Abstract
Summary
The examination of gene neighborhood is an integral part of comparative genomics but no tools to produce publication quality graphics of gene clusters are available. Gene Graphics is a straightforward web application for creating such visuals. Supported inputs include National Center for Biotechnology Information gene and protein identifiers with automatic fetching of neighboring information, GenBank files and data extracted from the SEED database. Gene representations can be customized for many parameters including gene and genome names, colors and sizes. Gene attributes can be copied and pasted for rapid and user-friendly customization of homologous genes between species. In addition to Portable Network Graphics and Scalable Vector Graphics, produced representations can be exported as Tagged Image File Format or Encapsulated PostScript, formats that are standard for publication. Hands-on tutorials with real life examples inspired from publications are available for training.
Availability and implementation
Gene Graphics is freely available at https://katlabs.cc/genegraphics/ and source code is hosted at https://github.com/katlabs/genegraphics.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Conserved physical clustering of genes (conserved genome context) in phylogenetically distant genomes is a strong suggestion of functional association (Overbeek et al., 1999). Thus, displaying and collating gene clusters is a common step in comparative genomics. Most of the specialized databases such as SEED (Overbeek et al., 2005), Integrated Microbial Genomes (Markowitz et al., 2012), Pathosystems Resource Integration Center (PATRIC) (Wattam et al., 2017), MicroScope (Vallenet et al., 2009) and Search Tool for the Retrieval of Interacting Genes/Proteins (Snel et al., 2000) allow users to display genes of interest and their neighbors. Except for genome selection and region window size, the display parameters are precomputed and fixed, and manipulation and export of representative image is impossible. Nevertheless, creating accurate and visually appealing representations of gene neighborhoods is frequently required.
The creation of gene neighborhoods representations using image editing software is time-consuming and prone to error. Open-source or vendor-supplied in silico cloning and sequence analysis programs, such as Clone Manager (Scientific and Educational Software), A Plasmid Editor, SnapGene (GSL Biotech), Vector NTI (Thermo Fisher Scientific) and pDRAW32 (AcaClone software) offer to create gene maps from sequence data, and are oriented towards record keeping purposes, not publication. In addition, these tools are neither user-friendly nor flexible, require installation, and can have costly licensing fees. Ultimately, the quality of the image produced may not be high enough for publications requirements.
Available within the bioinformatics environments BioPython and R, are GenomeDiagram (Pritchard et al., 2006) and genoPlotR (Guy et al., 2010), respectively. These tools aim at producing ready for publication, high quality figures in comparative genomics studies. Their utilization requires some programming skill, which may intimidate biologists.
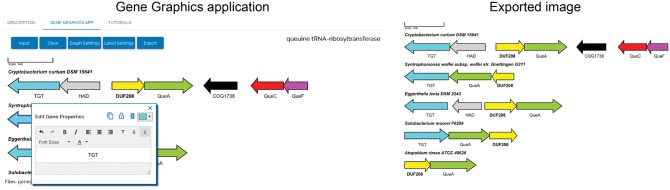
A free and easy to use application that allows for specific but accessible input and customization of the genomic regions to be displayed and can create high-quality graphics, ready for publication (Fig. 1) was clearly needed.
Fig. 1.
Gene Graphics application (left) and associated output (right). Information shown is accessible as ‘Tutorial 1; Final Result’ in Gene Graphics
Here, we present Gene Graphics https://katlabs.cc/genegraphics/, a web application that allows for consistent, visually appealing representations of physical gene neighborhoods with minimal effort. User-friendly tutorials based on the recreation of previously published figures are provided and illustrate how the conservation of genome context in phylogenetically distant organisms allows to identify functional association, and set within biological pathways, the previously uncharacterized genes duf208 and duf89 (Huang et al., 2016; Zallot et al., 2017), or identify an unsuspected relation between the genes folK and panB (Thiaville et al., 2016).
2 Data input
The information to be presented can be fetched from the National Center for Biotechnology Information (NCBI) or can be loaded as GenBank files or a tab separated values (TSV) files obtained from the SEED database. The user may incrementally upload multiple genomic regions from various sources into a single graphic. An input is interpreted and used for the graphic generation, and consolidated as a data object in a custom TSVs file that can be exported as a record. Previously exported records can be reimported. Supplementary Table S1 describes the structure of a typical data object in the TSV file.
We propose the ‘Fetch from NCBI’ as the primary input method. This option queries NCBI databases based on four different options: Gene ID, Protein ID, Gene symbol and genome or Genome and location. With all ‘Fetch from NCBI’ input, the user should specify a value corresponding to the region size (in number of base pairs) in which the gene of interest is centered within the graphic. Supplementary Figures S1–S4 describe in detail the data fetching process, the inputs allowed, and why RefSeq non-redundant protein IDs (‘WP_’ followed by nine numerical digits) cannot be used. Requests end with an eFetch to the nuccore database, and produce a GenBank file that is interpreted. In addition, GenBank files, conforming to the standard format (Benson et al., 2005), can be directly used as input. Information obtained is parsed and a data object is created. TSV files directly exported from the SEED database are also valid input.
3 Usage
The Gene Graphics application has an easy-to-use graphical interface which allows for full customization of the default layout: colors, fonts, sizes and positions of gene and genome names can be edited both individually and globally. Multiple genomes in a single view are supported to easily allow visualization of homologs. Images generated can be used for record keeping and publications.
Each genome has a label and is followed by arrows representing genes. Genomes can be displayed on multiple lines if they have overlapping genes. Gaps between genes can be shown or hidden. The length of each arrow is proportional to its gene size, in number of base pairs. The default display includes a scale for reference. Selecting a gene opens a multi-function editor in which the user can stylize and customize the size of the label and its position. It is possible to copy attributes from one gene to another to quickly homogenize the representation of homologous genes. By default, genes with the exact same annotation are displayed with the same color, which is accomplished using a hashing algorithm. Although customizing an individual gene, the user can ‘lock’ its formatting, so that any global options set has no effect.
Information is automatically saved within the browser, allowing the user to close the webpage and return to it later, with their information displayed as previously. This feature is automatic, requires no action on the part of the user, and uses the HyperText Markup Language 5 (HTML5) local storage object functionality.
The user can export their visualization as Tagged Image File Format or Encapsulated PostScript, formats that are standard for publication. Portable Network Graphic and Scalable Vector Graphic (SVG) formats are additional export options. TSV files can be exported and re-imported later.
4 Materials and methods
Gene Graphics is built with JavaScript, HTML5, and Cascading Style Sheets using AngularJS as the web framework. Information fetching from NCBI databases is done using the NCBI Entrez Programming Utilities: eFetch and eSearch. Data objects are rendered to SVG elements using D3.js. Path and text elements are positioned based on the start and stop locations of the genes, the strand, and user settings. Changes made by the user operating the application automatically update the data and re-render the view. The text editing features use TinyMCE editor. Gene colors are edited using the SpectrumJS colorpicker. The data are encoded into a Uniform Resource Identifier and linked on the export panel, so that the user can right click and save it as a file.
5 Conclusions
Gene Graphics is a web application for creating publication quality representations of gene neighborhoods for comparative genomics purposes. Genome regions are accurately represented. Options allow for customization of the information visualized. Tutorials are available. Gene Graphics only requirement is a modern web browser. Gene Graphics is available at https://katlabs.cc/genegraphics/.
Supplementary Material
Acknowledgements
We thank members of the de Crécy-Lagard (U. of Florida) and Gerlt (U. of Illinois at Urbana-Champaign) laboratories for testing and providing feedback.
Funding
This work was supported by the National Institutes of Health [grant R01 GM70641 to V.d.C.-L.].
Conflict of Interest: none declared.
References
- Benson D.A. et al. (2005) GenBank. Nucleic Acids Res., 33, D34–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy L. et al. (2010) genoPlotR: comparative gene and genome visualization in R. Bioinformatics, 26, 2334–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. et al. (2016) A family of metal-dependent phosphatases implicated in metabolite damage-control. Nat. Chem. Biol., 12, 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz V.M. et al. (2012) IMG: The integrated microbial genomes database and comparative analysis system. Nucleic Acids Res., 40, D115–D122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R. et al. (2005) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res, 33, 5691–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R. et al. (1999) Use of contiguity on the chromosome to predict functional coupling. In Silico Biol., 1, 93–108. [PubMed] [Google Scholar]
- Pritchard L. et al. (2006) GenomeDiagram: a python package for the visualization of large-scale genomic data. Bioinformatics, 22, 616–617. [DOI] [PubMed] [Google Scholar]
- Snel B. et al. (2000) STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res., 28, 3442–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiaville J.J. et al. (2016) Experimental and metabolic modeling evidence for a folate-cleaving side-activity of ketopantoate hydroxymethyltransferase (PanB). Front. Microbiol., 7, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet D. et al. (2009) MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford), 2009, bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam A.R. et al. (2017) Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res., 45, D535–D542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallot R. et al. (2017) Identification of a novel epoxyqueuosine reductase family by comparative genomics. ACS Chem. Biol., 12, 844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.