Summary
Detailed clinical and molecular evaluation of large cohorts of exceptional survivors provide an unprecedented opportunity to identify mechanisms underlying long term survival that can drive future therapeutic approaches and biomarker development. Exceptional survivors of high-grade serous ovarian cancer demonstrate concurrent disruption of homologous recombination DNA repair and retinoblastoma protein.
In this issue of CLINICAL CANCER RESEARCH, Garsed and colleagues use a novel and powerful approach of evaluating a large set of ovarian cancers patients to develop a cohort of unusual responders. This paradigm provides critical insight into mechanisms underlying long-term survival in high-grade serous ovarian cancer (HGSC) that could lead to therapeutic approaches designed to convert all HGSC patient into long-term survivors. Importantly, application of this paradigm could greatly improve our understanding of therapeutic liabilities in multiple cancer lineages (1).
HGSC is often diagnosed at an advanced stage with fewer than half of all HGSC patients surviving 5-years. However, a subset of HGSC patients have prolonged progression free survival, repeated responses to platinum-based therapy and prolonged survival. Most studies of exceptional responders focus on one or a small number of patients increasing the challenges of linking observations to causality. Through detailed clinical, pathological and molecular characterization of 96 exceptional HGSC survivors, Garsed and colleagues establish that patients who experience repeat responses to platinum based therapy are enriched for tumors with abnormalities in homologous recombination (HR)-mediated DNA repair. Patients with long progression free survival are more likely to have concurrent defective HR and retinoblastoma (Rb) protein loss. In contrast, long-term survivors are comprised of two subsets of patients, the set whose tumors have concurrent HR and Rb protein deficiency and a second subset with high proliferative rates and tumor infiltrating lymphocytes. This offers the intriguing possibility that inducing concurrent HR deficiency and Rb protein loss could markedly sensitize patients to platinum based chemotherapy.
In this study, three partially overlapping ‘exceptional responders/survivors’ were defined including: (1) long progression-free survival (PFS) of >36 months (Long-PFS); (2) Multiple Responders (MR) with three or more complete responses to chemotherapy; (3) long-term survivors (more than 10 years). Patterns of recurrent p53 mutations, which are almost universal in HGSC, did not predict survival benefit. Strikingly, 10 genes involved in HR repair (BRCA1, BRCA2, RAD51C, BRIP1, CDK12, PTEN, ATR, CHK1, CHK2 and RAD51D) were mutated in 73% of exceptional responders, which is significantly higher than the 41% observed in the unselected TCGA cohort. Notably, the highest portion of HR defects (88%) and BRCA1 mutations (59%) were found in MR reflecting a critical role for HR-deficiency in sensitivity to platinum-based chemotherapy. In contrast, RB1 loss was more frequent in long-FPS and long-term survivors than in MR. Importantly, co-occurrence of RB1 loss and HR-deficiency was highly enriched in long-PFS and long-term survivor groups. Surprisingly the majority of long-term survivors did not demonstrate BRCA1/2 abnormalities. Indeed a subset of long term survivors exhibited increased CD8 positive infiltrating T cells and, unexpectedly, high levels of the Ki67 proliferative marker. Taken together these observations offer the intriguing possibility that inducing concurrent defects in homologous recombination repair and RB1 protein could render “average” HGSC patients sensitive to platinum-based or potentially PARP inhibitor therapy increasing the number of long term survivors. Further, the likely long term outcomes for patients could better tailor therapy by embracing chemotherapy in patients likely to become long term responders and directing the remaining patients to alternative therapy approaches or focusing on palliation using regimens with more acceptable toxicity.
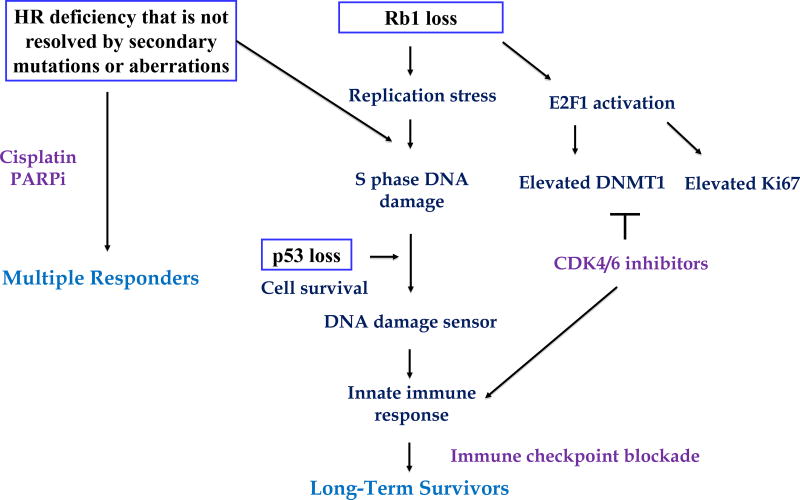
We propose a model that encompasses these important clinical and molecular observations that suggests intriguing therapeutic opportunities (Figure 1).
Figure 1.
A proposed model for concurrent deficiency of HR repair and Rb1 in exceptional survival in high-grade serous ovarian cancer.
HR represents an error-free repair mechanism to repair and relieve replication-associated DNA damage to maintain genomic integrity (2). Defective HR sensitizes cells to platinum-based therapy as well as to PARP inhibitors. Thus it is not surprising that HR deficiency defined by analysis of 32 genes implicated in DNA damage repair was enriched in exceptional responders and particularly in MR. A more extensive analysis of the broad set of genes involved in HR may further strengthen this association. It is intriguing to hypothesize that these MR patients are unable to engage mechanisms to reconstitute HR that are associated with both platinum and PARP inhibitor resistance such as “healing” of BRCA1 and Rad51. It also raises the intriguing question as to whether clinical trials of PARP inhibitor maintenance therapy in MR that led to approval of PARP inhibitors in HGSC were more markedly enriched for HR defects than was proposed by analysis of BRCA1/2 and HRD by loss of heterozygosity.
The RB1 tumor suppressor functions as a key regulator of the S-phase transition with loss leading to uncontrolled cell proliferation. Perhaps contributing to the interaction with HR deficiency seen in long-term survivors, loss of RB1 induces replication stress due to defects in replication fork progression and mitosis (3) that leads to increased DNA damage, which is most effectively repaired by HR (4). This increased DNA damage coupled with a decreased ability to repair DNA damage induced by platinum drugs could contribute to increased chemosensitivity and improved long-term survival.
In cells with normal p53 function, the DNA damage checkpoint is activated and if DNA damage is unresolved apoptosis eliminates damaged cells. However, in HGSC, the almost universal p53 mutations allow survival of cells with unresolved DNA damage by weakening the DNA damage checkpoint and apoptosis programs (5). It is possible that accumulation of DNA damage due to concurrent disruption of HR repair, Rb1 and p53 leads to activation of cytosolic DNA damage sensors that promote innate immune responses through mechanisms such as the STING pathway engaging anti-tumor immunity and contributing to long-term survival (6).
CDK4/6 inhibition triggers anti-tumor immunity by reducing expression of the E2F1 target gene DNMT1 thus activating innate immune responses through production of type III interferons (7). Based on this model, we speculate that immune checkpoint blockade in combination with CDK4/6 inhibitors may provide additional survival benefits in HGSC patients with concurrent deficiency in HR repair, Rb1 and p53 as well as in the population with CD8 T cell infiltration.
The HR repair network is composed of a number of proteins including DNA damage sensors, signaling transducers, repair effectors, and epigenetic regulators (2). It is extremely challenging to evaluate the complete spectrum of mutations, genomic alterations, methylation and protein levels for the large number of genes involved in HR repair. Moreover, post-translational protein modification and appropriate formation of molecular complexes are required for efficient HR repair. Unfortunately, functional HR repair assays such as RAD51 foci formation after DNA damage or HR reporter assays using DR-GFP and I-SceI plasmids are difficult to implement using clinical samples. Thus, a definitive assay for HR competency in patient samples remains elusive. To develop molecular markers of defective HR, recent studies have utilized genomic or transcriptomic changes that occur as consequence of HR repair deficiency, including patterns of loss of heterozygosity (8), transcriptomic alterations (9,10), and distinct somatic mutation patterns (11,12). This offers the exciting potential that emerging molecular signatures of HR deficiency might predict MR or long-term survival when assessed in combination with loss of Rb1.
In summary, as demonstrated by the current report, multiple molecular determinants of exceptional responders/survivors are associated with favorable outcomes. Distinct molecular pathways are identified in distinct subgroups such as HR deficiency in MR and Rb loss in Long-Term Survivors and Long-PFS. This information has the potential to lead to new therapeutic approaches as well methods to maximize patient benefit based on the likelihood of response to particular therapy approaches.
Acknowledgments
Funding: This research was supported by NCI Cancer Center Support Grant CA016672 to The University of Texas MD Anderson Cancer Center, Department of Defense grant OC140431, NIH R01 grant CA181663 to G.P., Cancer Prevention and Research Institute of Texas grant RP160242 to G.P.
Conflict of Interest: G. B. Mills is a consultant for AstraZeneca, Catena Pharmaceuticals, Critical Outcome Technologies, ImmunoMET, Ionis, Medimmune, Nuevolution, Pfizer, Precision Medicine, Signalchem Lifesciences, Symphogen, Takeda/Millennium Pharmaceuticals, Tarveda, has stock options with Catena Pharmaceuticals, ImmunoMet, SignalChem, Spindle Top Ventures, Tarveda, has licensed technology to Myriad Genetics and has sponsored research support from Abbvie, Adelson Medical Research Foundation, AstraZeneca, Breast Cancer Research Foundation, Critical Outcomes Technology, Horizon Diagnostics, Illumina, Ionis, Immunomet, Karus Therapeutics, Komen Research Foundation, Pfizer, Nanostring, Takeda/Millennium Pharmaceuticals, Tesaro
G. Peng has sponsored research support from Pfizer.
References
- 1.Garsed DW, Alsop K, Fereday S, Emmanuel C, Kennedy C, Etemadmoghadam D, et al. Homologous Recombination DNA Repair Pathway Disruption and Retinoblastoma Protein Loss are Associated with Exceptional Survival in High-Grade Serous Ovarian Cancer. Clin Cancer Res. doi: 10.1158/1078-0432.CCR-17-1621. [DOI] [PubMed] [Google Scholar]
- 2.Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol. 2013;5(11):a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coschi CH, Ishak CA, Gallo D, Marshall A, Talluri S, Wang J, et al. Haploinsufficiency of an RB-E2F1-Condensin II complex leads to aberrant replication and aneuploidy. Cancer Discov. 2014;4(7):840–53. doi: 10.1158/2159-8290.CD-14-0215. [DOI] [PubMed] [Google Scholar]
- 4.Gaillard H, Garcia-Muse T, Aguilera A. Replication stress and cancer. Nat Rev Cancer. 2015;15(5):276–89. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- 5.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408(6811):433–9. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 6.Parkes EE, Walker SM, Taggart LE, McCabe N, Knight LA, Wilkinson R, et al. Activation of STING-Dependent Innate Immune Signaling By S-Phase-Specific DNA Damage in Breast Cancer. J Natl Cancer Inst. 2017;109(1) doi: 10.1093/jnci/djw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goel S, DeCristo MJ, Watt AC, BrinJones H, Sceneay J, Li BB, et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature. 2017;548(7668):471–5. doi: 10.1038/nature23465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abkevich V, Timms KM, Hennessy BT, Potter J, Carey MS, Meyer LA, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer. 2012;107(10):1776–82. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng G, Chun-Jen Lin C, Mo W, Dai H, Park YY, Kim SM, et al. Genome-wide transcriptome profiling of homologous recombination DNA repair. Nat Commun. 2014;5:3361. doi: 10.1038/ncomms4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pitroda SP, Pashtan IM, Logan HL, Budke B, Darga TE, Weichselbaum RR, et al. DNA repair pathway gene expression score correlates with repair proficiency and tumor sensitivity to chemotherapy. Sci Transl Med. 2014;6(229):229ra42. doi: 10.1126/scitranslmed.3008291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3(1):246–59. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]