AND METHODS
Plasmids construction
Plasmids for the expression of WT- and 3xFLAG-TERT were previously described. The plasmid for overexpression of 3xFLAG-HaloTag TERT was generated by first ligating TERT (amplified with TERT for and TERT rev; see Table 1) into the pHTN HaloTag CMV-neo vector (Promega) cut with EcoRI and NotI (NEB). The sequence coding for the 3xFLAG-HaloTag was amplified from genomic DNA of a HeLa cell line stably expressing 3xFLAG-HaloTag-TERT from the endogenous TERT locus (amplified with 3xFLAG-HaloTag for and 3xFLAG-HaloTag rev; see Table 1) and ligated into pHTN containing TERT using NheI and EcoRI. To generate the exact coding sequence present in the HeLa cell line stably expressing 3xFLAG-HaloTag-TERT from the endogenous TERT locus, the EcoRI site was mutated to a KpnI site using quick change (amplified with EcoRI QC for and EcoRI QC rev; see Table 1). Plasmids for generation of retroviruses coding for various TERT alleles were generated by Gibson assembly, inserting a TERT (amplified with TERT Gibson for and TERT Gibson rev; see Table 1) or 3xFLAG-HaloTag-TERT (amplified with Halo TERT Gibson for and TERT Gibson rev; see Table 1) fragment into the pBABE PuroR vector (amplified with pBABE Gibson for and pBABE Gibson rev; see Table 1).
TABLE 1:
Primers used in this study.
| TERT for | GCGCGCGGAATTCATGCCGCGCGCTCCCCGCTGC |
| TERT rev | GCGCGCGGCGGCCGCTCAGTCCAGGATGGTCTTGAAG |
| 3xFLAG-HaloTag for | GCGCGCGCTAGCAAAGCCACCATGGACTACAAAGACCATGACGGTG |
| 3xFLAG-HaloTag rev | GCGCGCGAATTCGGAAGCGATCGCGTTATCGC |
| EcoRI QC for | TAACGCGATCGCTTCCGGTACCATGCCGCGCGCTCCC |
| EcoRI QC rev | GGGGAGCGCGCGGCATGGTACCGGAAGCGATCGCGTTA |
| Halo TERT Gibson for | ACGCTACCGGTCGCCACCATGGACTACAAAGACCATGACGGTG |
| TERT Gibson rev | TACCGTCGACTGCAGAATTCTCAGTCCAGGATGGTCTTGAAGTCTG |
| TERT Gibson for | ACGCTACCGGTCGCCACCATGCCGCGCGCTCCCCGC |
| pBABE Gibson for | GAATTCTGCAGTCGACGGTACCG |
| pBABE Gibson rev | GGTGGCGACCGGTAGCGTAC |
Cell culture and generation of stable cell lines
All cell lines were derivatives of Hela-EM2-11ht (Tet Systems Holding GmbH and Co. KG) and were grown in high glucose DMEM supplemented with 10% fetal bovine serum (FBS), 2 mM GlutaMAX-I (Life Technologies), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C with 5% CO2. Imaging experiments were carried out in CO2-independent media supplemented with 10% FBS, 2 mM GlutaMAX-I (Life Technologies), 100 U/ml penicillin, and 100 μg/ml streptomycin in a humidified imaging chamber heated to 37°C. For S-phase synchronization, cells were arrested in growth medium containing 2 mM thymidine for 16 h, released for 9 h, followed by a second thymidine 16 h arrest prior to release into S-phase. Puromycin selection was carried out at a concentration of 1 µg/ml (Sigma). Stable cell lines were generated by retroviral transduction as previously described (Schmidt et al., 2014). Single-cell clones were derived by diluting a suspension of the polyclonal cells to ∼6 cells/ml and plating 150 µl of this suspension into each well of a 96-well plate. Only clones that clearly formed a single colony of the appropriate size were used. Both polyclonal and clonal stable cell lines were continuously cultured in media containing puromycin.
Imetelstat and the mismatched control oligonucleotide were a gift of Janssen Research & Development, LLC (Raritan, NJ). Imetelstat and the mismatched control oligonucleotide were dissolved in PBS at concentrations between 1 and 2 mM and stored at –20 °C. Concentrations were verified using OD260. HeLa cells were incubated with 2 µM of imetelstat or the mismatched control oligonucleotide for 24 h prior to imaging or telomerase purification.
Transient transfections of BFP-coilin were carried out with the Nucleofector 2b device, using Kit R and the high-efficiency protocol for HeLa cells (Lonza).
Telomerase purification and activity assay
Telomerase was overexpressed as previously described (Sauerwald et al., 2013). Cell lysates were prepared using CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) lysis buffer (10 mM Tris-HCl, pH 7.5, 1 mM MgCl2, 1 mM ethylene glycol-bis(beta-aminoethyl ether)-tetraacetic acid, 0.5% CHAPS, 10% glycerol, 1 mM PMSF, 1 mM dithiothreitol [DTT]). Telomerase IP of endogenous and overexpressed TERT was carried out with a sheep polyclonal anti-TERT antibody, which was a gift from Scott Cohen (Children’s Medical Research Institute and University of Sydney, Westmead, Australia), from CHAPS lysates of ∼100 × 106 HeLa or HEK293T cells. For the comparison of telomerase in the presence and absence of POT1/TPP1, 3xFLAG- and 3xFLAG-HaloTag-TERT containing telomerases were purified from HEK293T cell lysates overexpressing telomerase with Anti-FLAG M2 Affinity Gel (Sigma-Aldrich; A2220), followed by elution with 3xFLAG peptide according to the manufacturer’s instructions. The HaloTag was labeled during the incubation with the resin using a concentration 0.5 µM JF646 (a kind gift from Luke Lavis, Howard Hughes Medical Institute [HHMI] Janelia Research Campus) or PEG-Biotin (Promega) HaloTag-ligand. The telomerase purifications and activity assays were carried out as previously described (Zaug et al., 2013), using indicated salt (KCl) and substrate concentrations. Purification of POT1/TPP1 and telomerase assays in the presence of POT1/TPP1 were carried out as previously described (Schmidt et al., 2014), using indicated salt (KCl), substrate, and POT1/TPP1 concentrations. Loading controls were phosphorylated telomeric DNA 18- and 21-mers. To analyze the effect of imetelstat on telomerase activity, the activity assay was initiated by adding primer, imetelstat, and nucleotides simultaneously. Telomerase activity was quantified by the total amount of radioactive counts incorporated into products and normalized to the sum of the loading controls. Telomerase processivity was determined by the decay method previously described (Latrick and Cech, 2010). The radioactive counts of each major telomeric repeat product were divided by the number of dG nucleotides incorporated during its synthesis (1 for the first product, 4 for the second product, 7 for the third product, etc.), which converts the radioactive signal into an estimate of the number of molecules present in each band. The number of molecules in each band represents products that have dissociated from telomerase after each round of repeat synthesis. Telomerase processivity was determined by fitting this decay from repeats (2–15), which contain greater than 95% of the measured molecules, excluding the first repeat, to a single exponential decay function Y = A*e(-k*Repeat). Processivity was calculated as ln(2)/k and has units of telomeric repeats.
Western blotting
The protein samples were separated on 4–12% Bis-Tris gels (Life Technologies), followed by standard Western blotting procedures. TERT was detected by a primary antibody anti-TERT (Rockland Immunochemicals; 600-401-252, 1:1000) and a secondary antibody peroxidase-AffiniPure donkey anti-rabbit immunoglobulin G (H+L) (Jackson; 711-035-152, 1:2000). The HaloTag modified with biotin was detected using Strepavidin-HRP (Pierce; 1:2000). SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific) was used to generate enhanced chemiluminescence signal, which was detected with a FluorChem HD2 imaging system (Alpha Innotech). JF646 fluorescence was detected directly on the acrylamide gel, prior to Western blotting, using a Typhoon Trio PhosphorImager (GE Healthcare).
RNA extraction and Northern blotting
To determine the level of TR contained in telomerase immunopurifications, telomerase elutions (∼50 µl) were subjected to Trizol (Invitrogen; 0.5 ml) extraction following the manufacturer’s instructions. In some instances, a loading and recovery control TR 34-328 (5 ng per sample) was included in the Trizol reagent. Precipitated RNA was resuspended in 50 µl of formamide loading buffer, and half of the sample was separated in a 6% Tris-borate-ethylenediaminetetraacetic acid (TBE) urea polyacrylamide gel (Life Technologies). RNA was transferred onto a Hybond N+ membrane (GE Healthcare) using a wet-blotting apparatus in 1× TBE for 1 h at 1 A of current. Following blotting, the RNA was UV-crosslinked to the membrane and incubated in Church buffer for 2 h at 50°C. TR was detected using three DNA oligos (CTTTTCCGCCCGCTGAAAGTCAGCGAG, CTCCAGGCGGGGTTCGGGGGCTGGGCAG, and CGTGCACCCAGGACTCGGCTCACACATG) that were radioactively labeled using T4 polynucleotide kinase (NEB). Probe (10 × 106 cpm) in Church buffer was incubated with the membrane for at least 2 h. The membranes were washed 3 times with 2xSSC, 0.1% SDS before exposure to a phosphor-imager screen overnight. Detection was carried out using a Typhoon Trio PhosphorImager (GE Healthcare).
Telomere length by Southern blotting
Telomere restriction fragment length analysis was carried out as previously described (Nandakumar and Cech, 2012; Schmidt et al., 2014), using 3 µg of genomic DNA prepared from HeLa cell lines using the GenElute Mammalian Genomic DNA Miniprep kit (Sigma-Aldrich). The rate of telomere length change was calculated by dividing the change in mean telomere length by the number of population doublings (PD), assuming 1.1 PD per day or 21.8 h per PD for the HeLa cells used in these experiments.
Single-molecule primer binding and telomerase activity assay
Surface-passivated Nexterion coverslips (22 × 22 mm, 170 ± 5 µm thickness, Schott) were prepared as follows. Coverslips were first cleaned with 3% Alconox (Alconox), which was brought to a boil in the microwave for 30 min in a sonicating water bath. After three washes with double-distilled water (ddH2O), the coverslips were treated with Piranha solution (3 parts conc. H2SO4, 1 part H2O2) followed by three ddH2O washes. The glass surface was activated in two steps. First, the coverslips were sonicated in 1 M fresh KOH for 30 min; after three ddH2O washes, the coverslips were dried and subjected to 60 min of UZ/Ozone cleaning (Novascan Technologies). Coverslips were silanized by vapor deposition of N-(2-aminoethyl)-3-aminopropyl-triethoxysilane (Gelest) in a desiccator for 4–16 h. After silanization, the coverslips were derivatized using PEG 5000–succinimidyl valerate (8% solution in fresh 0.1 M sodium bicarbonate) containing a small amount (∼3%) of Biotin-PEG 5000–succinimidyl valerate (Laysan Bio) for 24 h. Imaging chambers were assembled by taping a 18 × 18 mm coverslip onto a 22 × 22 mm coverslip using double-sided stick tape and mounted on the microscope using a custom-made holder as previously described (Schmidt et al., 2012). Each coverslip yielded three channels with a volume of 5–10 µl. To prepare the chambers for telomerase immobilization, the channels were incubated with NeutrAvidin (50 ng/ml in PBS, Thermo Scientific) for 5 min. Channels were then washed with five channel volumes of imaging buffer (50 mM Tris, pH 8.0, 50 mM KCl, 1 mM MgCl2, 0.5 mg/ml bovine serum albumin, 0.05% TWEEN-20, 2 mM TROLOX, 0.2 mg/ml glucose oxidase, 0.035 mg/ml catalase, 4.5 mg/ml glucose) and incubated in imaging buffer for 5 min. To analyze primer binding to telomerase, telomerase purified from HEK293T cells expressing 3xFLAG-HaloTag-TERT derivatized with Biotin-PEG-HaloTag-ligand was diluted 1:20 in imaging buffer and incubated with 20 nM of fluorescent primer A5 (Cy3-TTTTTAGGGTTAGCGTTAGGG, IDT) for 5 min. When analyzing the impact of imetelstat on primer binding, primer substrate and imetelstat were added simultaneously to telomerase. The telomerase solution was then loaded into the imaging channels and immobilized for 5 min before washing the channel with five channel volumes of imaging buffer. To visualize telomerase activity, immobilized telomerase-primer complexes were incubated with 500 µM each dATP, dTTP, and dGTP and 10 nM of detection oligonucleotide (Cy5-CCCTAACCCTAACCC, IDT) in imaging buffer for 5 min prior to imaging. The fraction of active telomerase RNPs was calculated as the ratio of primer molecules that colocalize with a product probe signal over the total number of primer molecules detected. TIRF imaging was carried out using a Nikon N-STORM microscope equipped with a TIRF illuminator; 405-nm (20-mW), 488-nm (50-mW), 561-nm (50-mW), and 647-nm (125-mW) laser lines; an environmental chamber to control humidity and temperature; two iXon Ultra 897 electron-multiplying charge-coupled device (EMCCD) cameras (Andor); a 100× oil-immersion objective (Nikon, NA = 1.49); two filter wheels; and the appropriate filter sets. To analyze primer binding, 10 frames of a given field of view were acquired at 20 frames per second. Average intensity projections of these short image sequences were analyzed using in-house MATLAB code, implementing particle detection code publicly available (https://site.physics.georgetown.edu/matlab/code.html). To determine the number of primer molecules in a given field of view, the number of particles with intensities corresponding to a single fluorophore was determined by fitting the intensity profiles of detected particles to a normal distribution and counting the number of particles within one SD from the mean of this distribution. To determine the fraction of primer molecules bound in the presence of imetelstat, the average particle number of five fields of view was divided by the number of particles detected in the absence of drug. One channel of each coverslip used for this experiment was a no drug control to account for coverslip surface variability. To determine the IC50 for inhibition of primer binding by imetelstat, the fraction of primer bound was plotted as a function of Imtelstat concentration and fit to a binding curve Fraction bound = 1 – [Drug]/([Drug] + IC50). To image telomerase product formation, primer substrate and product detection oligonucleotide were imaged simultaneously for 10 frames at 20 frames per second. The percentage of active telomerase RNPs was determined as the fraction of primer signals that colocalized with a product signal divided by the total number of primers detected.
Single-molecule live-cell imaging
Three-color single-molecule live-cell imaging was carried out as previously described (Schmidt et al., 2016), using the CRISPR genome-edited HeLa cell line stably expressing 3xFLAG-HaloTag TERT and mEOS3.2-TRF2 from their respective endogenous loci. Briefly, BFP-coilin was transfected into cells 48 h prior to imaging, followed by a double thymidine block. Imetelstat was added to cells 24 h before imaging, simultaneously with the release from the first thymidine block. Three to 4 hours after release into S-phase, FLAG-HaloTag-TERT was labeled by subjecting cells to a 2-min pulse of 100 nM JF646 HaloTag-ligand (a kind gift from Luke Lavis) in tissue culture medium (Grimm et al., 2015). BFP-coilin was imaged first for ∼1 s under continuous illumination. 3xFLAG-HaloTag-TERT and mEOS3.2-TRF2 (red state) were imaged simultaneously. Movies were acquired for 15 s on a Nikon N-STORM microscope under highly inclined and laminated optical sheet conditions (Tokunaga et al., 2008), with a 1.49 NA 100× oil-immersion TIRF objective (Nikon) at 46 frames per second. The two imaging channels were projected onto two iXon Ultra 897 EMCCD cameras (Andor) using TwinCam dual emission image splitter (Cairn). The channels were aligned prior to every imaging session using TetraSpeck microspheres (ThermoFisher).
Single-particle tracking
Single-particle trajectories were generated with MatLab 2011b (Mathworks) using SLIMfast, which implements the Multiple-Target-Tracing algorithm (Sergé et al., 2008; Liu et al., 2014) and evaluated using the script evalSPT (Normanno et al., 2015). Particle detection was carried out using 9 × 9 pixel detection, error rate of 10-6, and one deflation loop. Particle tracking for determining diffusion coefficients was carried out by setting the upper bound of the expected diffusion coefficient to D = 5 µm2/s. To determine the lifetime of short “probing” interactions, the maximal expected diffusion coefficient was set to D = 0.1 µm2/s, the maximal OFF-Time to three frames, and intensity fluctuation weight to 0.5. To analyze the binding properties of TERT particles at different nuclear locations, TERT tracks were assigned to telomeres, Cajal bodies, or other nuclear locations as previously described (Schmidt et al., 2016). To determine the dissociation rate of TERT interactions at these locations, the survival probabilities of TERT particles were fit to a double exponential decay function Y = A*e(-kfast*t) + B*e(-kslow*t) using tracks ranging from 0.044 to 1 s (2–46 frames), which encompassed >95% of the detected trajectories. The fraction of the particles that dissociate with the slower rate constant was calculated as Fractionslow = B/(A+B). Long, static interactions were identified by manual inspection of movies from HeLa cells expressing 3xFLAG-HaloTag and mEOS3.2-TRF2, as previously described (Schmidt et al., 2016). All analysis was carried out blinded to whether the particular movie was generated from cells treated with imetelstat, mismatched control oligonucleotide, or no drug.
Code availability
All Matlab scripts are available on request from the authors.
Supplementary Material
Acknowledgments
We thank Cech lab members Dan Youmans, Ci Ji Lim, and Yicheng Long for assistance with experiments and useful discussions; Joe Dragavon and the BioFrontiers Advanced Light Microscopy Core for assistance with imaging; Scott Cohen (Children’s Medical Research Institute and University of Syndey, Sydney, Australia) for providing the TERT antibody used for immunopurification of telomerase; and Luke Lavis and Zhe Liu (HHMI Janelia Research Campus) for providing HaloTag dyes and assistance with data analysis. We especially thank Janssen Research and Development, LLC, for the generous gift of imetelstat and the mismatched control oligonucleotide, which were essential for this study. This study was supported by grants from the National Institutes of Health to J.C.S. (K99GM120386) and T.R.C. (R01GM099705). R.K. was a Master student, University of Regensburg, under the direction of Herbert Tschochner and T.R.C. T.R.C. is an investigator of the Howard Hughes Medical Institute.
Abbreviations used:
- bp/PD
base pairs per population doubling
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- ddH2O
double-distilled water
- DTT
dithiothreitol
- EMCCD
electron-multiplying charge-coupled device
- FBS
fetal bovine serum
- HRP
horseradish peroxidase
- IC50
half-maximal inhibition
- IP
immunoprecipitation
- LC
loading controls
- MM
mismatch
- OB
oligonucleotide/oligosaccharide-binding
- PD
population doubling
- RNP
ribonucleoprotein
- ss
single-stranded
- TERT
telomerase reverse transcriptase
- TIRF
total internal reflection fluorescence
- TR
telomerase RNA
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-11-0637) on January 31, 2018.
REFERENCES
- Armanios M, Blackburn EH. (2012). The telomere syndromes. Nat Rev Genet , 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai A, Oshima Y, Yamamoto Y, Uochi TA, Kusaka H, Akinaga S, Yamashita Y, Pongracz K, Pruzan R, Wunder E. (2003). A novel telomerase template antagonist (GRN163) as a potential anticancer agent. Cancer Res , 3931–3939. [PubMed] [Google Scholar]
- Bell RJA, Rube HT, Kreig A, Mancini A, Fouse SD, Nagarajan RP, Choi S, Hong C, He D, Pekmezci M. (2015). Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science , 1036–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borah S, Xi L, Zaug AJ, Powell NM, Dancik GM, Cohen SB, Costello JC, Theodorescu D, Cech TR. (2015). Cancer. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science , 1006–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR. (2004). Beginning to understand the end of the chromosome. Cell , 273–279. [DOI] [PubMed] [Google Scholar]
- Chiba K, Lorbeer FK, Shain AH, McSwiggen DT, Schruf E, Oh A, Ryu J, Darzacq X, Bastian BC, Hockemeyer D. (2017). Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two-step mechanism. Science , 1416–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba K, Vogan JM, Wu RA, Gill MS, Zhang X, Collins K, Hockemeyer D. (2016). Endogenous TERT N-terminal tagging affects human telomerase function at telomeres in vivo. Mol Cell Biol , e00541–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, Lingner J. (2007). Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol Cell , 882–889. [DOI] [PubMed] [Google Scholar]
- de Lange T. (2005). Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev , 2100–2110. [DOI] [PubMed] [Google Scholar]
- Grimm JB, English BP, Chen J, Slaughter JP, Zhang Z, Revyakin A, Patel R, Macklin JJ, Normanno D, Singer RH. (2015). A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods , 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. (1990). Telomeres shorten during ageing of human fibroblasts. Nature , 458–460. [DOI] [PubMed] [Google Scholar]
- Herbert B-S, Gellert GC, Hochreiter A, Pongracz K, Wright WE, Zielinska D, Chin AC, Harley CB, Shay JW, Gryaznov SM. (2005). Lipid modification of GRN163, an N3“→P5” thio-phosphoramidate oligonucleotide, enhances the potency of telomerase inhibition. Oncogene , 5262–5268. [DOI] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fisher C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K. (2013). TERT promoter mutations in familial and sporadic melanoma. Science , 959–961. [DOI] [PubMed] [Google Scholar]
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. (2013). Highly recurrent TERT promoter mutations in human melanoma. Science , 957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H, Opresko P, Myong S. (2014). Single-molecule real-time detection of telomerase extension activity. Sci Rep , 6391–6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrick CM, Cech TR. (2010). POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J , 924–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Legant WR, Chen B-C, Li L, Grimm JB, Lavis LD, Betzig E, Tjian R. (2014). 3D imaging of Sox2 enhancer clusters in embryonic stem cells. Elife , e04236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Cech TR. (2012). DNA-induced dimerization of the single-stranded DNA binding telomeric protein Pot1 from Schizosaccharomyces pombe. Nucleic Acids Res , 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanno D, Boudarene L, Dugast-Darzacq C, Chen J, Richter C, Proux F, Bénichou O, Voituriez R, Darzacq X, Dahan M. (2015). Probing the target search of DNA-binding proteins in mammalian cells using TetR as model searcher. Nat Commun , 7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauerwald A, Sandin S, Cristofari G, Scheres SHW, Lingner J, Rhodes D. (2013). Structure of active dimeric human telomerase. Nat Struct Mol Biol , 454–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Arthanari H, Boeszoermenyi A, Dashkevich NM, Wilson-Kubalek EM, Monnier N, Markus M, Oberer M, Milligan RA, Bathe M. (2012). The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev Cell , 968–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Cech TR. (2015). Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev , 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Dalby AB, Cech TR. (2014). Identification of human TERT elements necessary for telomerase recruitment to telomeres. Elife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Zaug AJ, Cech TR. (2016). Live cell imaging reveals the dynamics of telomerase recruitment to telomeres. Cell , 1188–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergé A, Bertaux N, Rigneault H, Marguet D. (2008). Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat Methods , 687–694. [DOI] [PubMed] [Google Scholar]
- Sexton AN, Youmans DT, Collins K. (2012). Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J Biol Chem , 34455–34464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern JL, Theodorescu D, Vogelstein B, Papadopoulos N, Cech TR. (2015). Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes Dev , 2219–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Weinberg RA. (2006). Telomeres: cancer to human aging. Annu Rev Cell Dev Biol , 531–557. [DOI] [PubMed] [Google Scholar]
- Tokunaga M, Imamoto N, Sakata-Sogawa K. (2008). Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods , 159–161. [DOI] [PubMed] [Google Scholar]
- Wu RA, Tam J, Collins K. (2017a). DNA-binding determinants and cellular thresholds for human telomerase repeat addition processivity. Embo J , 1908–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RA, Upton HE, Vogan JM, Collins K. (2017b). Telomerase mechanism of telomere synthesis. Annu Rev Biochem , 439–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi L, Cech TR. (2014). Inventory of telomerase components in human cells reveals multiple subpopulations of hTR and hTERT. Nucleic Acids Res , 8565–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Crary SM, Jesse Fioravanti M, Campbell K, Cech TR. (2013). Many disease-associated variants of hTERT retain high telomerase enzymatic activity. Nucleic Acids Res , 8969–8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE. (2009). Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell , 463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong FL, Batista LFZ, Freund A, Pech MF, Venteicher AS, Artandi SE. (2012). TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell , 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.