Abstract
N-methyl-D-aspartate receptors (NMDARs) are glutamate-gated ion channels that play crucial roles in brain development and synaptic plasticity. They are also therapeutic targets of interest since their dysfunction is associated with multiple neurological and psychiatric disorders. In vivo, NMDARs exist as multiple subtypes that differ in their subunit composition, anatomical distribution, functional properties, as well as signaling capacities. While much is known about diheteromeric NMDARs composed of two GluN1 subunits and two identical GluN2 (or GluN3) subunits, the majority of native NMDARs are triheteromers containing two GluN1 and two different GluN2 (or a combination of GluN2 and GluN3). Knowledge about triheteromeric NMDARs has recently boomed, with the first decoding of their atomic structure and the development of a new methodology allowing selective expression of recombinant triheteromers at the cell-surface without confounding co-expression of diheteromers. Here we review these progresses and highlight the unique attributes of triheteromers. Particular emphasis is put on GluN1/GluN2A/GluN2B triheteromers, presumably the most abundant NMDARs in the adult forebrain and critical actors of synaptic plasticity. Better understanding triheteromeric NMDAR structure and function is of major interest for brain physiology and drug discovery.
Keywords: glutamate receptor, NMDA, triheteromeric receptor, synaptic transmission, LTP
Introduction
Glutamate is the major excitatory neurotransmitter in the vertebrate brain. It acts by binding to various transporters and receptors, including ionotropic glutamate receptors (iGluRs) that are responsible for fast neuronal communication at excitatory synapses. The iGluRs comprise three subfamilies, AMPA, kainate and NMDA receptors, that all share some degree of common molecular arrangement and mechanisms [1]. NMDARs stand out from other iGluRs by their capacity to act as coincidence detectors, converting specific patterns of neuronal activity into long-term changes in synaptic strength. NMDARs are also targets of therapeutic interest since their dysregulation is associated with a plethora of neuropathological conditions including ischemia, epilepsy, mental retardation, depression and schizophrenia. However, only very few NMDAR-based therapeutics have reached the clinics, usually because of poor side-effect profile [2–4].
NMDAR form massive molecular complexes that cluster at synaptic and extrasynaptic sites [5]. They are heterotetramers composed of two GluN1 subunits and two GluN2 or GluN3 subunits encoded by six different genes (GluN2A-D, GluN3A-B). This broad molecular heterogeneity translates into a wide variety of receptor subtypes, both diheteromers and triheteromers, each with distinct biophysical, pharmacological and signaling properties [3,6–9]. This is further diversified by their differential location between brain regions, developmental stages, and even subcellular localizations, supporting the idea that each receptor subpopulation is tailored to match the strict requirements of specific neuronal functions. Understanding the physiological relevance of NMDAR diversity on normal and diseased brain function is currently a major challenge. Studies on recombinant NMDARs have been extensively used as a conceptual framework to interpret functional diversity of native NMDARs. These studies, however, almost exclusively described diheteromers, leaving a gap in our knowledge of receptor plurality. With data on triheteromer structure and function becoming available, the landscape is fast evolving.
Widespread expression of triheteromeric NMDARs
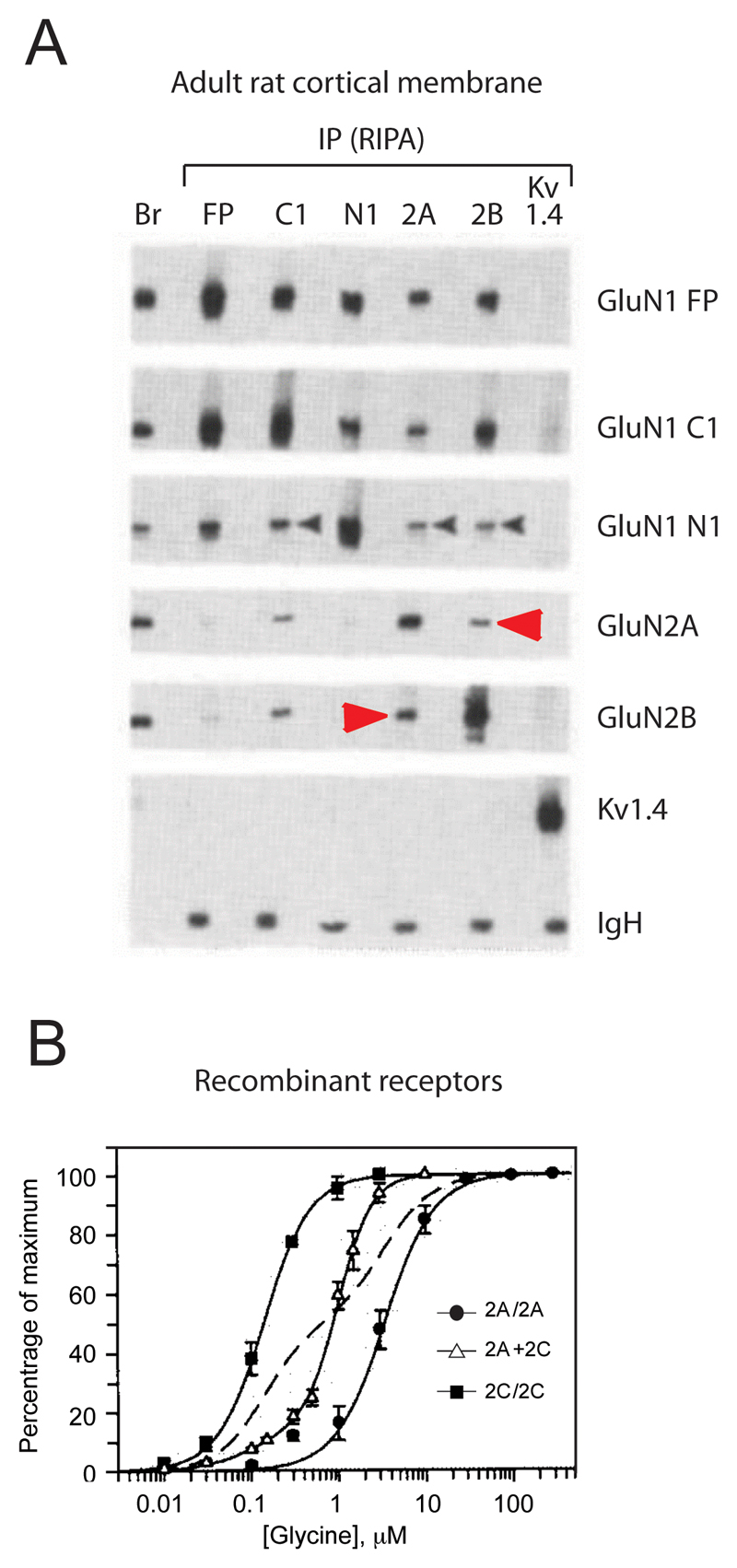
A compelling body of evidence shows that triheteromers are widespread in the CNS and represent a significant proportion, if not the majority, of native NMDARs [3]. The first hints for the presence for triheteromers in the CNS arose soon after the molecular cloning of NMDAR subunits from anatomical studies (i.e. in situ hybridization and immunolabelling) showing overlapping expression of GluN2 subunit in various brain regions, down to the single neuron level [10,11]. Definite proof for the existence of triheteromers in vivo came from biochemical co-immunoprecipitation studies on brain tissues (Figure 1A). Thus, in the rodent forebrain, GluN2A specific antibodies efficiently immunoprecipitate GluN2B, and vice versa, indicating the formation of GluN1/GluN2A/GluN2B ternary complexes [12–14] (see also [15] for the cerebellum). Similarly, triheteromeric GluN1/GluN2A/GluN2D and GluN1/GluN2B/GluN2D complexes populate the rat thalamus and midbrain [16], as well as the human spinal cord [17]. During this period, additional evidence for co-assembly of two different GluN2 subunits into the same receptor complex, including GluN1/GluN2A/GluN2C complexes, originated from studies on recombinant NMDARs expressed in heterologous systems [18–22]. It became evident that following co-expression of GluN1 with several GluN2 subunits, the properties of the resulting receptors could not be explained based on the sole properties of the diheteromers (Figure 1B). Functional studies on isolated neurons or brain slices further supported the existence of multiple co-existing NMDAR populations, both diheteromers and triheteromers [23–40]. Finally, GluN3A and GluN3B subunits can co-assemble with GluN2 subunits to form triheteromeric GluN1/GluN2/GluN3 receptors that are involved in synapse maturation during brain development. These receptors will not be discussed here but have been thoroughly reviewed recently [41].
Figure 1. Early evidence for triheteromeric NMDARs.
A. Biochemical evidence from native preparations. Co-immunoprecipitation experiments of NMDAR subunits from adult (P53) rat cortical membrane. GluN2A antibodies efficiently immunoprecipitate the GluN2B subunit and reciprocally, indicating that GluN2A and GluN2B subunits can assemble within the receptor complex in vivo. B. Functional evidence in a heterologous expression system. Glycine dose-response curves in the presence of 100 µM glutamate on oocytes co-expressing either two (GluN1 + GluN2A or GluN1 + GluN2C) or three (GluN1 + GluN2A + GluN2C) different NMDAR subunits. The dashed line indicates the theoretical fit for a glycine dose-response cure on a mixture of receptor populations consisting of 50% GluN1/GluN2A diheteromers (low glycine sensitivity) and 50% GluN1/GluN2C diheteromers (high glycine sensitivity). The monophasic, rather than biphasic, curve to describe the glycine sensitivity on oocytes co-expressing the three subunits suggests the presence of a single receptor population, GluN1/GluN2A/GluN2C triheteromers, with intermediate glycine sensitivity rather than a mixture of GluN1/GluN2A and GluN1/GluN2C diheteromers. Excerpt from [12] (A) and [18] (B).
The relative abundance of triheteromeric receptors in the CNS compared to their diheteromeric counterparts has been a subject of controversy. Based on immunoblot quantification, estimates of the fraction of GluN2A and GluN2B subunits co-assembling into triheteromers ranged from <10% [13,42], to 20-30% [43] and well above (>50%; [14,44]). Differences in animal species (mouse vs rat), brain region (hippocampus vs forebrain or cortex) and developmental stages may account, in part, for these discrepancies. More recent electrophysiological evidence strongly support a major contribution of GluN1/GluN2A/GluN2B triheteromers in the adult hippocampus, as the predominant NMDAR subtype at CA3-CA1 synapses [33,34,36]. Comparatively much less is known regarding the relative prevalence of other ternary combinations, incorporating GluN2C, GluN2D or GluN3 subunits.
Triheteromers display distinct functional properties
For a long time triheteromeric NMDARs have escaped proper functional characterization because of the difficulty of differentiating them from co-expressed diheteromers. The first isolation of triheteromers in heterologous systems was achieved in 2005 through combined mutagenesis and pharmacology allowing determination of the sensitivity of GluN1/GluN2A/GluN2B and GluN1/GluN2A/GluN2C receptors to the subunit-specific inhibitors zinc and ifenprodil [45]. The approach, however, had obvious limitations because of the introduced mutations in the receptor. Recently, a different approach exploiting the dual retention system of GABA-B receptors to selectively express a homogeneous population of triheteromers at the cell surface (while almost all diheteromers are retained intracellularly) enabled comprehensive evaluation of their functional attributes [46,47]. Using this method, and as detailed below, a range of biophysical and pharmacological properties of GluN1/GluN2A/GluN2B receptors, and to a lesser extent of GluN2C and GluN2D-containing triheteromers, were quantitatively determined. These studies also unveiled emerging principles on the influence of individual GluN2 subunits and domains in the tetrameric receptor complex.
Gating properties
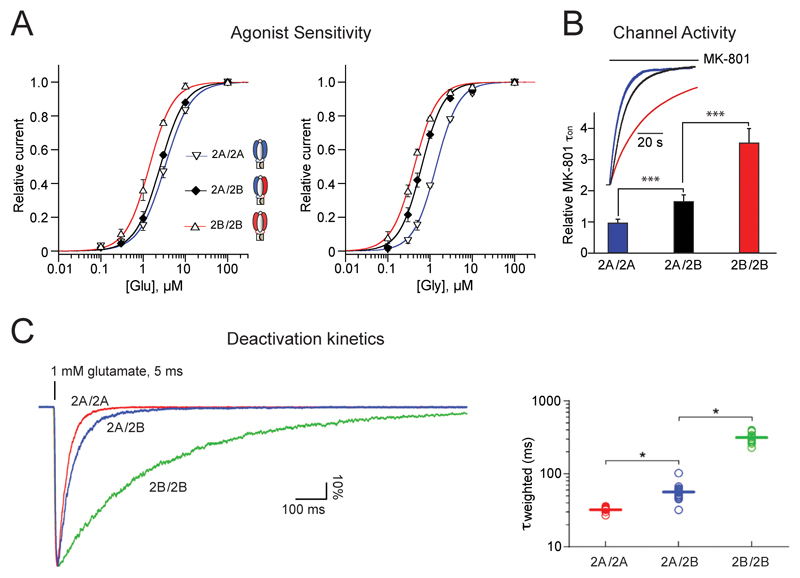
Compared to GluN1/GluN2A and GluN1/GluN2B diheteromers [6], GluN1/GluN2A/GluN2B triheteromers display distinct sensitivity to agonists, deactivation kinetics and channel activity (Figure 2). Differences in these parameters, together with specific coupling to intracellular partners (see below), likely confer unique charge transfer capacities and signaling properties on GluN1/GluN2A/GluN2B triheteromers.
Figure 2. Gating properties of GluN1/GluN2A/GluN2B triheteromers.
For comparison purposes, properties of GluN1/GluN2A/GluN2B triheteromers are displayed together with that of GluN1/GluN2A and GluN1/GluN2B diheteromers. A. Sensitivity to glutamate and glycine. B. Maximal channel open probability as assessed by MK-801 inhibition kinetics. C. Glutamate deactivation kinetics. Mean values of τweighted are 32, 57 and 314 ms for GluN1/GluN2A, GluN1/GluN2A/GluN2B and GluN1/GluN2B receptors, respectively. Excerpt from [47] (A,B) and [46] (C).
The sensitivity of GluN1/GluN2A/GluN2B triheteromers to the agonists glutamate and glycine is intermediate to that of diheteromers (Figure 2A). However, whereas the sensitivity to glycine, conferred by the GluN1 subunits, is close to that of GluN1/GluN2B receptors, the sensitivity to glutamate, conferred by the GluN2 subunits, is closer to that of GluN1/GluN2A receptors [47]. Similarly, the channel maximal open probability as assessed by the MK-801 inhibition kinetics is intermediate between that of the two parent diheteromers, although shifted towards the ‘high Po’ value of GluN1/GluN2A diheteromers [47,48] (Figure 2B). A ‘GluN2A-like’ phenotype is also observed for glutamate deactivation kinetics, a parameter of critical physiological importance that dictates the time window for NMDAR-mediated synaptic integration and plasticity. Thus, GluN1/GluN2A/GluN2B triheteromers deactivate much faster than GluN1/GluN2B diheteromers and nearly as fast as GluN1/GluN2A diheteromers [46–48]) (Figure 2C). Overall, it appears that the GluN2A subunit has a dominant role (over GluN2B) in setting the activation parameters of GluN1/GluN2A/GluN2B receptors. The sensitivity to glycine represents an exception however, the GluN2B ‘imposing’ high glycine affinity to the triheteromer. In GluN1/GluN2A/GluN2D triheteromers, the non-GluN2A subunit (i.e. GluN2D) also seems to dictate glycine sensitivity [22].
Pharmacology
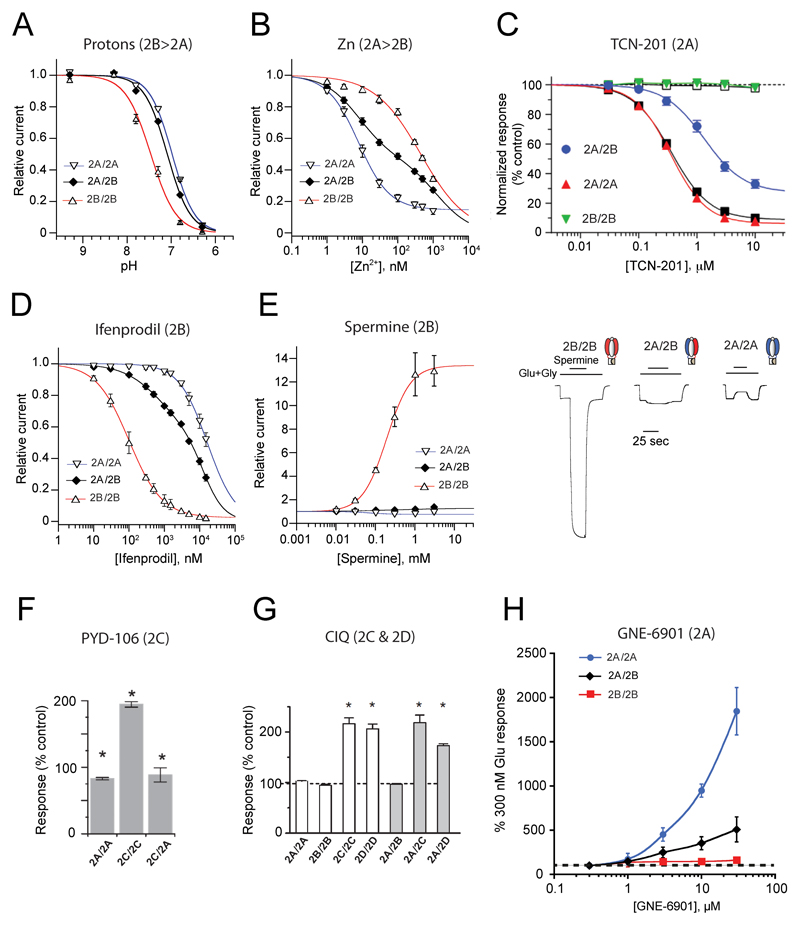
A distinctive feature of NMDARs is their strong allosteric capacity. NMDARs are studded with sites capable of binding small ligands, either exogenous or endogenous, which act as subunit-specific allosteric modulators [49]. Allosteric modulators are widely used as pharmacological tools to discriminate between receptor subtypes and hold great promise as therapeutics [2–4]. Accordingly, determining the sensitivity of triheteromers to allosteric modulators has been long sought after. Thanks to the methodology of subunit stoichiometry control (see above), recent years have witnessed great progress in this direction. The pharmacological properties of GluN1/GluN2A/GluN2B triheteromers have now been almost fully characterized, and application to other tiheteromeric combinations is emerging (Figure 3).
Figure 3. Pharmacological properties of triheteromeric NMDARs.
For comparison purposes, properties of GluN1/GluN2A/GluN2B triheteromers are displayed together with that of GluN1/GluN2A and GluN1/GluN2B diheteromers. Preferential GluN2 subunit targeting is specified for each modulator on top of ea. A. Sensitivity to protons. Values of pHIC50 are 7.0, 7.1 and 7.5, for GluN1/GluN2A, GluN1/GluN2A/GluN2B and GluN1/GluN2B receptors, respectively. B. Sensitivity to zinc. C. Sensitivity to TCN-201. D. Sensitivity to ifenprodil. E. Sensitivity to the polyamine spermine. F. Sensitivity to PYD-206. G. Sensitivity to CIQ. H. Sensitivity to GNE-6901. Excerpt from [47] (A,B, D,E), [46] (C), [64] (F), [65] (G) and [55] (H).
Ifenprodil and derivatives, such as Ro 25-6981 and CP-101,606, form a large family of synthetic allosteric inhibitors highly selective for GluN1/GluN2B over other GluN1/GluN2 diheteromers [2,50]. These compounds bind the N-terminal domain (NTD) region of the receptor and induce almost full inhibition of GluN1/GluN2B receptors through non-competitive antagonism. In contrast, at GluN1/GluN2A/GluN2B triheteromers, inhibition by ifenprodil is partial, with >60% of residual current at saturating inhibitor concentrations (1-10 µM). This decrease in efficacy is accompanied by a moderate decrease in potency (6-fold increase in IC50; [45–47]; Figure 3D). The reduced inhibition of the triheteromers by ifenprodil is also observed with CP-101,606 [46] and is likely shared by the entire family of GluN2B-selective antagonists [51]. Interestingly, GluN1/GluN2A/GluN2B receptors also display kinetics of ifenprodil inhibition that markedly differ from those of GluN1/GluN2B diheteromers, with both slower rates of inhibition and faster off relaxations [45,46]. This dependence of ifenprodil inhibition kinetics on GluN2B subunits copy number may prove useful for pharmacological profiling of NMDAR subunit composition in native systems.
The sensitivity of GluN1/GluN2A/GluN2B triheteromers to the GluN2A-selective antagonist TCN-201, a negative allosteric modulator (NAM) of glycine binding to GluN1 [52], provides another example of intermediate pharmacology, with reduced potency and efficacy [46,53] (Figure 3C). These effects are not as marked with GluN2B-selective antagonists however, highlighting the differential influence of the NTD region (where ifenprodil binds; [54]) and agonist-binding domain (ABD) region (where TCN binds; [55,56]) on triheteromer function (see below). Zinc and protons are two other allosteric inhibitors of NMDARs that have been thoroughly studied on triheteromers. Both modulators are endogenously present in the CNS and act as powerful regulators of NMDAR signaling and brain function [1,57–59]. Consistent with the strong influence of the GluN2A subunit on the channel maximal open probability, the proton sensitivity of GluN1/GluN2A/GluN2B triheteromers is significantly weaker than that of GluN1/GluN2B diheteromers but similar to that of GluN1/GluN2A diheteromers (Figure 3A) [47]. The zinc sensitivity of triheteromers appears somewhat more controversial. Through its binding to GluN2 NTDs, zinc inhibits both GluN1/GluN2A and GluN1/GluN2B diheteromers but with >100-fold higher affinity for the former [60–62]. At GluN1/GluN2A/GluN2B triheteromers, a ‘GluN2A-like’ high-affinity (low nM) component of zinc inhibition is also observed (Figure 3B), yet its relative amplitude compared to that at GluN1/GluN2A diheteromers varies substantially between studies (from similar [46], to weaker as for ifenprodil [45,47]).. The reason for this discrepancy remains unclear.
Several subunit-selective positive allosteric potentiators (PAMs) have been recently tested on triheteromers. Strikingly, the strong and specific potentiation of GluN1/GluN2B receptors by spermine is almost completely absent in GluN1/GluN2A/GluN2B receptors (Figure 3E), despite the expected presence of one remaining NTD polyamine-binding site [63]. Similarly, the GluN2C-specific pyrrolidinone PAM PYD-106 loses its effect at GluN1/GluN2A/GluN2C triheteromers [64] (Figure 3F), establishing the strict requirement for two copies of the GluN2C subunit. The situation differs for the GluN2C- and GluN2D-selective tetrahydroisoquinoline PAM CIQ, which potentiates GluN1/GluN2A/GluN2C and GluN1/GluN2A/GluN2D receptors to a similar extent than GluN1/GluN2C and Glu1N/GluN2D diheteromers [65] (Figure 3G). Finally, the recently identified GluN2A-PAM GNE-6901 produces an intermediate enhancement of GluN1/GluN2A/GluN2B responses when compared to GluN1/GluN2A diheteromers [55] (Figure 3H).
Several conclusions can be drawn by integrating all the available data on the functional properties of triheteromers, regarding their activation and pharmacology. First, triheteromers behave differently than diheteromers, forming receptors with unique properties. Second, it is unclear whether a simple general rule predicting triheteromer function based on the properties of the corresponding diheteromers exists. For triheteromeric GluN1/GluN2A/Glu2B receptors, it appears that, overall, the GluN2A subunit imposes its functional ‘signature’. Thus, sensitivity to glutamate, to subunit-specific allosteric modulators, channel maximal open probability, deactivation kinetics are all dominated by GluN2A (unlike glycine sensitivity, however). Third,pharmacological agents affecting diheteromers also affect triheteromers. Especially, no triheteromer-selective antagonist has been identified yet. Fourth, for most GluN2 subunit-selective allosteric modulators, two binding sites are ‘better’ than one. In other terms, subunit-specific PAMs or NAMs usually display stronger modulatory effect on diheteromers than triheteromers, only the former permitting two simultaneous binding events of the modulator. Interestingly, efficacy of the modulator rather than its potency appears to be primarily affected when comparing diheteromers and triheteromers. A single allosteric site is usually sufficient to maintain high potency but efficacy drops. This likely stems from the characteristic dimer-of-dimer arrangement of NMDARs, in which ligand binding occurs at the level of the individual GluN1/GluN2 dimer but transduction to the downstream gating machinery requires coordinated action of the two constitutive dimers.
Structure of GluN1/GluN2A/GluN2B triheteromers
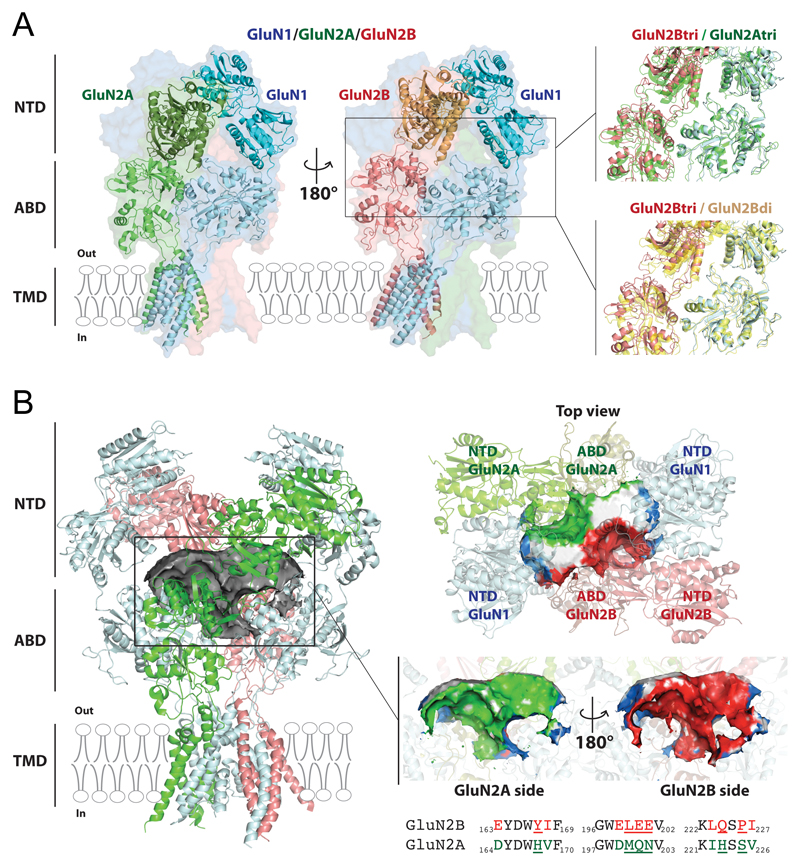
Recent years have witnessed major progress in our understanding of the structure of NMDARs, with several atomic structures of full-length GluN1/GluN2B receptors captured in different conformational states and complexed to various ligands [66–69]. These efforts culminated with the resolution of the first structure of an intact triheteromeric receptor, the CNS-abundant GluN1/GluN2A/GluN2B subtype, solved at 4.5 Å resolution using cryo-EM single particle reconstruction [70] (Figure 4). This newly solved structure provides an exceptional framework to correlate structure and function of triheteromeric NMDARs. It also opens interesting perspectives for the development of selective triheteromer pharmacology.
Figure 4. Atomic structure of GluN1/GluN2A/GluN2B triheteromers.
A. Cryo-EM structure of the GluN1/GluN2A/GluN2B triheteromer trapped in an inactive conformation [70]. Left: Two views of the triheteromer structure highlighting the GluN1/GluN2A dimer (B/A positions in green/cyan) and the pseudo-symmetric GluN1/GluN2B dimer (D/C positions in salmon/cyan). Right, top: Structural superimposition of the GluN1/GluN2B dimer (salmon/cyan) with the GluN1/GluN2A dimer (green) both extracted from the triheteromer (tri) structure. Right, bottom: Structural superimposition of the GluN1/GluN2B dimer (salmon/cyan) from the triheteromer structure with the GluN1/GluN2B (yellow) from an inactive GluN1/GluN2B diheteromer (di) structure (5fxi.pdb [69]). Note the looser packing between the GluN2B NTD and ABD region in the triheteromer. B. A large cavity at the core of the NMDAR extracellular region. Each subunit is colored as in panel A. Left: A tortuous cavity or pocket of 20 nm3 (grey surface; build using POVME [79]) lies in sandwich between the NTD and ABD layers in the triheteromer structure. Right, top: View of the triheteromer pocket from the top. The central part of the pocket is formed by the two GluN2 subunits. Right, middle: View of the pocket from towards the GluN2A site and towards the from GluN2B side. The pocket envelop is colored according to the contribution of the different subunits (blue for GluN1, green for GluN2A, red for GluN2B). The white surfaces correspond to solvent accessible entrances to the pocket. Right, bottom: Sequence alignment between NTD loops participating to the cavity of rat GluN2A and GluN2B subunits. Non-conserved residues are colored and those directly lining the cavity underlined. Because of subunit-specific contribution, the central cavity of triheteromers differs from that of corresponding diheteromers.
Molecular architecture and asymmetry
The GluN1/GluN2A/GluN2B triheteromer structure shows the typical hot-air balloon shape of NMDARs, with the transmembrane pore region forming the basket and the large extracellular domain or ectodomain forming the envelope (Figure 4A). The triheteromer retains the dimer-of-dimer assembly of the GluN1/GluN2B diheteromer, with one GluN1-GluN2A dimer and one GluN1-GluN2B dimer, and the typical alternating GluN1/GluN2/GluN1/GluN2 subunit arrangement around the central pore [66,67,71,72]. In the upper ectodomain, the two GluN1 subunits occupy a peripheral position (A/C positions) while the two GluN2 subunits are closer to the central axis where they contact each other (B/D position). The triheteromer also shows the layered organization previously observed in all iGluRs [73]: with the (ABDs sandwiched between the NTDs and the transmembrane domain (TMD). The triheteromer characterized is complexed to the agonists glycine and glutamate, yet its pore is closed, meaning that it represents an inactive (or desensitized) state. This is consistent with the presence of high concentrations of protons (purification at pH 6.5), powerful allosteric inhibitors of NMDARs (see above).
The main and most striking feature of the triheteromer structure is the asymmetry between the constitutive pairs (GluN1-GluN2A and GluN1-GluN2B; pdb 5UOW [70]) (Figure 4A). This departs from the structures of the GluN1/GluN2B diheteromer which shows an approximate two-fold symmetry in similar conditions (pdb 5IOU [68]; 5FXH and 5FXI [69]). Differences between the diheteromer and triheteromer spread throughout the receptor but are most salient in the extracellular domains.. Most strikingly, the GluN2A NTD tightly packs against the ABD layer, making multiple interactions with both GluN2A and GluN1 ABDs, while the GluN2B NTD detaches from the ABD layer by a solvent-filled gap absent in diheteromers (Figure 4A).
Importance of the NTD region
In AMPA and kainate receptors, direct interactions between the NTDs and ABDs are minimal, such that ABD and NTD motions appear uncoupled [73]. In contrast, in NMDARs, the ectodomain is much more compact, the NTDs literally sitting on the ABDs [66,67]. This tight coupling of the NTD and ABD layers is likely related to the well-established role of NMDAR NTDs in allosteric signaling and modulation of the downstream channel gating machinery. Thus, the pore-distal GluN2 NTDs control key gating properties, such as glutamate deactivation kinetics and channel maximal open probability [48,74,75]. NMDAR NTDs also harbor binding sites for small ligands acting as subunit-specific allosteric modulators that exert their effects through differential stabilization of NTD conformers [63,68–70,74].. In the triheteromer, the GluN1-GluN2A NTD dimer packing against the ABD layer is similar to that observed for the GluN1-GluN2B NTD dimer in GluN1/GluN2B diheteromers. On the contrary, the GluN1-GluN2B NTD dimer is less packed than in any known structures of GluN1/GluN2B diheteromers (Figure 4A). The tighter packing of the GluN1-GluN2A NTD dimer, as well as the more extensive interactions of GluN2A in the ABD layer, provide a structural basis for the dominant role of GluN2A in the triheteromer gating (see above and Figure 3). It is also consistent with the ‘biased’ pharmacology of GluN1/GluN2A/GluN2B triheteromers that display strongly attenuated sensitivity to GluN2B-specific modulators while retaining substantial modulation by GluN2A-specific agents (see Figure 3). Direct evidence that a single GluN2A NTD is sufficient to impose a GluN2A-like phenotype was recently obtained using chimeric subunits [48]. The presence of the GluN2A subunit in the triheteromer would relax the interaction of the GluN2B NTD with the rest of the receptor, allowing GluN2A NTD dominance [48,70].
Triheteromers in synaptic function and plasticity
Since triheteromeric NMDARs have distinct functional properties, their presence in neural tissue has been actively searched for. However, the lack of specific pharmacological tools has hampered their unequivocal identification. Often, the presence of triheteromers is inferred from properties that cannot be accounted by a simple mixture of diheteromers. This applies for partial effects of GluN2 subunit-selective antagonists, intermediate deactivation kinetics or intermediate level of Mg2+ block. Although such default criteria are prone to ambiguity, a rich body of information at several CNS synapses supports the existence of triheteromeric NMDARs in vivo. Evidence points to the existence of several subtypes of native triheteromers, including GluN1/GluN2A/GluN2B, GluN1/GluN2A/GluN2C and GluN1/GluN2B/GluN2D combinations.
One of the first indications for functional synaptic triheteromers was obtained in a study describing the developmental maturation of NMDAR-mediated excitatory postsynaptic currents (NMDA-EPSCs) at the mossy fiber to granule cell synapse in the cerebellum [25]. Granule cells in young animals express almost exclusively GluN1/GluN2B diheteromers. During maturation, GluN2B expression fades and is progressively replaced by GluN2A, then GluN2C, both strongly expressed in the adult [10,11]. During the first three postnatal weeks, NMDA-EPSC decay kinetics accelerate and ifenprodil sensitivity decreases, consistent with the replacement of GluN2B by GluN2A subunits. A slowing of decay kinetics is not seen before P40, likely due to the gradual incorporation of the ‘slow deactivating’ GluN2C subunit [6]. Sensitivity of NMDA-EPSCs to Mg2+ also evolves during maturation, yet the typical ‘low Mg2+ block’ conferred by GluN2C [6] precedes the slowing in decay kinetics. A possible explanation would be that GluN2C subunits are first incorporated into GluN1/GluN2A/GluN2C triheteromers, while slow-deactivating and low-Mg2+ sensitivity GluN1/GluN2C diheteromers appear later. This scenario assumes that GluN1/GluN2A/GluN2C receptors display fast deactivating kinetics (GluN2A signature) and low (or intermediate) Mg2+ block (GluN2C signature) (note that similar properties would also apply to GluN1/GluN2B/GluN2D receptors, see [76]). That the GluN2A subunit imposes its fast deactivation on GluN1/GluN2A/GluN2C receptors matches what has been observed with GluN1/GluN2A/GluN2B receptors [46,47]. In addition to GluN1/GluN2A/GluN2C triheteromers, GluN1/GluN2B/GluN2D triheteromers have also been described in cerebellar slices, more specifically in Golgi cells. These receptors, however, appear excluded from synaptic sites and populate extrasynaptic locations to control cellular excitability [27]. Thus, the presence of triheteromers in a given cell type does not guarantee their contribution to synaptic currents. Differential subcellular targeting of NMDAR subtypes likely enhances their signaling diversity and control of neuronal function.
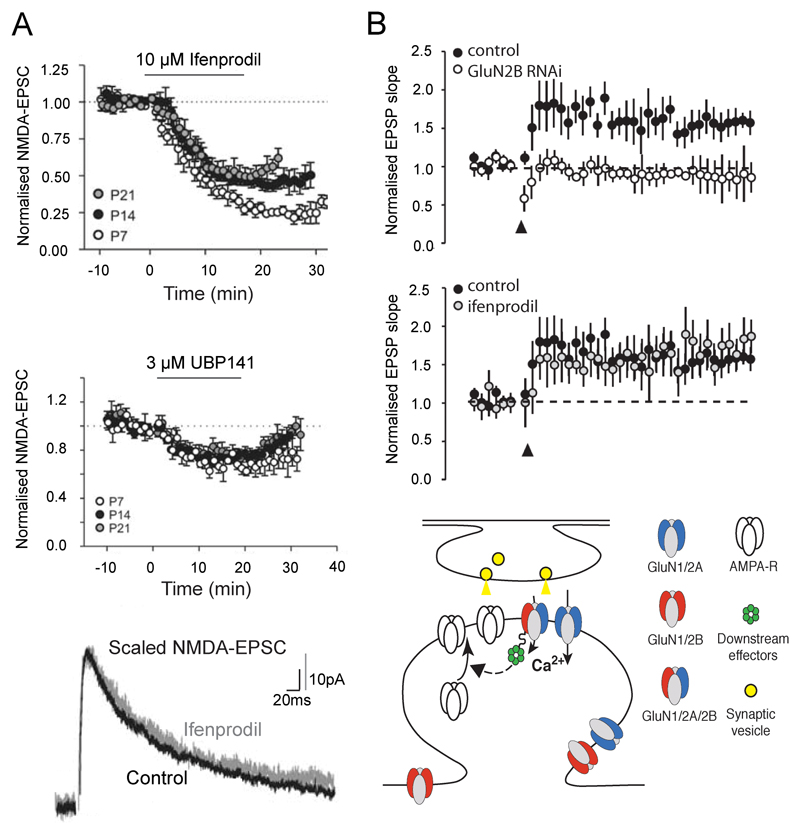
Substantia nigra [31] and hippocampus [37,39,40] are brain regions where GluN2D-containing triheteromers have been suggested to participate in synaptic transmission. During postnatal development, synaptic inputs onto rat substantia nigra pars compacta (SNc) dopaminergic neurons display characteristics compatible with a significant fraction of NMDARs being GluN1/GluN2B/GluN2D triheteromers [31]. Indeed, synaptic currents are partially inhibited by the GluN2B- and GluN2D-selective antagonist ifenprodil and UBP141. However, ifenprodil application does not alter NMDA-EPSC decay kinetics (Figure 5A), as expected if GluN1/GluN2D diheteromers, which display excessively slow deactivation kinetics [6], were predominant. The prevalence of GluN1/GluN2B/GluN2D triheteromers in SNc dopaminergic neurons increases with age [31]. In the hippocampus, CA1 interneurons may also bear GluN2D-containing triheteromers (of the GluN1/GluN2A/GluN2D and/or GluN1/GluN2B/GluN2D subtypes [39]). NMDA-EPSCs in these cells are potentiated by CIQ, a GluN2C- and GluN2D-selective PAM [65], yet NMDA-EPSC decay kinetics are neither affected by CIQ application nor GluN2D subunit deletion (GluN2D knockout). Again, the most parsimonious explanation of these data is the presence of postsynaptic GluN2D-containing triheteromers.
Figure 5. Triheteromeric NMDARs in synaptic transmission and plasticity.
A. Synaptic inputs onto substantia nigra pars compacta dopaminergic neurons activate NMDARs that display characteristics that are best explained by the presence of GluN1/GluN2B/GluN2D triheteromers. Top and middle panels: Inhibition of NMDA-EPSCs by ifenprodil, a GluN2B-selective antagonist, and by UBP141, a GluN2D-preferring antagonist. Sensitivity to both antagonists indicates the co-existence of GluN2B and GluN2D subunits. Bottom panel: Ifenprodil application does not alter NMDA-EPSC decay kinetics, indicating that it acts on a (relatively) homogeneous population of receptors (likely GluN1/GluN2B/GluN2D triheteromers) rather than a mix of GluN1/GluN2B and GluN1/GluN2D diheteromers. B. Activation of triheteromeric GluN1/GluN2A/GluN2B NMDARs is necessary for LTP induction in basolateral amygdala principal neurons. Top and middle panels: Knocking-down the GluN2B subunit impairs LTP induction, while acute application of the GluN2B-selective antagonist ifenprodil does not. Bottom panel: Schematic representation of an adult glutamatergic synapse. Triheteromeric GluN1/GluN2A/GluN2B receptors cluster at postsynaptic sites and anchored to downstream effectors necessary for LTP induction through the C-terminus of the GluN2B subunit. Excerpt from [31] (A) and [35] (B).
Numerous evidence exists for native triheteromeric GluN1/GluN2A/GluN2B receptors, in cultured neurons [23,24,30,32,36] or brain slices [28,33–35,37,38,40,77]. The consensus is that this receptor subtype is widespread in the adult forebrain, where it represents a major, if not the most abundant, synaptic NMDAR population. GluN1/GluN2A/GluN2B triheteromers are present at various synapses, including corticostriatal synapses [28], synapses on principal neurons of the basolateral amygdala [35] and on hippocampal CA1 pyramidal cells [33,34,37,38,40,77]. Do GluN1/GluN2A/GluN2B receptors play specific roles in synapse function? A discrete, and essential, task that these triheteromers have been proposed to play is triggering long-term potentiation (LTP), a form of synaptic plasticity that underlies memory formation and storage. Studying synaptic plasticity, investigators have faced an apparent paradox when comparing molecular pharmacology and gene silencing (RNA interference) approaches [35,77] (Figure 5B): no impairment of LTP by GluN2B-selective antagonists on one hand, suppression of LTP after GluN2B subunit removal on the other. Interestingly, after GluN2B silencing, LTP is rescued following overexpression of the GluN2B subunit or a chimeric GluN2A subunit with GluN2B C-terminal cytoplasmic tail, but not using wild-type GluN2A [77]. The ‘hybrid’ nature of GluN1/GluN2A/GluN2B triheteromers together with their unique properties provides an explanation (Figure 5B). LTP induction is insensitive to GluN2B-selective antagonists because it relies on GluN1/GluN2A/GluN2B triheteromers that are poorly sensitive to these antagonists (in contrast to GluN1/GluN2B diheteromers; see Figure 3D). In contrast, LTP requires expression of the GluN2B subunit because its C-terminus tail recruits molecules necessary for LTP induction, such as CamKII [8]. GluN2B tail also controls intracellular trafficking of GluN1/GluN2A/GluN2B receptors through preferential recycling to the cell surface [32]. Combined with their fast ‘GluN2A-like’ deactivation kinetics (see Figure 2C), GluN1/GluN2A/GluN2B triheteromers may thus offer a unique mix of precise coincidence detection and specific anchoring to essential LTP effectors within a single receptor complex.
Conclusions and perspectives
Through a combination of pharmacological, physiological and structural approaches, major advances in our understanding of triheteromeric NMDAR structure and function have been made recently. The intricate workings of these molecular machines as well as their impact on neuronal function are progressively revealed. Many questions remain open however. For instance, it is still unclear if, in vivo, triheteromers are preferentially assembled (compared to diheteromers). And what are the mechanisms targeting triheteromers to specific subcellular compartments? Another key challenge concerns pharmacology. The combinatorial diversity and widespread occurrence of NMDARs require highly selective and precise tools for investigating NMDAR-mediated signaling in native tissue. This is particularly true for triheteromers, for which no selective pharmacology exists. Designing pharmacological agents able to distinguish between diheteromers and triheteromers is not an easy task however. At the structural level, triheteromers are fundamentally arranged as two distinct halves, each found in the two related GluN1/GluN2 diheteromers. Any compound that binds within one GluN1-GluN2 dimer or the other is thus unlikely to be fully specific for triheteromers. Interfaces involving the two constitutive GluN2 subunits are the only target regions to offer possibilities for heterophilic interactions unique to triheteromers. According to the GluN1/GluN2A/GluN2B triheteromer structure [70], regions where the GluN2A and GluN2B subunits come in close proximity are scarce. A cavity at the center of the ectodomain provides interesting opportunities however. It is lined, in its upper part, by the lower lobes of the two GluN2 NTDs, and in its lower part, by the upper lobes of the two GluN2 ABDs (Figure 4B). Interestingly, the homologous region in AMPARs is a site of allosteric modulation, binding the polypeptide con-ikot-ikot toxin [78]. We speculate that targeting this cavity in NMDARs, although smaller in size due to the NTD-ABD compaction, may provide a path forward for selective pharmacology and offer novel tool compounds to explore triheteromer physiology. Given the critical role of triheteromers in brain function (e.g. GluN1/GluN2A/GluN2B receptors in LTP), drugs specifically targeting these receptors also hold promise in neuroprotection and cognitive restoration.
Highlights.
Triheteromers are NMDA receptors containing three distinct subunits in the tetramer
Triheteromers are abundantly expressed in the developing and adult brain
Triheteromers display unique atomic structure and functional properties
Triheteromers are essential mediators of synapse maturation and plasticity
Strategies to selectively target triheteromers remain to be developed
Acknowledgements
Research of authors is supported by the French government under the program ‘Investissements d’Avenir’ (ANR-10-LABX-54 MEMO LIFE and ANR-11-IDEX-0001-02 PSL* Research University), and the European Research Council (ERC Advanced Grant #693021 to PP).
References and recommended reading
- 1.Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mony L, Kew JN, Gunthorpe MJ, Paoletti P. Allosteric modulators of NR2B-containing NMDA receptors: molecular mechanisms and therapeutic potential. Br J Pharmacol. 2009;157:1301–1317. doi: 10.1111/j.1476-5381.2009.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- 4.Hackos DH, Hanson JE. Diverse modes of NMDA receptor positive allosteric modulation: Mechanisms and consequences. Neuropharmacology. 2017;112:34–45. doi: 10.1016/j.neuropharm.2016.07.037. [DOI] [PubMed] [Google Scholar]
- 5.Frank RA, Komiyama NH, Ryan TJ, Zhu F, O'Dell TJ, Grant SG. NMDA receptors are selectively partitioned into complexes and supercomplexes during synapse maturation. Nat Commun. 2016;7:11264. doi: 10.1038/ncomms11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paoletti P. Molecular basis of NMDA receptor functional diversity. Eur J Neurosci. 2011;33:1351–1365. doi: 10.1111/j.1460-9568.2011.07628.x. [DOI] [PubMed] [Google Scholar]
- 7.Wyllie DJ, Livesey MR, Hardingham GE. Influence of GluN2 subunit identity on NMDA receptor function. Neuropharmacology. 2013;74:4–17. doi: 10.1016/j.neuropharm.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanz-Clemente A, Nicoll RA, Roche KW. Diversity in NMDA receptor composition: many regulators, many consequences. Neuroscientist. 2013;19:62–75. doi: 10.1177/1073858411435129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glasgow NG, Siegler Retchless B, Johnson JW. Molecular bases of NMDA receptor subtype-dependent properties. J Physiol. 2015;593:83–95. doi: 10.1113/jphysiol.2014.273763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akazawa C, Shigemoto R, Bessho Y, Nakanishi S, Mizuno N. Differential expression of five N-methyl-D-aspartate receptor subunit mRNAs in the cerebellum of developing and adult rats. J Comp Neurol. 1994;347:150–160. doi: 10.1002/cne.903470112. [DOI] [PubMed] [Google Scholar]
- 11.Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 12.Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature. 1994;368:144–147. doi: 10.1038/368144a0. [• A classic paper providing the first evidence for triheteromeric NMDARs in the mammalian brain.] [DOI] [PubMed] [Google Scholar]
- 13.Chazot PL, Stephenson FA. Molecular dissection of native mammalian forebrain NMDA receptors containing the NR1 C2 exon: direct demonstration of NMDA receptors comprising NR1, NR2A, and NR2B subunits within the same complex. J Neurochem. 1997;69:2138–2144. doi: 10.1046/j.1471-4159.1997.69052138.x. [DOI] [PubMed] [Google Scholar]
- 14.Luo J, Wang Y, Yasuda RP, Dunah AW, Wolfe BB. The majority of N-methyl-D-aspartate receptor complexes in adult rat cerebral cortex contain at least three different subunits (NR1/NR2A/NR2B) Mol Pharmacol. 1997;51:79–86. doi: 10.1124/mol.51.1.79. [DOI] [PubMed] [Google Scholar]
- 15.Didier M, Xu M, Berman SA, Bursztajn S. Differential expression and co-assembly of NMDA zeta 1 and epsilon subunits in the mouse cerebellum during postnatal development. Neuroreport. 1995;6:2255–2259. doi: 10.1097/00001756-199511000-00036. [DOI] [PubMed] [Google Scholar]
- 16.Dunah AW, Luo J, Wang YH, Yasuda RP, Wolfe BB. Subunit composition of N-methyl-D-aspartate receptors in the central nervous system that contain the NR2D subunit. Mol Pharmacol. 1998;53:429–437. doi: 10.1124/mol.53.3.429. [DOI] [PubMed] [Google Scholar]
- 17.Sundstrom E, Whittemore S, Mo LL, Seiger A. Analysis of NMDA receptors in the human spinal cord. Exp Neurol. 1997;148:407–413. doi: 10.1006/exnr.1997.6691. [DOI] [PubMed] [Google Scholar]
- 18.Wafford KA, Bain CJ, Le Bourdelles B, Whiting PJ, Kemp JA. Preferential co-assembly of recombinant NMDA receptors composed of three different subunits. Neuroreport. 1993;4:1347–1349. doi: 10.1097/00001756-199309150-00015. [DOI] [PubMed] [Google Scholar]
- 19.Chazot PL, Coleman SK, Cik M, Stephenson FA. Molecular characterization of N-methyl-D-aspartate receptors expressed in mammalian cells yields evidence for the coexistence of three subunit types within a discrete receptor molecule. J Biol Chem. 1994;269:24403–24409. [PubMed] [Google Scholar]
- 20.Brimecombe JC, Boeckman FA, Aizenman E. Functional consequences of NR2 subunit composition in single recombinant N-methyl-D-aspartate receptors. Proc Natl Acad Sci U S A. 1997;94:11019–11024. doi: 10.1073/pnas.94.20.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawkins LM, Chazot PL, Stephenson FA. Biochemical evidence for the co-association of three N-methyl-D-aspartate (NMDA) R2 subunits in recombinant NMDA receptors. J Biol Chem. 1999;274:27211–27218. doi: 10.1074/jbc.274.38.27211. [DOI] [PubMed] [Google Scholar]
- 22.Cheffings CM, Colquhoun D. Single channel analysis of a novel NMDA channel from Xenopus oocytes expressing recombinant NR1a, NR2A and NR2D subunits. J Physiol. 2000;526(Pt 3):481–491. [PubMed] [Google Scholar]
- 23.Kew JN, Richards JG, Mutel V, Kemp JA. Developmental changes in NMDA receptor glycine affinity and ifenprodil sensitivity reveal three distinct populations of NMDA receptors in individual rat cortical neurons. J Neurosci. 1998;18:1935–1943. doi: 10.1523/JNEUROSCI.18-06-01935.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tovar KR, Westbrook GL. The incorporation of NMDA receptors with a distinct subunit composition at nascent hippocampal synapses in vitro. J Neurosci. 1999;19:4180–4188. doi: 10.1523/JNEUROSCI.19-10-04180.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cathala L, Misra C, Cull-Candy S. Developmental profile of the changing properties of NMDA receptors at cerebellar mossy fiber-granule cell synapses. J Neurosci. 2000;20:5899–5905. doi: 10.1523/JNEUROSCI.20-16-05899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pina-Crespo JC, Gibb AJ. Subtypes of NMDA receptors in new-born rat hippocampal granule cells. J Physiol. 2002;541:41–64. doi: 10.1113/jphysiol.2001.014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brickley SG, Misra C, Mok MH, Mishina M, Cull-Candy SG. NR2B and NR2D subunits coassemble in cerebellar Golgi cells to form a distinct NMDA receptor subtype restricted to extrasynaptic sites. J Neurosci. 2003;23:4958–4966. doi: 10.1523/JNEUROSCI.23-12-04958.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, Murphy TH, Hayden MR, Raymond LA. Enhanced striatal NR2B-containing N-methyl-D-aspartate receptor-mediated synaptic currents in a mouse model of Huntington disease. J Neurophysiol. 2004;92:2738–2746. doi: 10.1152/jn.00308.2004. [DOI] [PubMed] [Google Scholar]
- 29.Jones S, Gibb AJ. Functional NR2B- and NR2D-containing NMDA receptor channels in rat substantia nigra dopaminergic neurones. J Physiol. 2005;569:209–221. doi: 10.1113/jphysiol.2005.095554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu C, Fu Z, Karavanov I, Yasuda RP, Wolfe BB, Buonanno A, Vicini S. NMDA receptor subtypes at autaptic synapses of cerebellar granule neurons. J Neurophysiol. 2006;96:2282–2294. doi: 10.1152/jn.00078.2006. [DOI] [PubMed] [Google Scholar]
- 31.Brothwell SL, Barber JL, Monaghan DT, Jane DE, Gibb AJ, Jones S. NR2B- and NR2D-containing synaptic NMDA receptors in developing rat substantia nigra pars compacta dopaminergic neurones. J Physiol. 2008;586:739–750. doi: 10.1113/jphysiol.2007.144618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang TT, Badger JD, 2nd, Roche PA, Roche KW. Novel approach to probe subunit-specific contributions to N-methyl-D-aspartate (NMDA) receptor trafficking reveals a dominant role for NR2B in receptor recycling. J Biol Chem. 2010;285:20975–20981. doi: 10.1074/jbc.M110.102210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rauner C, Kohr G. Triheteromeric NR1/NR2A/NR2B receptors constitute the major N-methyl-D-aspartate receptor population in adult hippocampal synapses. J Biol Chem. 2011;286:7558–7566. doi: 10.1074/jbc.M110.182600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray JA, Shi Y, Usui H, During MJ, Sakimura K, Nicoll RA. Distinct modes of AMPA receptor suppression at developing synapses by GluN2A and GluN2B: single-cell NMDA receptor subunit deletion in vivo. Neuron. 2011;71:1085–1101. doi: 10.1016/j.neuron.2011.08.007. [• Using single GluN2 subunit genetic deletion in mouse hippocampal slices, this study shows that a large proportion of synaptic NMDARs in CA1 pyramidal cells are GluN1/GluN2A/GluN2B triheteromers.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delaney AJ, Sedlak PL, Autuori E, Power JM, Sah P. Synaptic NMDA receptors in basolateral amygdala principal neurons are triheteromeric proteins: physiological role of GluN2B subunits. J Neurophysiol. 2013;109:1391–1402. doi: 10.1152/jn.00176.2012. [• In this study combining electrophysiology, genetics and pharmacology, the authors reveal that triheteromeric GluN1/GluN2A/GluN2B receptors are essential for triggering LTP at amygdalar synapses. The GluN2A subunit would have an essential ionotropic role by controlling the receptor kinetics while the GluN2B subunit would serve as a structural scaffold for important LTP effectors. See also ref. [77] for a related study.] [DOI] [PubMed] [Google Scholar]
- 36.Tovar KR, McGinley MJ, Westbrook GL. Triheteromeric NMDA receptors at hippocampal synapses. J Neurosci. 2013;33:9150–9160. doi: 10.1523/JNEUROSCI.0829-13.2013. [• A thorough study showing that triheteromeric GluN1/GluN2A/GluN2B receptors are preferentially assembled (or preferentially localized) at mature hippocampal synapses.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volianskis A, Bannister N, Collett VJ, Irvine MW, Monaghan DT, Fitzjohn SM, Jensen MS, Jane DE, Collingridge GL. Different NMDA receptor subtypes mediate induction of long-term potentiation and two forms of short-term potentiation at CA1 synapses in rat hippocampus in vitro. J Physiol. 2013;591:955–972. doi: 10.1113/jphysiol.2012.247296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izumi Y, Zorumski CF. Sensitivity of N-methyl-D-aspartate receptor-mediated excitatory postsynaptic potentials and synaptic plasticity to TCN 201 and TCN 213 in rat hippocampal slices. J Pharmacol Exp Ther. 2015;352:267–273. doi: 10.1124/jpet.114.220582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perszyk RE, DiRaddo JO, Strong KL, Low CM, Ogden KK, Khatri A, Vargish GA, Pelkey KA, Tricoire L, Liotta DC, et al. GluN2D-Containing N-methyl-d-Aspartate Receptors Mediate Synaptic Transmission in Hippocampal Interneurons and Regulate Interneuron Activity. Mol Pharmacol. 2016;90:689–702. doi: 10.1124/mol.116.105130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.France G, Fernandez-Fernandez D, Burnell ES, Irvine MW, Monaghan DT, Jane DE, Bortolotto ZA, Collingridge GL, Volianskis A. Multiple roles of GluN2B-containing NMDA receptors in synaptic plasticity in juvenile hippocampus. Neuropharmacology. 2017;112:76–83. doi: 10.1016/j.neuropharm.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez-Otano I, Larsen RS, Wesseling JF. Emerging roles of GluN3-containing NMDA receptors in the CNS. Nat Rev Neurosci. 2016;17:623–635. doi: 10.1038/nrn.2016.92. [• A comprehensive review on the 'non-conventional' GluN3-containing NMDARs. GluN1/GluN2/GluN3A triheteromeric receptors appear to have important roles in brain development by influencing the time course of synapse maturation.] [DOI] [PubMed] [Google Scholar]
- 42.Blahos J, 2nd, Wenthold RJ. Relationship between N-methyl-D-aspartate receptor NR1 splice variants and NR2 subunits. J Biol Chem. 1996;271:15669–15674. doi: 10.1074/jbc.271.26.15669. [DOI] [PubMed] [Google Scholar]
- 43.Al-Hallaq RA, Conrads TP, Veenstra TD, Wenthold RJ. NMDA di-heteromeric receptor populations and associated proteins in rat hippocampus. J Neurosci. 2007;27:8334–8343. doi: 10.1523/JNEUROSCI.2155-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunah AW, Standaert DG. Subcellular segregation of distinct heteromeric NMDA glutamate receptors in the striatum. J Neurochem. 2003;85:935–943. doi: 10.1046/j.1471-4159.2003.01744.x. [DOI] [PubMed] [Google Scholar]
- 45.Hatton CJ, Paoletti P. Modulation of triheteromeric NMDA receptors by N-terminal domain ligands. Neuron. 2005;46:261–274. doi: 10.1016/j.neuron.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 46.Hansen KB, Ogden KK, Yuan H, Traynelis SF. Distinct functional and pharmacological properties of Triheteromeric GluN1/GluN2A/GluN2B NMDA receptors. Neuron. 2014;81:1084–1096. doi: 10.1016/j.neuron.2014.01.035. [• This paper, together with ref. 47, presents an original strategy to heterologously express in heterologous systems 'pure' triheteromeric NMDARs enabling detailed analysis of their biophysical and pharmacological properties.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stroebel D, Carvalho S, Grand T, Zhu S, Paoletti P. Controlling NMDA receptor subunit composition using ectopic retention signals. J Neurosci. 2014;34:16630–16636. doi: 10.1523/JNEUROSCI.2736-14.2014. [• See annotation [46].] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun W, Hansen KB, Jahr CE. Allosteric Interactions between NMDA Receptor Subunits Shape the Developmental Shift in Channel Properties. Neuron. 2017;94:58–64 e53. doi: 10.1016/j.neuron.2017.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu S, Paoletti P. Allosteric modulators of NMDA receptors: multiple sites and mechanisms. Curr Opin Pharmacol. 2015;20:14–23. doi: 10.1016/j.coph.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 50.Williams K. Ifenprodil discriminates subtypes of the N-methyl-D-aspartate receptor: selectivity and mechanisms at recombinant heteromeric receptors. Mol Pharmacol. 1993;44:851–859. [PubMed] [Google Scholar]
- 51.Stroebel D, Buhl DL, Knafels JD, Chanda PK, Green M, Sciabola S, Mony L, Paoletti P, Pandit J. A Novel Binding Mode Reveals Two Distinct Classes of NMDA Receptor GluN2B-selective Antagonists. Mol Pharmacol. 2016;89:541–551. doi: 10.1124/mol.115.103036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hansen KB, Ogden KK, Traynelis SF. Subunit-selective allosteric inhibition of glycine binding to NMDA receptors. J Neurosci. 2012;32:6197–6208. doi: 10.1523/JNEUROSCI.5757-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheriyan J, Balsara RD, Hansen KB, Castellino FJ. Pharmacology of triheteromeric N-Methyl-D-Aspartate Receptors. Neurosci Lett. 2016;617:240–246. doi: 10.1016/j.neulet.2016.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karakas E, Simorowski N, Furukawa H. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature. 2011;475:249–253. doi: 10.1038/nature10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hackos DH, Lupardus PJ, Grand T, Chen Y, Wang TM, Reynen P, Gustafson A, Wallweber HJ, Volgraf M, Sellers BD, et al. Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function. Neuron. 2016;89:983–999. doi: 10.1016/j.neuron.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 56.Yi F, Mou TC, Dorsett KN, Volkmann RA, Menniti FS, Sprang SR, Hansen KB. Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors. Neuron. 2016;91:1316–1329. doi: 10.1016/j.neuron.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nozaki C, Vergnano AM, Filliol D, Ouagazzal AM, Le Goff A, Carvalho S, Reiss D, Gaveriaux-Ruff C, Neyton J, Paoletti P, et al. Zinc alleviates pain through high-affinity binding to the NMDA receptor NR2A subunit. Nat Neurosci. 2011;14:1017–1022. doi: 10.1038/nn.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vergnano AM, Rebola N, Savtchenko LP, Pinheiro PS, Casado M, Kieffer BL, Rusakov DA, Mulle C, Paoletti P. Zinc dynamics and action at excitatory synapses. Neuron. 2014;82:1101–1114. doi: 10.1016/j.neuron.2014.04.034. [DOI] [PubMed] [Google Scholar]
- 59.Anderson CT, Radford RJ, Zastrow ML, Zhang DY, Apfel UP, Lippard SJ, Tzounopoulos T. Modulation of extrasynaptic NMDA receptors by synaptic and tonic zinc. Proc Natl Acad Sci U S A. 2015;112:E2705–2714. doi: 10.1073/pnas.1503348112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paoletti P, Ascher P, Neyton J. High-affinity zinc inhibition of NMDA NR1-NR2A receptors. J Neurosci. 1997;17:5711–5725. doi: 10.1523/JNEUROSCI.17-15-05711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karakas E, Simorowski N, Furukawa H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J. 2009;28:3910–3920. doi: 10.1038/emboj.2009.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Romero-Hernandez A, Simorowski N, Karakas E, Furukawa H. Molecular Basis for Subtype Specificity and High-Affinity Zinc Inhibition in the GluN1-GluN2A NMDA Receptor Amino-Terminal Domain. Neuron. 2016;92:1324–1336. doi: 10.1016/j.neuron.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mony L, Zhu S, Carvalho S, Paoletti P. Molecular basis of positive allosteric modulation of GluN2B NMDA receptors by polyamines. EMBO J. 2011;30:3134–3146. doi: 10.1038/emboj.2011.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khatri A, Burger PB, Swanger SA, Hansen KB, Zimmerman S, Karakas E, Liotta DC, Furukawa H, Snyder JP, Traynelis SF. Structural determinants and mechanism of action of a GluN2C-selective NMDA receptor positive allosteric modulator. Mol Pharmacol. 2014;86:548–560. doi: 10.1124/mol.114.094516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mullasseril P, Hansen KB, Vance KM, Ogden KK, Yuan H, Kurtkaya NL, Santangelo R, Orr AG, Le P, Vellano KM, et al. A subunit-selective potentiator of NR2C- and NR2D-containing NMDA receptors. Nat Commun. 2010;1:90. doi: 10.1038/ncomms1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karakas E, Furukawa H. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science. 2014;344:992–997. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee CH, Lu W, Michel JC, Goehring A, Du J, Song X, Gouaux E. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 2014;511:191–197. doi: 10.1038/nature13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu S, Stein RA, Yoshioka C, Lee CH, Goehring A, McHaourab HS, Gouaux E. Mechanism of NMDA Receptor Inhibition and Activation. Cell. 2016;165:704–714. doi: 10.1016/j.cell.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tajima N, Karakas E, Grant T, Simorowski N, Diaz-Avalos R, Grigorieff N, Furukawa H. Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature. 2016;534:63–68. doi: 10.1038/nature17679. [• In this paper, combining X-ray crystallography and single-particle cryo-EM, the authors reveal critical aspects of NMDAR structure and mechanisms.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu W, Du J, Goehring A, Gouaux E. Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation. Science. 2017;355 doi: 10.1126/science.aal3729. [• Using cyro-EM, this important paper reports the first atomic map of a triheteromeric NMDAR (GluN1/GluN2A/GluN2B). It shows how incorporation of two different GluN2 subunits results into structural asymmetry between the two constitutive GluN1/GluN2 dimers, allowing each subunit to uniquely influence receptor structure and function.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salussolia CL, Prodromou ML, Borker P, Wollmuth LP. Arrangement of subunits in functional NMDA receptors. J Neurosci. 2011;31:11295–11304. doi: 10.1523/JNEUROSCI.5612-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Riou M, Stroebel D, Edwardson JM, Paoletti P. An alternating GluN1-2-1-2 subunit arrangement in mature NMDA receptors. PLoS One. 2012;7:e35134. doi: 10.1371/journal.pone.0035134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459:703–707. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci. 2009;29:12045–12058. doi: 10.1523/JNEUROSCI.1365-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang Z, Gibb AJ. Mg2+ block properties of triheteromeric GluN1-GluN2B-GluN2D NMDA receptors on neonatal rat substantia nigra pars compacta dopaminergic neurones. J Physiol. 2014;592:2059–2078. doi: 10.1113/jphysiol.2013.267864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Foster KA, McLaughlin N, Edbauer D, Phillips M, Bolton A, Constantine-Paton M, Sheng M. Distinct roles of NR2A and NR2B cytoplasmic tails in long-term potentiation. J Neurosci. 2010;30:2676–2685. doi: 10.1523/JNEUROSCI.4022-09.2010. [• This study reveals a critical role for triheteromeric GluN1/GluN2A/GluN2B receptors in LTP induction. In the triheteromeric complex, the GluN2B subunit cytoplasmic tail has an essential role by recruiting signaling molecules necessary for synaptic plasticity. See also ref. 35 for a related study.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen L, Durr KL, Gouaux E. X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism. Science. 2014;345:1021–1026. doi: 10.1126/science.1258409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Durrant JD, de Oliveira CA, McCammon JA. POVME: an algorithm for measuring binding-pocket volumes. J Mol Graph Model. 2011;29:773–776. doi: 10.1016/j.jmgm.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]