Abstract
A major obstacle in understanding the complex biology of the malaria parasite remains to discover how gene transcription is controlled during its life cycle. Accumulating evidence indicates that the parasite’s epigenetic state plays a fundamental role in gene expression and virulence. Using a comprehensive and quantitative mass spectrometry approach, we determined the global and dynamic abundance of histones and their covalent post-transcriptional modifications throughout the intra-erythrocytic developmental cycle of Plasmodium falciparum. We detected a total of 232 distinct modifications, of which 160 had never been detected in Plasmodium and 88 had never been identified in any other species. We further validated over 10% of the detected modifications and their expression patterns by multiple reaction monitoring assays. In addition, we uncovered an unusual chromatin organization with parasite-specific histone modifications and combinatorial dynamics that may be directly related to transcriptional activity, DNA replication, and cell cycle progression. Overall, our data suggest that the malaria parasite has a unique histone modification signature that correlates with parasite virulence.
Keywords: Cell cycle, Epigenetics, Histones, Label-free quantification, Malaria, Multiple reaction monitoring, Parasite, Post-translational modifications, Tandem Mass Spectrometry
Table of Contents Graphic
Introduction
In all eukaryotic genomes, DNA is packed with histone proteins to form chromatin. The dynamic structure of these chromatin fibers impacts several processes, including replication, transcription and repair, by modulating DNA accessibility. Each core histone contains two functional domains: a histone-fold motif, which mediates histone/histone and histone/DNA interactions, and an N-terminal tail (NT) that projects from the core and can undergo a large number of post-translational modifications (PTMs). Known histone PTMs in model eukaryotes include acetylation, methylation, phosphorylation, ubiquitylation, sumoylation, ADP-ribosylation, propionylation, butyrylation, formylation, citrullination, proline isomerization1, 2, and the recently reported crotonylation3. These modifications can be found in complex combinations and are primarily localized in the accessible N-terminal tails. Variant histones, found in subsets of nucleosomes, can provide additional complexity. The combined occurrence of histone PTMs and histone variants at a particular chromosomal locus is thought to play a key role in gene regulation. Some histone PTMs have been associated with specific cellular functions such as transcriptional activation (e.g., H3 and H4 lysine acetylations), transcriptional elongation (e.g., H3-K36 methylation), or transcriptional repression (e.g., H3-K9, H3-K27, and H4-K20 methylations). During DNA duplication, chromatin not only undergoes destabilization and reassembly on the two daughter strands but histones recycle during the formation of the new nucleosomes and histone PTMs are transferred to the daughter chromatids. This process needs to be tightly regulated as uncoordinated or imperfect replication can cause genomic instability and developmental abnormalities. As with transcriptional activation, elongation or repression, several lines of evidence suggest that histone PTMs play a role in coordinating DNA replication. Histone lysine methylation such as H4-K20me2 and H3-K79me2 or the euchromatin markers, H3-K4me3 and H3-K9ac, are often found at replication origins, while H3-K27me1 is also often detected at elongation sites4, 5. As a whole, dynamic changes of histone PTMs involved in replication and transcription correlate generally well with the genetic activity of the cell6.
The protozoan parasite Plasmodium falciparum causes the most severe form of human malaria. In humans, symptomatic disease is associated with intensive parasite replication in red blood cells. During this intra-erythrocytic developmental cycle (IDC), the parasite progresses through three distinct stages, ring, trophozoite, and schizont, of which the trophozoite stage in particular is characterized by high transcriptional activity. It is also at this stage that the parasite starts to replicate its DNA. Between the trophozoite and the schizont stages, the parasite undergoes multiple rounds of asynchronous nuclear division (up to 16) without chromosomes condensation or loss of the nuclear membrane. To better understand the DNA replication process in the malaria parasite, we not only need to determine DNA replication origins, the molecular components of the pre-replication complex, but also the chromatin landscape that participates in genome replication and how histone post-translational modification patterns are transferred to the daughter chromatids. Furthermore, in addition to these high transcriptional and DNA replication activities, Plasmodium has the capacity to develop phenotypic diversity by the selection of clonally variant parasites. The exact molecular processes regulating cell cycle progression and clonal variant selection at the transcriptional level are still poorly understood, but emerging evidence indicates that chromatin structure plays a critical role in regulating transcription7–14.
Most canonical and structural variant histones (H2A, H2A.Z, H2B, H2B.Z, H3, H3.3, H4, and the centromeric-specific CenH3) have been identified in the malaria parasite, but no clear homolog of the linker histone H1 has been found15. Initial genome-wide chromatin studies demonstrated an abnormal distribution of histone marks. H3-K36me3 and the silencing H3-K9me3 mark are uniquely associated with repression of genes involved in antigenic variation within restricted subtelomeric and chromosome internal regions10, 16, 17. On the contrary, active marks such as H3-K4me3 and H3-K9ac have a broad distribution across the genome. Finally, preliminary mass spectrometry studies identified a limited number of additional histone PTMs18–20. However, the detailed role of chromatin and histone PTMs in DNA replication, control of transcription and pathogenicity of the parasite is incompletely understood and deserves a deeper investigation.
Histone PTMs have been identified with a variety of techniques, including immunohistochemistry, chromatography, spectroscopy and mass spectrometry. The characterization of all combinatorial histone PTMs is a great analytical challenge. Antibodies can only measure known PTMs independently and are not designed to achieve a complete combinatorial data set. Furthermore, adjacent modifications can occlude the epitope that the antibody is designed to recognize, leading to biased results21, 22. The broad understanding of the role of epigenetic mechanisms in controlling a eukaryotic cell requires an extensive characterization of all histone modifications in a combinatorial manner. Recently, the ability to determine which PTMs co-exist in a particular cell on a genome-wide scale has emerged by the development of High-Throughput Histone Code Analysis (HT-HCA) methods using mass spectrometry (reviewed in ref.23). While not the most accurate to conserve the combinatorial information, bottom up mass spectrometry, which sequences small peptides from enzymatic digestions, provides sufficient throughput and quantitative analysis for biologically meaningful information. Similar to how the current high-throughput next-generation DNA sequencers can rapidly analyze genomes, HT-HCA using mass spectrometry can significantly contribute to a better understanding of the epigenome in the malaria parasite.
Using the comprehensive and quantitative Multidimensional Protein Identification Technology (MudPIT), we report an extensive atlas of histone PTMs across the parasite IDC. Overall, we detected 232 distinct histone PTMs, of which 160 had never been identified in apicomplexan parasites and 88 had never been discovered in other species. Furthermore, the dynamic and combinatorial analyses of these histone modifications across the IDC suggests that Plasmodium falciparum has a unique histone modification signature that correlates with parasite virulence.
Experimental Procedures
Dataset generation and processing (see Supporting Information SI-1)
Synchronized Plasmodium falciparum strain 3D7-infected human blood cultures24 were harvested every 6 hours at seven time points along the asexual intra-erythrocytic cycle from 0 hours post-invasion (hpi) to 36-hpi. Two biological replicates of acid-extracted histone preparations were digested with enzymes of diverse specificities25, 26: endoproteinase LysC + trypsin, endoproteinase LysC + GluC, and endoproteinase ArgC. The resulting peptide mixtures were analyzed by MudPIT25, 27 on a Thermo Scientific LTQ Orbitrap (SI-1.1).
After offline recalibration by RawDistiller v. 1.028, the MS/MS spectra were searched using SEQUEST v.27 (rev.9)29 with a mass tolerance of 10 ppm for precursor and +/− 0.5 amu for fragment ions. No enzyme specificity was imposed for the search against a protein database combining 5,439 non-redundant Plasmodium falciparum (PlasmoDB release 5.5) and 30,552 human proteins (NCBI 2008-03–04 release), as well as 162 usual contaminants. To estimate false discovery rates (FDR), each protein sequence was randomized leading to a total search space of 72,306 sequences. To account for alkylation, 57.02146 Da were added statically to cysteines.
As described previously30, the MS/MS dataset was searched for modified peptides in a recursive manner (SI-1.2). A total of 44 differential modification searches were set up to query for various combinations methylated lysines and arginines, oxidized methionines and hydroxylated lysines, prolines, aspartates, histidines, and tyrosines, formylated lysines and arginines, dimethylated lysines and arginines, trimethylated lysines, acetylated lysines, serines and threonines, crotonylated lysines, phosphorylated serines, threonines, and tyrosines, ubiquitinated lysines, as well as potentially N-terminally acetylations on methionines (oxidized or not), serines, alanines, and valines (SI-1.3).
Data assembly and statistical analysis (see Supporting Information SI-2)
Results from different digestions were merged for each of the duplicates and time-points were compared using CONTRAST31. NSAF732 was used to create the final reports on all detected peptides and non-redundant proteins identified (SI-2.1). Spectral and protein level FDRs were, on average, 0.25 ± 0.14% and 1.6 ± 0.5%, respectively (SI-2.1). To assess the similarities in relative protein abundance (dNSAF) between all 14 samples, pairwise Pearson correlation coefficients were calculated (SI-2.2.A), while independent analyses were hierarchically clustered (SI-2.2.B). To assess relative enrichment over all detected proteins, the normalized spectral counts values of specific group of proteins (Plasmodium proteins vs. human proteins in SI-2.2.C, and histones in SI-2.2.D) were summed (ΣdNSAF) as described in33. The sequence coverages obtained for each of the eight Plasmodium histones were compared across the seven time-points (SI-2.2.E). To assess variations in histone abundances, a multiple comparison analysis was performed using a Bonferroni correction on dNSAF values (SI-2.2.D and SI-2.2.F).
Identification of histones modifications (see Supporting Information SI-3)
Spectra/peptide matches (PSMs) were filtered using conservative criteria using DTASelect31. PSMs were only retained if they had a DeltCn of at least 0.08. Minimum XCorr values were set at 1.8 for singly-, 2.0 for doubly-, and 3.0 for triply-charged spectra for the Arg-C and trypsin-digested samples, and at 2.5 and 3.5 for +2 and +3 spectra in the Lys-C+Glu-C digested samples. In addition, the peptides had to be at least seven amino acids long and comply with the specificity of the enzymes used to digest samples. All PSMs assigned by SEQUEST29 to a modified peptide from Plasmodium histones were sorted into three categories: i) peptides containing modifications that had been previously reported in the literature (SI-3.1 and Fig. 1A/S1) in Plasmodium and/or other organisms (based on sequence alignments in SI-3.2 and tridimensional structures in SI-3.3); ii) peptides containing at least one newly reported modification (Table 1); iii) peptides containing only oxidized methionines. Any PSM with at least one newly described modification was cross-validated against OMSSA34 search results for the same spectrum (SI-1.2 and SI-1.3). Spectra mapped to the exact same peptide by both search engines were flagged yes (Y) using SQTMASSFixer28, while spectra matching the same sequence but with the modification located on different residues were flagged maybe (M) (SI-3.4). Spectra matched to different sequences by the two search engines were removed from the final data. All spectra matching the reported modified peptides were visually assessed (SI-3.5).
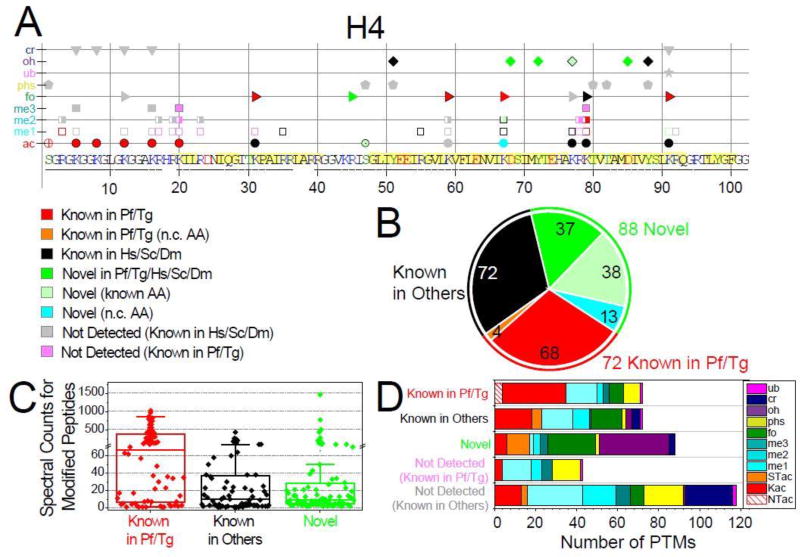
Figure 1. Qualitative overview of Plasmodium histones modifications.
A. Detected PTMs on Plasmodium histone H4 are plotted along its sequence, while the plots for H2A, H2A.Z, H2B, H2B.Z, H3, and H3.3 are provided in Fig. S1. PTMs are sorted into three main categories based on their novelty status (see SI-3.1): 1) PTMs previously reported in Plasmodium falciparum and/or Toxoplasma gondii (red and orange symbols for non-conserved (n.c.) residues); 2) PTMs previously reported in species other than apicomplexans (black symbols); 3) novel PTMs on conserved residues are in bright green symbols, a subset of those are in lighter green symbols drawn with a black edge to denote that the amino acid had been previously reported as modified by other types of PTMs, while novel modifications on n.c. residues are in cyan. In addition, PTMs previously reported in other organisms but were not detected (n.d.) in our analysis are plotted as grey symbols, while PTMs previously detected in apicomplexans are colored in light pink (SI-3.1). Underlined amino acid residues are covered by detected peptides. Amino acids predicted to be within structured domains (SI-3.2) are highlighted in yellow. B. Total numbers of PTMs for each of the novelty categories. C. Spectral counts for peptides bearing the 232 detected PTMs within each of the novelty categories. D. Tally of the PTMs types falling into each of the novelty categories and the known PTMs not detected in our analysis.
Table 1.
Overview of the novel modifications identified in P. falciparum
| PTM | H2A | H2A.Z | H2B | H2B.Z | H3 | H3.3 | CenH3 | H4 |
|---|---|---|---|---|---|---|---|---|
| Kac | K35c | K23nc | K67a, nc | |||||
| K64nc | K26nc | |||||||
| K60a, nc | ||||||||
|
| ||||||||
| STac | T101a | S24 | T30nc | T11 | S47a | |||
| S28a | S54 | S86 | ||||||
| S50nc | T107 | |||||||
| T61nc | ||||||||
|
| ||||||||
| me1 | K75 | K91a | ||||||
|
| ||||||||
| me2 | K64 | K115 | K67 | |||||
|
| ||||||||
| me3 | K35 | K115 | ||||||
| K49 | ||||||||
| K64 | ||||||||
|
| ||||||||
| fo | K20 | R115 | R25 | R29 | K14 | R108 | R45 | |
| K41nc | K49 | K53 | K27 | |||||
| R42 | K64 | R95 | K36 | K36 | ||||
| R42b | R42b | |||||||
| R53 | K53 | |||||||
| R72 | R72 | |||||||
| R83 | ||||||||
| R116b | R116b | |||||||
|
| ||||||||
| oh | P48 | Y77 | Y32 | K53 | Y54 | D68 | ||
| Y50 | H138 | K35 | D64 | K79 | Y72 | |||
| Y57 | K139 | K38 | D67nc | K53 | K77 | |||
| K118 | P42 | Y99 | D85 | |||||
| K49 | D106 | D106 | ||||||
| D60 | H113 | H113 | ||||||
| K64 | K115 | K115 | ||||||
| P95 | P121b | P121b | ||||||
| K100 | ||||||||
|
| ||||||||
| phs | S56a | Y78nc | ||||||
|
| ||||||||
| cr | K49a | K115a | K115 | |||||
Label-free quantitation of modification levels (see Supporting Information SI-4)
NSAF732 was used to extract total and modified label-free features for each amino acid within Plasmodium histones (SI-4.1.A) and calculate modification levels based on local spectral counts30 or MS1 peak intensities35. The correlation between modifications levels calculated using these two features was evaluated by linear regression through the relative levels of the 202 quantified PTMs (SI-4.1.B). Modifications levels were also calculated for each replicate analysis (SI-4.2 and Fig. 2/S2 for spectral counts and SI-4.3 for sum of MS1 peak intensities).
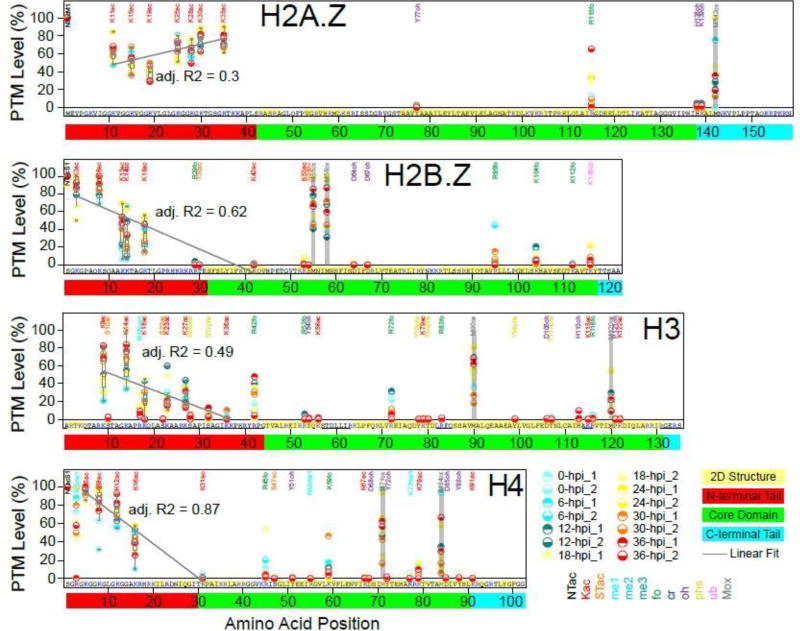
Figure 2. Quantitative profiles of modification levels on Plasmodium histones.
Percentages of modification occupancy calculated based on local spectral counts (SI-4.1) from all 14 analyses (“Mod/Total (%)” columns in SI-4.2) are plotted along the whole sequences of Plasmodium H2A.Z, H2B.Z, H3, and H4, while the plots for the other histones are provided in Fig. S2. Boundaries for the structural domains (N-terminal tail, core domain, and C-terminus) are shown by the red, green, and cyan boxes around the sequence numbering. When multiple PTMs are observed on one residue (SI-4.1), the modification with the highest levels is reported here. The measured levels of methionine oxidations are plotted for reference (grey bars). Box-plots are overlaid above the data points for the K acetylations within the N-terminal tails. Linear regressions through the data points within these boxes are plotted as a grey line and the adjusted R2 statistics calculated for each regression are provided.
Dynamic and combinatorial profiles of modification levels across the erythrocyte cycle (see Supporting Information SI-5)
Modifications levels calculated based on spectral counts (SpC) or MS1 peak intensities (PI) were hierarchically clustered (SI-5.1) using PermutMatrix software with Euclidean as the distance metric and Ward’s method36. The trees were generated using the multiple-fragment heuristic algorithm (MF) as a seriation rule (Fig. 3A/S3 and SI-5.2).
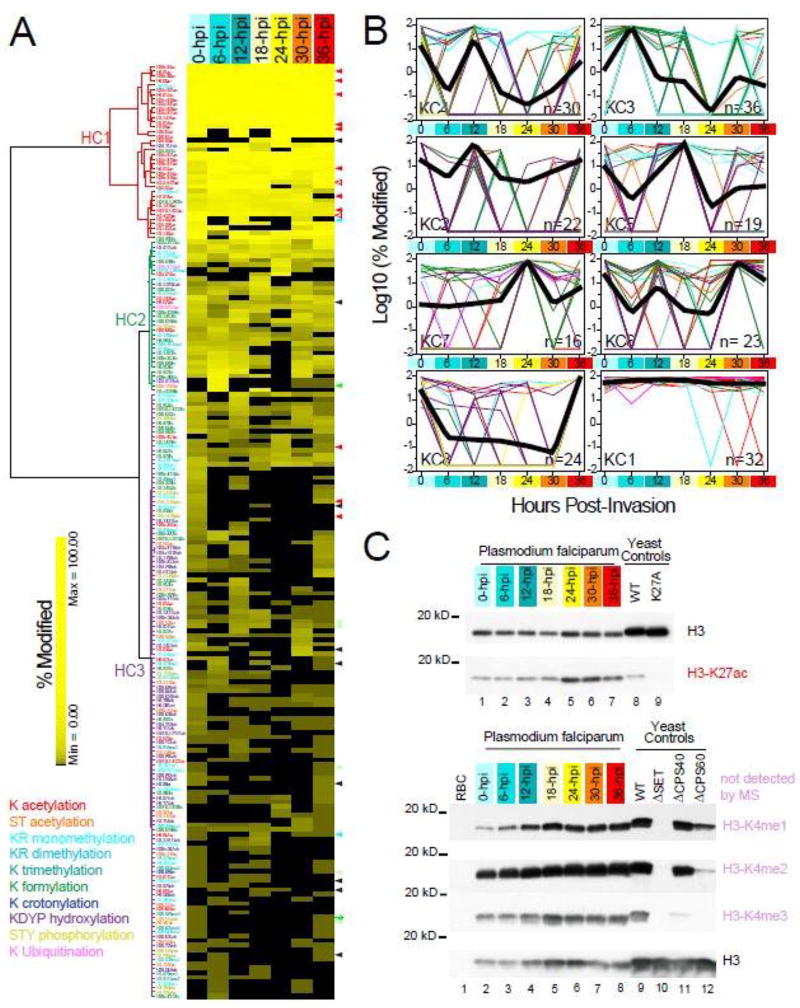
Figure 3. Dynamic landscape of PTM levels across seven erythrocytic stages of P. falciparum.
A. Unsupervised hierarchical clustering analyses of histone PTMs. SpC-based modification levels (SI-4.2) are hierarchically clustered (SI-5.1) with Euclidean as the distance metric and Ward’s method with multiple-fragment heuristic algorithm (MF) as a seriation rule. For each of the 202 quantified histone PTMs (rows), the color intensity is proportional to the modification level (%) measured in each stage (columns). PTM types are color-coded as in Fig. 2 and the dendrogram line colors denote the modification type that is the most represented in each cluster. An expanded version of this heat map is provided in Fig. S3.
B. k-means clustering of histone PTMs. Histone modifications optimally separate into eight k-means clusters using the Hartigan-Wong algorithm on their spectral count-based abundance during the IDC (SI-5.2). For each histone PTM, abundance values for the seven time points are expressed as the percentage of the maximum measured value for this PTM at any time point during the IDC. For each k-means cluster, log10 values of the average normalized modification level are plotted as a function of hours post-invasion (SI-5.1). Clusters are ordered based on the time point at which their maximum modification levels are measured (from 0 to 36-hpi), with cluster KC1 containing PTMs detected at near maximum levels across all seven time points. The number of PTMs in each cluster in indicated in the bottom right corner. The temporal profile of each individual PTMs is represented by lines color-coded by modification type, while thick black lines represent the result of multiple curves averaging.
C. Validation of mass-spectrometry results by Western blot. Acetylation at H3-K27 and the three methylation states of H3-K4 are detected at seven time points of the parasite cell cycle using antibodies against the conserved modifications in S. cerevisiae. Whole cell extracts from wild-type yeast, a strain expressing the H3-K27A mutant, and yeast strains deficient in the H3-K4 methylating machinery are analyzed in parallel to ensure proper behavior of each antibody.
Loading amounts in each lane are normalized using an antibody against Saccharomyces cerevisiae histone H3. K27ac levels in both H3 isoforms as measured by our MS analysis of the seven time points are indicated by the open red arrowheads on the right side of the heat map in A. While the peptide bearing H3-K4 was not detected by MS, mono-, di- ant tri-methylation of this residue are present at all stages of the parasite cell cycle, hence behaving similarly to H3-K27ac. An additional 30 PTMs are validated by MRM assays (SI-6.2), as indicated by arrowheads on the right side of the heat map in A. These arrowheads are color-coded by novelty status as in Fig.1A/S1.
To group histone modifications based on their SpC- or PI-based abundance profiles across the seven time points, we first normalized each individual modification in each time point to the highest percent of modification across the seven stages (i.e. the highest value equals to 100%). We then applied k-means clustering to these normalized matrices using the Hartigan-Wong algorithm37. The optimal number of clusters was obtained when k=8 for both quantitation methods (Fig. 3B and SI-5.2). All computations were run using R environment using k-means function for the partition and daisy function to compute all the pairwise dissimilarities (Euclidean distances) between observations in the dataset for the silhouette. To compare the hierarchical and k-means clustering results between the SpC- and PI-based modification levels, Spearman’s rank correlation and Jaccard indexes were calculated (SI-5.1).
NSAF732 was used to implement the Apriori algorithm as described in38 on the spectral counts of modified peptides (SI-5.3) to obtain the probabilities for combinatorial associations between modified residues (SI-5.4). Frequencies of co-occurrence were calculated considering either the merged dataset (“ALL”, Fig. 4A/S4A) or each of the seven time-points analyzed (0 to 36-hpi, Fig. 4B/S4B). For each of the seven canonical histones, probabilities of combinatorial associations between histone PTMs were plotted as cord diagrams using Circos (http://www.circos.ca/)39.
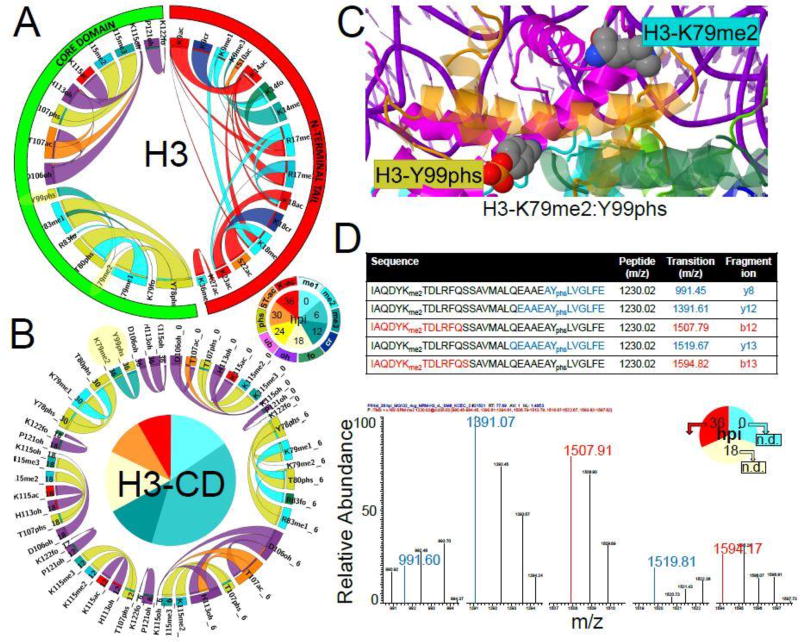
Figure 4. Combinatorial associations between modifications on Plasmodium histone H3.
The Apriori algorithm is used on the spectral counts of modified peptides (SI-5.3) to derive the probabilities of combinatorial associations between individual modified residues (SI-5.4). Frequencies of co-occurrence are plotted as cord diagrams using Circos. The ratio layout is used such as probabilities of PTMs XY given PTM X and of PTMs XY given PTM Y are depicted by one ribbon with ends of variable sizes corresponding to P(XY|X) and P(XY|Y) values. The order of the segments in the chord diagrams is imposed such as the PTMs locations are from N-terminus to C-terminus (segment starting at 12:00, clockwise), while the segments and ribbons are colored based on modification types as in Fig. 2.
A. Frequencies of co-occurrence derived from the merged dataset (“ALL”). The outer segments reflect the location of each residue along H3 sequence (N-terminal tail or core domain). Full-sized Circos output images for all histones are provided in Fig. S4A.
B. Frequencies of co-occurrence calculated for each of the seven time-points (0 to 36-hpi). The dynamic combinatorial patterns between residues located within H3 core domain (CD) are shown. The outer segments and ribbons are colored based on the type of PTMs, while the inner quadrants are colored to denote the time points at which PTM combinations are observed. For all histones, full-sized Circos output images for the dynamic combinatorial patterns between residues located within the N-terminal tails or core domains are provided in Fig. S4B.
C. Mapping of the combinatorial peptide bearing K79me2 and Y99phs on histone H3 in the tridimensional structure of the human nucleosome. Dimethyl and phosphate groups were added to the side-chains of H3-K79 and Y99, respectively, using the Vienna-PTM 2.0 web application. Histones backbones are displayed as ribbon cartoons: H2A in blue/cyan; H2B in green/bright green; H3 in magenta/light pink; H4 in orange/yellow. Modified residues side-chains are displayed as space-filled atoms color-coded using the CPK convention and their labels are cyan for methylation and dark yellow for phosphorylation. Additional structural analyses of combinatorial peptides on H2B and H3 are provided in SI-3.3.C.
D. MRM assay to validate the co-occurrence of H3-K79me2 and H3-Y99phs. Five transitions of the peptide bearing both H3-K79 and Y99 (LysC + GluC digested) are assayed for the presence of dimethylation on K79 and phosphorylation on Y99 at 0-, 18- and 36-hpi. The only positive identifications are reproducibly observed at 36-hpi (see SI-6.2), with a representative MRM spectrum shown here, confirming the MudPIT results (see combination highlighted in yellow in A and B panels).
Validation of histones modifications (see Supporting Information SI-6)
The transitions of 24 modified peptides identified in the MudPIT dataset (SI-3.4) were assayed by multiple reactions monitoring (MRM) on a LTQ-Velos-Orbitrap in low resolution mode, as adapted from40, 41. The MRM method was built by including the parent mass, parent mass selection window, product mass and mass selection windows for these three to seven product ions (SI-6.1). The criteria for selecting the fragment ions were that the fragments included the amino acid residue(s) modified in the peptide. We collected MRM data and the MS/MS spectra for the same parent ion within the same analysis on three of the seven previously analyzed samples (0-hpi_2, 18-hpi_1 and 36-hpi_2) digested with endoproteinase ArgC or endoproteinases LysC followed by GluC. The datasets were searched for acetylation, crotonylation, methylations, and phosphorylation using SEQUEST. The data was validated manually to confirm the presence of the relevant transitions followed by their corresponding MS/MS spectrum (SI-6.2), with representative annotated SRM transitions and MS2 spectra provided in SI-6.3 and Fig. 4D.
Western blot analyses were performed using an antibody against H3-K27 acetylation, which was detected by MS, and antibodies directed against each of the three methylation states of Saccharomyces cerevisiae H3-K4, which were not detected by MS analysis (Fig. 3C).
Data accessibility
The mass spectrometry data can be obtained from MassIVE (ftp://massive.ucsd.edu/) or ProteomeXchange (http://www.proteomexchange.org/) using the following accession numbers with password (ANS01146): MSV000079225/PXD002709 to MSV000079231/PXD002715 for the 0- to 36-hpi MudPIT datasets (SI-1.1) and MSV000079792/PXD004268 for the MRM dataset (SI-6.1). In addition, all of the original data (unprocessed Western Blots images) and processing steps (OriginPro files as well as R and jmol scripts) may be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/LIBPB-536.
Results
Dataset overview
Synchronized P. falciparum-infected blood cultures were harvested at seven time points corresponding to the early ring morphological stage at 0-hpi, mid-ring at 6-hpi, late ring at 12-hpi, early trophozoite at 18-hpi, late trophozoite at 24-hpi, early schizont at 30-hpi, and late schizont at 36-hpi (SI-1.1). The MS/MS datasets acquired for these histone samples were searched in a recursive manner (SI-1.2) for oxidation/hydroxylation, formylation, methylation, dimethylation, trimethylation, acetylation, crotonylation, ubiquitination, phosphorylation, as well as N-terminal acetylation (SI-1.3).
Plasmodium proteins represented between 41% and 86% of all proteins identified in this study (SI-2.1). The biological duplicate analyses were overall reproducible at the qualitative and quantitative levels (SI-2.2.A-C). The high enrichment in Plasmodium histones (between 24 and 52%, SI-2.2.D) combined with the three types of digestion performed on each replicate resulted in reproducibly high sequence coverage, ranging from 95% to 100% for the canonical histones (SI-2.2.E). H4 was the most abundant as estimated by its dNSAF values32, while both H2B/H2B.Z and H3/H3.3 isoforms added up to H4 levels (SI-2.2.F, inset). CenH3 was the least abundant isoform, consistent with its centromere-restricted localization. Most interestingly, the transcriptionally permissive histone variant H2A.Z, which has been characterized to be at least one order of magnitude less abundant than H2A in human and budding yeast, was recovered at levels similar to H2A in Plasmodium.
Benchmarking of post-translational modifications detected on Plasmodium histones
We identified a total of 232 amino acid/PTM combinations on the eight Plasmodium histones (Fig. 1A/S1), which fell into three main categories (SI-3.1): (1) 72 modifications that had been previously described in Plasmodium and/or Toxoplasma (62 of which were also known in other species)18–20, 42–44; (2) 72 modifications that had not been observed in apicomplexans but that have been reported on evolutionary conserved residues (SI-3.2 and SI-3.3) in other organisms; and (3) 88 completely novel modifications (Table 1). All spectra matching peptides (PSMs) with at least one novel PTM were cross-validated by two different search engines (SI-3.4) and visually assessed (SI-3.5). Of these 88 new PTMs, 38 modifications were detected on residues that had previously been shown to be marked by other modifications, 37 modifications were detected on conserved residues that have never been reported as modified, while another 13 PTMs were detected on non-conserved or semi-conserved residues (Fig. 1B). The majority of the novel sites were located within the histone fold domains (Fig. 1A/S1), suggesting that many of these may play a role in histone stability. The location of 67 residues out of the 95 sites detected as modified in Plasmodium canonical histones was assessed in the context of the human nucleosome core particle (SI-3.3). Six residues located within the long α2 helix of the histone-fold domains were deemed to be buried in the nucleosome tridimensional structure (SI-3.3.A). Five of these buried residues carried novel modifications, four of which were hydroxylations. Over half of the surface accessible residues were located at the DNA interface, where they might be involved in regulating protein DNA interactions and chromatin assembly.
At the quantitative level, the peptides bearing the 72 PTMs previously reported in Plasmodium and/or Toxoplasma were detected with large numbers of spectral counts (Fig. 1C) and mostly consisted of lysine acetylations and methylations (Fig. 1D) on the N-terminal tails (Fig. 1A/S1), while the peptides bearing the 72 PTMs known in other species were overall less abundant. The 88 entirely novel PTMs were detected within peptides with the lowest spectral counts (Fig. 1C), with the exceptions of a few formylations and hydroxylations (SI-3.1), indicative of the high sensitivity of our methodology. O-acetylations on serine and threonine, formylations, and hydroxylations contributed the majority of the newly discovered PTMs (Fig. 1D and Table 1).
A total of 158 PTMs that had been previously reported in other organisms on residues conserved in Plasmodium were not detected in our dataset (SI-3.1). About 60% of these missing PTMs were on residues that we detected as modified by another type of modification (Fig. 1A/S1). It is likely that these previously reported PTMs are not present in P. falciparum, but we cannot exclude that they were present below our detection limit. For residues in regions of low spectral coverage (not covered by peptides or with spectral counts below 50 when combining all 42 analyses; SI-3.1), we could not definitively conclude whether these PTMs were indeed absent in the Plasmodium samples we analyzed. The three largest classes of PTMs missing from our dataset were methylations, phosphorylations, and crotonylations (Fig. 1D).
Of the 115 PTMs previously reported in Plasmodium and/or Toxoplasma, 42 were not detected in this dataset (SI-3.1). 44% of these were phosphorylated sites (Fig. 1D) that were detected in two mass spectrometry studies using phosphopeptide enrichment18, 19, which may explain their absence in our dataset since we did not perform such enrichment steps. For other previously observed modifications, such as H3-K4 methylation20, technical limitations in our analysis pipeline may have resulted in low spectral coverage of peptides bearing the respective residues (e.g., H3/H3.3-K4, H4-K20, and H2B.Z-T84, SI-3.1). Due to the hydrophobic nature of the small tryptic peptide bearing H3-K4, we also had trouble detecting this H3-K4 methylation in other organisms that have relatively high levels of this PTM, such as S. cerevisiae45. Conflicting results have been obtained in the past about H4-R17 methylation20, 46. H4 peptides bearing R17 were detected as unmodified with high spectral coverage in all of the 14 analyzed samples (SI-3.1), suggesting that H4-R17 is not methylated during the IDC. Trimethylation at H3-K36 has recently been shown to be essential for silencing of the var virulence gene family17. In our dataset, K36 was detected as acetylated and formylated on both H3 isoforms and as mono- and dimethylated on only H3 (Fig. S1). The restricted chromosomal localization and thus low abundance of the H3-K36me3 mark likely prevented detection in our study.
Quantitative landscape of modification levels along histone sequences
To gain more insight into global histone modification patterns, we evaluated the abundance of histone marks. First, we estimated to what extend a particular residue was modified, by dividing the local label-free features matching a particular modification by the total label-free features for this amino acid position (SI-4.1)30. We estimated modification levels using both spectral counts (SpC) and sum of MS peak intensities (PI) as label-free values (SI-4.1.A). A total of 202 PTMs were quantified in this study (previously reported PTMs detected by single spectrum and/or in only one run were not used in the quantitative analyses, neither were N-terminal acetylations). The correlation between SpC- and PI-based modification levels was high with an adjusted R2 of 0.9 when merging all time-points (SI-4.1.B). Modifications levels were also calculated for each replicate analysis of the seven time-points (Fig. 2/S2 and SI-4.2 for SpC-based values and SI-4.3 for PI-based values).
A clear dichotomy in modification levels was observed for all Plasmodium histones analyzed (Fig. 2/S2): accessible N-terminal tails were heavily modified (mostly acetylated) at high levels, while most PTMs detected within the structured domains were present at much lower levels. The exception was methionine oxidations (positions shaded in grey in Fig. 2/S2), which occur on unfolded peptides during sample processing, leading to high oxidation levels no matter their position within the histone structures. A few sites located within the core domains were observed to be modified at similarly high levels (Fig. 2/S2), in particular formylations, which raises the question as to whether such modifications were also the artefactual result of sample processing47.
Within the N-terminal tails, acetylation levels appeared to be lower for residues located more internally. To investigate this trend further, we plotted all of the SpC-based quantitative data-points for each acetylated lysine within N-terminal tails (Fig. 2/S2). Overall, the N-terminal tails of H2A.Z, H2B.Z and H4 were the most heavily acetylated. Linear decreases in lysine acetylation levels over the length of the N-terminal tail were observed for all canonical histones and were statistically significant for H4 (R2 = 0.87) and H2B.Z (R2= 0.62). Interestingly, high levels of lysine acetylations were observed to be stable across the long H2A.Z N-terminal tail, even for lysine residues close to its core domain (CD).
Histones PTMs dynamics across the intra-erythrocytic developmental cycle
Next, we investigated changes in histone modification levels during the different stages of the IDC. We used both SpC- and PI-based modification levels measured for the 202 PTMs quantified across the IDC (SI-5.1). The hierarchical and k-means clustering results obtained using the SpC-and PI-based quantitative values as input were compared by Spearman’s rank correlation and Jaccard similarity statistics (SI-5.1) and were shown to be very similar (SI-5.2). The SpC-based clustering results were plotted in Fig. 3A–B and discussed below.
Unsupervised hierarchical clustering of the modification levels revealed three major groups, each exhibiting different patterns (Fig. 3A/S3). The first cluster (HC1) contained mainly known acetylated lysine residues (32 out of the 37 PTMs in HC1) located in N-terminal tails that were present at high abundance in all seven stages (SI-5.1). The second cluster (HC2) contained formylation as the most predominant modification (14 sites) in a group of 32 PTMs present at intermediate levels and frequently observed, yet not always present. The third cluster (HC3) contained PTMs that were only sporadically present in the seven stages and at very low levels. This group was the largest and contained all possible types of PTMs, with hydroxylations being the most represented (33 out of 133 PTMs). This last cluster is potentially of great interest, since it encompasses histone PTMs that undergo dramatic changes during the Plasmodium IDC and may therefore reflect changes in chromatin organization associated with DNA replication and transcriptional activity.
To further analyze the variations in relative histone PTM abundances during the IDC, in particular for the PTMs detected in a limited number of time-points (i.e. belonging to HC3), we clustered abundance profiles with the k-means algorithm (Fig. 3B). Each of the eight k-means clusters had similar silhouette widths above 0.6 and contained similar numbers of PTMs (SI-5.2). PTMs within each cluster were sorted based on modification type, novelty status, and location of the modified residue within the histone structures (SI-5.1). KC1 “all” cluster mostly overlapped with HC1 and contained the large majority of the histone acetylations previously reported in Plasmodium (SI-5.1). Most of these PTMs (93%) were located on the N-terminal tails (SI-5.1) and abundance levels did not vary along the IDC (Fig. 3B). Many PTMs detected on histone variants belonged to this group including all seven lysine acetylations on H2A.Z-NT and all five lysine acetylations detected on H2B.Z-NT (vs. none for H2A and H2B; SI-5.1). On average, 59% of the PTMs found in the k-means clusters with maximum relative abundances at the early, mid-, and late ring and early trophozoite stages (0 to 18-hpi) were novel and were largely located within the core domains (SI-5.1). KC7, with maximum levels at 24-hpi, mostly contained PTMs previously described in other organisms. Most of the PTMs found in KC6 and KC8, with maximum levels at early and late schizont stages (30- and 36-hpi), were also detected at fairly high levels at early ring stage (Fig. 3B). This is in agreement with the fact that the 36-hpi k-means clusters diverged the most between the SpC- and PI-based quantitation methods, with over half of PTMs within the 36-hpi k-means cluster based on spectral counts falling in the 0-hpi cluster based on peak intensities (SI-5.1). This highlights the cyclic nature in the modification dynamics, with events at the end of one IDC bleeding into the beginning of a new cycle.
As a validation of our quantitative analysis, we confirmed that the combined abundance of H3-K27 and H3.3-K27 acetylation in all seven-time points as detected by Western blot (Fig. 3C) correlated well with our quantitation of H3-K27 and H3.3-K27 acetylation levels by local spectral counts (Fig. 3A/S3, open red arrowheads). We also used highly characterized antibodies recognizing specific methylation states of H3-K4 to confirm that H3-K4me1, H3-K4me2 and H3-K4me3 were present across the erythrocytic cycle, at levels comparable to what is found in yeast cell extracts (Fig. 3C). The presence of three methylation states on H3-K4 was in agreement with previously analyzed histones isolated from an asynchronous mixture of Plasmodium asexual stages20. While we could not detect these modification by mass spectrometry (Fig. S1), here we showed that H3/H3.3-K4 mono-, di- and tri-methylations behave similarly to H3-K27ac and the other lysine acetylations within the N-terminal tails and would fall in the first hierarchical cluster and k-means cluster 1, i.e. observed at high abundance in all time points.
The stage-specific detection of an additional 30 PTMs was further validated by specifically targeting the MRM transitions of 24 peptides bearing these modified residues by themselves or in combination (SI-6.1). We tested three of the seven time-points analyzed by MudPIT at the beginning (0-hpi), middle (18-hpi), and end (36-hpi) of the IDC. The assayed peptides included nine novel PTMs (underlined in Table 1), 10 PTMs known in other organisms, as well as 11 PTMs previously described in Plasmodium and/or Toxoplasma to ensure that our method was working properly (see color-coded arrowheads in Fig. 3A/S3). Overall the MRM results were in good agreement with the MudPIT dataset: 83% of the modified peptides tested at 0-hpi and 74% of the peptides assayed at 18- and 36-hpi showed the same pattern of detection/not detection by both methods (SI-6.2 and SI-6.3). In particular, the four phosphorylated peptides we assayed (H2A–S120phs, H2A–T126phs, H3-Y99phs and the novel H2B–S56phs) were only detected at the beginning and/or the end of the IDC, but never in the 18-hpi sample, confirming the quantitative observations derived from the MudPIT dataset.
Combinatorial associations between histone modifications
To investigate potential cross-regulation between histone modifications, we next used the spectral counts of modified peptides (SI-5.3) to derive the frequencies of co-occurrence between modified residues (SI-5.4) using the Apriori association rule algorithm as described in38. We calculated these probabilities of combinatorial associations between individual residues on the merged dataset (Fig. 4A/S4A) and for the seven time points (Fig. 4B/S4B).
In all histones, the strongest associations were observed between acetylated residues within the N-terminal tails (Fig. 4A/S4A) that are associated with transcriptionally permissive chromatin. In particular the H2A.Z and H2B.Z variants showed very similar combinatorial profiles (Fig. S4A) with exceptionally strong associations between multiple acetylated lysines across all seven time points (Fig. S4B). On the other hand, only acetylated H2A–K3/K5, H2A–K9/K10 and H2B–K3/K7 were considered combinatorial (Fig. S4A), with the H2A peptides bearing modified residues only detected at 18- and 24-hpi, while the modified H2B–K3/K7 peptides were more consistently detected in five of the seven time points (Fig. S4B). H4 had many different combinatorial peptides mapped to its N-terminal tail (SI-5.3), leading to strong and stable association probabilities between R3me1 and K5/K8/K12/K16ac (Fig. S4A) during the entire IDC (Fig. S4B).
The H3 and H3.3 isoforms had the most diverse batch of modified peptides combining many different types of modifications (SI-5.3), both within their N-terminal tails and core domains (Fig. 4A/S4A). Overall, the association probabilities observed were low (SI-5.4), with the exception of combinations involving acetylated K9, K14, K18, and K23 and methylated R17. Acetylations of H3-K9 and H3-K14 in P. falciparum are carried out by PfGCN546, 48, 49 and are known to occur together in active regulatory gene elements in other eukaryotes50. H3-K14ac was found in combination with three other modifications on H3-K9 besides acetylation (crotonylation, mono-and trimethylation). While the associations between H3-K9ac or K9me1 with K14ac were stably observed at all seven time points (Fig. S4B), K9cr was only associated with K14ac at the beginning (0-hpi) and the end of the IDC (36-hpi). The temporally-restricted co-occurrence of K9cr with K14ac was also observed at 36-hpi on an orthologous peptide from H3.3 that is distinct from the H3 peptide (Fig. S4B). Similarly, H3-K23ac was associated with K18ac and/or K18me1 throughout the cycle, while K18cr was only found in combination with K23ac at the late schizont stage. Interestingly, modifications at residues located further away from the N-terminus (K27, S28, S32, K36, and K37) were almost exclusively present by themselves in both H3 and H3.3 (SI-5.3).
Some of these combinatorial patterns showed interesting changes during the IDC. While associations within H3.3 core domain (CD) mostly involved hydroxylations (Fig. S4A), a more diverse combinatorial landscape was observed for H3-CD (Fig. 4B). In particular phosphorylations on H3-Y78, T80, and Y99 were detected in combination with mono- and/or di-methylations on K79 or R83, while phosphorylation on H3-T107 co-occurred with K115 di- and tri-methylations. These combinatorial events were again restricted to specific stages of the IDC (Fig. 4B). The combination of H3-Y78/Y80 phosphorylations with K79/R83 methylations was detected at 6- and 30-hpi, while T107phs in combination with K115 methylations was detected from 0 to 18-hpi. A peptide bearing both H3-Y99phs and H3-K79me2 (Fig. 4C) was only detected at 36-hpi (Fig. 4B, residues highlighted in yellow). MRM assays (SI-6.2 and SI6.3) confirmed that this combinatorial peptide was only detected in the late schizont sample (Fig. 4D).
Collectively, the co-occurrence of many histone modifications, in particular acetylations, as observed here suggests that these modifications are likely to be co-regulated. In addition, variations in combinatorial patterns during the IDC may indicate that such histone modifications are involved in cell cycle progression, DNA replication, and/or transcriptional regulation.
Discussion
In this study, we have analyzed histone modifications at seven time points during the IDC of Plasmodium falciparum, detecting 232 unique amino acid position/PTM combinations (Fig. 1A/S1). All peptides bearing the 88 novel modifications we report have been cross-validated by two independent search engines, significantly increasing the confidence in their identification51. Most (85%) novel PTMs and another 72 PTMs not previously identified in apicomplexans occur on residues conserved across species, demonstrating that our approach has gained access to previously unknown and potentially critical and conserved PTMs. In addition, we have performed an in-depth characterization of 202 histone PTM dynamics using two label-free quantitative methods, leading to identifying changes in the occurrence and abundance of single histone modifications as well as PTM combinations. We have also systematically characterized the detected PTMs in the context of the human nucleosome tridimensional structure and the possible functional significance for the residues within the core domains that were found to be formylated, hydroxylated, O-acetylated or phosphorylated are discussed below. Overall, our data indicate that histone modifications are controlled by an intricate regulatory network that affects global and local chromatin structure and plays an important role in parasite development and virulence.
Classical epigenetics marks detected on Plasmodium histones
As previously observed in an asynchronous mixture of Plasmodium asexual stages20, a high abundance of acetylations and monomethylations, which are associated with transcriptionally permissive states of chromatin, is observed in our dataset (37.5% of all detected PTMs). In contrast, dimethylations and trimethylations, which are usually associated with transcriptional silencing, only constitute 6 and 3% of the detected PTMs, respectively (SI-3.1), and are often present at low levels or at a limited number of time points (SI-4.2). In particular, 12 of the 19 detected me2 and me3 marks show highest abundance at the beginning of the cycle (0- and 6-hpi k-means clusters; SI-5.1), while another four me2 and me3 marks are detected at highest abundance in the last two time points (30- and 36-hpi k-means clusters). Methylations are mostly found within the histone cores and only histone H3 and its variant H3.3 have multiple methylation sites on their N-terminal tails (Fig. S1). In addition to the classical methylations usually observed in eukaryotic cells, nine novel methylation marks are identified (Table 1), many of which are located on residues that have been previously described to be marked with different modifications or different methylation states, e.g. H2B–K35, H2B–K49, H3-K115, and H4-K91 (SI-3.1). These classical silencing marks may be restricted to sub-telomeric regions associated with silencing of gene families involved in antigenic variations10, 16, 17. Overall, the high abundance of activation marks and the low number of known repressive modifications are consistent with the prevalence of euchromatin observed in the human malaria parasite IDC11, 52.
While a large number of the identified PTMs consisted of well-known lysine acetylations (Fig. 1A/S1), a total of six new lysine residues are found to be acetylated (Table 1). However five of them are on lysines not conserved or semi-conserved across species (SI-3.1). H2B–K64, and H4-K67 are both substituted for arginines in other species as well as in the Plasmodium H2B.Z isoform (SI-3.2). The presence of lysines at these positions in both Plasmodium and Toxoplasma histones allows for a larger array of modifications (acetylations and hydroxylations) that are not possible on an arginine side chain. In addition, the CenH3 N-terminal tail is highly divergent across species (SI-3.2) and the three acetylated lysines we detected on Plasmodium CenH3 (K23ac, K26ac, K60ac) are hence not conserved (SI-3.1). Peptides bearing H4-K67ac and CenH3-K60ac have been independently validated at all three of the time-points we assayed by MRM (SI-6.1) for the H4 peptide and at 0-hpi for the CenH3 peptide, in agreement with the MudPIT results (SI-6.2). To our knowledge, these are the only modifications ever detected on centromeric histone H3 from any species.
Ubiquitylation of H2B–K112 has been previously reported in Plasmodium20 and is detected with an average occupancy of 5% in all stages, leading this modification to fall into hierarchical cluster 2 and the k-means “all” cluster (SI-5.1). On the other hand, its homologous residue H2B.Z-K116 is ubiquitylated at 6 out of the 7 time points (SI-4.2) with an average level of about 3%, except in 24-hpi where its level reaches 18%. H2B.Z-K116 hence is grouped in the k-means “24-hpi” cluster (SI-5.1) using both SpC- (SI-4.2) and PI-based quantitative values (SI-4.3). Ubiquitylation of H2B–K112 is a well-characterized event associated with active transcription in both yeast (H2B–K123ub) 53 and human (H2B–K120ub)54. In Plasmodium, this signal might be carried out by the H2B.Z variant, whose K116 residue reaches maximum ubiquitylation during the transcriptionally-active late trophozoite stage.
A total of twelve phosphorylations are confidently identified (SI-3.1), seven of which have previously been reported in Plasmodium19 and/or Toxoplasma44, one in yeast55, and one in mouse38. Phosphorylations on H2B–S56 and H3-Y78 are novel (Table 1), with H2B–S56 being conserved and H3-Y78 aligning with a phenylalanine in other species (SI-3.2). The abundance of all phosphorylation marks is extremely low with the exception of the conserved H3-S32phs that is present at slightly higher levels (on average 3% of all histones H3) and detected at all stages but 18-hpi. The twelve detected phosphorylations neatly segregate into four k-means clusters with maximum relative abundance at 0-, 6-, 30-, and 36-hpi, indicating that histone phosphorylations are mainly present at the beginning and the end of the IDC (SI-5.1). The temporally-restricted expression patterns of four of these phosphopeptides (H2A–S120phs, H2A–T126phs, H30Y99phs and the newly-described H2B–S56phs) have been validated by MRM assays (SI-6.1) on three of the seven stages analyzed by MudPIT (0-, 18- and 36-hpi). Specific transitions for all four phosphopeptides are detected in the 36-hpi sample (Fig. 4D and SI-6.2), but none are detected at 18-hpi. Pfcrk-3, a kinase that phosphorylates histones, has recently been characterized in P. falciparum56 and is thought to play a crucial role in parasite proliferation. In other eukaryotes, phosphorylation is an abundant histone modification, potentially connecting chromatin remodeling and intracellular signaling pathways. Given the limited ability of the malaria parasite to respond to environmental stress57, 58, it may not be surprising that phosphorylation of P. falciparum histones was observed at low levels.
The location of five of these twelve phosphorylated sites could be assessed using the tridimensional structure of the human nucleosome (Fig. 4C and SI-3.3). Phosphorylations on H3-Y78 and T80 are detected in combinations with mono- and/or di-methylations on K79 or R83 (Fig. 4A) and all four of these residues are located within Loop 1 at the surface of H3 (SI-3.3.C). On the other hand, H3-T107, whose phosphorylation has been previously reported in Toxoplasma and other organisms (SI-3.1) and co-occurs with K115 di- and tri-methylations (Fig. 4A), and H3-Y99, whose phosphorylation is detected in combination with H3-K79me2, are both located within the long α2 helix and are buried residues at the interface of the H3-H4 heterodimer (Fig. 4C and SI-3.3.C). The methylated lysines co-occurring with these four phosphorylations are interestingly all located at the interface with the DNA strands (Fig. 4C and SI-3.3.C). While H3-Y99 and H3-T107 are not likely to be phosphorylated in a post-translational manner on a fully assembled histone octamer or even on a H3-H4 dimer55, the temporally-restricted occurrence of these combinatorial modifications (validated by MRM for the H3-K79me2:Y99phs peptide, Fig. 4D) along with their close structural proximity suggest that they might be involved in a co-regulated event. Modifications of buried residues have been reported before, for example H4-K91ac that has been involved in the docking of the H2A–H2B dimer with the H3-H4 dimer and the stabilization of the octamer, and associated with chromatin assembly59. On the other hand, phosphorylation on H3-Y99 has been associated with disrupting and/or preventing the formation of the H3-H4 dimer and targeting free H3 for degradation55. In this context, H3-T107 and H2B–S56, which is located at the interface with H4 in close proximity with its very C-terminal glycines (SI-3.3.C), could play a similar role in octamer stabilization when not phosphorylated.
Other types of modifications detected on Plasmodium histones
Hydroxylation is less common than other types of PTMs60, yet hydroxylations of lysine, tyrosine, aspartate, proline and histidine residues constitute the largest class of novel PTMs in our dataset (Fig. 1D). A total of four tyrosine hydroxylations have recently been reported on H2B–Y29, H4-Y51, H4-Y88, and H2A–Y393, all but the last being detected in our dataset (SI-3.1). Among the novel hydroxylated residues are H2A–Y57, H3-H113, and H4-Y72, which are conserved and have been shown by alanine substitution scanning to be essential for viability under normal growth conditions in S. cerevisiae61 but for which no modification has been reported so far. Interestingly, all but three of the 37 hydroxylated sites detected in this study are located within the histone globular domains (Fig. 1A/S1), and 30 are involved in alpha-helices (SI-3.3.B). Proline and lysine hydroxylation have been shown to stabilize the triple helix structure of collagen 62. The clear enrichment of hydroxylations on residues involved in secondary structures hence hints at a stabilizing conformational role. Four of the 18 hydroxylated sites mapped to the nucleosome structure (SI-3.3.A) are not surface-accessible, while the reactive groups of another three residues are somewhat buried. With a few exceptions, most (33 out of 37) of the hydroxylations are detected at low levels (Fig. 2) and fall within the third hierarchical cluster (Fig. 3A). Hydroxylation is hence not likely a prominent post-translational modification occurring while the histones are part of the nucleosome, but might rather occur co-translationally while histones are being synthetized or post-translationally before octamer assembly.
While some formylation of lysines has previously been associated with histones (see references in SI-3.1) and other nuclear proteins63, lysine and arginine formylations are the second largest class of novel PTMs identified in this study (Fig. 1D): in total, formylation represents 20% of our dataset, on par with acetylation and methylation (SI-3.1). Most of the formylated residues (80%) are located within the core domains (Fig. 1A/S1) and are reproducibly observed at high levels (Fig. 2 and SI-4.2). High levels of formylations could be the artefactual result of the presence of formic acid during sample processing and acquisition47, in particular for arginine residues, which are not known to be formylated in vivo. However, if a modification happens chemically in vitro at the unfolded peptide level, as opposed to enzymatically on a folded native protein, one should expect to see an equal frequency of formylation events on all K/R residues, even those that are not accessible in vivo to histone modifiers. In contrast to methionine oxidations that are observed on average in 13.2 samples out of the 14 analyzed and on 12 of the 13 methionines in Plasmodium histones (SI-3.1), our data shows that not all possible K/R residues are formylated (24.4 % of all lysines and 12.9% of all arginines; Fig. 1A/S1) with an average frequency of 7.2 out of 14 samples (SI-3.1). Furthermore all formylated sites located within the core domains are accessible within the nucleosome tridimensional structure (SI-3.3.A), suggesting that these modifications could have happened in vivo at the protein level. Half of these formylations are newly described, yet the majority (73%) are on modification hot-spots, i.e. are detected on residues observed here or known to be otherwise acetylated, methylated, hydroxylated, and/or ubiquitylated (Fig. 1A/S1 and SI-3.1). In the seven cases where the potentially differentially-modified residues are located within the N-terminal tail, the acetylation or methylation events are usually the ones occurring with the highest frequency and levels (Fig. 2/S2), with the exception of H3/H3.3-R42 which was detected in all time-points at higher formylation levels than dimethylation. In the 26 instances where the modification hot-spots are located within the core domains, formylation is the most reproducibly detected modification with the highest spectral counts (SI-4.2), with the exception of the well-known ubiquitylations on H2B–K112 and H2B.Z-K116 and the acetylation on H4-K79. Formylation of modification hot-stops may yet happen post-translationally on the assembled nucleosome and play a competing role for other modifications on Plasmodium histones.
Most of the novel acetylation sites were on 11 serines and threonines (Fig. 1A/S1). All are on conserved residues, except H2B.Z-T30, which aligns with a lysine residue (SI-3.2) that is known to be acetylated, formylated and/or crotonylated in other species (see references in SI-3.1). All but a couple of the peptides bearing S/T acetylations also contain lysine residues, which could indicate wrongly assigned modification sites. For example, H2B–S28 acetylation is detected on a peptide bearing two unmodified lysines at positions 35 and 38 (SI-3.4). However both SEQUEST and OMSSA search engines are in agreement with S/T acetylation assignments (SI-3.4). Furthermore the acetylation of H2B–S28 and H4-S47 are confirmed by MRM assays (SI-6.1) that detected specific SMR transitions from fragments containing these modified serines (SI-6.2) in all three of the tested time-points (0-, 18- and 36-hpi), while H2A–T101 acetylation was observed at 18- and 36-hpi (SI-6.3). Nonetheless, most of the acetylated serines and threonines are located in close proximity to acetylated lysines either at the level of the primary or secondary structures within each histone (Fig. 1A/S1) or at the level of the quaternary structure within the nucleosome (H2A–T101 and H4-S47 are within 10Å of acetylated lysines from other histones; SI-3.3.C). In addition, all S/T acetylations are detected at low levels (Fig. 2/S2) and all but one fall in the third hierarchical cluster (SI-5.1). This indicates that these S/T acetylations could be the by-products of permissive reactions by lysine acetyltransferases. On the other hand, while acetylations on serine and threonine side-chains are not widely known38, H3-S10ac has been linked to cell cycle progression and cellular pluripotency64. In addition, two studies have suggested that such modifications may be a way to inhibit or prevent phosphorylation on these residues65, 66. In this context, it is interesting to note that all but three of these S/T acetylated sites (H2A–T101, H2B–T61 and H2B.Z-S54) have been also detected as phosphorylated in this study (H2B–S50, H3-S28, H3-T107) or others (see references in SI-3.1).
Crotonylation of lysine side chain has been recently reported on mammalian histones3. While crotonylated lysines are present in our dataset (H2B–K49, H3-K115, H3.3-K115 are novel, and H3-K9, H3/H3.3-K18, H3-K27, H3.3-K9 are known, see references in SI-3.1), crotonylation constitutes one of the main classes of PTMs that was notably absent from our list (Fig. 1D). Most of the missing crotonylations are reported on the N-terminal tail of H2B, H3/H3.3 (Fig. S1) and H4 (Fig. 1A) on lysine residues that are acetylated at high levels in the seven Plasmodium stages we analyzed (SI-4.2 and SI-4.3). In addition, lysine crotonylations are enriched on sex chromosomes3, which are very different cellular conditions than the Plasmodium IDC and it is therefore likely that these modifications are indeed absent in the malaria parasite. With the exception of H3-K115cr, which reaches maximum abundance at 18-hpi (SI-5.1), the other two crotonylation events detected in this study are mainly observed at the early ring and late schizont stage. We confirmed the presence and expression pattern of crotonylation on H2B–K49, H3-K9 and H3-K115 by MRM assays (SI-6.1). This type of modification may play a role in gene activation during the process of parasite egress and re-invasion that is characterized by global transcriptional silencing, similar to enrichment of this histone mark in post-meiotic spermatids in mice3.
Plasmodium-specific epigenetics features
Based on our results, we can highlight several ways in which P. falciparum differs from other eukaryotes. First, histone variants H2A.Z and H2B.Z are highly abundant (SI-2.2.F) and carry stable lysine acetylations in their N-terminal tails during the complete cell cycle (Fig. 4B). In particular, the long N-terminal H2A.Z shows reproducibly high acetylation levels, even for residues located next to the first putative alpha helix (Fig. 2/S2). In contrast to the restricted localization of H2A.Z around promoter-associated nucleosome-depleted regions in other eukaryotes, H2A.Z and H2B.Z have recently been shown to be located throughout the highly AT-rich intergenic regions of the P. falciparum genome67. P. falciparum may have evolved these histone variants to promote nucleosome binding in AT-rich DNA sequence, and the high acetylation levels of H2A.Z and H2B.Z could play a key role in nucleosome stability and chromatin organization in these intergenic regions.
Second, the observation that acetylation levels on N-terminal tails decrease linearly as lysines become more internally located (Fig. 2/S2) is also unique to P. falciparum, and is in stark contrast to results in human and mouse. For example, acetylation occupancy of H4 in mammals is higher on K16, followed by K12, then K8/K538, 68–71, supporting the zip model suggested in earlier MS70 and Edman sequencing72 analyses. This model proposes H4 acetylation to occur from the C-to N-terminus of the N-terminal tail, while deacetylation would occur in the opposite direction. The reverse occupancy pattern observed in Plasmodium H4 has been previously raised at the qualitative level for Plasmodium H4-K8 and K1220 and our comprehensive quantitative data supports this contradictive pattern in Plasmodium. Similar discrepancies are observed for other histones38, 73, highlighting strong differences in the mechanisms regulating histone PTMs between the malaria parasite and other eukaryotes.
Finally, the relative abundance of histone proteins in our samples (SI-2.2.D) correlates well with chromatin dynamics observed in a previous study11, 74 and confirms the depletion of histone proteins at 18-hpi. Based on these observations, chromatin remodeling seems to be particularly prevalent during this time point of the IDC and is likely to be closely linked to initiation of DNA replication and transcriptional activity. Histone modifications that are highly abundant during the middle point of the asexual cycle may thus play important roles in initiation of DNA replication and regulation of gene expression. K-means clustering grouped 17 PTMs mostly located within core domains, whose maximum abundance levels peak at 18-hpi based on both spectral counts and peak intensities values (SI-5.1). These include known acetylations on H3-K115 and H4-K91, which has been shown to be involved in nucleosome stability59, a novel acetylation on an apicomplexan-specific lysine at position 67 on H4, known methylations on H3.3-R40 and H3-R17, a novel methylation on H2A-75, and a newly described crotonylation on H3-K115. Overall, the presence of complex histone modification patterns that vary over the course of the IDC indicates that many different regulatory cues, involving a multitude of chromatin remodeling enzymes, are likely to be at work.
In conclusion, this study has generated a detailed atlas of histone modifications throughout the IDC of the human malaria parasite. We have greatly increased the number of histone PTMs detected in any eukaryotic organism. In addition, we describe unique histone modification combinatorial signatures and changes in histone modification patterns during the parasite asexual cycle, many of which may be related to parasite-specific cell cycle control and virulence. Understanding the role of each of these modifications and molecular mechanisms that control these atypical features may offer novel therapeutic strategies.
Supplementary Material
Acknowledgments
We thank Edwin Smith and Ali Shilatifard for the gift of the H3-K4 me1/me2/me3 and H3-K27ac antibodies and comments on our manuscript.
Funding Sources
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) (grant R01 AI85077-01A1 to KLR), the National Institutes of Health (fellowship F31 AI096840-01 to SC), the Human Frontier Science Program (grant LT000507/2011-L to EMB) and the Stowers Institute for Medical Research (AS, MES, WZ, JMV, MPW, LF).
ABBREVIATIONS
- PTM
post-translational modification
- MudPIT
Multidimensional Protein Identification Technology
- IDC
intra-erythrocytic developmental cycle
- TCEP
Tris(2-Carboxylethyl)-Phosphine Hydrochloride
- CAM
chloroacetamide
- FDR
false discovery rate
- ASFP
All Spectra vs. Few Proteins
- ASAP
All Spectra vs. All Proteins
- MSAP
Modified Spectra vs. All Proteins
- hpi
hours post-invasion
- ac
acetylation
- me1
monomethylation
- me2
dimethylation
- me3
trimethylation
- fo
formylation
- oh
hydroxylation
- phs
phosphorylation
- ub
ubiquitination
- cr
crotonylation
- ox
oxidation
- MS
Mass Spectrometry
- PSMs
Spectra/Peptide Matches
- SRM
Single Reaction Monitoring
- MRM
Multiple Reaction Monitoring
- n.c
non-conserved
- NT
N-terminal Tail
- CD
Core Domain
Footnotes
“This material is available free of charge via the Internet at http://pubs.acs.org.”
Author Contributions
The manuscript was written through contributions of all authors. KLR, AS, and LF conceived and designed the experiments. SC, AS, JP, and DWDC performed the experiments. MS, ZW, MPW, JMV, contributed reagents, materials, and analysis tools. LF, AS, EMB, NP and KLR analyzed the data. LF, EMB and KLR wrote the paper. All authors edited and approved the manuscript. All authors have given approval to the final version of the manuscript.
The authors report no competing financial interests.
References
- 1.Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr Opin Cell Biol. 2007;19(3):266–72. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8(12):983–94. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, Lu Z, Ye Z, Zhu Q, Wysocka J, Ye Y, Khochbin S, Ren B, Zhao Y. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–28. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rivera C, Gurard-Levin ZA, Almouzni G, Loyola A. Histone lysine methylation and chromatin replication. Biochim Biophys Acta. 2014;1839(12):1433–9. doi: 10.1016/j.bbagrm.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Smith OK, Kim R, Fu H, Martin MM, Lin CM, Utani K, Zhang Y, Marks AB, Lalande M, Chamberlain S, Libbrecht MW, Bouhassira EE, Ryan MC, Noble WS, Aladjem MI. Distinct epigenetic features of differentiation-regulated replication origins. Epigenetics Chromatin. 2016;9:18. doi: 10.1186/s13072-016-0067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 7.Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, Freitas-Junior LH, Scherf A, Crabb BS, Cowman AF. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121(1):13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 8.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mancio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, Scherf A. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121(1):25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 9.Issar N, Ralph SA, Mancio-Silva L, Keeling C, Scherf A. Differential sub-nuclear localisation of repressive and activating histone methyl modifications in P. falciparum. Microbes and infection / Institut Pasteur. 2009;11(3):403–7. doi: 10.1016/j.micinf.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 10.Lopez-Rubio JJ, Mancio-Silva L, Scherf A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell host & microbe. 2009;5(2):179–90. doi: 10.1016/j.chom.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Ponts N, Harris EY, Prudhomme J, Wick I, Eckhardt-Ludka C, Hicks GR, Hardiman G, Lonardi S, Le Roch KG. Nucleosome landscape and control of transcription in the human malaria parasite. Genome Res. 2010;20(2):228–38. doi: 10.1101/gr.101063.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ralph SA, Scheidig-Benatar C, Scherf A. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(15):5414–9. doi: 10.1073/pnas.0408883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stubbs J, Simpson KM, Triglia T, Plouffe D, Tonkin CJ, Duraisingh MT, Maier AG, Winzeler EA, Cowman AF. Molecular mechanism for switching of P. falciparum invasion pathways into human erythrocytes. Science. 2005;309(5739):1384–7. doi: 10.1126/science.1115257. [DOI] [PubMed] [Google Scholar]
- 14.Tonkin CJ, Carret CK, Duraisingh MT, Voss TS, Ralph SA, Hommel M, Duffy MF, Silva LM, Scherf A, Ivens A, Speed TP, Beeson JG, Cowman AF. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS biology. 2009;7(4):e84. doi: 10.1371/journal.pbio.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419(6906):498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flueck C, Bartfai R, Volz J, Niederwieser I, Salcedo-Amaya AM, Alako BT, Ehlgen F, Ralph SA, Cowman AF, Bozdech Z, Stunnenberg HG, Voss TS. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog. 2009;5(9):e1000569. doi: 10.1371/journal.ppat.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang L, Mu J, Zhang Q, Ni T, Srinivasan P, Rayavara K, Yang W, Turner L, Lavstsen T, Theander TG, Peng W, Wei G, Jing Q, Wakabayashi Y, Bansal A, Luo Y, Ribeiro JM, Scherf A, Aravind L, Zhu J, Zhao K, Miller LH. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature. 2013;499(7457):223–7. doi: 10.1038/nature12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dastidar EG, Dzeyk K, Krijgsveld J, Malmquist NA, Doerig C, Scherf A, Lopez-Rubio JJ. Comprehensive histone phosphorylation analysis and identification of pf14-3-3 protein as a histone h3 phosphorylation reader in malaria parasites. PloS one. 2013;8(1):e53179. doi: 10.1371/journal.pone.0053179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Treeck M, Sanders JL, Elias JE, Boothroyd JC. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites’ boundaries. Cell host & microbe. 2011;10(4):410–9. doi: 10.1016/j.chom.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trelle MB, Salcedo-Amaya AM, Cohen AM, Stunnenberg HG, Jensen ON. Global histone analysis by mass spectrometry reveals a high content of acetylated lysine residues in the malaria parasite Plasmodium falciparum. J Proteome Res. 2009;8(7):3439–50. doi: 10.1021/pr9000898. [DOI] [PubMed] [Google Scholar]
- 21.Egelhofer TA, Minoda A, Klugman S, Lee K, Kolasinska-Zwierz P, Alekseyenko AA, Cheung MS, Day DS, Gadel S, Gorchakov AA, Gu T, Kharchenko PV, Kuan S, Latorre I, Linder-Basso D, Luu Y, Ngo Q, Perry M, Rechtsteiner A, Riddle NC, Schwartz YB, Shanower GA, Vielle A, Ahringer J, Elgin SC, Kuroda MI, Pirrotta V, Ren B, Strome S, Park PJ, Karpen GH, Hawkins RD, Lieb JD. An assessment of histone-modification antibody quality. Nature structural & molecular biology. 2011;18(1):91–3. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of combinatorial histone modifications on antibody and effector protein recognition. Current biology : CB. 2011;21(1):53–8. doi: 10.1016/j.cub.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. High throughput characterization of combinatorial histone codes. Mol Cell Proteomics. 2009;8(10):2266–84. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De La Vega P, Holder AA, Batalov S, Carucci DJ, Winzeler EA. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301(5639):1503–8. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 25.Florens L, Washburn MP. Proteomic analysis by multidimensional protein identification technology. Methods Mol Biol. 2006;328:159–75. doi: 10.1385/1-59745-026-X:159. [DOI] [PubMed] [Google Scholar]
- 26.MacCoss MJ, McDonald WH, Saraf A, Sadygov R, Clark JM, Tasto JJ, Gould KL, Wolters D, Washburn M, Weiss A, Clark JI, Yates JR. 3rd, Shotgun identification of protein modifications from protein complexes and lens tissue. Proc Natl Acad Sci U S A. 2002;99(12):7900–5. doi: 10.1073/pnas.122231399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Washburn MP, Wolters D, Yates JR. 3rd, Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Wen Z, Washburn MP, Florens L. Improving proteomics mass accuracy by dynamic offline lock mass. Anal Chem. 2011;83(24):9344–51. doi: 10.1021/ac201867h. [DOI] [PubMed] [Google Scholar]
- 29.Eng J, McCormack AL, Yates JR. III, An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Amer. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 30.Xiang Y, Takeo S, Florens L, Hughes SE, Huo LJ, Gilliland WD, Swanson SK, Teeter K, Schwartz JW, Washburn MP, Jaspersen SL, Hawley RS. The inhibition of polo kinase by matrimony maintains G2 arrest in the meiotic cell cycle. PLoS Biol. 2007;5(12):pe323. doi: 10.1371/journal.pbio.0050323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tabb DL, McDonald WH, Yates JR. 3rd, DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1(1):21–6. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal Chem. 2010;82(6):2272–81. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- 33.Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, Washburn MP. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40(4):303–11. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J Proteome Res. 2004;3(5):958–64. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Wen Z, Washburn MP, Florens L. Improving label-free quantitative proteomics strategies by distributing shared peptides and stabilizing variance. Anal Chem. 2015;87(9):4749–56. doi: 10.1021/ac504740p. [DOI] [PubMed] [Google Scholar]
- 36.Caraux G, Pinloche S. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics. 2005;21(7):1280–1. doi: 10.1093/bioinformatics/bti141. [DOI] [PubMed] [Google Scholar]
- 37.Wong JAHaMA. Algorithm AS 136: A K-Means Clustering Algorithm. The Journal of the Royal Statistical Society. Series C (Applied Statistics) 1979;28(1):100–108. [Google Scholar]
- 38.Tweedie-Cullen RY, Brunner AM, Grossmann J, Mohanna S, Sichau D, Nanni P, Panse C, Mansuy IM. Identification of combinatorial patterns of post-translational modifications on individual histones in the mouse brain. PloS one. 2012;7(5):e36980. doi: 10.1371/journal.pone.0036980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Research. 2009;19(9):1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka N, Nagasaka K, Komatsu Y. Selected reaction monitoring by linear ion-trap mass spectrometry can effectively be applicable to simultaneous quantification of multiple peptides. Biol Pharm Bull. 2011;34(1):135–41. doi: 10.1248/bpb.34.135. [DOI] [PubMed] [Google Scholar]
- 41.Yang X, Lazar IM. MRM screening/biomarker discovery with linear ion trap MS: a library of human cancer-specific peptides. BMC Cancer. 2009;9:96. doi: 10.1186/1471-2407-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cui L, Fan Q, Miao J. Histone lysine methyltransferases and demethylases in Plasmodium falciparum. International journal for parasitology. 2008;38(10):1083–97. doi: 10.1016/j.ijpara.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui L, Miao J. Chromatin-mediated epigenetic regulation in the malaria parasite Plasmodium falciparum. Eukaryot Cell. 2010;9(8):1138–49. doi: 10.1128/EC.00036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nardelli SC, Che FY, Silmon de Monerri NC, Xiao H, Nieves E, Madrid-Aliste C, Angel SO, Sullivan WJ, Jr, Angeletti RH, Kim K, Weiss LM. The histone code of Toxoplasma gondii comprises conserved and unique posttranslational modifications. MBio. 2013;4(6):e00922–13. doi: 10.1128/mBio.00922-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, Recht J, Shabanowitz J, Mishra N, Strahl BD, Allis CD, Hunt DF. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282(10):7641–55. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- 46.Miao J, Fan Q, Cui L, Li J. The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation. Gene. 2006;369:53–65. doi: 10.1016/j.gene.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 47.Parker CE, Mocanu V, Mocanu M, Dicheva N, Warren MR. Mass Spectrometry for Post-Translational Modifications. In: Alzate O, editor. Neuroproteomics. Boca Raton (FL): 2010. [PubMed] [Google Scholar]
- 48.Fan Q, An L, Cui L. Plasmodium falciparum histone acetyltransferase, a yeast GCN5 homologue involved in chromatin remodeling. Eukaryotic cell. 2004;3(2):264–76. doi: 10.1128/EC.3.2.264-276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cui L, Miao J. Cytotoxic effect of curcumin on malaria parasite Plasmodium falciparum: inhibition of histone acetylation and generation of reactive oxygen species. Antimicrobial agents and chemotherapy. 2007;51(2):488–94. doi: 10.1128/AAC.01238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, Tora L. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC genomics. 2012;13:424. doi: 10.1186/1471-2164-13-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan ZF, Lin S, Molden RC, Garcia BA. Evaluation of proteomic search engines for the analysis of histone modifications. J Proteome Res. 2014;13(10):4470–8. doi: 10.1021/pr5008015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westenberger SJ, Cui L, Dharia N, Winzeler E. Genome-wide nucleosome mapping of Plasmodium falciparum reveals histone-rich coding and histone-poor intergenic regions and chromatin remodeling of core and subtelomeric genes. BMC Genomics. 2009;10:610. doi: 10.1186/1471-2164-10-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, Kaiser P. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivocross-linking. Mol Cell Proteomics. 2006;5(4):737–48. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125(4):703–17. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 55.Singh RK, Kabbaj M-HM, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol. 2009;11(8):925–933. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Halbert J, Ayong L, Equinet L, Le Roch K, Hardy M, Goldring D, Reininger L, Waters N, Chakrabarti D, Doerig C. A Plasmodium falciparum transcriptional cyclin-dependent kinase-related kinase with a crucial role in parasite proliferation associates with histone deacetylase activity. Eukaryot Cell. 2010;9(6):952–9. doi: 10.1128/EC.00005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ganesan K, Ponmee N, Jiang L, Fowble JW, White J, Kamchonwongpaisan S, Yuthavong Y, Wilairat P, Rathod PK. A genetically hard-wired metabolic transcriptome in Plasmodium falciparum fails to mount protective responses to lethal antifolates. PLoS pathogens. 2008;4(11):e1000214. doi: 10.1371/journal.ppat.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Le Roch KG, Johnson JR, Ahiboh H, Chung DW, Prudhomme J, Plouffe D, Henson K, Zhou Y, Witola W, Yates JR, Mamoun CB, Winzeler EA, Vial H. A systematic approach to understand the mechanism of action of the bisthiazolium compound T4 on the human malaria parasite, Plasmodium falciparum. BMC genomics. 2008;9:513. doi: 10.1186/1471-2164-9-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye J, Ai X, Eugeni EE, Zhang L, Carpenter LR, Jelinek MA, Freitas MA, Parthun MR. Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Mol Cell. 2005;18(1):123–30. doi: 10.1016/j.molcel.2005.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaelin WG. Proline hydroxylation and gene expression. Annu Rev Biochem. 2005;74:115–28. doi: 10.1146/annurev.biochem.74.082803.133142. [DOI] [PubMed] [Google Scholar]
- 61.Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehling-Hampton K, Shilatifard A. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat Struct Mol Biol. 2008;15(8):881–8. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chopra RK, Ananthanarayanan VS. Conformational implications of enzymatic proline hydroxylation in collagen. Proc Natl Acad Sci U S A. 1982;79(23):7180–4. doi: 10.1073/pnas.79.23.7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang T, Zhou X, Taghizadeh K, Dong M, Dedon PC. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc Natl Acad Sci U S A. 2007;104(1):60–5. doi: 10.1073/pnas.0606775103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Britton LM, Newhart A, Bhanu NV, Sridharan R, Gonzales-Cope M, Plath K, Janicki SM, Garcia BA. Initial characterization of histone H3 serine 10 O-acetylation. Epigenetics. 2013;8(10):1101–13. doi: 10.4161/epi.26025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mittal R, Peak-Chew SY, McMahon HT. Acetylation of MEK2 and I kappa B kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc Natl Acad Sci U S A. 2006;103(49):18574–9. doi: 10.1073/pnas.0608995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, Goldsmith EJ, Orth K. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312(5777):1211–4. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 67.Hoeijmakers WA, Salcedo-Amaya AM, Smits AH, Francoijs KJ, Treeck M, Gilberger TW, Stunnenberg HG, Bartfai R. H2A.Z/H2B.Z double-variant nucleosomes inhabit the AT-rich promoter regions of the Plasmodium falciparum genome. Molecular microbiology. 2013;87(5):1061–73. doi: 10.1111/mmi.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol Cell Biol. 2008;28(1):468–86. doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith CM, Gafken PR, Zhang Z, Gottschling DE, Smith JB, Smith DL. Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4. Analytical biochemistry. 2003;316(1):23–33. doi: 10.1016/s0003-2697(03)00032-0. [DOI] [PubMed] [Google Scholar]
- 70.Zhang K, Williams KE, Huang L, Yau P, Siino JS, Bradbury EM, Jones PR, Minch MJ, Burlingame AL. Histone acetylation and deacetylation: identification of acetylation and methylation sites of HeLa histone H4 by mass spectrometry. Molecular & cellular proteomics : MCP. 2002;1(7):500–8. doi: 10.1074/mcp.m200031-mcp200. [DOI] [PubMed] [Google Scholar]
- 71.Keen JC, Yan L, Mack KM, Pettit C, Smith D, Sharma D, Davidson NE. A novel histone deacetylase inhibitor, scriptaid, enhances expression of functional estrogen receptor alpha (ER) in ER negative human breast cancer cells in combination with 5-aza 2’-deoxycytidine. Breast cancer research and treatment. 2003;81(3):177–86. doi: 10.1023/A:1026146524737. [DOI] [PubMed] [Google Scholar]
- 72.Thorne AW, Kmiciek D, Mitchelson K, Sautiere P, Crane-Robinson C. Patterns of histone acetylation. European journal of biochemistry / FEBS. 1990;193(3):701–13. doi: 10.1111/j.1432-1033.1990.tb19390.x. [DOI] [PubMed] [Google Scholar]
- 73.Jiang L, Smith JN, Anderson SL, Ma P, Mizzen CA, Kelleher NL. Global assessment of combinatorial post-translational modification of core histones in yeast using contemporary mass spectrometry. LYS4 trimethylation correlates with degree of acetylation on the same H3 tail. The Journal of biological chemistry. 2007;282(38):27923–34. doi: 10.1074/jbc.M704194200. [DOI] [PubMed] [Google Scholar]
- 74.Ay F, Bunnik EM, Varoquaux N, Bol SM, Prudhomme J, Vert JP, Noble WS, Le Roch KG. Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Res. 2014;24(6):974–88. doi: 10.1101/gr.169417.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wakamori M, Fujii Y, Suka N, Shirouzu M, Sakamoto K, Umehara T, Yokoyama S. Intra- and inter-nucleosomal interactions of the histone H4 tail revealed with a human nucleosome core particle with genetically-incorporated H4 tetra-acetylation. Sci Rep. 2015;5:17204. doi: 10.1038/srep17204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data can be obtained from MassIVE (ftp://massive.ucsd.edu/) or ProteomeXchange (http://www.proteomexchange.org/) using the following accession numbers with password (ANS01146): MSV000079225/PXD002709 to MSV000079231/PXD002715 for the 0- to 36-hpi MudPIT datasets (SI-1.1) and MSV000079792/PXD004268 for the MRM dataset (SI-6.1). In addition, all of the original data (unprocessed Western Blots images) and processing steps (OriginPro files as well as R and jmol scripts) may be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/LIBPB-536.