Abstract
Mutations in bestrophin-1 (BEST1) are associated with distinct retinopathies, notably three forms with autosomal dominant inheritance and one condition with an autosomal recessive mode of transmission. The molecular mechanisms underlying their distinct retinal phenotypes are mostly unknown. Although heterozygous missense mutations in BEST1 reveal dominant-negative effects in patients with autosomal dominant Best disease (BD), heterozygous mutations associated with autosomal recessive bestrophinopathy (ARB) display no disease phenotype. Here we show that the recessive mutations trigger a strong and fast protein degradation process in the endoplasmic reticulum (ER), thereby favoring a decreased stoichiometry of mutant versus normal BEST1 subunits in the assembly of the homo-pentameric BEST1 chloride channel. In contrast, dominant mutations escape ER-associated degradation and are subjected to a slightly delayed post-ER degradation via the endo-lysosomal degradation pathway. As a result, increased formation of a non-functional BEST1 channel occurs due to a roughly equimolar incorporation of normal and mutant BEST1 subunits into the channel complex. Taken together, our data provide insight into the molecular pathways of dominantly and recessively acting BEST1 missense mutations suggesting that the site of subcellular protein quality control as well as the rate and degree of mutant protein degradation are ultimately responsible for the distinct retinal disease phenotypes in BD and ARB.
Introduction
In human, bestrophin-1 (BEST1) is highly expressed in the retinal pigment epithelium (RPE) (1,2), where it localizes to the basolateral aspect (3) by forming a homo-pentameric (4,5), calcium-activated (6–8) and volume-regulated (9) chloride channel. Mutations in the BEST1 gene are associated with distinct retinopathies, including the autosomal dominant forms of Best vitelliforme macular dystrophy or Best disease (BD) (MIM 153700) (1), adult-onset vitelliforme macular dystrophy (AVMD) (MIM 608161) (10) and the vitreoretinochoroidopathy (ADVIRC) (MIM 193220) (11). In addition, there is an autosomal recessive bestrophinopathy (ARB) (MIM 611809) with heterozygous BEST1 mutation carriers free of retinal manifestations (12). So far, more than 250 independent pathologic BEST1 mutations have been deposited in the Human Gene Mutation Database (http://www-huge.uni-regensburg.de/BEST1_database/home.php; date last accessed December 2017), the vast majority of these mutations affecting the evolutionarily highly conserved N-terminal part of the protein. Of the known mutations, 90% are of the missense type, although with no apparent correlation between BEST1 genotypes and clinical phenotypes.
BD is the most common pathology of the bestrophinopathies with an estimated prevalence of ∼1:50 000 (13). It affects primarily the macular area of the posterior pole of the retina and is initially characterized by prominent deposits of lipofuscin-like material beneath the neurosensory retina and an abnormal Arden ratio (light peak/dark trough ratio) in the electrooculogram highly suggestive of an impaired RPE as the primary site of pathology (14). Later, disintegration of the yellowish lesions progressively leads to atrophy of the RPE/photoreceptor complex and consequently to vision loss although disease expression in BD varies widely (15).
An autosomal recessive mode of inheritance of BEST1 mutations is estimated at a prevalence of <1:1 000 000. Affected individuals usually are compound heterozygous (12) or less often homozygous (16,17) carriers of pathogenic BEST1 mutations while heterozygous parents generally show no retinal symptoms. Unlike BD, ARB is not associated with the classic macular ‘egg-yolk’ lesion; rather, the main characteristics of ARB are multifocal subretinal deposits, abnormal autofluorescence and subretinal fluid accumulation or macular edema (12,18).
So far, the molecular mechanisms underlying the individual manifestations of the BEST1-associated pathologies have not been well defined. We and others have shown that protein mislocalization and thus loss of chloride channel function is a consequence not only for several BD- but also for some ARB-associated mutations (7,9,19–23). These findings suggest that simply a failure to traffic to the plasma membrane (PM) is not sufficient to explain the distinct pathologies of the two disease entities. There is also evidence that regardless of their clinical expression mutant BEST1 protein still has the capacity to oligomerize and thus form a homo-pentameric BEST1 channel (24). Again, this provides no explanation as to why the various missense mutations result in distinct clinical phenotypes.
Recently, we demonstrated that BD-associated mutations exert a dominant-negative effect (9), which appears intuitive by assuming incorporation of normal and mutant BEST1 subunits into the homo-pentameric structure of the mature chloride channel (4,5). For the autosomal recessive BEST1 mutations, loss of function of both BEST1 alleles seems likely. Although ARB-associated nonsense and frameshift mutations predict a truncated and thus likely inactive protein, the mechanisms by which missense mutations result in protein dysfunction is less straightforward. Uggenti et al. (25) suggested that ARB-associated BEST1 protein may be less stable due to degradation via the ubiquitin-proteasome pathway. Consequently, they demonstrated rescue of mutant BEST1 by chemical chaperons as was similarly shown for other endoplasmic reticulum (ER)-retained mutant proteins (26,27).
Taken together, a number of preliminary findings suggest that specifics in the degradation process of mutant BEST1 could play a crucial role in phenotypic expression of the defect. We therefore aimed to clarify the cellular/molecular mechanisms underlying the distinct pathologies of BEST1-driven BD and ARB phenotypes. Understanding these differences may be relevant when searching for appropriate treatment options in these sight-threatening retinal disease entities.
Results
Algorithm-based prediction of protein stability
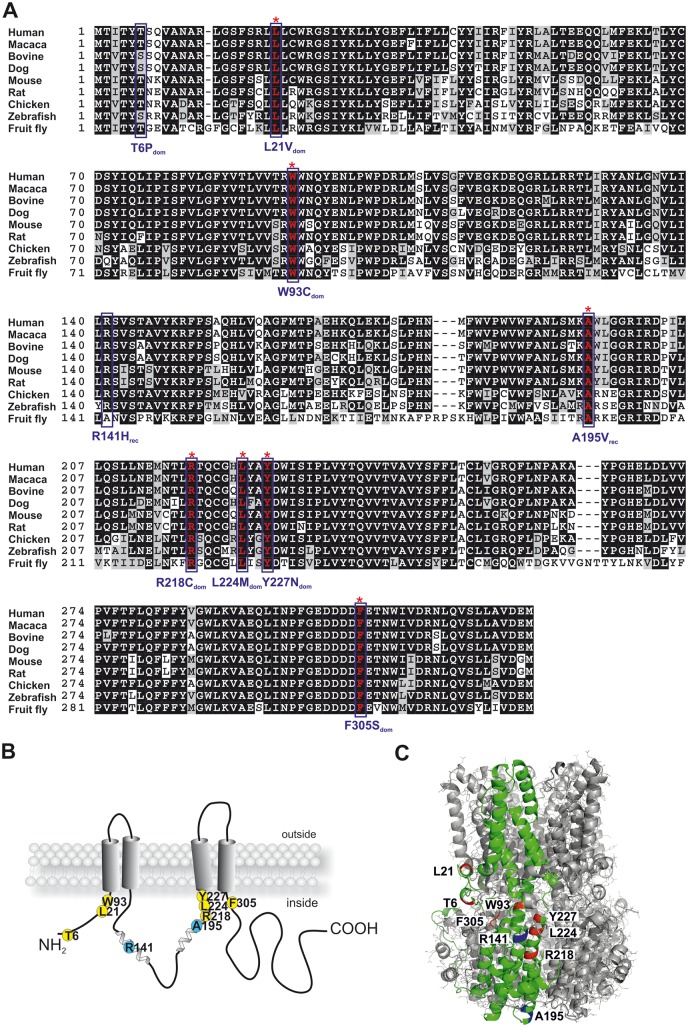
To predict effects of pathologic BEST1 missense mutations on thermodynamic protein stability, we selected BD- (termed T6Pdom, L21Vdom, W93Cdom, R218Cdom, L224Mdom, Y227Ndom and F305Sdom) and ARB- (termed R141Hrec and A195Vrec) associated amino acid changes, all affecting the highly conserved N-terminal half of the protein (2) (Fig. 1A–C). Conservation analysis was performed with nine homologous bestrophin sequences across vertebrate and insect species. We found 100% sequence identity each for residue L21, W93, A195, R218, L224, Y227 and F305, while R141 revealed 90% and T6 70% amino acid identity (Fig. 1A). The high conservation values suggest an essential contribution of these amino acids to function or structural BEST1 stability. For computational stability analysis we relied on the recently solved 3D protein structure of chicken BEST1 (cBESTcryst) (4), which was implemented into the structure-based prediction tools mutation cutoff scanning matrix (mCSM), site-directed mutator (SDM), DUET and the three-state predictor I-mutant 2.0. Accordingly, the analyzed pathologic mutations were all classified as destabilizing noticeable by a large decline in Gibbs free energy, as indicated by a negative ΔΔG value (Table 1).
Figure 1.
Localization of selected BD- and ARB-associated mutations in the BEST1 gene. (A) Multiple sequence alignment of the first N-terminal 317 amino acid residues of BEST1 from human (NP_004174), macaca (BAE02471), bovine (XP_585778), dog (XP_540912), mouse (NP_036043), rat (XP_574621), chicken (XP_421055), zebrafish (XP_689098) and fruit fly (NP_652603). Identical residues are given on black background and conservative amino acid substitutions are shaded. BEST1 mutations analyzed in this study are denoted by a blue box. Red asterisks above the aligned sequences indicate 100% conservation of amino acid residues in the species analyzed. (B) Topology model of human BEST1 as given in reference (57) and localization of codons for BD- and ARB-associated mutations are shown in yellow and blue, respectively. (C) Crystal structure model of the homo-pentameric chicken BEST1 chloride channel. Homologous positions of the BD- and ARB-associated residues are indicated in red and blue, respectively, in the superimposed monomer (green).
Table 1.
Prediction of protein stability changes due to single amino acid substitutions in BEST1
| AA change | Inheritance | I-Mutant2.0 | mCSM | SDM | DUET | Prediction |
|---|---|---|---|---|---|---|
| T6P | AD | –1.58 | –0.58 | –0.19 | –0.76 | Destabilizing |
| L21V | AD | –0.75 | –0.86 | –1.06 | –0.88 | Destabilizing |
| W93C | AD | –2.44 | –1.71 | –1.44 | –1.56 | Destabilizing |
| R218C | AD | –1.55 | –1.81 | 0.39 | –1.90 | Destabilizing |
| L224M | AD | –0.82 | –1.26 | –0.8 | –1.37 | Destabilizing |
| Y227N | AD | –3.32 | –2.33 | –4.69 | –2.5 | Destabilizing |
| F305S | AD | –2.71 | –3.0 | –3.88 | –3.1 | Destabilizing |
| R141H | AR | –2.69 | –2.95 | –0.05 | –3.12 | Destabilizing |
| A195V | AR | –1.39 | –0.45 | –0.18 | –0.32 | Destabilizing |
Two-stage prediction classification: destabilizing (<0 kcal/mol), stabilizing(>0 kcal/mol); AD, autosomal dominant; AR, autosomal recessive.
Protein expression of mutant BEST1
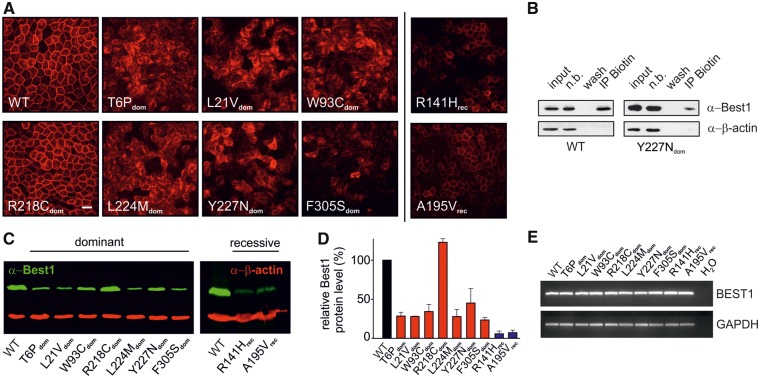
To further explore the impact of the selected BEST1 mutations on function and protein stability in vitro, we chose a non-viral, non-integrating single cell cloning strategy with untagged vector constructs to obtain stably transfected monoclonal MDCKII cell lines (WT, T6Pdom, L21Vdom, W93Cdom, R218Cdom, L224Mdom, Y227Ndom and F305Sdom, R141Hrec and A195Vrec). All lines were tested for 100% clonal purity. Consistent with an earlier study (28), we confirmed that untransfected MDCKII cells revealed no detectable Best1 expression (Supplementary Material, Fig. S1). Each transfected cell line was analyzed for BEST1 protein localization and quantified for RNA and protein expression (Fig. 2A–E). The polarized MDCKII cell monolayers were immunostained for BEST1 and examined by confocal microscopy. BEST1 WT and R218Cdom, and to a lesser extent R141Hrec and A195Vrec revealed localization of BEST1 to the basolateral PM in contrast to predominantly intracellular localizations for mutant proteins T6Pdom, L21Vdom, W93Cdom, L224Mdom, Y227Ndom and F305Sdom (Fig. 2A). Furthermore, cell surface biotinylation experiments revealed a substantial staining of WT in the biotinylated fraction confirming cell surface localization of the normal protein, whereas only ∼10% of biotinylated Y227Ndom was detectable (Fig. 2B), further demonstrating intracellular retention of the mutant protein. By quantitative Western blot analysis of MDCKII cell protein extracts, we observed reduced expression for the mislocalized proteins expressed by the autosomal dominant BEST1 mutations, except for mutant R218Cdom (Fig. 2C and D). Protein expression of the autosomal recessive mutants R141Hrec and A195Vrec were dramatically reduced (<7%), despite their obvious PM localization (Fig. 2A, C and D). Of note, RNA expression was similar in all ten cell lines analyzed (Fig. 2E). To control for clonal effects, protein expression of four independent clonal cell lines with BEST1 WT, and mutations L224Mdom, A195Vrec and R141Hrec, was measured demonstrating similar expression for each group of clonal cell lines (Supplementary Material, Fig. S2).
Figure 2.
Localization, RNA and protein expression of normal and mutant BEST1 stably expressed in MDCKII cells. (A) Confocal immunofluorescence images of MDCKII cells stably expressing wildtype, seven BD- and two ARB-associated mutants after 5 days growth on coverslips. Scale bars: 20 µm. (B) Wildtype and Y227Ndom were subjected to surface protein biotinylation. Labeled cells were precipitated with streptavidin, transferred to nylon membranes and probed as indicated. (n.b., fraction not bound). Immunoblot analysis revealed a substantial staining in the biotinylated fraction. Beta-actin served as negative control. (C) Western blot images of whole cell lysates of wildtype and BD- (left) and ARB-associated mutants (right). Also see Supplementary Material, Figure S1 for immunostaining of untransfected MDCKII cells. (D) Quantification of BEST1 protein expression from (C). Data from mutants are given relative to wildtype and normalized against beta-actin. For each sample, the mean ± SD is given (n = 2–4). Also see Supplementary Material, Figure S2 for individual variations within different clonal cell lines from the same genotype. (E) BEST1 mRNA expression of wildtype, BD- and ARB-associated mutants by semi-quantitative RT-PCR. GAPDH served as a control for RNA integrity. Immunostaining of BEST1 protein was performed using antibody hBEST1–334, targeting the last 15 amino acids of human BEST1 (9).
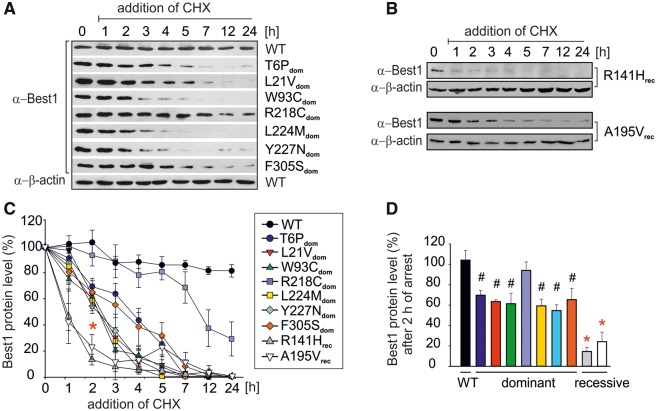
Half-life of mutant BEST1 protein
Next, we tested protein stability in the MDCKII cell lines by quantifying the levels of remaining BEST1 protein at different time points after treating cells with cycloheximide (CHX), a known inhibitor of eukaryotic translation. Protein quantity of BEST1 WT remained stable even after 24 h CHX treatment, whereas six out of seven autosomal dominant mutant proteins showed significant degradation within 3 h (Fig. 3A). Notably, mutant R218Cdom was stable for over 7 h and only showed signs of degradation at time points 12 and 24 h. Overall, the findings suggest that PM mislocalization of autosomal dominant BEST1 mutant proteins is accompanied by enhanced protein instability.
Figure 3.
Autosomal dominant and recessive mutations result in different biochemical half-life of BEST1 mutant protein. Representative Western blots of wildtype and stable mutant cell lines associated with autosomal dominant (A) or autosomal recessive (B) mutant BEST1 after inhibition of protein synthesis. Cells were treated with CHX (20 µg/ml) and harvested at the indicated time points. After determination of protein concentration in the cell lysates, equal amounts of protein were loaded on SDS-gels. (C) Scans of Western blots from (A) and (B) were analyzed densitometrically. The amount of BEST1 protein at different time points was quantified as the percentage of initial BEST1 protein level (0 h of CHX treatment). For each time point, the mean ± SD is given (n = 4 from at least 2 independent experiments). (D) Bar graph showing BEST1 protein expression of autosomal dominant and recessive mutants relative to wildtype after 2 h of CHX treatment. Data were extracted from (C). Values are given as mean ± SD. Two-sided paired Student’s t-test; #, significant relative to wildtype P < 0.05; *, significant relative to wildtype and dominant mutants P < 0.05. Also see Supplementary Material, Figure S3A–D for individual variations within different clonal cell lines from the same genotype.
Proteins of autosomal recessive mutants R141Hrec and A195Vrec revealed a strong degradation with a biochemical half-life of only 1–2 h (Fig. 3B–D). Degradation rates of independent clonal cell lines generated for BEST1 WT, R218Cdom, L224Mdom and R141Hrec revealed no clonal effects and confirmed interclonal comparability (Supplementary Material, Fig. S3A–D). Despite their PM localization, autosomal recessive mutations encode proteins that undergo a significant faster rate of degradation than those of autosomal dominant mutations or of wildtype (Fig. 3C and D).
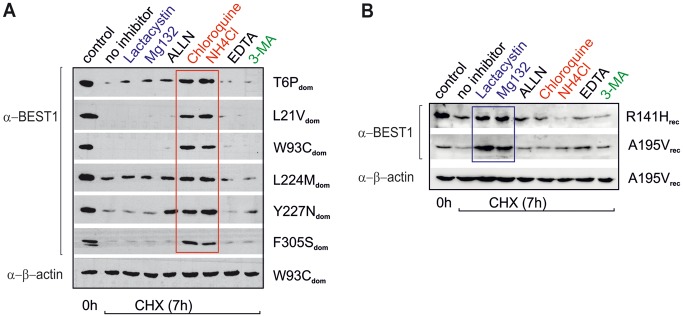
Degradation pathways
We then investigated characteristics of the degradation pathway underlying the autosomal dominant and recessive BEST1 gene mutations. Potent and selective inhibitors for the three major degradation systems were used, namely the proteasomal [lactacystin, MG132 and ALLN (N-Acetyl-Leu-Leu-Nle-al)], endo-lysosomal [the two weak bases chloroquine and ammonium chloride (NH4Cl)] or autophagy pathway [3-methyladenine (3-MA)] and the metal chelator ethylenediamine tetraacetic acid (EDTA). Our results revealed that protein levels encoded by the six BD-associated mutations showing mislocalization remained unaltered after incubation with NH4Cl (50 mM) and chloroquine (50 µM) (Fig. 4A). In contrast, degradation of the two ARB-associated mutant proteins R141Hrec and A195Vrec was inhibited only by the potent 26S proteasomal inhibitors lactacystin (10 µM) and MG132 (20 µM) well in agreement with earlier reports (20,25). Of note, the ARB-associated proteins were not affected by the lysosomal inhibitors NH4Cl and chloroquine (Fig. 4B), suggesting distinct degradation mechanisms for dominant and recessive mutant BEST1 proteins.
Figure 4.
Inhibition of lysosomal hydrolases leads to increased levels of BEST1 protein. Representative Western blots of MDCKII cells expressing wildtype and mutant BEST1 carrying autosomal dominant (A) and recessive mutations (B) after treatment with CHX. BEST1 protein was determined after incubation for 7 h in the absence or presence of indicated inhibitors. Beta-actin served as control. Inhibitors: proteasomal (blue), lactacystin (10 µM), MG132 (20 µM), ALLN (10 µM); lysosomal (red), chloroquine (50 µM), NH4Cl (50 mM); autophagy (green), 3-MA (5 mM) and EDTA (2 mM).
Rescue of autosomal dominant and recessive mutant BEST1 by low temperature and chemical chaperon 4-phenylbutyrate
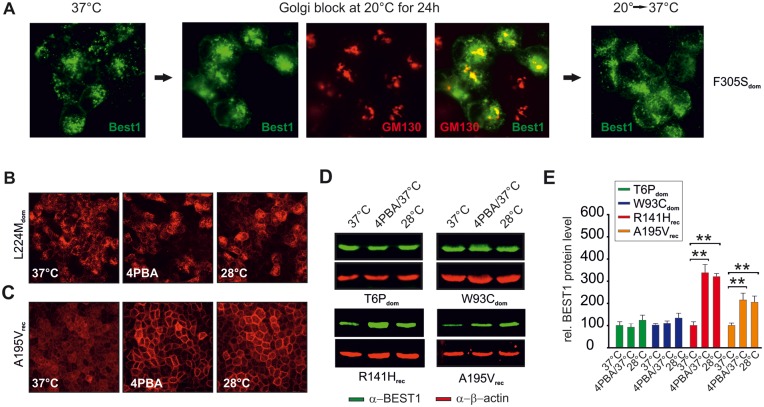
Next, we aimed to characterize trafficking routes of dominant BEST1 mutant protein from ER to the PM in more detail. Cell lines were incubated at 20°C, a condition that inhibits the exit of proteins from the Golgi complex. Under these conditions, mutant BEST1 proteins that are capable to exit the ER are expected to co-localize with Golgi marker GM130. After the 20°C temperature block, cells were shifted back to 37°C. This allowed differentiation between mutants retained in the Golgi complex and those capable to exit the Golgi complex ultimately localizing to the lysosome. As expected, mutant F305Sdom, a representative for the six intracellularly retained BD-associated mutant proteins, clearly co-localized with GM130 after 20°C incubation and regained a vesicle-like staining pattern after release from the temperature block (Fig. 5A). A behavior similar to F305Sdom is also shown for mutants L21Vdom, W93Cdom and L224Mdom (Supplementary Material, Fig. S4). Together, these data suggest that degradation of BD-mutant proteins occur after exit from the Golgi apparatus, thereby excluding an ER-associated degradation.
Figure 5.
Expression, localization and rescue of intracellularly retained autosomal dominant BEST1 mutant protein by 4-PBA or low temperature. (A) Immunofluorescence staining shows F305Sdom localization at 37°C (left), after a 20°C block for 24 h (middle) and after release of the temperature block and transfer to 37°C (right). After 20°C block co-localization of BEST1 with Golgi marker GM130 was examined. Also see Supplementary Material, Figure S4 for co-localization analysis of mutant L21Vdom, W93Cdom and L224Mdom after a 20°C block. (B and C) Confocal immunofluorescence images of L224Mdom and A195Vrec at 37°C in the presence or absence of 2 mM 4-PBA and at 28°C. (D) Representative Western blots of mutants T6Pdom, L224Mdom, R141Hrec and A195Vrec at 37°C in the presence or absence of 4-PBA (2 mM) and at 28°C. (E) Quantification of BEST1 protein expression relative to wildtype levels from (D). Scans of Western blots were analyzed densitometrically and normalized to beta-actin on the same blot. Data were represented as the mean ± SD (n = 6 from at least three independent experiments). Two-sided paired Student’s t-test; **,P < 0.01.
To corroborate these findings and to extend the study to recessive BEST1 mutant protein, cell lines L224Mdom and A195Vrec were incubated for 24 h with the commonly used pharmacological chaperon 4-phenylbutyrate (4-PBA) (2 mM) or at low temperature (28°C), two conditions that result in the enhancement of the ER folding capacity by modulating the expression of molecular chaperones. As a consequence, misfolded/instable proteins are able to escape endoplasmic-reticulum-associated protein degradation (ERAD) and traffic to the PM. Subsequently, cell surface expression of BEST1 was monitored by immunofluorescence. As a result, L224Mdom mutant was not detected at the PM after treatment (Fig. 5B). As recessive mutants showed surface expression at steady state, treatment with 4-PBA or 28°C revealed no effect on the localization although BEST1 labeling appeared more pronounced in treated than untreated cells at 37°C (Fig. 5C). To analyze to what extent 4-PBA or 28°C temperature condition enhances BEST1 expression in BD and ARB mutants, we performed Western blot analysis from mutant proteins T6Pdom, W93Cdom, R141Hrec and A195Vrec in treated and untreated cells (Fig. 5D and E). BEST1 expression in the T6Pdom and W93Cdom cell lines remained unaffected upon 4-PBA treatment. In contrast, BEST1 expression increased 3- to 4-fold in the two recessive mutant cell lines upon treatment with 4-PBA and low temperature, consistent with the known cellular processes of enhancing chaperone expression for clearance of misfolded proteins (29). These results argue further against a contribution of the ER in the degradation of misfolded/instable mutant protein carrying autosomal dominant mutations.
Effects of mutant BEST1 processing on ER homeostasis
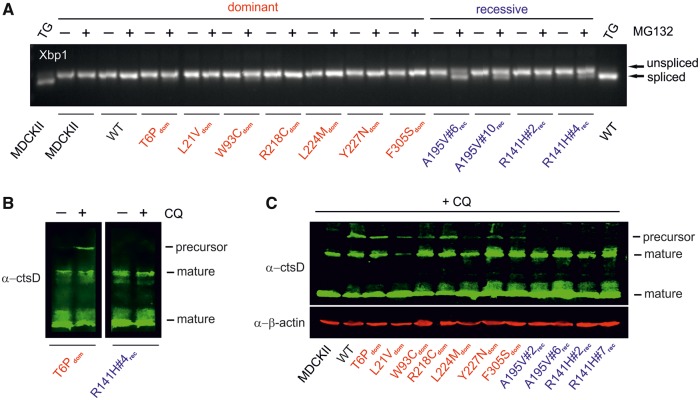
Next, we examined whether accumulation of misfolded/unfolded mutant BEST1 protein in the ER triggers the unfolded protein response (UPR), a condition known to reflect ER stress. Since UPR initiates a cascade of signaling pathways by activating downstream transcription factors which counteract ER stress, we analyzed the mRNA expression of X-box binding protein 1 (XBP1) in all cell lines. In response to ER stress, XBP1 is spliced to produce a highly active transcription factor, XBP1active (30). As expected, treatment of untransfected MDCKII cells and WT with ER stress-inducing agent thapsigargin (TG) (1 µM) results in the detection of a 26 bp smaller (spliced) XBP1 product indicative of successfully induced ER stress (Fig. 6A). In contrast, all untreated cell lines showed the unspliced XBP1 product implying that the capacity of the proteasome for processing misfolded/unfolded protein is sufficient to prevent the cells from ER stress at steady state (Fig. 6A).
Figure 6.
Effects of protein degradation of dominant and recessive mutants on ER and lysosome homeostasis. (A) XBP1 mRNA expression of untransfected MDCKII cells, wildtype (WT) control, BD- and ARB-associated mutants in the presence (+) or absence (−) of MG132 (20 µM). Semi-quantitative RT-PCR results in an unspliced 287 bp mRNA fragment. In response to ER stress a 26 bp XBP1 fragment is removed by splicing from the Xbp1 mRNA. TG was used to induce ER stress in untransfected MDCKII cells and WT. (B) Western blot images of whole cell lysates from a representative BD- and ARB-associated mutant cell line before (−) and after (+) treatment with 50 µM chloroquine (CQ) for 5 h. Also see Supplementary Material, Figure S5 for Western blot results of untreated and treated mutant lines L21Vdom, W93Cdom, L224Mdom, Y227Ndom and F305Sdom. (C) Western blot image of whole cell lysates from untransfected MDCKII cells, WT and BD- and ARB-associated mutant lines after treatment with 50 µM CQ. An antibody against α-ctsD was used to detect two mature (∼34 and ∼14 kDa) and a short-lived (∼55 kDa) precursor form of ctsD. Beta-actin was used as loading control.
To precondition for weak ER stress, we treated the cells with MG132 (20 µM) resulting in an increased protein load in the ER. Two independent clonal cell lines of the ARB-associated mutants A195Vrec and to a weaker extent R141Hrec clearly show both, spliced and unspliced XBP1. In contrast, in the seven BD-associated dominant mutants only the unspliced product of the transcription factor XBP1 is detectable (Fig. 6A). These findings suggest that proteasome-associated degradation of autosomal recessive BEST1 mutants increases susceptibility for ER stress.
Effects of mutant BEST1 processing on lysosomal homeostasis
To explore effects of enhanced degradation of the autosomal dominant BEST1 mutant proteins on lysosomal homeostasis, we analyzed the proteolytic maturation of cathepsin D (ctsD), a lysosomal enzyme representing a major protease type of lysosomal activity (31). CtsD is synthesized in the ER as an inactive and short-lived proenzyme of molecular weight ∼55 kDa that is subsequently proteolytically processed to a 48 kDa intermediate form in the endosome. Final processing in acidic lysosomes yields an active mature enzyme composed of a larger (∼34 kDa) and a smaller (∼4 kDa) chain (32). In Western blot images of protein lysates from all BD- and ARB-associated mutations solely the two 34 and 14 kDa mature forms of ctsD but not the proenzyme (∼55 kDa) were detectable (Fig. 6B and Supplementary Material, Fig. S5). This indicates that overexpression of BEST1 mutants does not interfere with ctsD processing in the lysosome at steady state.
We then preconditioned the cell lines with chloroquine (50 µM) to induce protein accumulation in the lysosome and thus lysosomal stress. Although chloroquine-treated ARB-associated cell lines remained unaffected, the presence of the ∼55 kDa proenzyme in cell lysates from dominant BEST1 mutants is suggestive of lysosomal dysfunction (Fig. 6B and C and Supplementary Material, Fig. S5). Interestingly, the ∼55 kDa proenzyme is also present in WT BEST1 indicating that normal protein is recognized for endo-lysosomal degradation by a quality control checkpoint at the PM, a well-established mechanism to preserve cellular protein homeostasis (33). Together, these findings indicate that enhanced degradation in the lysosome and accumulation of autosomal dominant but not recessive BEST mutant protein affect the integrity of endo-lysosomal compartments.
Discussion
Here we show that BEST1 missense mutations leading to the autosomal dominant BD phenotype escape the ER quality check and instead are recognized by a post-ER quality control mechanism at the Golgi complex (34). Several findings strongly support this conclusion. First, six out of seven BEST1 mutations analyzed revealed a cellular mislocalization into vesicle-like structures accompanied by a strikingly reduced steady state level of BEST1 protein and a shortened half-life that could not be rescued by low temperature or the chemical chaperon 4-PBA. Pore-forming mutation R218Cdom, which is an exception to the above findings, could be recognized by a quality control mechanism at the PM, a known alternative in cases where quality control at the Golgi complex does not prove effective (35). Second, protein degradation was inhibited by two lysosomal inhibitors, chloroquine and NH4Cl, but not by other potent and selective inhibitors of the proteasomal or the autophagy pathway. Third, at 20°C the dominant BEST1 mutants exited the ER and accumulated in the Golgi complex while release of the temperature block resulted in their vesicular redetection. Finally, dominant BEST1 mutants triggered abnormal processing and maturation of catalytic enzymes in the endo-lysosome after chloroquine treatment. In contrast, mutant BEST1 associated with the autosomal recessive ARB is recognized by the ER and subsequently prone to ERAD as shown before by other groups (20,25). These cellular events are well established as a regular route of protein degradation for many other mutated membrane proteins (36,37).
The rate of protein degradation of misfolded BEST1 protein could have a significant impact on partial functionality of the homo-pentameric BEST1 chloride channel. For example, as mutant subunits in heterozygous carriers of autosomal recessive mutations would be rapidly removed by ERAD from the process of proper channel formation, stoichiometry would shift to include rather normal BEST1 subunits. This way, the likelihood of forming a channel solely by regular non-mutated BEST1 subunits should increase and ERAD would be beneficial for disease outcome over post-ER protein degradation as is observed for the autosomal dominant BEST1 missense mutations. We suggest that only BEST1 mutations that escape ERAD can act upon their dominant-negative potential.
Although the surveillance mechanism at the Golgi apparatus is a common post-ER checkpoint for a number of misfolded proteins in yeast (38,39), little is known about this degradation mechanism in mammalian cells. There are only few reports on Golgi quality control-induced endo-lysosomal degradation of mutant proteins. For example, the gap-junction protein connexin 43 (40) and the T-cell receptor (41) require the Golgi complex for their final assembly. Incomplete complexes have been shown to be retained in the Golgi apparatus and targeted for lysosomal degradation. There is also the prion protein where a lysine-based motif in the N-terminus together with a misfolded C-terminal domain influences the destination for degradation of mutant protein (42,43). In addition, studies indicate that membrane proteins with limited conformational defects may escape ERAD while recognized by the more stringent Golgi quality control suggesting that Golgi surveillance is more sensitive to subtle structural changes than ERAD (38,44). At present, the recognition motif for autosomal recessive mutant BEST1 to discriminate folded from misfolded protein in the ER is unknown and it is difficult to predict differences between dominant and recessive mutations based on their linear or conformational 3D locations. Autosomal dominant BEST1 may allow multimerization without grossly affecting the overall protein fold. This complex may then be targeted for endo-lysosomal degradation by post-translational modifications, such as mono- or multi-ubiquitination, as described previously for a variety of integral proteins (45,46). Alternatively, as misfolded BEST1 is capable of exiting ER due to a strong cytosolic ER exit signal (47), this signal may be weakened by autosomal recessive but not autosomal dominant mutations. At present, it is unclear why dominant BEST1 mutant proteins are not retained in the ER but instead traffic to the Golgi system.
The channeling of dominant BEST1 mutant proteins into the endo-lysosomal degradation pathway could have important implications for BD pathogenesis. One of the consequences could be an inefficient maturation of lysosomal proteases due to an accumulation of mutant protein in post-ER compartments. This, in turn, could result in saturation of the Golgi quality control machinery and negatively influence the degradation capacity of lysosomes. In BD, the striking lipofuscin accumulation in the RPE may be an expression of an impaired lysosomal function with the known adverse effects on lysosomal degradation of photoreceptor outer segment membranes (48–50). Conversely, the observed increased sensitivity to MG132-induced ER stress in the ARB-associated mutants may be caused by enhanced accumulation of mutant protein in the ER, thereby exhausting chaperone activity over time. This may then impair the degradation capacity of the proteasomal machinery. Only recently, work by Hamdan et al. (51) showed that limitations in chaperone availability account for increased protein aggregation in ER stress mutants. Thus, in contrast to BD, multifocal subretinal deposits and abnormal autofluorescence in the ARB phenotype may not originate from accumulated photoreceptor cell waste but, instead, from ER stress-induced accumulation of cytosolic aggregation-prone proteins (51). Together, our divergent findings on degradation of the autosomal dominant versus the autosomal recessive BEST1 mutant proteins suggest that activation of quality control mechanisms at separate subcellular compartments likely underlie the phenotypic differences in BEST1-associated disease. It will be of interest to investigate additional autosomal dominant BEST1 mutations to explore whether the well-known phenotypic heterogeneity in BD could at least in part be due to missense mutations favoring the retention in or escaping from the ER. It will also be attractive to test mutant BEST1 protein associated with other phenotypic manifestations such as ADVIRC.
In conclusion, this study has identified distinct degradation mechanisms underlying selected autosomal dominant and recessive BEST1 missense mutations. We hypothesize that activation of ERAD may be triggered by specific missense mutations and this may determine whether the mutated protein is retained in the ER or is targeted for post-ER degradation. This, in turn, has major implications for BEST1-associated disease manifestations and possibly for therapeutic intervention. For example, a pharmacological intervention targeted at post-ER quality control mechanisms may be beneficial for treatment of the autosomal dominant but not the autosomal recessive bestrophinopathies.
Materials and methods
Predicting the structural effects of BEST1 missense mutations
Sequence alignments were accomplished using ClustalW (http://www.ch.embnet.org/software/ClustalW.html; date last accessed December 2017) and BoxShade (http://www.ch.embnet.org/software/BOX_form.html; date last accessed December 2017). PyMOL was used for the cBEST structure figure (http://www.pymol.org/; date last accessed December 2017). For prediction analysis, the 3D structure of cBESTcryst was obtained from protein data bank (PDB) (52). Given a PDB file (cBESTcryst: PBB ID 4rdq) and a point mutation, the server calculates the stability difference score between the wildtype and mutant protein. The predicted free energy change is derived from the unfolding Gibbs free energy change of the mutated protein minus the unfolding free energy value of the native protein (kcal/mol) (53). I-Mutant2.0 is a support vector machine-(SVM) based web server for the automatic prediction of protein stability changes upon single-site variations from the protein structure or sequence (http://folding.biofold.org/i-mutant/i-mutant2.0.html; date last accessed December 2017) (53). mCSM relies on graph-based signatures. Distance patterns between atoms are used to train predictive models and to represent the protein residue environment (54). SDM is based on knowledge of observed substitutions within homologous protein families to calculate a stability score which is analogous to the free energy difference between a wildtype and mutant protein (55). DUET consolidates two complementary approaches (mCSM and SDM) in a consensus prediction, by combining the results of the two methods in an optimized predictor using SVM (56). The predicted results from I-Mutant 2.0, SDM, mCSM and DUET (http://biosig.unimelb.edu.au/duet/stability; date last accessed December 2017) are expressed as the variation in ΔΔG: destabilizing (<0 kcal/mol) and stabilizing (>0 kcal/mol).
Chemicals and antibodies
CHX, chloroquine phosphate, lactacystin, MG132, 3-MA, calpain inhibitor I (ALLN), 4-PBA and TG (Biomol, Hamburg, Germany) were obtained commercially. Substances were prepared as stock solutions in dimethyl sulfoxide: lactacystin (10 mM), MG132 (40 mM), ALLN (5 mg/ml) and TG (2.5 mM).
Rabbit polyclonal antibody hBEST1–334 was described previously (9), and used in 1:2500 (WB) or 1:250 (ICC) dilution. Rabbit polyclonal antibody ctsD (ab75852, Abcam, Cambridge, UK; dilution 1: 1500), mouse monoclonal antibodies β-actin (no. 5441, Sigma-Aldrich, Munich, Germany; dilution 1:10 000) and GM130 (ab 52649, Abcam, Cambridge, UK; dilution 1:1000), are commercially available. Secondary antibodies for immunofluorescence were goat Alexa 594- and Alexa 488-conjugated anti-rabbit (Thermofisher Scientific, Waltham, USA 1:500). Western blot experiments were performed with goat near-infrared fluorescent dyes (IRDye 1:10 000) (Lycor, Bad Homburg, Germany) or horseradish peroxidase-conjugated secondary antibodies (Calbiochem/Merck Millipore, Darmstadt, Germany; 1:10 000).
Plasmid constructs
Generation of BEST1 constructs of wildtype and mutants T6P, L21V, R218C, L224M, Y227N and F305S in a pCDNA3 vector has been described previously in (22). Mutations W93C, R141H and A195V were generated via site-directed mutagenesis.
Cell culture and generation of stable cell lines
MDCKII (ATCC, cat no. CCL-34) cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and penicillin/streptomycin solution. Cells were maintained at 37 or 28°C with a 5% CO2 environment. Transfection of MDCKII cells was performed with Lipofectamine 3000 transfection reagent following the manufacturer’s instructions. MDCKII cells expressing wildtype and mutant BEST1 were cultured for 2 weeks in a selection medium containing 500 µg/ml G418 before single cell seeding in 96-well plates. Two to nine single clones were selected for each cell line. For all immunofluorescence and Western blot experiments cells were seeded on coverslips or 6-well plates, respectively, to reach confluency the next day and maintained for another 5 days in media containing 1% FBS to prevent overgrowth. For half-life and rescue studies, cells were subjected to the same protocol followed by the addition of CHX, 4-PBA or growth at 28°C in media containing 1% FBS. For biotinylation experiments cells were grown for 6 days on Transwell-filters (Corning Costar by Sigma-Aldrich, Munich, Germany). For Golgi block experiments cells were exposed to 20°C for 3 h in a CO2-free incubator in MEM media with reduced NaHCO3 concentration supplemented with 10 mM Hepes. All cell culture supplements and transfection reagent were provided from Thermofisher Scientific, Waltham, USA.
Protein sample preparation, SDS page and quantitative Western blot analysis
Whole cell protein sample preparation was performed as previously described in (9). Protein samples were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis on 10% gels and subsequently transferred onto Immobilon-FL (LI-COR Bioscience, Bad Homburg, Germany) or Immobilon-P PVDF membrane (Millipore, Bedford, MA, USA). Incubation of primary and secondary antibodies was carried out at 4°C ON, respectively. Protein labeling was visualized by fluorescence detection using the Odyssey Fc Imaging System and signal intensities were quantified with the Image Studio software and normalized against beta-actin from the same blot.
RNA isolation, reverse transcription and semi-quantitative reverse transcriptase-PCR
Using the RNeasy Mini Kit total RNA was extracted after DNAse treatment (Roche, Mannheim, Germany) according to the manufactureŕs instructions (Qiagen, Hilden, Germany). First strand cDNA synthesis from 1 μg of total RNA was performed with RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas, Thermofisher Scientific, Waltham, USA) and random hexamer oligonucleotide primers. Human BEST1 and GAPDH was analyzed by reverse transcriptase (RT)-PCR with primer pairs TU15newF2 (5′-cag tac gag aac ctg ccg tg -3′)/TU15newR1 (5′- ggt agg ctc agt ttc tcc aaa-3′), GAPDH-F (5′-atc gtg gaa gga ctc atg acc-3′)/GAPDH-R (5′-agc gcc agt aga ggc agg gat-3′), respectively.
Analysis of ER stress
Cells were seeded onto 5 cm dishes to confluency and cultured for 5 consecutive days with media containing 1% FBS. Cells were treated in the presence or absence of MG132 (20 µM) or TG (1 µM) and collected after 6 h. 1 µg of total RNA was reverse transcribed and RT-PCR was performed from cDNA using primer pair huXBP1-F (5′-tta cga gag aaa act cat ggc c-3′)/dogXbp1 (5′-gga tcc aag ttg aac aga atg c-3′).
Plasma membrane surface biotinylation
Surface proteins of MDCKII cells, grown on Transwell inserts in a 6-well plate, were biotinylated according to the manufacturer’s instructions (#89881, Pierce, Thermo Fisher Scientific, Waltham, USA). Homogenization of cell lysates was performed in 1 ml lysis buffer (50 mM Tris HCl, pH 7, 5, 5 mM EDTA, 15 mM NaCl, 1% Triton, 1 × Protease inhibitor cocktail [#04693116001, Roche, Switzerland]) by passing the lysate six times through a 27 gauge needle. Biotinylated proteins were eluted, subjected to SDS-PAGE and immunostained for BEST1.
CHX treatment
Cells were seeded onto 6-well plates cultured for 5 consecutive days with media containing 1% FBS. Cells were treated with CHX (20 µg/ml) in the presence or absence of various inhibitors and collected at various time points up to 24 h. Whole cell lysates were subjected to quantitative Western blot analysis using antibodies against BEST1 and normalized against beta-actin.
Statistical analysis
Statistical analysis was performed applying two-paired Student’s t-test. P < 0.05 was considered statistically significant.
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
Funding
This work was supported in part by a grant from the Deutsche Forschungsgemeinschaft (DFG) to BHFW [WE1259/29–1]. Funding to pay the Open Access publication charges for this article was provided by institutional resources.
Supplementary Material
References
- 1. Marquardt A., Stohr H., Passmore L.A., Kramer F., Rivera A., Weber B.H. (1998) Mutations in a novel gene, VMD2, encoding a protein of unknown properties cause juvenile-onset vitelliform macular dystrophy (Best's disease). Hum. Mol. Genet., 7, 1517–1525. [DOI] [PubMed] [Google Scholar]
- 2. Stohr H., Marquardt A., Nanda I., Schmid M., Weber B.H. (2002) Three novel human VMD2-like genes are members of the evolutionary highly conserved RFP-TM family. Eur. J. Hum. Genet. , 10, 281–284. [DOI] [PubMed] [Google Scholar]
- 3. Marmorstein A.D., Marmorstein L.Y., Rayborn M., Wang X., Hollyfield J.G., Petrukhin K. (2000) Bestrophin, the product of the Best vitelliform macular dystrophy gene (VMD2), localizes to the basolateral plasma membrane of the retinal pigment epithelium. Proc. Natl. Acad. Sci. U.S.A., 97, 12758–12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kane Dickson V., Pedi L., Long S.B. (2014) Structure and insights into the function of a Ca(2+)-activated Cl(-) channel. Nature, 516, 213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang T., Liu Q., Kloss B., Bruni R., Kalathur R.C., Guo Y., Kloppmann E., Rost B., Colecraft H.M., Hendrickson W.A. (2014) Structure and selectivity in bestrophin ion channels. Science (New York, N.Y.), 346, 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sun H., Tsunenari T., Yau K.W., Nathans J. (2002) The vitelliform macular dystrophy protein defines a new family of chloride channels. Proc. Natl. Acad. Sci. U.S.A., 99, 4008–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hartzell H.C., Qu Z., Yu K., Xiao Q., Chien L.T. (2008) Molecular physiology of bestrophins: multifunctional membrane proteins linked to best disease and other retinopathies. Physiol. Rev., 88, 639–672. [DOI] [PubMed] [Google Scholar]
- 8. Xiao Q., Prussia A., Yu K., Cui Y.Y., Hartzell H.C. (2008) Regulation of bestrophin Cl channels by calcium: role of the C terminus. J. Gen. Physiol., 132, 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Milenkovic A., Brandl C., Milenkovic V.M., Jendryke T., Sirianant L., Wanitchakool P., Zimmermann S., Reiff C.M., Horling F., Schrewe H.. et al. (2015) Bestrophin 1 is indispensable for volume regulation in human retinal pigment epithelium cells. Proc. Natl. Acad. Sci. U.S.A., 112, E2630–E2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kramer F., White K., Pauleikhoff D., Gehrig A., Passmore L., Rivera A., Rudolph G., Kellner U., Andrassi M., Lorenz B.. et al. (2000) Mutations in the VMD2 gene are associated with juvenile-onset vitelliform macular dystrophy (Best disease) and adult vitelliform macular dystrophy but not age-related macular degeneration. Eur. J. Hum. Genet., 8, 286–292. [DOI] [PubMed] [Google Scholar]
- 11. Yardley J., Leroy B.P., Hart-Holden N., Lafaut B.A., Loeys B., Messiaen L.M., Perveen R., Reddy M.A., Bhattacharya S.S., Traboulsi E.. et al. (2004) Mutations of VMD2 splicing regulators cause nanophthalmos and autosomal dominant vitreoretinochoroidopathy (ADVIRC). Investig. Ophthalmol. Vis. Sci., 45, 3683–3689. [DOI] [PubMed] [Google Scholar]
- 12. Burgess R., Millar I.D., Leroy B.P., Urquhart J.E., Fearon I.M., De Baere E., Brown P.D., Robson A.G., Wright G.A., Kestelyn P.. et al. (2008) Biallelic mutation of BEST1 causes a distinct retinopathy in humans. Am. J. Hum. Genet., 82, 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bitner H., Schatz P., Mizrahi-Meissonnier L., Sharon D., Rosenberg T. (2012) Frequency, genotype, and clinical spectrum of best vitelliform macular dystrophy: data from a national center in Denmark. Am. J. Ophthalmol., 154, 403–412 e404. [DOI] [PubMed] [Google Scholar]
- 14. Cross H.E., Bard L. (1974) Electro-oculography in Best’s macular dystrophy. Am. J. Ophthalmol., 77, 46–50. [DOI] [PubMed] [Google Scholar]
- 15. Mohler C.W., Fine S.L. (1981) Long-term evaluation of patients with Best’s vitelliform dystrophy. Ophthalmology, 88, 688–692. [DOI] [PubMed] [Google Scholar]
- 16. Bitner H., Mizrahi-Meissonnier L., Griefner G., Erdinest I., Sharon D., Banin E. (2011) A homozygous frameshift mutation in BEST1 causes the classical form of Best disease in an autosomal recessive mode. Investig. Ophthalmol. Vis. Sci., 52, 5332–5338. [DOI] [PubMed] [Google Scholar]
- 17. Wong R.L., Hou P., Choy K.W., Chiang S.W., Tam P.O., Li H., Chan W.M., Lam D.S., Pang C.P., Lai T.Y. (2010) Novel and homozygous BEST1 mutations in Chinese patients with Best vitelliform macular dystrophy. Retina (Philadelphia, Pa.), 30, 820–827. [DOI] [PubMed] [Google Scholar]
- 18. Boon C.J., Klevering B.J., Leroy B.P., Hoyng C.B., Keunen J.E., den Hollander A.I. (2009) The spectrum of ocular phenotypes caused by mutations in the BEST1 gene. Prog. Retin. Eye Res., 28, 187–205. [DOI] [PubMed] [Google Scholar]
- 19. Marmorstein A.D., Cross H.E., Peachey N.S. (2009) Functional roles of bestrophins in ocular epithelia. Prog. Retin. Eye Res., 28, 206–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Davidson A.E., Millar I.D., Burgess-Mullan R., Maher G.J., Urquhart J.E., Brown P.D., Black G.C., Manson F.D. (2011) Functional characterization of bestrophin-1 missense mutations associated with autosomal recessive bestrophinopathy. Investig. Ophthalmol. Vis. Sci., 52, 3730–3736. [DOI] [PubMed] [Google Scholar]
- 21. Moshfegh Y., Velez G., Li Y., Bassuk A.G., Mahajan V.B., Tsang S.H. (2016) BESTROPHIN1 mutations cause defective chloride conductance in patient stem cell-derived RPE. Hum. Mol. Genet., 25, 2672–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Milenkovic V.M., Rohrl E., Weber B.H., Strauss O. (2011) Disease-associated missense mutations in bestrophin-1 affect cellular trafficking and anion conductance. J. Cell Sci., 124, 2988–2996. [DOI] [PubMed] [Google Scholar]
- 23. Johnson A.A., Bachman L.A., Gilles B.J., Cross S.D., Stelzig K.E., Resch Z.T., Marmorstein L.Y., Pulido J.S., Marmorstein A.D. (2015) Autosomal recessive bestrophinopathy is not associated with the loss of bestrophin-1 anion channel function in a patient with a novel BEST1 mutation. Investig. Ophthalmol. Vis. Sci., 56, 4619–4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson A.A., Lee Y.S., Chadburn A.J., Tammaro P., Manson F.D., Marmorstein L.Y., Marmorstein A.D. (2014) Disease-causing mutations associated with four bestrophinopathies exhibit disparate effects on the localization, but not the oligomerization, of Bestrophin-1. Exp. Eye Res., 121, 74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Uggenti C., Briant K., Streit A.K., Thomson S., Koay Y.H., Baines R.A., Swanton E., Manson F.D. (2016) Restoration of mutant bestrophin-1 expression, localisation and function in a polarised epithelial cell model. Dis. Models Mech., 9, 1317–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zode G.S., Bugge K.E., Mohan K., Grozdanic S.D., Peters J.C., Koehn D.R., Anderson M.G., Kardon R.H., Stone E.M., Sheffield V.C. (2012) Topical ocular sodium 4-phenylbutyrate rescues glaucoma in a myocilin mouse model of primary open-angle glaucoma. Investig. Ophthalmol. Vis. Sci., 53, 1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rubenstein R.C., Egan M.E., Zeitlin P.L. (1997) In vitro pharmacologic restoration of CFTR-mediated chloride transport with sodium 4-phenylbutyrate in cystic fibrosis epithelial cells containing delta F508-CFTR. The J. Clin. Investig., 100, 2457–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Doumanov J., Zeitz C., Gimenez P., Audo I., Krishna A., Alfano G., Diaz M., Moskova-Doumanova V., Lancelot M.-E., Sahel J.-A.. et al. (2013) Disease-causing mutations in BEST1 gene are associated with altered sorting of bestrophin-1 protein. Int. J. Mol. Sci., 14, 15121–15140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cummings C.J., Mancini M.A., Antalffy B., DeFranco D.B., Orr H.T., Zoghbi H.Y. (1998) Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat. Genet., 19, 148–154. [DOI] [PubMed] [Google Scholar]
- 30. Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell, 107, 881–891. [DOI] [PubMed] [Google Scholar]
- 31. Turk V., Stoka V., Vasiljeva O., Renko M., Sun T., Turk B., Turk D. (2012) Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim. Biophys. Acta, 1824, 68–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gieselmann V., Hasilik A., von Figura K. (1985) Processing of human cathepsin D in lysosomes in vitro. J. Biol. Chem., 260, 3215–3220. [PubMed] [Google Scholar]
- 33. Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. (2006) Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell, 21, 737–748. [DOI] [PubMed] [Google Scholar]
- 34. Okiyoneda T., Apaja P.M., Lukacs G.L. (2011) Protein quality control at the plasma membrane. Curr. Opin. Cell Biol., 23, 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Babst M. (2014) Quality control: quality control at the plasma membrane: one mechanism does not fit all. J. Cell Biol., 205, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hayashi H., Sugiyama Y. (2009) Short-chain ubiquitination is associated with the degradation rate of a cell-surface-resident bile salt export pump (BSEP/ABCB11). Mol. Pharmacol., 75, 143–150. [DOI] [PubMed] [Google Scholar]
- 37. Sharma M., Pampinella F., Nemes C., Benharouga M., So J., Du K., Bache K.G., Papsin B., Zerangue N., Stenmark H.. et al. (2004) Misfolding diverts CFTR from recycling to degradation: quality control at early endosomes. J. Cell Biol., 164, 923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang S., Ng D.T. (2010) Evasion of endoplasmic reticulum surveillance makes Wsc1p an obligate substrate of Golgi quality control. Mol. Biol. Cell, 21, 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chang A., Fink G.R. (1995) Targeting of the yeast plasma membrane [H+]ATPase: a novel gene AST1 prevents mislocalization of mutant ATPase to the vacuole. J. Cell Biol., 128, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vanslyke J.K., Naus C.C., Musil L.S. (2009) Conformational maturation and post-ER multisubunit assembly of gap junction proteins. Mol. Biol. Cell, 20, 2451–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sussman J.J., Bonifacino J.S., Lippincott-Schwartz J., Weissman A.M., Saito T., Klausner R.D., Ashwell J.D. (1988) Failure to synthesize the T cell CD3-zeta chain: structure and function of a partial T cell receptor complex. Cell, 52, 85–95. [DOI] [PubMed] [Google Scholar]
- 42. Ashok A., Hegde R.S. (2009) Selective processing and metabolism of disease-causing mutant prion proteins. PLoS Pathogens, 5, e1000479.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Heske J., Heller U., Winklhofer K.F., Tatzelt J. (2004) The C-terminal globular domain of the prion protein is necessary and sufficient for import into the endoplasmic reticulum. J. Biol. Chem., 279, 5435–5443. [DOI] [PubMed] [Google Scholar]
- 44. Coughlan C.M., Walker J.L., Cochran J.C., Wittrup K.D., Brodsky J.L. (2004) Degradation of mutated bovine pancreatic trypsin inhibitor in the yeast vacuole suggests post-endoplasmic reticulum protein quality control. J. Biol. Chem., 279, 15289–15297. [DOI] [PubMed] [Google Scholar]
- 45. Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P.P., Dikic I. (2003) Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol., 5, 461–466. [DOI] [PubMed] [Google Scholar]
- 46. Kolling R., Hollenberg C.P. (1994) The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J., 13, 3261–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kincaid M.M., Cooper A.A. (2006) Misfolded proteins traffic from the endoplasmic reticulum (ER) due to ER export signals. Mol. Biol. Cell, 18, 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Katz M.L., Shanker M.J. (1989) Development of lipofuscin-like fluorescence in the retinal pigment epithelium in response to protease inhibitor treatment. Mech. Ageing Dev., 49, 23–40. [DOI] [PubMed] [Google Scholar]
- 49. Rakoczy P., Kennedy C., Thompson-Wallis D., Mann K., Constable I. (1992) Changes in retinal pigment epithelial cell autofluorescence and protein expression associated with phagocytosis of rod outer segments in vitro. Biol. Cell, 76, 49–54. [DOI] [PubMed] [Google Scholar]
- 50. Wassell J., Ellis S., Burke J., Boulton M. (1998) Fluorescence properties of autofluorescent granules generated by cultured human RPE cells. Investig. Ophthalmol. Vis. Sci., 39, 1487–1492. [PubMed] [Google Scholar]
- 51. Hamdan N., Kritsiligkou P., Grant C.M. (2017) ER stress causes widespread protein aggregation and prion formation. J. Cell Biol., 216, 2295–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. (2000) The protein data bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Capriotti E., Fariselli P., Calabrese R., Casadio R. (2005) Predicting protein stability changes from sequences using support vector machines. Bioinformatics (Oxford, England), 21, ii54–ii58. [DOI] [PubMed] [Google Scholar]
- 54. Pires D.E., Ascher D.B., Blundell T.L. (2014) mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics (Oxford, England), 30, 335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pandurangan A.P., Ochoa-Montano B., Ascher D.B., Blundell T.L. (2017) SDM: a server for predicting effects of mutations on protein stability. Nucleic Acids Res., 45, W229–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pires D.E., Ascher D.B., Blundell T.L. (2014) DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res., 42, W314–W319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Milenkovic V.M., Rivera A., Horling F., Weber B.H. (2007) Insertion and topology of normal and mutant bestrophin-1 in the endoplasmic reticulum membrane. The J. Biol. Chem., 282, 1313–1321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.