Abstract
Motivation
Mapping bias causes preferential alignment to the reference allele, forming a major obstacle in allele-specific expression (ASE) analysis. The existing methods, such as simulation and SNP-aware alignment, are either inaccurate or relatively slow. To fast and accurately count allelic reads for ASE analysis, we developed a novel approach, ASElux, which utilizes the personal SNP information and counts allelic reads directly from unmapped RNA-sequence (RNA-seq) data. ASElux significantly reduces runtime by disregarding reads outside single nucleotide polymorphisms (SNPs) during the alignment.
Results
When compared to other tools on simulated and experimental data, ASElux achieves a higher accuracy on ASE estimation than non-SNP-aware aligners and requires a much shorter time than the benchmark SNP-aware aligner, GSNAP with just a slight loss in performance. ASElux can process 40 million read-pairs from an RNA-sequence (RNA-seq) sample and count allelic reads within 10 min, which is comparable to directly counting the allelic reads from alignments based on other tools. Furthermore, processing an RNA-seq sample using ASElux in conjunction with a general aligner, such as STAR, is more accurate and still ∼4× faster than STAR + WASP, and ∼33× faster than the lead SNP-aware aligner, GSNAP, making ASElux ideal for ASE analysis of large-scale transcriptomic studies. We applied ASElux to 273 lung RNA-seq samples from GTEx and identified a splice-QTL rs11078928 in lung which explains the mechanism underlying an asthma GWAS SNP rs11078927. Thus, our analysis demonstrated ASE as a highly powerful complementary tool to cis-expression quantitative trait locus (eQTL) analysis.
Availability and implementation
The software can be downloaded from https://github.com/abl0719/ASElux.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Allele specific expression (ASE) denotes the preferential allelic expression of a gene in the diploid genome. Integrating ASE with expression quantitative trait locus (eQTL) analysis improves fine-mapping accuracy and sensitivity (Kumasaka et al., 2015), thus helping identify biologically meaningful regulatory signals such as imprinting and cis regulation. Although several methods have been developed to identify ASE events from RNA-sequencing (RNA-seq) data (Castel et al., 2015; León-novelo et al., 2014; Liu et al., 2014; Li et al, 2012), mapping bias remains a major obstacle in ASE analysis (Degner et al., 2009; Panousis et al., 2014; Stevenson et al., 2013). Therefore, there is an important scientific knowledge gap that motivates the development of fast and accurate allelic expression analysis tools.
Previously, simulations have been used to identify variant sites showing bias towards one allele (Buil et al., 2015). However, simulations perform sub-optimally in practice since they are largely based on single-end reads whereas most RNA-seq data are now paired-end reads. There are also methods that utilize available genotype information and build personal allelic reference genomes for an allele-aware alignment that are implemented in programs such as SNP-o-matic (Manske and Kwiatkowski, 2009) and GSNAP (Wu and Nacu, 2010). In these approaches, the aligners are aware of single nucleotide polymorphisms (SNP) and align reads against both alleles. Even though the SNP-aware methods are more accurate than simulation-based approaches, they are more time consuming and computationally intensive, which makes them impractical for large RNA-seq datasets. A recently developed allele-specific analysis method (WASP) (van de Geijn et al., 2015) substitutes the SNP base with the alternative genotype in allelic reads and re-aligns those reads to correct for the reference bias. By excluding the allelic reads that are affected by different genotypes, WASP obtains extremely low false positive rate when identifying ASE SNPs. However, the process of generating reads with alternative genotypes in WASP takes a relatively long time (∼3.5 h) and many reads are excluded due to its stringent requirements.
To this end, we developed a new and more efficient approach, ASElux, which focuses on SNP-overlapping reads and combines the alignment and estimation of allelic expression into one step. Since accurate genotyping is essential for ASE analysis, the genotype information is usually obtained separately from the RNA-seq data using SNP array or genome/exome sequencing (Lonsdale et al., 2013). ASElux builds a personal allelic reference genome by using the individual’s existing genotype information to generate all possible ASE reads and pre-screen the RNA-seq data. This allows us to perform SNP-aware alignment and to efficiently identify only the reads that cover the unique set of SNPs present in each individual. Compared to all of the tested tools, ASElux is ultra-fast while achieving the closest allelic mapping accuracy to the benchmark SNP-aware aligner, GSNAP. Adding the time consumption to analyze an RNA-seq sample using a general-purpose aligner, such as STAR, the overall runtime of ASE analysis using ASElux is still ∼4 times faster than STAR (Dobin et al., 2013) followed by WASP (STAR + WASP), which re-aligns the reads with SNPs to decrease the reference bias (van de Geijn et al., 2015). We applied ASElux to 273 lung transcriptomes from the Genotype-Tissue Expression Project (GTEx) (Lonsdale et al., 2013) to demonstrate the increased power of ASE analysis in detecting local gene regulation. The high speed and accuracy of this novel ASE software makes it possible to analyze ASE in large datasets, helping efficient transformative interrogation of variants.
2 Materials and methods
2.1 Workflow of ASElux
Since only ∼10% of sequencing reads can be identified as SNP-overlapping, ASElux saves time by focusing on aligning reads that overlap with an individual’s SNPs obtained either from a genotype array, imputed SNPs based on a reference panel, or DNA sequencing. To implement this new alignment, we designed a hybrid index system that performs both genome-wide alignment and personal SNP-aware alignment (Supplementary Fig. S1A). The hybrid index system contains a static index that is built once for each reference genome. ASElux aggregates the genic regions in the reference genome to form a trimmed genome and uses a suffix array (Nong et al., 2009; Manber and Myers, 1990) as the static index for a fast alignment. The other part of our hybrid index system is the dynamic index. We extract the flanking sequence on both sides of the exonic SNP and store that in the dynamic index. The dynamic index is generated before alignment and it takes only ∼3 min to build it for each individual. Supplementary Figure S1B shows the workflow of aligning paired-end reads. For a pair of reads, we follow the workflow twice to treat each read first as the main read and then as the mate read. To accommodate sequencing errors, ASElux by default allows up to two mismatches elsewhere than at the SNP site. The user can set the number of allowed mismatches to fit various read lengths. We first use the dynamic index to identify the allelic reads during the alignment. Only the reads that match the dynamic index would be mapped to the genome with the static index to locate their multi-alignment loci. Then we try to align the other read, known as the mate read, near the identified multi-alignment loci. Thus, we only align the mate read if the main read matches the dynamic index. If both reads are uniquely aligned to one gene, we count the reads for the allele they originated from.
2.2 Filtering candidate SNPs
Since exonic reads can provide the best estimation of gene expression, ASElux disregards non-exonic SNPs and alignments for ASE analysis. Within ASElux, we provide a fast and useful tool to select exonic SNPs using genome annotation. A standard genome annotation contains overlapped exons and transcripts due to alternative splicing, and the overlapping information is redundant for pruning SNPs. To facilitate the pruning process, we merge all overlapping exons from different transcripts within the same gene into one. Small indels are another mechanism of allelic expression, but they tend to cause an alignment error leading to bias in ASE estimation (Heap et al., 2010; Stevenson et al., 2013). Thus, most ASE analyses focus on SNPs alone rather than the combination of SNPs and indels for the better accuracy (David et al., 2017). Therefore, we load the SNP and indel information from the vcf file and disregard all SNPs within one read length of an indel. The distance allowed between SNPs and indels varies according to the read length of the particular set of RNA-seq data. For example, if the read length is 50 bp, all SNPs within 50 bp of any indels in each individual would be disregarded from the further alignment. As shown in the 20 GTEx samples, only ∼0.9% of the SNPs were excluded by this process (Supplementary Table S1).
2.3 Hybrid index system
To perform a personalized SNP-aware alignment and maintain a high speed, we designed a hybrid index system that contains both static and dynamic indices. The static indices are built only once for each reference genome. Since only a small proportion of RNA-seq reads consist of intergenic reads (Mortazavi et al., 2008), ASElux uses the genic regions as the reference genome to achieve the least compromised balance between the alignment accuracy and speed and relatively low memory usage. We locate the start and the end of each gene so that the sequence between them covers all the components of a gene (exons, introns, UTRs etc.). Then we aggregate the sequences of all genes to form a trimmed genome. In the human genome (hg19), ASElux generates a new genome that contains ∼1.5 billion bp out of the ∼3 billion bp. For a genome that contains N genes, we construct N suffix arrays for the N genes and 1 more general suffix array for the trimmed genome as the static index using the sais algorithm (Nong et al., 2011). Although in theory searching globally in one suffix array is faster than using N suffix arrays for N genes, in practice combining local and global indices is faster due to the low-level memory management strategy in a modern computer (Kim et al., 2015). Briefly, the static index is built based on the trimmed reference genome and accordingly, the global alignment is not allele-specific. The suffix array indices of the trimmed genome and genes costs ∼30 GB of RAM (10 bytes for each base). We only use ∼15 GB with the trimmed genome, thus keeping our overall RAM usage at ∼ 20GB.
For each individual, we build a personalized dynamic index for SNP-aware alignment. We first prune the non-exonic SNPs to make sure that ASElux focuses on aligning only the expression-related reads. For each exonic SNP, we extract N-1 bp flanking sequence on both sides of the exonic SNP from the reference transcripts, where N is the read length, and replace the allele at the SNP location to generate reference sequences for all possible exonic reads that overlap with the SNP. To cover the SNPs adjacent to various splicing junctions, we extract the SNP flanking regions from all transcripts in each gene. Thus, each SNP has two 2N-1 bp long sequences for the reference and alternative alleles from each transcript. If the individual has additional known SNPs within the flanking sequence, we generate all possible haplotypes with alternative alleles of these adjacent SNPs to avoid misaligning reads with multiple variants. As there are regions with extremely high SNP density, ASElux only counts the first 10 heterozygous SNPs in each read. Noteworthy, as most indices are unique, we do not expect ambiguous indices to substantially bias the alignment of the ASE reads. To quickly locate SNP-overlapping reads, we aggregate all of the generated sequences as the dynamic index and build a suffix array for it. Then we save the generated sequences, SNPs and gene names for the dynamic index to query.
2.4 Alignment
Aligning only to the SNP-overlapping regions of the genome to identify the allelic reads is the key to the high speed of ASElux. For paired-end reads, we treat one read as the main read and the other as the mate read to help alignment. As shown in Supplementary Figure S1B and Algorithm 1 of Supplementary Methods, we check if the main read can be identified as an allelic read and use the mate read to properly align the whole read fragment. Only the ASE reads that are aligned to the dynamic index with up to two mismatches by default (not counting the SNP locus) will be aligned against the static index built on the trimmed genome (global alignment) to identify all of the multi-alignment loci. During the local alignment step, ASElux tries to locally align each main read’s mate read to the static index of the same gene. Thus, both the global alignment and the local alignment are against the static index. Since the read fragment should come from the same gene, we require the read mates to be aligned to the same gene. In the case where the major read is multi-aligned, we count the major read towards the ASE estimate only if both the main and mate reads are aligned to the same gene and at least one of them is uniquely aligned. Finally, we exchange the roles of the main and mate read and align the main read again to identify all possible alignments for the read pair. Thus, each read is treated once as the main read of the paired end reads.
2.4.1 Alignment against the dynamic index
Similarly to STAR, ASElux uses a binary search strategy to identify the Maximal Mappable Prefix (MMP) of a read in a suffix array index. The alignment of the main read starts from the left end of the read and identifies the longest common sequence with the dynamic index. Since the suffix array is built in the forward genome direction, we also align the reverse complement of the read to cover both directions. Algorithm 2 in the Supplementary Methods shows the process of aligning reads against the dynamic index. For the reads with mismatches, the alignment process stops at the mismatched locus and restarts at the base after the mismatched locus. Thus, several regions divided by the mismatched loci in the main reads are aligned to different loci in the dynamic index. For the regions aligned by no less than 20 bp, ASElux compares the whole read against the sequence around the mapped loci to check if the main read can be aligned to the locus with no more than 2 mismatches (using the ASElux default setting) while not counting indels (Supplementary Fig. S2). Since ASElux aligns the main read while being aware of the individual’s SNP loci, the mismatches are mainly caused by sequencing errors or unknown adjacent variants. Furthermore, we calculated that for a 100-bp read, allowing for up to 2 mismatches (using the default setting) covers 99.985% of the reads with the typical sequencing error rate of 0.1% per base expected for the Illumina platform (Schirmer et al., 2016). Although ASElux allows 2 mismatches for ASE reads by default, users can adjust the number of allowed mismatches to fit for the various read lengths.
2.4.2 Local alignment
Using the static index, ASElux aligns the mate read against the same gene region that the main read aligns to. Therefore, the reads without mismatches, indels, or splice junctions are perfectly mapped to the reference genome in this step. Supplementary Figure S3 shows an example of aligning a junction read. For reads that are not identical to the reference genome, the MMP is a substring of the read that stops before a variant or splice site. As shown in Algorithm 3 of the Supplementary Methods, we skip eight bases to avoid mapping indels or SNPs and search the MMP again for the unmapped part of the read. We chose to skip 8 bp in line with STAR because in practice most indels would safely be skipped with this set-up and it still allows us to utilize the remaining read for alignment. Separate MMPs of a read indicate that mismatches or splicing occurs between MMPs. We repeatedly search for the MMP until all parts of the read are mapped or we have searched more than the default of four times, indicating that the read should not be mapped to the reference due to too many mismatches or splicing loci. We selected the default of four times since it provided the best balance between the alignment accuracy and speed. After identifying all MMPs, we reassemble the read and only accept the read alignment if the read was properly reconstructed such that the MMPs are in the same order in the query read and the reference.
2.4.3 Global alignment
The global alignment is similar to the local alignment but extends to the trimmed genome in the static index. Hence, the MMP can originate from multiple local indices, indicating that the read is aligned to multiple genes. Since the lengths of the perfectly aligned prefixes in multi-aligned reads vary and searching for the MMP requires a perfect alignment, the multi-aligned reads will only be aligned to the locus that has the longest prefix shared with the reference genome. Thus, if we only align a read once to the trimmed genome, the multi-aligned loci that have the shorter perfectly aligned prefixes would be missed. Since it is crucial to find all possible multi-alignment targets for the ASE reads, we developed a masked binary search strategy to align the read to additional possible loci by masking off the known alignment results (Algorithm 4 of the Supplementary Methods). To globally fast align the ASE read, we utilize the fact that the information about the one perfectly mapped locus is available for the ASE reads. To find all possible genes where the read may be mapped to, we skip the locus that the read is already aligned to when searching for the MMP for the read. Since smaller MMPs have too many matches to the trimmed genome by chance alone, we only use MMPs longer than 20 bases and record the genes they reside in. Then we locally align the main read and the mate read in those genes to finish the alignment. ASElux can repeatedly align the reads with more and more masked genes. Therefore, the loci with smaller MMPs will not be missed due to the existence of the other loci with longer MMPs. In more detail, to find all MMPs within the read, we will start from the beginning of the read and search for the longest shared sequence between the particular read and the trimmed genome. We move along the read to find all MMPs longer than 20 bp in the read. Accordingly, there can be several MMPs which all must be longer than 21 bp. After the global alignment, we still locally align the mate read (Supplementary Fig. S1B), which ensures that a locus with only 20 bp match will not be identified as a properly aligned locus. As the next step, since the static index contains no SNP information, we align not only the ASE read but also the read that resides in the same locus with a different genotype. ASElux combines the alignment results of the two reads to make sure that we have the most comprehensive multi-alignment result.
The details of alignment with existing methods as well as the simulation data are described in the Supplementary Methods.
2.5 ASE and splice-QTL analyses in the GTEx project
We processed 273 RNA-seq samples from the GTEx project (Lonsdale et al., 2013) with ASElux. We downloaded the RNA-seq data and the imputed genotype data from the dbGaP accession phs000424.v6.p1. We randomly selected 20 samples for the comparisons in this study. The samples have on average 40 million 50-bp paired-end reads. Reads were aligned to the human genome (hg19) with the four tested aligners. We used the default alignment parameters of all the tested methods. The uniquely aligned reads were then kept for the subsequent analyses.
The results of the cis-eQTL analysis (version 6) were obtained from the GTEx portal (Ardlie et al., 2015). For each individual, we pruned out all SNPs aligned with less than 30 reads or less than 6 reads from one allele. To identify ASE SNPs across the population, we picked all SNPs that were heterozygous and passed the read count threshold in at least 30 individuals. We performed a paired t-test with the read counts of the reference allele and alternative allele from all individuals. The SNPs with Bonferroni corrected P-values less than 0.05 were identified as ASE SNPs. The GWAS SNPs (P ≤ 5 × 10−8, two-sided) were obtained from the NHGRI GWAS Catalog (Welter et al., 2014). We calculated the linkage disequilibrium (LD) between the ASE SNPs and GWAS SNPs within 1 Mb distance and obtained all of the SNPs in LD (R2 ≥ 0.8) with the ASE SNPs using PLINK (Purcell et al., 2007). Then we annotated the SNPs in LD with the ASE SNPs using ANNOVAR (Wang et al., 2010).
For the splice-QTL analysis, we aligned the 273 GTEx lung samples with STAR and identified all the splice events using LeafCutter. Following the analytical guideline of LeafCutter, we then used MatrixeQTL (Shabalin, 2012) to identify whether rs11078928 is a significant splice-QTL of GSDMB. For the isoform level eQTL analysis, we used RSEM (Li and Dewey, 2011) to estimate the isoform expression of the 273 GTEx lung samples and calculated the proportional transcript expression as the transcript expression level over the total gene expression as the phenotype in the eQTL analysis performed by MatrixeQTL (Shabalin, 2012).
3 Results
3.1 Test on simulated RNA-seq dataset
We first tested ASElux and other alignment methods on a simulated RNA-seq dataset with ∼180 M 2× 50 bp paired-end reads (see Supplementary Methods). Since comprehensively testing the alignment bias is important, we generated a high coverage simulated dataset. SNPs and junction reads were introduced to mimic real RNA-seq data. We added alternative alleles to the simulated reads based on imputed genotypes from a random GTEx sample and set both alleles to be equally expressed, which allowed us to accurately calculate the mapping bias of all methods. Besides ASElux, we also tested STAR 2.4.2a (Dobin et al., 2013), GSNAP 2015-6-23 (Wu and Nacu, 2010), HISAT2 2.0.4 (Kim et al., 2015) and WASP (van de Geijn et al., 2015) on the simulated dataset using the default parameters during the alignment (see Supplementary Methods). Since we focus on the alignment bias, we only tested the mapping function of WASP. We used the reference genome hg19 for all aligners and the GENCODE v19 annotation if the gene annotation could be supplied. To utilize the power of SNP-aware alignment, we used GSNAP to build a SNP-integrated alignment index for GSNAP. The HISAT2 alignment index was downloaded from its website along with the SNPs and transcript information. We used the default parameters for each aligner.
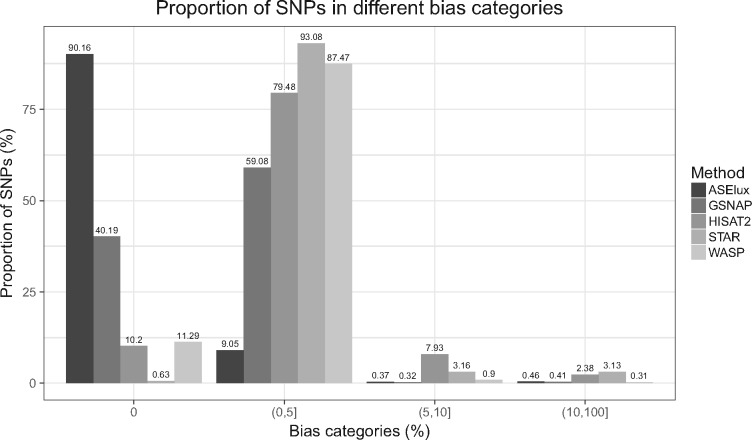
Using the genome-wide SNP data (genotyped and imputed) from GTEx (Lonsdale et al., 2013), we calculated read counts of each allele at exonic SNP sites to estimate ASE. The proportion of reference allele read counts when compared to the total read counts indicates the imbalance of allelic expression. Since the two alleles were equally expressed in the simulated dataset, the expected RACR of each SNP is 0.5. Accordingly, we measured the reference bias as the deviation of RACR from 0.5. Since each method aligns allelic reads differently, we performed the reference bias analysis using SNPs with enough aligned reads in all methods. ASElux, GSNAP, STAR and HISAT2 uniquely aligned ∼10 M allelic reads whereas WASP aligned ∼24% less reads than the other tested methods using the same simulated dataset (Supplementary Table S2). Figure 1 shows the proportion of SNPs in different bias categories. Although the majority of the SNPs displayed a bias less than 5% using all methods, ASElux achieved the highest accuracy by properly accounting for allele imbalance for ∼90% of the SNPs. Among the biased SNPs identified by each method, ASElux and the SNP-aware GSNAP showed substantially fewer SNPs with reference allele bias (∼70%) when compared to HISAT2 and STAR (∼99%). Even though STAR + WASP identified the fewest SNPs with a bias more than 5%, still the majority (88%) of the SNPs identified by WASP showed a bias in the range of more than 0% but less than 5% (Fig. 1). STAR alone performed worst since no SNP information was used during the alignment. Even though HISAT2 considers all common SNPs (MAF > 1%), it performs better than STAR but not as well as WASP, GSNAP and ASElux.
Fig. 1.
Proportions of SNPs in different bias categories show that ASElux performs better than general RNA-seq aligners. Y axis shows the proportion of SNPs, and X axis shows the different bias categories. The bias is the absolute difference between the predicted proportion of the reference allele reads and the real proportion of reference allele reads (0.5)
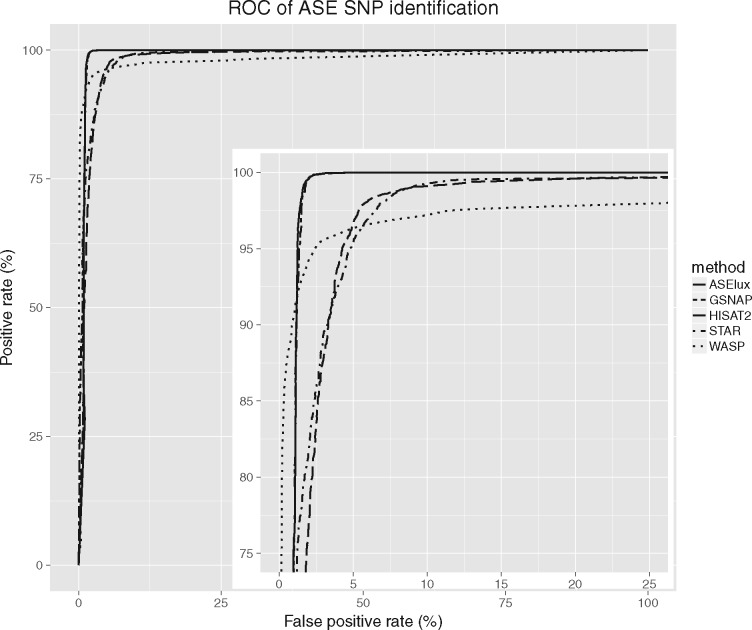
To test the ability of identifying ASE SNPs by ASElux and other methods, we generated another simulated dataset with 20% of genes exhibiting imbalanced allelic expression. These imbalanced genes were randomly selected and one random allele from the selected genes was overexpressed. Compared to the less expressed allele, we generated 1.5–3.5× more reads from the overexpressed allele. To mimic real RNA-seq data, we introduced sequencing error in addition to SNPs and junction reads. To ensure a 50× coverage for each allele, we overexpress one allele by generating more reads when simulating the imbalanced allelic expression. Using the binomial test, we identified a SNP as an ASE SNP if the Bonferroni corrected P-value is less than 0.05. To ensure that the tested methods are fairly compared, we used the intersection of the SNPs identified by all of the tested methods. The receiver operating characteristic (ROC) curve (Fig. 2) indicates that ASElux outperforms HISAT2 and STAR alone on identifying ASE SNPs. Since GSNAP and ASElux both utilize personal SNP information, they identified all of the ASE SNPs (true positive rate = 100%) while maintaining a low false positive rate of ∼5%. Although ASElux performed better than GSNAP in the first simulation test (Fig. 1), ASElux and GSNAP showed a comparable number of SNPs showing more than 5% bias. Thus, GSNAP and ASElux performed similarly based on the ROC curve (Fig. 2). The false positive rate of WASP is the lowest among all the tested methods while the true positive is below 92%. In the first simulation test under the null condition (Fig. 1), WASP showed the smallest number of SNPs that have bias more than 5%, suggesting that WASP tends to be highly conservative in order to achieve a low false positive rate. However, since WASP filters out potentially falsely aligned reads by STAR, some SNPs might have insufficient coverage to pass a stringent threshold, which may contribute to the low positive rate of WASP.
Fig. 2.
The receiver operating characteristic (ROC) of ASE SNP identification shows that ASElux performs as well as GSNAP, and outperforms HISAT2 and STAR in a simulated dataset. The X axis is the false positive rate and the Y axis is the positive rate
3.2 Speed benchmarks
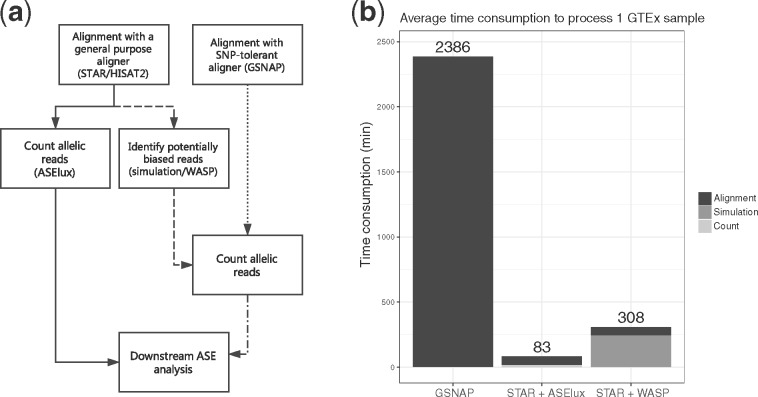
We performed the speed benchmark on a server with 64-bit Intel CPUs @2.66 GHz with ∼95GB RAM. Figure 3A shows the common workflow of ASE analysis using different methods. Researchers can first map reads using a RNA-seq aligner (STAR/HISAT2) and then count allelic reads with specialized tools, such as ASElux or WASP, or alternatively use a SNP-aware aligner (GSNAP) and count allelic reads directly based on the alignment. Figure 3B shows the average time consumption of a single thread to perform ASE analysis on 10 samples from the GTEx project using STAR + ASElux, STAR + WASP and GSNAP, respectively. Among the tested methods, GSNAP used ∼12 GB RAM, HISAT2 ∼8 GB RAM and ASElux ∼22 GB of RAM, respectively. WASP itself requires no more than 1GB of RAM but the actual RAM requirement of WASP depends on the alignment tool it uses, e.g. STAR would need additional ∼30 GB RAM. Counting allelic reads with ASElux is, however, ultra-fast since it only takes ∼20 min to process a GTEx RNA-seq sample. Therefore, STAR + ASElux can has a ∼33× faster processing speed than GSNAP. WASP requires ∼4 CPU hours for each GTEx sample, which makes STAR + WASP ∼4× slower than STAR + ASElux. The tests shown in Figure 3 were based on single thread mode. ASElux, HISAT2, STAR and GSNAP all have a multi-thread mode, however, WASP does not support multi-thread computing. Thus, we also tested the multithread mode of each tool except for WASP (Supplementary Fig. S4), which resulted in similar relative alignment speeds across the tools as in the single thread mode. As I/O often plays a significant factor in runtime, the system cache was cleared before each alignment run to avoid any bias due to pre-loaded reference index in the memory during the benchmarking. On average, index loading contributes up to 25% of the overall runtime of ASElux without caching.
Fig. 3.
ASElux is faster than the other tested methods (WASP and GSNAP). (a) The workflow of ASE analysis using different programs. (b) The estimated average time consumptions to count allelic reads from 10 GTEx RNA-seq samples. The X axis shows the tested method. The Y axis is the time needed for processing the dataset
3.3 Comparing GSNAP, ASElux, HISAT2 and STAR on 20 experimental samples
To evaluate the performance of ASElux, GSNAP, STAR, HISAT2 and WASP on real RNA-seq data, we processed 20 lung RNA-seq samples from the GTEx (Lonsdale et al., 2013) cohort with the five methods. Each sample contains ∼40 million pairs of 76 bp reads. The genotype data of each sample consists of the imputed and genome-wide SNP array data from the GTEx study. For each sample, ∼120 000 exonic SNPs were obtained from the VCF file. We built a personalized index for each sample for ASElux (see methods) and GSNAP, and provided the same indices as in the simulated analysis to STAR and HISAT2. After alignment, we extracted the allelic read counts on each heterozygous SNP for further analyses.
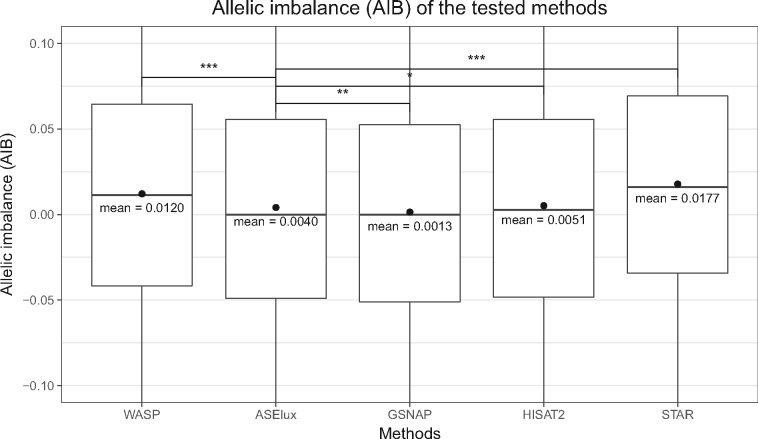
The level of imbalanced allelic expression represented by the RACR provides more information on ASE than the statistics by the binomial test. In an ASE analysis, the proportion of reference allelic reads closer to 0 or 1 often indicates stronger allele-specific gene expression. Therefore, we compared the allelic imbalance (AIB), which is the difference between 0.5 and RACR, derived by ASElux to AIBs by the other methods. Under the null hypothesis that most SNPs will not have an ASE effect, we expect equal expression from both the reference and alternative haplotypes. Consequently, the theoretical distribution of AIB should be centered at zero with a few outliers towards the two tails. If the reference bias hampers the alignment, the mean and median of AIB of all SNPs would shift up from 0, which is shown in Figure 4. GSNAP shows a minimal reference bias in the test. Although ASElux shows a higher average AIB when compared to GSNAP (Fig. 4), its average AIB is significantly lower than the AIBs obtained using WASP, HISAT2 and STAR. WASP, HISAT2 and STAR aligned significantly more reads to the reference allele, indicating a higher reference bias. Although WASP showed the lowest false positive rate in the simulation test, the majority of the WASP SNPs still had a bias more than 0% and less than 5%, which is similar to HISAT2 (Fig. 1). The AIBs derived from the 20 GTEx samples confirmed this similarity between WASP and HISAT2.
Fig. 4.
For ASE analysis, ASElux has less reference bias than WASP, and the general aligners, STAR and HISAT, when testing the 20 real RNA-seq samples. * indicates a P-value of 1.05 × 10−3 (two-sided); ** indicates a P-value of 7.44 × 10−14 (two-sided); and *** indicates a P-value <2.2 × 10−16 (two-sided). Y axis displays the allele frequency differences between the tested methods and 0.5 (i.e. the two alleles are expressed equally). The reference bias of ASElux is significantly smaller than that of HISAT2, STAR and WASP, but higher than GSNAP. Y axis was limited from -0.1 to 0.1 to show the distribution of most SNPs. The red dots indicate the mean values of each method
ASElux uniquely aligned ∼1.3 M allelic reads for each sample; whereas WASP aligned ∼1.5 M allelic reads; GSNAP and HISAT2∼1.7 M allelic reads; and STAR ∼2.8 M allelic reads for each sample, respectively (Supplementary Fig. S5a, Table S2). ASElux identified ∼15% fewer SNPs than GSNAP but ∼37% more than WASP (Supplementary Fig. S5b). It is worth noting, however, that not all SNPs identified by STAR and HISAT2 are suitable for downstream ASE analysis. Previous studies show that more than 10% of the heterozygous SNPs would be excluded when employing a simulation procedure to correct for the reference alignment bias, (Kukurba et al., 2014; Panousis et al., 2014) while using a general purpose aligner. Thus, overall ASElux would identify a similar number of heterozygous SNPs that are suitable for the downstream ASE analysis when compared to STAR, and HISAT2.
Although STAR uniquely aligned more reads than the other tested tools, it identified a similar number of SNPs with a coverage of ≥30 reads when compared to HISAT2 and GSNAP (Supplementary Table S3, Fig. S4b). The extra allelic reads aligned by STAR mainly overlap with the low coverage SNPs that do not contribute to the ASE analysis (Supplementary Fig. S6). Since WASP depends on STAR for the alignment, a large amount of reads in WASP also overlap with the low coverage SNPs (Supplementary Fig. S6). Thus, WASP identified less SNPs than the other tested tools with similar number of reads aligned (Supplementary Fig. S5).
3.4 ASE analysis strengthens cis-eQTL analysis in identifying local regulation of gene expression
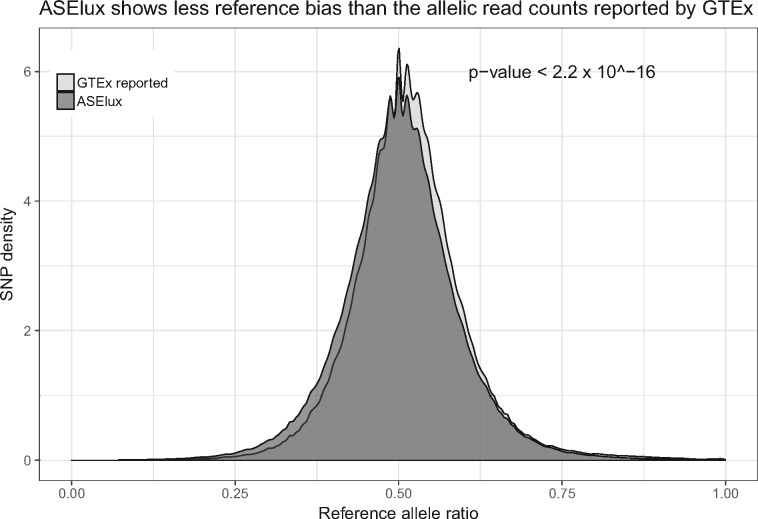
Utilizing the ultra-fast speed of ASElux, we applied ASElux to a dataset of 273 lung RNA-seq samples and imputed SNP array data from the GTEx study. Figure 5 shows that ASElux has significantly less reference bias when compared to the allelic read counts reported by GTEx using their ASE analysis pipeline (Ardlie et al., 2015) (P-value< 2.2 × 10−16, two-sided t-test). The distribution of RACR from ASElux is centered at 0.5 whereas the distribution reported by GTEx displays an upward bias. To verify whether ASElux has identified enough heterozygous SNPs for the ASE analysis, we compared the number of SNPs identified by ASElux and the GTEx study. In both analysis by ASElux and the GTEx study a heterozygous SNP must be covered by ≥30 reads to be counted for the downstream ASE analysis. A median of 6385 SNPs passed the simulation correction in the GTEx study (Panousis et al., 2014) for each sample which is ∼20% less than the median SNP number identified by ASElux in the 273 GTEx lung samples, indicating that ASElux can identify more ASE SNPs than the GTEx ASE protocol.
Fig. 5.
Compared to the allelic read counts reported by GTEx, ASElux shows less reference bias in 273 lung samples (P-value < 2.2 × 10−16, two-sided). The X axis shows the reference allele ratio, and the y axis shows the density of all SNPs
In addition to ASE analysis, a cis-eQTL analysis is also widely used to detect allele-specific regulation of gene expression. We compared the ASE results from ASElux to the cis-eQTL results on the same set of SNPs publicly available at the GTEx website. Among the 273 lung samples we aligned with ASElux, we identified 21 550 heterozygous exonic SNPs covered by at least 30 reads in no less than 30 samples. Using a paired t-test, we identified 2765 SNPs residing in 1790 genes that showed ASE (P < 2.32 × 10-6, two-sided). Although not all ASE events are caused by exonic SNPs, the paired t-test of the exonic SNPs should identify either the causal exonic ASE SNPs or the exonic SNPs tagged by non-coding causal variants. Next, we investigated whether these 2765 ASE SNPs were also identified as cis-eQTLs by GTEx. Using the gene-specific permutation threshold used by GTEx (Ardlie et al., 2015), 1421 of the ASE SNPs were significant cis-eQTLs in lung for the genes they are located in. Overall, 1344 (48.61%) of the ASE SNPs were missed by the cis-eQTL analysis, and 965 (53.91%) of the ASE genes were not identified as cis-eQTL genes. Accordingly, the combination of ASE and cis-eQTL analysis increased the power to identify variants associated with local regulation of gene expression when compared to cis-eQTL analysis alone.
To further investigate the association between ASE and lung disorders, we calculated the linkage disequilibrium (LD) between ASE SNPs and 67 GWAS SNPs (Ardlie et al., 2015) of lung disorders, such as smoking; asthma; lung cancer; chronic obstructive pulmonary disease (COPD); and pulmonary hypertension. There are 11 ASE SNPs in strong LD (r2 > 0.8) with the GWAS SNPs (Supplementary Table S4). Of the 11 ASE SNPs, 10 are identified as cis-eQTLs of the genes in which they reside. Both the cis-eQTL and ASE analysis indicate that the alternative genotype of the 10 ASE SNPs is associated with a lower gene expression. It is worth noting, however, that the ASE SNP rs2305480 located in Gasdermin B (GSDMB) is in LD (R2 = 1) with a GWAS SNP rs11078927 which is associated with the increased risk of asthma (Bouzigon et al., 2008; Bønnelykke et al., 2014). This SNP rs11078927 has never been identified as a significant cis-eQTL of GSDMB in the lung tissue before. Moreover, rs2305480 has also been identified as a GWAS SNP of another inflammatory disorder, ulcerative colitis (McGovern et al., 2010), supporting the role of the GSDMB gene in several disorders with a known inflammatory component.
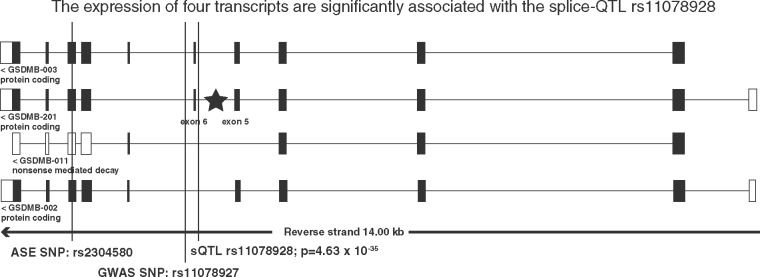
We further investigated the potential mechanism of the GWAS SNP rs11078927 and discovered rs11078928, which is a splice donor site variant previously identified in the whole blood and suggested to be involved in asthma (Morrison et al., 2013). It is in tight LD (R2 = 0.99) with two asthma GWAS hits, rs2305480 (the ASE SNP) and rs11078927. To examine the splicing effect in a human tissue highly relevant for asthma, we performed a splice-QTL analysis in 273 GTEx lung RNA-seq samples using LeafCutter and identified rs11078928 as a significant splice-QTL of GSDMB in the lung (Fig. 6). The genotype of rs11078928 is significantly associated (P-value = 4.63 × 10−35, two-sided) with the proportional expression level of the junction reads overlapping exon 5 and exon 6 of GSDMB, which is consistent with the splicing event identified previously in the whole blood (Morrison et al., 2013).
Fig. 6.
Using the proportional transcript expression as the phenotype, four transcripts GSDMB-003, GSDMB-201, GSDMB-011 and GSDMB-002 are significantly associated with the genotype of the splice-QTL SNP, rs11078928 (P-value < 9.43 × 10−4, two-sided). The stars indicate that genotypes of rs11078928 are significantly associated with the splice junction reads between the exon 5 and exon 6 of GSDMB (P-value = 4.63 × 10−35, two-sided)
To determine which isoform expression of GSDMB is impacted by the splice variant, we used RSEM (Li and Dewey, 2011) to estimate the expression of isoforms in 273 GTEx lung samples and used the proportional transcript expression as the phenotype for an isoform eQTL analysis. The relative expression of four transcripts GSDMB-003, GSDMB-201, GSDMB-011 and GSDMB-002 are significantly associated with the genotype of the splice-QTL rs11078928 [P-value < 9.43 × 10−4 using linear regression via MatrixeQTL (Shabalin, 2012) with Bonferroni correction] (Fig. 6, Supplementary Table S5). Thus, the biological mechanism underlying the asthma risk SNPs, rs2305480 and rs11078927, is likely mediated by the SNP rs11078928 via splicing regulation on GSDMB in the human lungs.
We further functionally annotated the 52 460 SNPs (R2 > 0.8) tagged by the ASE SNPs, identified by ASElux, using ANNOVAR (Wang et al., 2010) (Supplementary Fig. S7). There are 19 additional SNPs identified as splice variants by ANNOVAR and 7 of them were missed by the GTEx cis-eQTL analysis. Taken together, an ASE analysis provides substantially more power for analysis of local gene expression, complementing the regular cis-eQTL analysis.
4 Discussion
With growing interest in ASE analysis, mapping bias remains a critical barrier that hinders the accuracy of ASE analysis in RNA-seq. We provide a novel approach, ASElux that focuses solely on SNP-overlapping reads, allowing a fast and accurate SNP-aware alignment for ASE analysis. To ensure a high alignment accuracy, we used the whole gene body (50% of the reference genome) to build the alignment index. It is worth noting that this speed gain is largely due to the fact that ASElux first aligns all reads to the very small dynamic index to identify the allelic reads and then only aligns them to the large static index. The size of the static index will not affect the speed substantially because the time complexity of searching through suffix array is O(mlog(n)), where the n is the size of the reference and m is the size of the pattern. In addition, ASElux shows a minimal reference bias when compared with other methods based on both simulated and experimental RNA-seq data. ASElux aligns against both alleles by employing personal dynamic indices to minimize the reference bias. We demonstrated that ASElux works optimally with short reads currently generated by most RNA-seq studies.
Due to the complexity of RNA-seq alignment and variable expression of genes across tissues, SNP-calling from RNA-seq is often less accurate than from DNA-sequencing data (Quinn et al., 2013). Thus, external genotype information from whole exome sequencing (WES), whole genome sequencing (WGS), or SNP-arrays are preferred for ASE or eQTL analysis (Ardlie et al., 2015). ASElux and all of the tools tested here do not directly identify SNPs from RNA-seq reads and are therefore only applicable to RNA-seq cohorts that have genotype data available. Simultaneously calling SNPs and ASE from RNA-seq data will enable ASE analyses in additional RNA-seq cohorts, but it will require development of new methods in the future.
Multi-alignment also presents a serious challenge in ASE analysis. Reads generated from different regions might be falsely identified as ASE reads due to their similar sequences. ASElux tries to find all possible multi-alignment loci in addition to the optimal alignment even if the read has the best alignment quality as an ASE read to stringently remove possible false ASE reads. As ambiguously aligned reads are more stringently excluded, ASElux tends to align less allelic reads than the other tested tools. However, not all SNPs are reliable for the ASE analysis due to the reference alignment bias when using a general-purpose aligner such as STAR and HISAT2, and in fact, the previous studies show a ∼10% loss in the number of SNPs during the simulation correction (Kukurba et al., 2014; Panousis et al., 2014). We have shown here that the high accuracy of ASElux has provided more reliable SNPs for the downstream ASE analysis than STAR did in the analyzed GTEx lung samples.
As an alignment tool exclusively designed for ASE analysis, ASElux outperforms most existing methods in speed and provides a better accuracy than the existing non-SNP-aware aligners for correcting the reference bias in alignment while also achieving the closest accuracy to GSNAP. ASElux is ultra-fast: it is able to process 40 million 2× 50 bp reads in 16 min. Combined with a general purpose aligner, such as STAR, STAR + ASElux is ∼33 times faster than the golden standard SNP-aware aligner GSNAP, and ∼4 times faster than the popular combination of STAR + WASP. The high speed and accuracy make ASElux an ideal tool to perform ASE analysis in large-scale RNA-seq studies. We demonstrated the usefulness of ASElux by performing the ASE analysis in lung RNA-seq data from 273 individuals of the GTEx project in two days (∼70 CPU hours using multi-CPU). By comparing the ASE SNPs and eQTLs from the same dataset, we also demonstrated that the combination of ASE and cis-eQTL analysis provides more power to detect local regulation of gene expression.
Supplementary Material
Acknowledgements
The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH and NINDS. The data used for the analyses described in this manuscript were obtained from: the GTEx Portal on 08/14/2016 and dbGaP accession number phs000424.v6.p1 on 08/11/2016.
Funding
This work was supported by the National Institutes of Health (NIH) [grant numbers HL-095056, HL-28481]. A.K. was supported by the NIH [grant number F31HL127921] and M.A. was supported by the NIH [grant number T32HG002536].
Conflict of Interest: none declared.
References
- Ardlie K.G. et al. (2015) The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science, 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bønnelykke K. et al. (2014) A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat. Genet., 46, 51–55. [DOI] [PubMed] [Google Scholar]
- Bouzigon E. et al. (2008) Effect of 17q21 variants and smoking exposure in early-onset asthma. N. Engl. J. Med., 359, 1985–1994. [DOI] [PubMed] [Google Scholar]
- Buil A. et al. (2015) Gene-gene and gene-environment interactions detected by transcriptome sequence analysis in twins. Nat. Genet., 47, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel S.E. et al. (2015) Tools and best practices for allelic expression analysis. Genome Biol., 16, 195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David A.K. et al. (2017) Allele-specific expression reveals interactions between genetic variation and environment. Nat. Methods, 14, 699–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner J.F. et al. (2009) Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics, 25, 3207–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A. et al. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap G.A. et al. (2010) Genome-wide analysis of allelic expression imbalance in human primary cells by high-throughput transcriptome resequencing. Hum. Mol. Genet., 19, 122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. et al. (2015) HISAT: a fast spliced aligner with low memory requirements. Nat. Methods, 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukurba K. et al. (2014) Allelic expression of deleterious protein-coding variants across human tissues. PLoS Genet., 10, e1004304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumasaka N. et al. (2015) Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nat. Genet., 48, 206–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- León-Novelo L.G. et al. (2014) A flexible Bayesian method for detecting allelic imbalance in RNA-seq data. BMC Genomics, 15, 920.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12, 323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. et al. (2012) Identification of allele-specific alternative mRNA processing via transcriptome sequencing. Nucleic Acids Res., 40, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. et al. (2014) Comparing computational methods for identification of allele-specific expression based on next generation sequencing data. Genet. Epidemiol., 38, 591–598. [DOI] [PubMed] [Google Scholar]
- Lonsdale J. et al. (2013) The Genotype-Tissue Expression (GTEx) project. Nat. Genet., 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manber U., Myers G. (1990) Suffix string arrays: a new searches method for on-line. Proc. first Annu. ACM-SIAM Symp. Discret. Algorithms, 319–327. [Google Scholar]
- Manske H.M., Kwiatkowski D.P. (2009) SNP-o-matic. Bioinformatics, 25, 2434–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern D. et al. (2010) Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat. Genet., 42, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison F.S. et al. (2013) The splice site variant rs11078928 may be associated with a genotype-dependent alteration in expression of GSDMB transcripts. BMC Genomics, 14, 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A. et al. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods, 5, 621–628. [DOI] [PubMed] [Google Scholar]
- Nong G. et al. (2009) Linear suffix array construction by almost pure induced-sorting. In: 2009 Data Compression Conference, pp. 193–202.
- Nong G. et al. (2011) Two efficient algorithms for linear time suffix array construction. IEEE Trans. Comput., 60, 1471–1484. [Google Scholar]
- Panousis N.I. et al. (2014) Allelic mapping bias in RNA-sequencing is not a major confounder in eQTL studies. 15, 467.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn E.M. et al. (2013) Development of strategies for SNP detection in RNA-Seq data: application to lymphoblastoid cell lines and evaluation using 1000 genomes data. PLoS One, 8, e58815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M. et al. (2016) Illumina error profiles: resolving fine-scale variation in metagenomic sequencing data. BMC Bioinformatics, 17, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin A.A. (2012) Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics, 28, 1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson K.R. et al. (2013) Sources of bias in measures of allele-specific expression derived from RNA-sequence data aligned to a single reference genome. BMC Genomics, 14, 536.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Geijn B. et al. (2015) WASP: allele-specific software for robust discovery of molecular quantitative trait loci. Nat. Methods, 12, 1061–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. et al. (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res., 38, e164.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D. et al. (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res., 42, 1001–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.D., Nacu S. (2010) Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics, 26, 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.