Summary
We have reported altered choline and tryptophan metabolism with malnutrition in our murine model and in children. Here, we report the additional metabolitic alterations with murine cryptosporidiosis, including trimethylamine N-oxide (TMAO), which is largely independent of changes in microbiota.
Keywords: choline, Cryptosporidium, malnutrition, metabonome, microbiome, .
Abstract
Cryptosporidium infections have been associated with growth stunting, even in the absence of diarrhea. Having previously detailed the effects of protein deficiency on both microbiome and metabolome in this model, we now describe the specific gut microbial and biochemical effects of Cryptosporidium infection. Protein-deficient mice were infected with Cryptosporidium parvum oocysts for 6–13 days and compared with uninfected controls. Following infection, there was an increase in the urinary excretion of choline- and amino-acid-derived metabolites. Conversely, infection reduced the excretion of the microbial–host cometabolite (3-hydroxyphenyl)propionate-sulfate and disrupted metabolites involved in the tricarboxylic acid (TCA) cycle. Correlation analysis of microbial and biochemical profiles resulted in associations between various microbiota members and TCA cycle metabolites, as well as some microbial-specific degradation products. However, no correlation was observed between the majority of the infection-associated metabolites and the fecal bacteria, suggesting that these biochemical perturbations are independent of concurrent changes in the relative abundance of members of the microbiota. We conclude that cryptosporidial infection in protein-deficient mice can mimic some metabolic changes seen in malnourished children and may help elucidate our understanding of long-term metabolic consequences of early childhood enteric infections.
Malnutrition is a global health epidemic. Undernutrition results in growth and cognitive impairment, a host of inflammatory markers and an increased susceptibility to enteric infections [1]. The consequences of these infections, such as diarrhea and malabsorption from intestinal damage, further deprive the host of nutrients and lead to a vicious reciprocal cycle of undernutrition and enteric infections. The cumulative effects of this chronic cycle result in developmental shortfalls and impaired responses to vaccines [2–6]. Understanding how nutritional deficiencies prolong enteric infections and combine to the detriment of the host is essential to break this cycle.
Cryptosporidiosis, a protozoan infection, is prevalent in malnourished populations, areas with limited access to clean water and sanitation, as well as immunocompromised individuals [2, 7–13]. Cryptosporidium infections with or without overt diarrhea have been associated with significant growth shortfalls, and we have previously demonstrated that, as seen in children, malnutrition substantially worsens Cryptosporidium infection in a murine model [14–16]. Following 3 days of Cryptosporidium infection, significant growth impairment was observed in protein-malnourished mice, accompanied by increased intestinal injury and inflammation. Unlike nourished equivalents, protein-malnourished mice were unable to rapidly clear Cryptosporidium and continue to have detectable organisms in the stool for more than 7 days [17, 18].
A previous study found strong effects of C. parvum on the murine microbiota, although the authors acknowledge that small treatment group size across multiple experiments may have amplified differences between uninfected and infected groups [19]. Studies in children and rodents have indicated that undernutrition results in both compositional and functional modulations in the resident gut microbiota [19]. In a study examining the effect of various diets on the mouse microbiota and metabolome, we observed that mice fed a low-protein diet retained a fecal microbiota more similar in composition to newly weaned mice than those on any other diet [20–22]. In the current study, we examined the effects of cryptosporidiosis on the fecal microbiome and urinary metabolic phenotypes of mice on the same protein-deficient diets. We disentangled the infection-specific metabolic alterations from those related to the diet. This approach reveals that several metabolic shifts after infection occur independent of changes in specific microbiota.
METHODS
Animal Husbandry
Mice used in this study were male, 22 days old, C67BL/6 strain, and ordered from Jackson Laboratories (Bar Harbor, ME). Mice weighed approximately 11 g on arrival and were cohoused in groups up to 5 animals per cage. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of the University of Virginia (protocol no. 3315).
Rodent Diet
Weaned mice (22 days old) were fed a defined protein-deficient diet (dPD; 2% protein; Research Diets, New Brunswick, NJ) as previously published [23].
Cryptosporidium Challenge
Mice were infected with a single inoculum of 5 × 106 purified C. parvum oocysts (Iowa Strain, Waterborne, Inc) by orogastric gavage as previously described [17]. Following challenge with C. parvum oocysts, mice were weighed daily. Feces and urine samples were collected every other day.
Lipocalin-2 and Myeloperoxidase Measurements
Lipocalin-2 (Lcn2) and myeloperoxidase (MPO) were measured in fecal samples collected 5 days after Cryptosporidium infection. Samples were homogenized in a radioimmunoprecipitation assay buffer with protease inhibitors, centrifuged at 8000 rpm for 10 minutes, and supernatant collected. Stool supernatant was assayed for total protein (BCA), Lcn2 (R&D Systems), and MPO (R&D Systems), according to manufacturer’s instructions. Data were reported as pg of Lcn2 or MPO per µg total protein.
DNA Isolation
DNA was isolated from fecal pellets using the QIAamp DNA stool mini kit as previously described [24]. Stool DNA was analyzed for the C. parvum–specific 18S ribosomal ribonucleic acid (rRNA) gene to determine shedding of organism in the stool. Polymerase chain reaction conditions and primer sequences were as previously published [18]. The V3-V4 hypervariable regions of the 16S rRNA gene from fecal DNA samples were amplified using Illumina specific primers: (Illumina; forward: 5'-TCGTCGGCAGCGTCAGATGTGTATA-AGAG ACAGCCTACGGGNGGCWGCAG–3', reverse: 5'–GT- CTCGTGGGCTCGGAGATGTGTATAAGAGACAGG ACTAC- HVGGGTATCTAATCC–3'). For detailed methods of 16S rRNA gene sequencing and 1H NMR spectroscopy-based metabolic profiling, please see Supplementary Methods.
RESULTS
We have previously shown significant weight loss, colonization, fecal shedding of the parasite, and inflammation in mice maintained on a protein-deficient diet infected with C. parvum [15, 17, 18, 24]. A protein-malnourished state was necessary for the infection to establish, as C. parvum infections could not be sustained in nourished mice. Peak shedding of oocysts usually occurs approximately 5 days postinfection, which coincides with up to 20% weight loss in infected mice. Using this same model of protein malnutrition, we infected mice with 5 × 106C. parvum oocysts. As in previous studies, infected mice had significant weight loss and shedding of oocysts in stool (Supplementary Figure 1).
Lcn2 Is Significantly Upregulated by Cryptosporidiosis
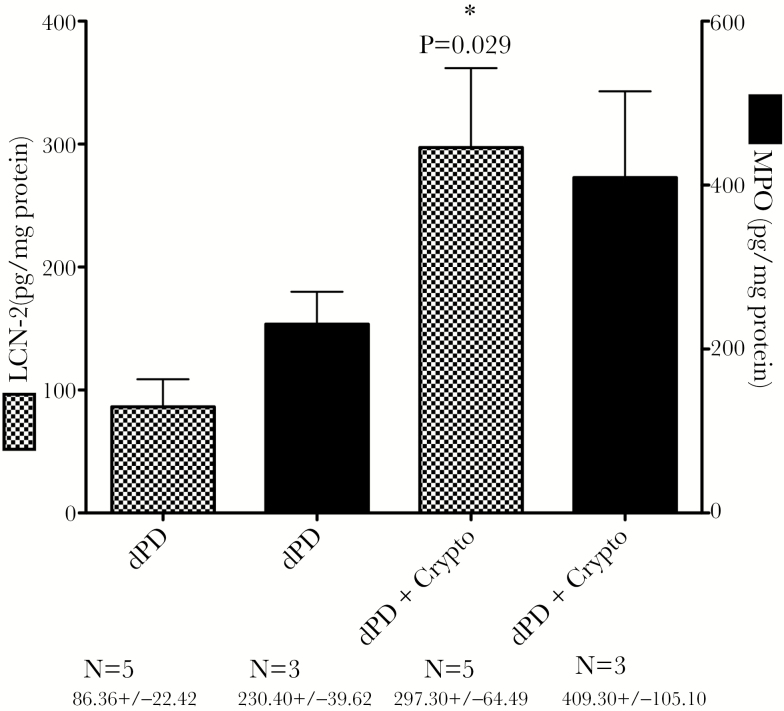
We have previously shown that protein deficiency alone is capable of disrupting intestinal epithelial tight junctions [18] and elevating fecal biomarkers of inflammation [23]. As shown in Figure 1, C. parvum infection with protein deficiency further increases myeloperoxidase and significantly increased Lcn2 compared with protein deficiency alone.
Figure 1.
Lcn2 and myeloperoxidase levels in stools from C. parvum–infected mice 3 days postinfection. *Protein Deficient (dPD) vs dPD + Crypto; P = .029. Abbreviations: Crypto, C. parvum; Lcn2, lipocalin-2.
Urinary Metabolic Alterations Induced by C. parvum Infection
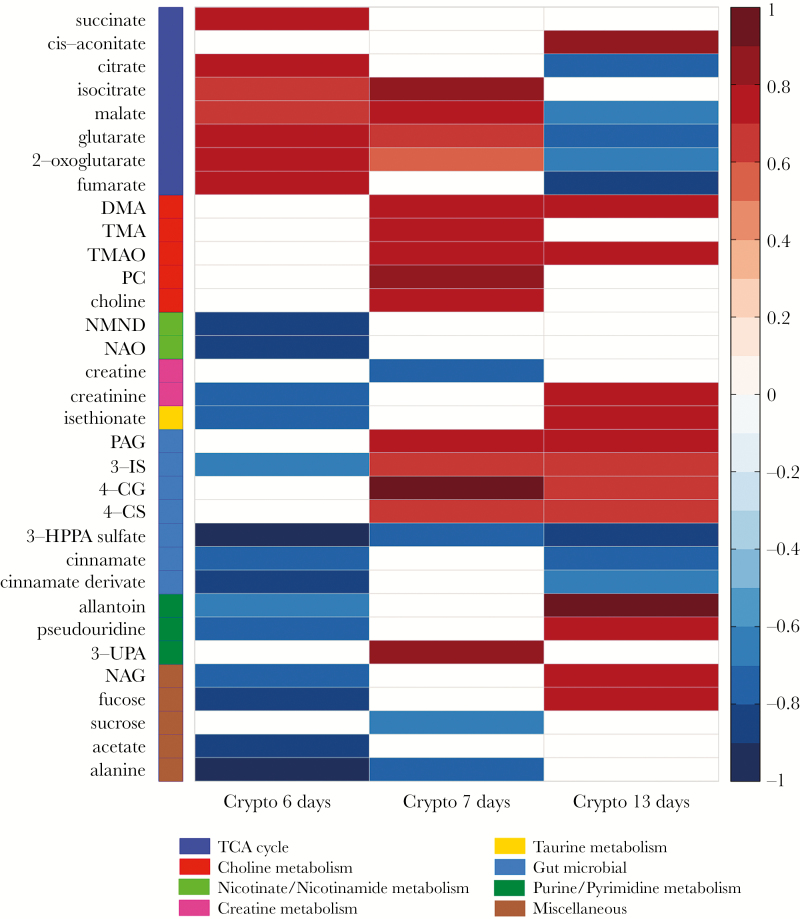
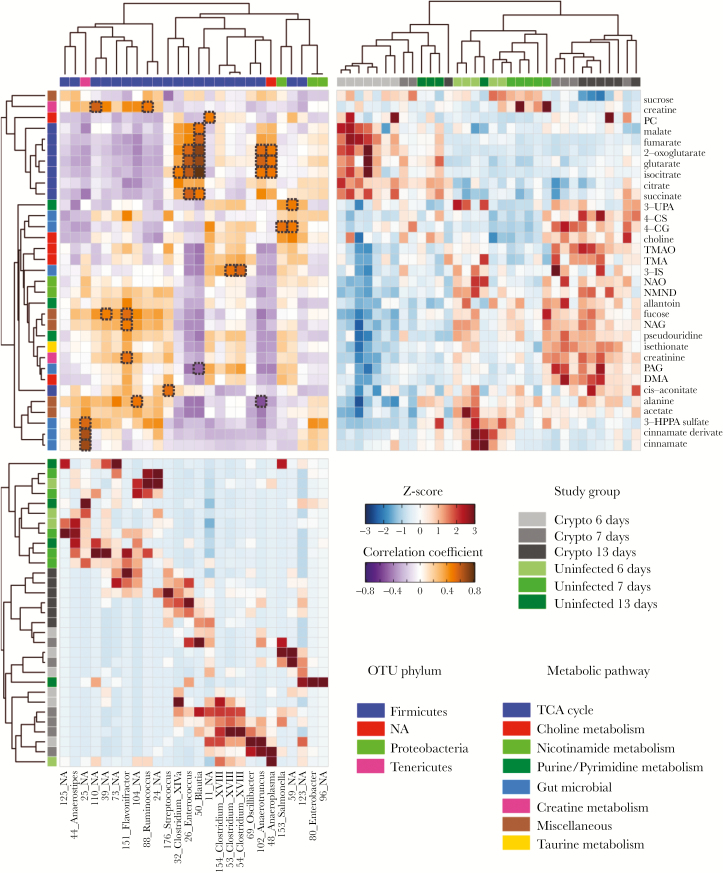
The urinary metabolic profiles of control and infected mice were compared at 6, 7, and 13 days postinfection using an orthogonal partial least squares–discriminant analysis (OPLS-DA) approach to assess the biochemical response to C. parvum exposure. Significant OPLS-DA models with good predictive abilities were obtained for this comparison at all time points. Following 7 days of C. parvum infection (OPLS-DA model: Q2Y = 0.66; P = .025), the excretion of choline, phosphocholine, and the gut microbial metabolites of choline, dimethylamine (DMA) and trimethylamine (TMA), as well as the host oxidation product of TMA, trimethylamine N-oxide (TMAO), were increased compared to the uninfected controls (Figure 2). At 13 days of infection (OPLS-DA model: Q2Y = 0.71; P = .001) DMA and TMAO excretion remained higher in the infected mice compared with the control mice, but the excretion of the other choline-related metabolites was comparable. Products of the combinatorial metabolism between the gut microbiota and host were also increased 7 and 13 days postinfection, respectively. This included metabolites of the amino acids phenylalanine (phenylacetylglycine [PAG]), tryptophan (3-indoxyl-sulfate [3-IS]), and tyrosine (4-cresyl glucuronide [4-CG] and 4-cresyl-sulfate [4-CS]). However, the excretion of 3-hydroxy-phenyl propionic acid sulfate (3-HPPA sulfate) and cinnamate and a cinnamate derivate, all gut microbial–host cometabolites of polyphenols and/or phenylalanine, was reduced 6 and/or 7 and 13 days postinfection, respectively. In addition, following 6 days of C. parvum infection (OPLS-DA model: Q2Y = 0.82; P = .001), several TCA cycle metabolites (succinate, citrate, isocitrate, malate, 2-oxoglutarate, and fumarate) were increased in urine compared with uninfected controls. However, some of these changes (succinate, citrate, and fumarate) were not observed at 7 days, and by 13 days postinfection most alterations were observed in the opposite direction (decreased), with the exception of cis-aconitate, which was excreted in greater amounts compared with uninfected controls. Similar trends were observed in the excretion of metabolites related to inflammation and oxidative stress (allantoin, pseudouridine, N-acetyl glycoproteins [NAG], and fucose). These metabolites were excreted in lower amounts 6 days postinfection but were excreted in higher amounts 13 days postinfection compared with uninfected controls. No difference was observed in their excretion 7 days postinfection. Other notable changes in the urinary metabolic profiles included a decrease in sucrose, acetate, and alanine at 6 and/or 7 days postinfection, which was not observed after 13 days of infection.
Figure 2.
Heatmap summary of the Crypto-induced urinary metabolic alterations identified by the OPLS-DA models. The color (ranging from –1 dark blue to +1 dark red) indicates the correlation coefficient (R) of Crypto at 6, 7, and 13 days postinfection compared with diet-(protein deficient) and age-matched uninfected controls. Metabolic grouping are indicated by color code on the left with the group color key below. n = 5/7 at day 6; n = 5/6 at day 7; and n = 4/7 at day 13 postinfection (uninfected/infected). Abbreviations: 3-IS, 3-indoxyl sulfate; 3-UPA, 3-ureidopropionic acid; 4-CG, 4-cresol glucuronide; 4-CS, 4-cresyl sulfate; DMA, dimethylamine; 3-HPPA sulfate, 3-hydroxy-phenyl propionic acid sulfate; NAG, N-acetyl glycoproteins; NAO, nicotinamide-N-oxide; NMND, N-methylnicotinamide; PAG, phenylacetylglycine; PC, phosphocholine; TMA, trimethylamine; TMAO, trimethylamine-N-oxide.
Effects of Protein Deficiency and Cryptosporidium on Gut Resident Microbiota
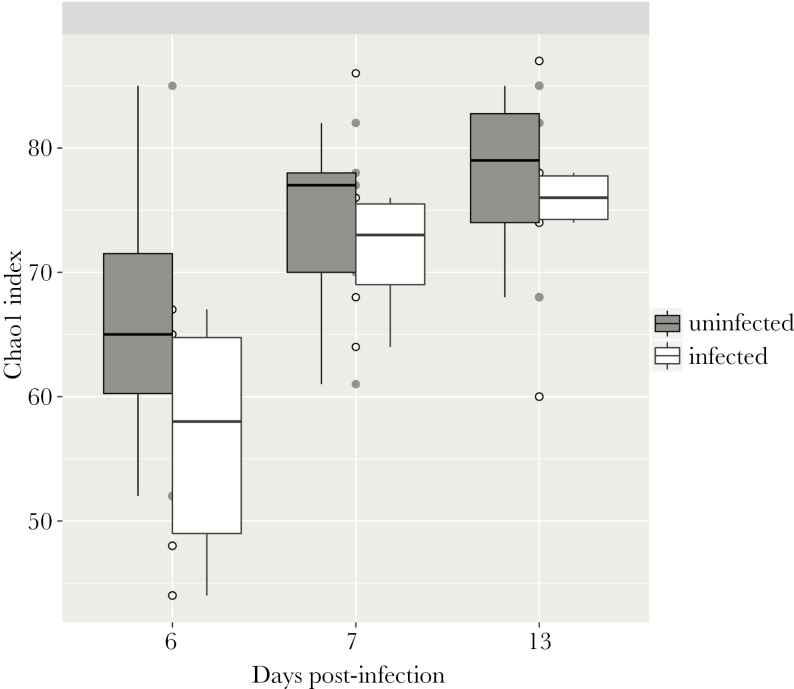
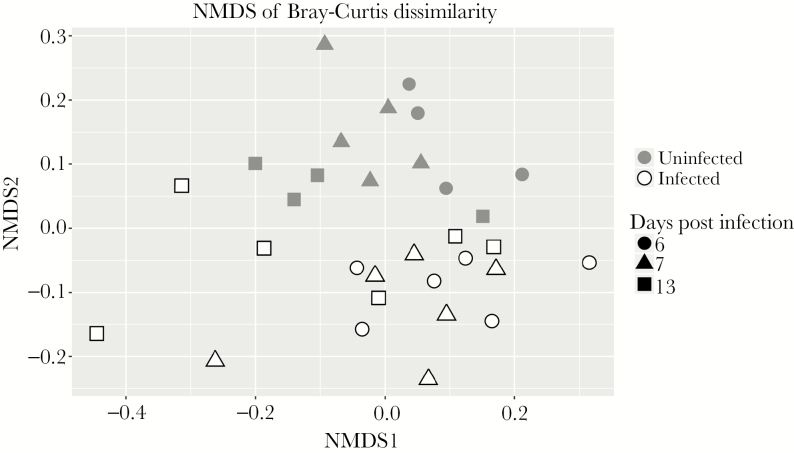
We have recently shown that protein deficiency retards the aging-induced diversification of the fecal microbiome relative to a defined nourished diet [23]. Although cryptosporidiosis caused major changes in urinary metabolites, there was surprisingly little effect of C. parvum infection on the bacterial composition of the feces. Only 26 microbial sequence variants (SVs) were found to be differentially abundant between infected and uninfected groups at any time point, and these 26 SVs stratified infected and uninfected groups independent of time (Supplementary Figure 2). At all time points, infection slightly reduced α-diversity, consistent across a variety of α-diversity metrics (Figure 3, Supplementary Figure 2). Considering all SVs that met filtering criteria (see Methods), β-diversity calculations and nonmetric multidimensional scaling led to delineation of infected and uninfected samples (Figure 4).
Figure 3.
Alpha diversity of infected and uninfected fecal microbiota. Alpha diversity, as calculated by the Chao 1 Index, is slightly higher in uninfected mice at all time points. All samples are shown as circles, whiskers extend to maximum and minimum, and boxplot shows median (middle line) and 25th and 75th percentiles (lower and upper hinge, respectively). n = 4/6 at day 6; n = 5/6 at day 7; and n = 4/6 at day 13 postinfection (uninfected/infected).
Figure 4.
Beta diversity of fecal samples from infected and uninfected mice. Nonmetric multidimensional scaling was performed on the Bray-Curtis dissimilarity matrix to visualize the similarity of samples. n = 4/6 at day 6; n = 5/6 at day 7; and n = 4/6 at day 13 post infection (uninfected/infected).
Microbial–Metabolic Interactions in Response to Malnutrition
Unsupervised hierarchical clustering analysis (HCA) of the metabolites identified to contribute to the C. parvum infection at days 6, 7, and 13 is shown in Figure 5 (upper-right quadrant). The interpretation that 7 days postinfection represents a transition in the metabolic shift from 6 to 13 days postinfection is more evident from the heatmap. C. parvum–infected samples at 6 days appear on the left, whereas all 13-day postinfected samples are located on the right, with some 7-day infected samples clustering with either 6- or 13-day infected samples. The clustergram, constructed using relative abundances of the bacteria identified to be significantly different between any age-matched infected and uninfected mice, resulted in distinct clusters of uninfected and infected mice (Figure 5, lower-left quadrant). In addition, there seems to be a transition in the SVs from 6 to 13 days, with some 7-day samples clustering with either 6- or 13-day infected samples.
Figure 5.
Unsupervised hierarchical clustering of the metabonome and microbiome for all mice. Upper right quadrant: Metabolite clustergram displaying z scores of metabolites for each mouse. A metabolite z-score transformation was performed on the intensity of each metabolite across samples. Metabolites shown are those that were identified in orthogonal projection to latent structures discriminant analysis models as varying between study groups. Study-group assignments are indicated by color bars. Lower-left quadrant: Bacterial clustergram displaying z scores of specific bacteria abundance for each mouse. Each row corresponds to an individual mouse, and each column corresponds to a specific OTU. OTUs selected were those that passed the filtering criteria as stated in Methods. Metabolites, OTUs, and samples are clustered by correlation distance and average-linkage hierarchical clustering. Upper-left quadrant: Pearson’s correlation heatmap between OTUs and metabolites. The intensity of the colors represents the degree of correlation. Significant correlations after adjustment of P values (with the use of the Storey method for 5% false-discovery rate) are shown in a dotted black frame. n = 4/6 at day 6; n = 5/6 at day 7; and n = 4/6 at day 13 post infection (uninfected/infected). For the correlations we only used the animals for which we had both microbiome and metabonomic data. Abbreviations: 3-IS, 3-indoxyl sulfate; 3-UPA, 3-ureidopropionic acid; 4-CG, 4-cresol glucuronide; 4-CS, 4-cresyl sulfate; DMA, dimethylamine; 3-HPPA sulfate, 3-hydroxy-phenyl propionic acid sulfate; NAO, nicotinamide-N-oxide; NMND, N-methylnicotinamide; PAG, phenylacetylglycine; PC, phosphocholine; TMA, trimethylamine; TMAO, trimethylamine-N-oxide.
The correlation heatmap (Figure 5, upper-left quadrant) identified significant correlations between SVs and metabolites. The most notable metabolite–bacterial correlations included those between TCA cycle metabolites and 4 SVs identified as members of the genera Enterococcus, Blautia, Anaeroplasma, and Anaerotruncus. Additionally, an unknown bacterial group (25_NA) was strongly correlated with 3 products of microbial-specific metabolism (cinnamate, a cinnamate derivative, and 3-HPPA sulfate). The bacterial group 25_NA was not detected in any infected samples, and cinnamate and the cinnamate derivative are present at very low amounts in infected samples.
DISCUSSION
These findings of metabolites and biomarkers seen in experimentally controlled dietary (protein)–deficient mice with and without cryptosporidial infection with additional microbiome studies extend studies of cryptosporidial infection and their potential impact in field studies. In particular, we identify that modulations in choline, protein, and tryptophan metabolism seen with protein deficiency alone are magnified by cryptosporidial infection. Furthermore, these choline pathway metabolites, such as TMAO, may well have long-term risk consequences, as they have been strongly associated with cardiovascular disease risk in humans in later life [25]. Indeed, we and others have shown that early-life malnutrition, enteric infections, diarrhea, or enteropathy may have such long-term consequences as metabolic syndrome and cardiovascular disease, as well as stunting and cognitive effects [26–28]. Thus, these are the first specific metabolic findings with a common enteric infection, cryptosporidiosis, in a protein-deficient state that provide experimental evidence to support such potential consequences of the common dietary and enteric infections seen in young children in resource-limited settings.
Regarding fecal biomarkers that are altered in malnourished children and in our murine cryptosporidial infections, we find significant increases in the epithelial and inflammatory marker, Lcn2, as well as the inflammatory marker MPO. Consistently, cryptosporidiosis in malnourished children living in endemic areas (Haiti and Northeast Brazil) has also been associated with evidence of mild to moderate intestinal or systemic inflammation, including elevated fecal lactoferrin, and fecal or systemic interleukin-8 and tumor necrosis factor–α, more than that seen in healthy adult volunteers [29–31]. Similarly, we see effects on intestinal and inflammatory biomarkers in our protein-deficient murine model of experimental cryptosporidial infection that exceed those seen in protein deficiency alone, again suggesting a compounding of modest diet-induced changes [23] with added cryptosporidial infection, thus potentially modeling (and dissecting) both diet and infection components of environmental enteropathy. Also of interest is the greater increase in Lcn2 relative to MPO in our model. Because Lcn2 is seen in epithelial cells as well as in inflammatory cells, while MPO is more specifically associated with inflammation, these effects likely reflect a more direct disruption of epithelial-cell integrity by cryptosporidial infection, opposite to the greater relative increases in MPO we see in our murine models of enteroaggregative Escherichia coli or Campylobacter infections (unpublished data, submitted).
As expected after the introduction of an enteric pathogen, cryptosporidiosis impacted the composition of the microbiota. Infected mice were most notably enriched with SVs from Clostridium XVIII and had reduced levels of several SVs that could not be assigned identity at the genus level. Given the limited taxonomic resolution of 16S rRNA gene sequencing analysis, as well as the small region we used (180 base pairs), the cause of this shift requires further investigation. Overall, it is unclear whether modulation of the microbiota is a result of interaction between C. parvum and the microbiota, or an indirect effect in which cryptosporidiosis alters the intestinal environment, thereby altering the microbiota. Consistent with our observations the study of murine C. parvum infection by Ras et al. [19] found clear differences in the fecal microbial profiles between infected and uninfected mice using beta diversity analyses. In their study, 4 Bacteroidetes operational taxonomic units (OTUs) were identified as enriched during C. parvum oocyst shedding relative to uninfected controls, while 4 other Bacteroidetes OTUs were depleted. These results contrast with the shifts we identified, which were almost exclusively within the Firmicutes phylum. However, Ras and colleagues performed 3 other mouse experiments with slightly altered infection protocols, and a meta-analysis of 16S rRNA sequencing from all 4 experiments indicated strong clustering by experiment in beta diversity analysis. The inconsistencies in specific taxonomic shifts across these experiments imply that C. parvum infection does not induce a specific alteration of the microbiota, or that the effect cannot be identified using taxonomic analyses.
A previous murine study found that cryptosporidiosis induced by C. parvum was enhanced in germ-free immunodeficient mice relative to immunodeficient mice harboring the altered Schaedler flora, a model microbiota composed of 8 bacterial species [32]. This finding indicates that C. parvum colonization is affected by interactions between the intestinal microbiota and the parasite or host, even with a reduced microbiota. C. parvum growth is also inhibited to a greater extent in mature mice compared to freshly weaned mice, a finding originally thought to be due to increasing diversity of the gut microbiota with age [33]. The protein-deficient model in our study results in lower alpha diversity than age-matched mice on other diets, reinforcing that the result shown in Harp et al. [33] may be due to age-related differences in microbiota composition. However, colonization resistance conferred by the altered Schaedler flora rules out diversity as a determinant of colonization resistance; instead, indicating that specific functionalities carried out by 8 or fewer bacterial species are sufficient. The potential for individual bacterial species to influence colonization of C. parvum suggests that direct interaction between C. parvum and the intestinal microbiota is likely during cryptosporidiosis. Studies using mouse models with varying degrees of colonization resistance, coupled with metagenomic and/or metatranscriptomic analyses, are required to further characterize interactions between C. parvum and the microbiota.
At 6 days postinfection, we observed an increase in the urinary excretion of TCA cycle intermediates, with the exception of cis-aconitate. This initial stimulation of the TCA cycle may reflect the high energy demands required to fight the infectious process, resulting from the recruitment of inflammatory cells and elevated proliferation rates among lymphocyte populations [34]. However, C. parvum lacks both the TCA cycle and cytochrome-based respiratory chain, thus relying mainly on glycolysis for adenosine triphosphate production. This could lead to a depletion of the TCA cycle intermediates as the infection progresses and explains the lower levels of TCA cycle intermediates in the urine of C. parvum–infected mice at 13 days compared with uninfected controls. However, we did not observe any difference in glycolysis metabolites, such as pyruvate, lactate, or acetate.
Thus, the suppression of TCA cycle may not be due to a stimulation of glycolysis. Alternatively, it could be related to the higher excretion of cis-aconitate, a metabolite produced from the dehydration of citrate by aconitase, an iron-sulfur protein. Successful iron acquisition from host cells is almost entirely necessary for any protozoan pathogen to survive, grow, and replicate. C. parvum lacks a sulfur assimilation pathway, which is expected to be substituted from the host cells. This can lead to an aconitase deficiency and depletion of TCA cycle intermediates. In fact, aconitase knockdown flies contained high levels of cis-aconitate, while most other TCA cycle metabolites, including 2- oxoglutarate, succinate, fumarate, and malate, as well as metabolites of glycolysis, were significantly decreased [35]. In addition, several studies have shown that aconitases are inhibited by iron deficiency or oxidative stress [36].
Decreased aconitase activity may also lead to decreased production of nicotinamide adenine dinucleotide phosphate, an important defense against oxidative stress [36]. Thus, oxidative stress seems to play an important role in the development of C. parvum infection in mice [37].
Cryptosporidiosis is also associated with a persistent systemic inflammatory response. Consistently, the unsupervised HCA showed a cluster comprising NAG, fucose, pseudouridine, and allantoin. These metabolites are related to inflammation and oxidative stress. Thus, allantoin appears to be a useful biomarker of gut inflammation, and both allantoin and pseudouridine have been associated with inflammation and oxidative stress. Elevated levels of NAG signals from acute-phase reactive proteins have been associated with inflammatory conditions [38]. Finally, fucose is a sugar that can be found in the glycosylated part of the glycoproteins and is also found abundantly in the mammalian gut. Systemic invasion of pathogens or microbial products in the circulation has been shown to induce fucosylation of the epithelium in the small intestine, where it seems to have a protective role in both gut-centered and systemic infection and inflammation [39].
Interestingly, choline and phosphocholine (PC) were excreted in higher amounts in infected mice. PC was identified as the predominant lipid in C. parvum–infected bovine kidney cells and, interestingly, C. parvum has been suggested to be capable of sequestering phospholipids from the host to incorporate them into the membranes [40]. Thus, the greater abundance of choline and PC can be related to the degradation of host membranes by the parasite. In addition, choline-derived metabolites (DMA, TMA, and TMAO) were also excreted in higher amounts by infected mice.
Our results also suggested a change in microbial activity, in some cases coupled to the relative abundance of specific SVs. Gut microbial–derived metabolites such as PAG, 4-CG, 4-CS, 3-IS, DMA, TMA, and TMAO were excreted in higher amounts, whereas 3-HPPA sulfate, cinnamate, and a cinnamate derivate were excreted in lower amounts by infected mice. This was reinforced by the fact that TMA, TMAO, choline, 4-CS, 4-CG, and 3-IS cluster together, as did 3-HPPA sulfate, cinnamate, and the cinnamate derivate. PAG, 4-CG and 4-CS, and 3-IS are derived from phenylalanine, tyrosine, and tryptophan, respectively. On the other hand, DMA, TMA, and TMAO are choline-derived metabolites, while 3-HPPA sulfate and cinnamate result from polyphenol metabolism. Similar to our findings, elevations in TMA, PAG, 4-CG and reductions in hippurate and/or 3-HPPA sulfate have been previously identified in hosts infected with helminths (such as Schistosoma mansoni [41, 42] and S. japonicum [43–46]) and intestinal nematodes [47]. Similar results have also been observed in protozoan parasites such as Plasmodium berghei [48] and Trypanosoma brucei brucei [49].
The correlation between these metabolites and the SVs confirmed their microbial origin. For example, 4-CG is derived from microbial degradation of tyrosine by species such as Clostridium difficile, C. scatalogens, and certain Lactobacillus species. In accordance, 4-CG correlated with 59_NA and 123_NA, both clostridia SVs. However, choline-derived metabolites showed no significant correlations with the microbiome. Alternatively, these changes could be due to renal damage. For example, DMA and TMAO have been associated with renal cortex damage in methanol intoxication patients [50]. An increase in DMA and TMAO was observed at 13 days, when TCA cycle intermediates were depleted. These changes could be the direct products of parasite metabolism. Future studies should also address whether altered renal function might contribute to these findings as well.
In conclusion, these metabolomic, biomarker, and microbiome studies of cryptosporidial infections in protein-deficient mice help dissect common major dietary and pathogen elements in enteropathy commonly seen in children in developing areas. They also elucidate metabolic and inflammatory pathways that have direct relevance to growth failure and even potential long-term metabolic disease consequences. They also provide a powerful experimental model to help elucidate key pathways involved, and also to assess innovative interventions to ameliorate environmental enteropathy and its triple burden of potentially lasting consequences in impoverished children.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Financial support. This work was supported by the Bill and Melinda Gates Foundation Biomarker Grant no. OPP1066140 entitled, “Novel metabonomic biomarkers of gut function and health: Modeling enteropathy (EE) and field validation” (to R. L. G.), and by the following National Institutes of Health (NIH) grants: National Institute of Allergy and Infectious Diseases (NIAID) Centers for Excellence for Translational Research U19 AI109776 (to R.L.G.; Myron Levine University of Maryland, Principal Investigator), NIH Grant RO1 GM108501 (to J.A.P.), and NIH grant T32LM012416.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Guerrant RL, Oriá RB, Moore SR, Oriá MO, Lima AA. Malnutrition as an enteric infectious disease with long-term effects on child development. Nutr Rev 2008; 66:487–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guerrant DI, Moore SR, Lima AA, Patrick PD, Schorling JB, Guerrant RL. Association of early childhood diarrhea and cryptosporidiosis with impaired physical fitness and cognitive function four-seven years later in a poor urban community in northeast Brazil. Am J Trop Med Hyg 1999; 61:707–13. [DOI] [PubMed] [Google Scholar]
- 3. Guerrant RL, DeBoer MD, Moore SR, Scharf RJ, Lima AA. The impoverished gut–a triple burden of diarrhoea, stunting and chronic disease. Nat Rev Gastroenterol Hepatol 2013; 10:220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Niehaus MD, Moore SR, Patrick PD, et al. Early childhood diarrhea is associated with diminished cognitive function 4 to 7 years later in children in a northeast Brazilian shantytown. Am J Trop Med Hyg 2002; 66:590–3. [DOI] [PubMed] [Google Scholar]
- 5. Pinkerton R, Oriá RB, Lima AA, et al. Early childhood diarrhea predicts cognitive delays in later childhood independently of malnutrition. Am J Trop Med Hyg 2016; 95:1004–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Taniuchi M, Platts-Mills JA, Begum S, et al. Impact of enterovirus and other enteric pathogens on oral polio and rotavirus vaccine performance in Bangladeshi infants. Vaccine 2016; 34:3068–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Checkley W, Epstein LD, Gilman RH, Black RE, Cabrera L, Sterling CR. Effects of Cryptosporidium parvum infection in Peruvian children: growth faltering and subsequent catch-up growth. Am J Epidemiol 1998; 148:497–506. [DOI] [PubMed] [Google Scholar]
- 8. Checkley W, Gilman RH, Epstein LD, et al. Asymptomatic and symptomatic cryptosporidiosis: their acute effect on weight gain in Peruvian children. Am J Epidemiol 1997; 145:156–63. [DOI] [PubMed] [Google Scholar]
- 9. Mølbak K, Andersen M, Aaby P, et al. Cryptosporidium infection in infancy as a cause of malnutrition: a community study from Guinea-Bissau, West Africa. Am J Clin Nutr 1997; 65:149–52. [DOI] [PubMed] [Google Scholar]
- 10. Agnew DG, Lima AA, Newman RD, et al. Cryptosporidiosis in northeastern Brazilian children: association with increased diarrhea morbidity. J Infect Dis 1998; 177:754–60. [DOI] [PubMed] [Google Scholar]
- 11. Mondal D, Haque R, Sack RB, Kirkpatrick BD, Petri WA., Jr Attribution of malnutrition to cause-specific diarrheal illness: evidence from a prospective study of preschool children in Mirpur, Dhaka, Bangladesh. Am J Trop Med Hyg 2009; 80:824–6. [PMC free article] [PubMed] [Google Scholar]
- 12. Checkley W, White AC, Jr, Jaganath D, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect Dis 2015; 15:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Korpe PS, Haque R, Gilchrist C, et al. Natural history of cryptosporidiosis in a longitudinal study of slum-dwelling Bangladeshi children: association with severe malnutrition. PLOS Negl Trop Dis 2016; 10:e0004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Costa LB, JohnBull EA, Reeves JT, et al. Cryptosporidium-malnutrition interactions: mucosal disruption, cytokines, and TLR signaling in a weaned murine model. J Parasitol 2011; 97:1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coutinho BP, Oriá RB, Vieira CM, et al. Cryptosporidium infection causes undernutrition and, conversely, weanling undernutrition intensifies infection. J Parasitol 2008; 94:1225–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bartelt LA, Sevilleja JE, Barrett LJ, et al. High anti-Cryptosporidium parvum IgG seroprevalence in HIV-infected adults in Limpopo, South Africa. Am J Trop Med Hyg 2013; 89:531–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Costa LB, Noronha FJ, Roche JK, et al. Novel in vitro and in vivo models and potential new therapeutics to break the vicious cycle of Cryptosporidium infection and malnutrition. J Infect Dis 2012; 205:1464–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bartelt LA, Bolick DT, Kolling GL, et al. Cryptosporidium priming is more effective than vaccine for protection against cryptosporidiosis in a murine protein malnutrition model. PLOS Negl Trop Dis 2016; 10:e0004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ras R, Huynh K, Desoky E, Badawy A, Widmer G. Perturbation of the intestinal microbiota of mice infected with Cryptosporidium parvum. Int J Parasitol 2015; 45:567–73. [DOI] [PubMed] [Google Scholar]
- 20. Preidis GA, Ajami NJ, Wong MC, Bessard BC, Conner ME, Petrosino JF. Composition and function of the undernourished neonatal mouse intestinal microbiome. J Nutr Biochem 2015; 26:1050–7. [DOI] [PubMed] [Google Scholar]
- 21. Subramanian S, Huq S, Yatsunenko T, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014; 510:417–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smith MI, Yatsunenko T, Manary MJ, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 2013; 339:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mayneris-Perxachs J, Bolick DT, Leng J, et al. Protein- and zinc-deficient diets modulate the murine microbiome and metabolic phenotype. Am J Clin Nutr 2016; 104:1253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu J, Bolick DT, Kolling GL, Fu Z, Guerrant RL. Protein malnutrition impairs intestinal epithelial cell turnover, a potential mechanism of increased cryptosporidiosis in a murine model. Infect Immun 2016; 84:3542–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011; 472:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guerrant RL, DeBoer MD, Moore SR, Scharf RJ, Lima AA. The impoverished gut–a triple burden of diarrhoea, stunting and chronic disease. Nat Rev Gastroenterol Hepatol 2013; 10:220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. DeBoer MD, Chen D, Burt DR, et al. Early childhood diarrhea and cardiometabolic risk factors in adulthood: the Institute of Nutrition of Central America and Panama Nutritional Supplementation Longitudinal Study. Ann Epidemiol 2013; 23:314–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schroeder DG, Martorell R, Flores R. Infant and child growth and fatness and fat distribution in Guatemalan adults. Am J Epidemiol 1999; 149:177–85. [DOI] [PubMed] [Google Scholar]
- 29. Kirkpatrick BD, Daniels MM, Jean SS, et al. Cryptosporidiosis stimulates an inflammatory intestinal response in malnourished Haitian children. J Infect Dis 2002; 186:94–101. [DOI] [PubMed] [Google Scholar]
- 30. Kirkpatrick BD, Noel F, Rouzier PD, et al. Childhood cryptosporidiosis is associated with a persistent systemic inflammatory response. Clin Infect Dis 2006; 43:604–8. [DOI] [PubMed] [Google Scholar]
- 31. Alcantara CS, Yang CH, Steiner TS, et al. Interleukin-8, tumor necrosis factor-alpha, and lactoferrin in immunocompetent hosts with experimental and Brazilian children with acquired cryptosporidiosis. Am J Trop Med Hyg 2003; 68:325–8. [PubMed] [Google Scholar]
- 32. Harp JA, Chen W, Harmsen AG. Resistance of severe combined immunodeficient mice to infection with Cryptosporidium parvum: the importance of intestinal microflora. Infect Immun 1992; 60:3509–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harp JA, Wannemuehler MW, Woodmansee DB, Moon HW. Susceptibility of germfree or antibiotic-treated adult mice to Cryptosporidium parvum. Infect Immun 1988; 56:2006–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kominsky DJ, Campbell EL, Colgan SP. Metabolic shifts in immunity and inflammation. J Immunol 2010; 184:4062–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheng Z, Tsuda M, Kishita Y, Sato Y, Aigaki T. Impaired energy metabolism in a Drosophila model of mitochondrial aconitase deficiency. Biochem Biophys Res Commun 2013; 433:145–50. [DOI] [PubMed] [Google Scholar]
- 36. Tong WH, Rouault TA. Metabolic regulation of citrate and iron by aconitases: role of iron-sulfur cluster biogenesis. Biometals 2007; 20:549–64. [DOI] [PubMed] [Google Scholar]
- 37. Bhagat M, Sood S, Yadav A, et al. Alterations in oxidative stress parameters and its associated correlation with clinical disease on experimental Cryptosporidium parvum infection in Swiss albino mice. J Parasit Dis 2016. doi:10.1007/s12639-016-0871-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lawler PR, Akinkuolie AO, Chandler PD, et al. Circulating N-linked glycoprotein acetyls and longitudinal mortality risk. Circ Res 2016; 118:1106–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pickard JM, Chervonsky AV. Intestinal fucose as a mediator of host-microbe symbiosis. J Immunol 2015; 194:5588–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mitschler RR, Welti R, Upton SJ. A comparative study of lipid compositions of Cryptosporidium parvum (Apicomplexa) and Madin-Darby bovine kidney cells. J Eukaryot Microbiol 1994; 41:8–12. [DOI] [PubMed] [Google Scholar]
- 41. Wang Y, Holmes E, Nicholson JK, et al. Metabonomic investigations in mice infected with Schistosoma mansoni: an approach for biomarker identification. Proc Natl Acad Sci U S A 2004; 101:12676–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Garcia-Perez I, Earll ME, Angulo S, Barbas C, Legido-Quigley C. Chemometric analysis of urine fingerprints acquired by liquid chromatography-mass spectrometry and capillary electrophoresis: application to the Schistosomiasis mouse model. Electrophoresis 2010; 31:2349–55. [DOI] [PubMed] [Google Scholar]
- 43. Wang Y, Utzinger J, Xiao SH, et al. System level metabolic effects of a Schistosoma japonicum infection in the Syrian hamster. Mol Biochem Parasitol 2006; 146:1–9. [DOI] [PubMed] [Google Scholar]
- 44. Wu JF, Holmes E, Xue J, et al. Metabolic alterations in the hamster co-infected with Schistosoma japonicum and Necator americanus. Int J Parasitol 2010; 40:695–703. [DOI] [PubMed] [Google Scholar]
- 45. Saric J, Li JV, Wang Y, et al. Metabolic profiling of an Echinostoma caproni infection in the mouse for biomarker discovery. PLOS Negl Trop Dis 2008; 2:e254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saric J, Li JV, Wang Y, et al. Panorganismal metabolic response modeling of an experimental Echinostoma caproni infection in the mouse. J Proteome Res 2009; 8:3899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Y, Xiao SH, Xue J, Singer BH, Utzinger J, Holmes E. Systems metabolic effects of a Necator americanus infection in Syrian hamster. J Proteome Res 2009; 8:5442–50. [DOI] [PubMed] [Google Scholar]
- 48. Li JV, Wang Y, Saric J, et al. Global metabolic responses of NMRI mice to an experimental Plasmodium berghei infection. J Proteome Res 2008; 7:3948–56. [DOI] [PubMed] [Google Scholar]
- 49. Wang Y, Utzinger J, Saric J, et al. Global metabolic responses of mice to Trypanosoma brucei brucei infection. Proc Natl Acad Sci USA 2008; 105:6127–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Janus T, Borowiak KS, Pabisiak K, Machoy-Mokrzyńska A, Swiniarski A, Rozwadowski Z. H nuclear magnetic resonance spectroscopic investigation of urine for diagnostic and clinical assessment of methanol intoxication. Basic Clin Pharmacol Toxicol 2005; 97:257–60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.