Abstract
It is emerging that the pathways which process newly transcribed RNA molecules also regulate the response to DNA damage at multiple levels. Here, we discuss recent insights into how RNA processing pathways participate in DNA damage recognition, signalling and repair, selectively influence the expression of genome-stabilizing proteins, and resolve deleterious DNA/RNA hybrids (R-loops) formed during transcription and RNA processing. The importance of these pathways for the DNA damage response (DDR) is underscored by the growing appreciation that defects in these regulatory connections may be connected to the genome instability involved in several human diseases, including cancer.
Introduction
After their transcription from genomic DNA, RNA molecules that encode cellular proteins, or perform non-coding cellular functions, are extensively processed into their mature functional forms by complex cellular pathways. For instance, protein-coding mRNA undergoes a series of interconnected processing steps. In human cells, these steps are predominantly co-transcriptional and include the addition of a 5’ methylguanosine cap on nascent transcripts, splicing to remove intronic sequences, addition of a 3’ poly(A) tail, and the export of spliced, polyadenylated transcripts from the nucleus into the cytoplasm (Bentley, 2014; Muller-McNicoll and Neugebauer, 2013; Wickramasinghe and Laskey, 2015). It is emerging that RNA processing pathways such as these play an unexpectedly intimate role in the cellular response to DNA damage (or DNA damage response, DDR). In this review, we will discuss how RNA processing pathways participate in DNA damage recognition, signalling and repair, selectively influence the expression of genome-stabilizing proteins, and resolve deleterious DNA/RNA hybrids (R-loops) formed during transcription and RNA processing.
Transcription and RNA processing - an integral part of the DDR
It has long been appreciated that the transcriptional machinery itself periodically surveys the integrity of transcribed genomic DNA, and triggers the repair of DNA lesions that engage or impede it, as exemplified in the intimate connections between transcription and nucleotide excision repair (Friedberg, 1996; Hanawalt and Spivak, 2008). Conversely, genome- and proteome-wide screens to identify factors involved in the DDR have shown that proteins involved in RNA processing are modified by enzymes of the DDR such as ATM, ATR and PARP as well as the MAPKAP kinase-2, MK2 (e.g Adamson et al., 2012; Beli et al., 2012; Blasius et al., 2014; Matsuoka et al., 2007; Reinhardt et al., 2010). These studies have led to an understanding that proteins involved in transcription and RNA processing are part of the first response to DNA damage, and conversely, DNA damage responses are part of the feedback that regulates transcription and RNA processing. The sections that follow explain this concept in more detail.
Selective control by RNA processing factors of DDR gene expression against background changes in transcription and translation
One effect of DNA damage is to transiently repress transcription (Mayne and Lehmann, 1982), polyadenylation of pre-mRNA (Kleiman and Manley, 2001) and translation (Deng et al., 2002). This is generally assumed to occur to limit the collisions between the repair and transcription machineries. Global alterations of this kind may be accompanied by site-specific changes around the DNA lesions. For example, at nucleotide lesions induced by UV light, elongating RNA polymerase II stalls and is locally poly-ubiquitylated for degradation by the ubiquitin ligase Nedd4 and the Elongin complex (Harreman et al., 2009), thus facilitating repair. The tumour suppressor BRCA1 and its interacting partner BARD1 have been implicated in RNA pol II ubiquitylation after UV exposure (Kleiman et al., 2005), but this remains controversial (Anindya et al., 2007).
Varying global effects of DNA damage on RNA transcription and processing are also lesion-dependent (Fry et al., 2005; Rieger and Chu, 2004; Workman et al., 2006), and exhibit dose dependencies (Albrecht et al., 2012). Thus, the transcriptional response to various nucleotide lesions caused by UV light and ionizing radiation differ (Rieger and Chu, 2004). The dose-dependent nature of the response to damage raises the question of whether global changes in transcription after high-dose damage arise in part simply from the summation of multiple local changes. Arguing against this, while the majority of transcription is repressed after high-dose DNA damage, the transcription of genes required for the DDR itself is increased (Rieger and Chu, 2004), and p53 is likely to be a key player in this event (Riley et al., 2008). For instance, following IR, alterations in the genomic binding of transcription factors p53 and NF-kB is observed, triggered by ATM kinase (Rashi-Elkeles et al., 2014).
Indeed, against the background of global changes in gene expression, many RNA processing factors selectively regulate the expression of DDR genes both positively and negatively (Figure 1). The selective nature of these regulatory influences should be emphasized: they affect a relatively small pool of genes, highly enriched for factors involved in DNA replication, repair or the cell cycle. Regulation may be exerted either directly or indirectly at each step of the gene expression pathway (Table 1). For instance, DDR gene expression is regulated during transcription elongation (Blazek et al., 2011), mRNA splicing (Paronetto et al., 2011; Savage et al., 2014), mRNA export (Wickramasinghe et al., 2013), miRNA regulation (Moskwa et al., 2011), or translation (Powley et al., 2009). Although the mechanisms responsible for these regulatory effects and their apparent selectivity remain to be fully defined, the available information suggests that certain steps depend on recognition by RNA binding proteins (RBPs) of conserved RNA sequence motifs shared between mRNAs encoding genome maintenance factors that may have a common secondary structure (Blackinton and Keene, 2014; Keene, 2007; Mazan-Mamczarz et al., 2011; Wickramasinghe et al., 2013). Not all of these mechanisms are induced after DNA damage. Some may also be constitutive. For example, transcription of DDR genes such as ATR, BRCA1 or FANCD2 is regulated through the formation of a specific cyclin/CDK complex that promotes transcript elongation (Blazek et al., 2011). CyclinK/Cdk12 protects cells from genomic instability by promoting the phosphorylation of Ser2 in the C-terminal domain of RNA polymerase II on a subset of long transcripts enriched for those that function in the DDR and its depletion results in increased DNA damage and sensitivity to DNA damaging agents (Blazek et al., 2011). Another constitutive mechanism is linked to second messengers involved in growth signaling such as phosphatidylinositol (3,4,5)-trisphosphate (PIP3), which along with its catalytic enzyme inositol polyphosphate multi-kinase (IPMK), regulates mRNA export (Wickramasinghe et al., 2013).
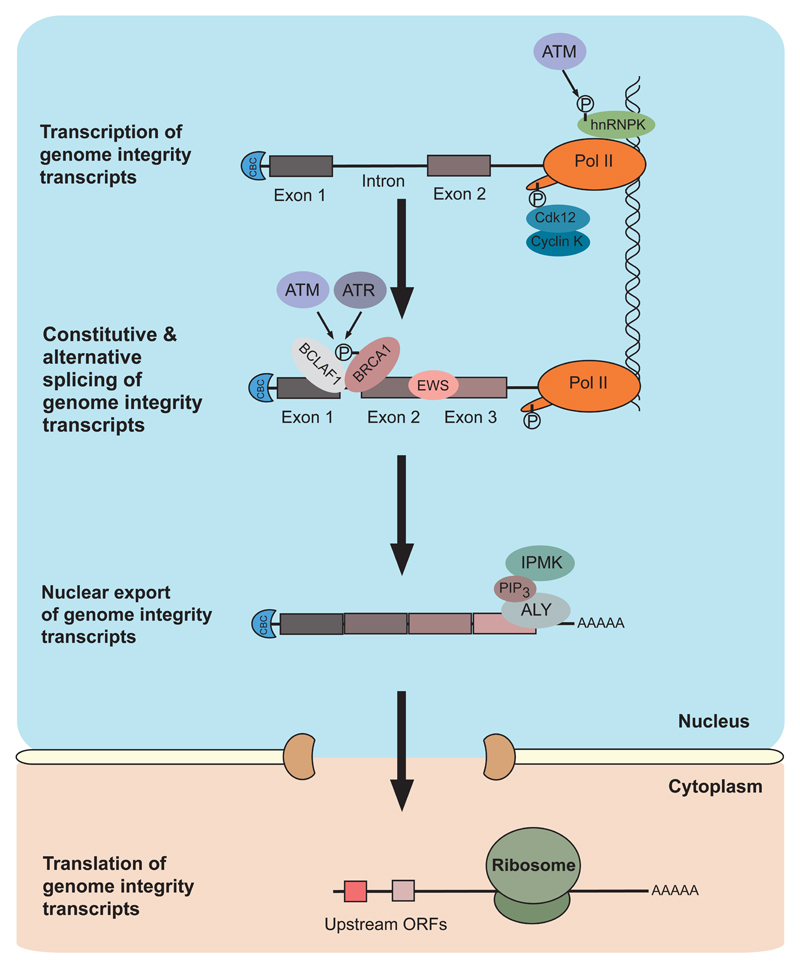
Figure 1. Expression of genome integrity transcripts.
This figure represents the gene expression pathway for transcripts that encode various genome maintenance factors, and the proteins that contribute to their proper transcription, splicing and export. Specific examples are indicated here or in the text, and referenced in Table 1. ATM-mediated phosphorylation of hnRNPK promotes transcriptional activation of p53. CyclinK/Cdk2 promotes transcriptional elongation of genome integrity transcripts. Constitutive and alternative splicing of genome integrity transcripts is regulated by the BRCA1/BCLAF1 splicing complex and the Ewing sarcoma protein EWS. Nuclear export of genome integrity transcripts is promoted by inositol polyphosphate multi-kinase (IPMK), its catalytic product PIP3, and TREX mRNA export complex component ALY. Different upstream open reading frames (ORFs), may be used by the translation machinery to ensure efficient translation of genome integrity proteins following DNA damage.
Table 1. RNA processing factors involved in maintaining genome integrity.
| Protein Name/s | Role in Gene Expression Pathway | Function in RNA processing | Function in DNA damage response | References |
|---|---|---|---|---|
| Selective control by RNA processing factors of DDR gene expression at different levels | ||||
| Cyclin K | Transcription elongation | Cyclin subunit of CDK12 that associates with RNA pol II | Promotes transcription of DNA repair genes (BRCA1, ATR, FANCI and FANCD2) | (Blazek et al., 2011) |
| EWS | Transcription and mRNA splicing | RNA and DNA binding protein which interacts with RNA pol II and splicing factors | Functions in alternative splicing of DNA repair genes (CHEK2); its association with target transcripts is reduced after DNA damage | (Paronetto et al., 2011) |
| BCLAF1 | mRNA splicing | Factor that interacts with core spliceosome components | Interacts with phosphorylated BRCA1 to promote splicing of DNA repair transcripts after DNA damage (ATRIP, EXO1, BACH1) | (Savage et al., 2014) |
| IPMK | mRNA export | Inositol and PI3-kinase that promotes binding of RNA export factor ALY to target transcripts | Functions constitutively in the nuclear export of DNA repair transcripts (RAD51, CHK1, FANCD2) | (Wickramasinghe et al., 2013) |
| Active roles for RNA processing factors in the DDR | ||||
| TAF15 | Transcription and mRNA splicing | hnRNP that is part of TFIID transcription factor complex and can also regulate alternative splicing | Binds to damaged chromatin in a PARP-dependent manner | (Izhar et al., 2015) |
| THRAP3 | Transcription and mRNA splicing | Factor that interacts with core spliceosome components | Phosphorylated in response to DNA damage and excluded from DNA damage sites | (Beli et al., 2012) |
| hnRNPU | mRNA splicing | hnRNP implicated in transcription elongation, alternative splicing and RNA stability | Free form accumulates at sites of DNA damage, chromatin-bound form is excluded from sites of DNA damage | (Britton et al., 2014) |
| RBMX | mRNA splicing | hnRNP that regulates splice site selection | Accumulates at sites of DNA damage and may regulate expression/splicing of BRCA2 | (Adamson et al., 2012) |
| PRP19 | mRNA splicing | Forms PRP19 complex that associates with and remodels spliceosome Complex B | Promotes ubiquitylation of RPA following DNA damage and facilitates accumulation of ATRIP at sites of DNA damage | (Marechal et al., 2014) |
| hnRNPU L1 and 2 | mRNA splicing | hnRNP like proteins | Recruited to DNA double strand breaks and stimulates DNA-end resection through CtIP binding and BLM recruitment | (Polo et al., 2012) |
| RNA processing factors involved in R-loop prevention and resolution | ||||
| Senataxin | Transcription termination | Functions in pause-dependent RNA pol II transcription termination and recruits exonuclease Xrn2 | Resolves R-loops at termination elements, releasing RNA for degradation by Xrn2 prior to termination | (Skourti-Stathaki et al., 2011) |
| ASF/SF2 | mRNA splicing | SR splicing factor | Suppresses R-loop formation in a Top1-dependent manner | (Li and Manley, 2005) |
| PCID2 | mRNA export | Component of TREX-2 mRNA export complex | May interact with BRCA2 to suppress R-loop formation | (Bhatia et al., 2014) |
| THOC1 | mRNA export | Component of TREX mRNA export complex | Along with other TREX components, suppresses R-loop formation | (Dominguez-Sanchez et al., 2011) |
Finally, we note in passing that DNA damage also triggers a global decrease in protein synthesis, accompanied by preferential translation of a subset of mRNAs dependent on DNA-PK activity (Powley et al., 2009). This subset is enriched for genes required during the DDR, such as ERCC1, ERCC5 and DDB1 (Powley et al., 2009). These forms of selective enhancement of gene expression at the level of transcription, mRNA processing or translation may exist to ensure that a sufficient pool of DNA repair proteins is maintained against the background of global decreases in gene expression after DNA damage.
Altered RNA splicing
RNA splicing patterns are also altered following DNA damage. For example, co-transcriptional exon skipping occurs in response to various forms of genotoxic stress (Dutertre et al., 2010). The formation of a damage-induced mRNA splicing complex is promoted by BRCA1 phosphorylation-dependent binding to the mRNA splicing factor, BCLAF1 (Savage et al., 2014). This complex facilitates splicing of a number of transcripts encoding genome maintenance factors such as ATRIP, EXO1 and BACH1 (Savage et al., 2014). Another example involves the Ewing sarcoma protein (EWS), which functions constitutively in the alternative splicing of a subset of transcripts encoding genome maintenance factors (Paronetto et al., 2011). Interestingly, DNA damage and EWS depletion cause similar changes in alternative splicing, due to reduced interaction of EWS with its splicing targets such as CHK2, following damage (Paronetto et al., 2011). EWS also controls skipping of several exons of the MDM2 gene, which may in turn contribute to p53 regulation and the transcriptional regulation of DDR factors (Dutertre et al., 2010). Importantly, depletion of either BCLAF1 or EWS results in sensitivity to DNA damage. BCLAF1 depleted cells are also defective for DNA repair and have genomic instability, thus demonstrating the powerful effect that modulating RNA splicing activity can have on the DDR. Depletion of some RNA processing factors such as hnRNPC and RBMX have also been reported to reduce the expression of key regulators of homologous recombination (HR) such as BRCA2, presumably due to aberrant mRNA splicing (Adamson et al., 2012; Anantha et al., 2013). Thus, selective enhancement of the expression of DDR factors through altered splicing may facilitate the recovery of cells from damage-induced cellular stress.
The maintenance of genome stability also relies on proper splicing of proteins involved in mitotic progression and chromosome segregation. Depletion of a number of core spliceosome components results in mitotic catastrophe (Sundaramoorthy et al., 2014; van der Lelij et al., 2014), owing to the altered splicing of essential mitotic transcripts, which in the case of the core splicing factor PRPF8 has recently been linked to 5’ splice site strength (Wickramasinghe et al., 2015).
Altered RNA transport
RNA processing factors may also function constitutively to sustain the synthesis or nuclear export of transcripts encoding genome maintenance factors. For example, the export of RAD51, CHK1 and FANCD2 mRNAs is selectively regulated by inositol polyphosphate multi-kinase (IPMK), its catalytic product, phosphatidylinositol (3,4,5)-trisphosphate (PIP3), and ALY, a component of the TREX mRNA export complex (Wickramasinghe et al., 2013). IPMK depleted cells are sensitive to various genotoxic lesions that result in DSBs and accumulate structural chromosomal aberrations such as chromatid breaks and radial structures, typical of defective DNA repair by HR (Wickramasinghe et al., 2013). Transcripts affected by this mechanism appear to be enriched for a degenerate sequence motif responsible for regulation. This again underscores the question of whether common regulatory motifs are shared amongst RNA species that are constitutively maintained, or coordinately regulated after DNA damage, by RNA processing mechanisms.
DDR proteins - an integral part of transcription and RNA processing
A number of DNA repair factors can also moonlight as transcription factors themselves. For example, FANCD2 can activate transcription of the tumour suppressor gene TAp53 (Park et al., 2013), and XPC can activate transcription of hormone-inducible genes such as RARβ2 (Le May et al., 2010). In certain contexts, transcription can be activated by DNA damage itself. Transcription of estrogen receptor responsive genes is facilitated by DNA topoisomerase IIβ-dependent, DSB formation (Ju et al., 2006). This results in recruitment of PARP, which may induce a permissive chromatin architecture for transcription initiation (Ju et al., 2006). Collectively, these considerations suggest that DNA damage, transcription and repair are functionally and physically intertwined (Fong et al., 2013).
Connecting damage sensing to signaling to RNA processing
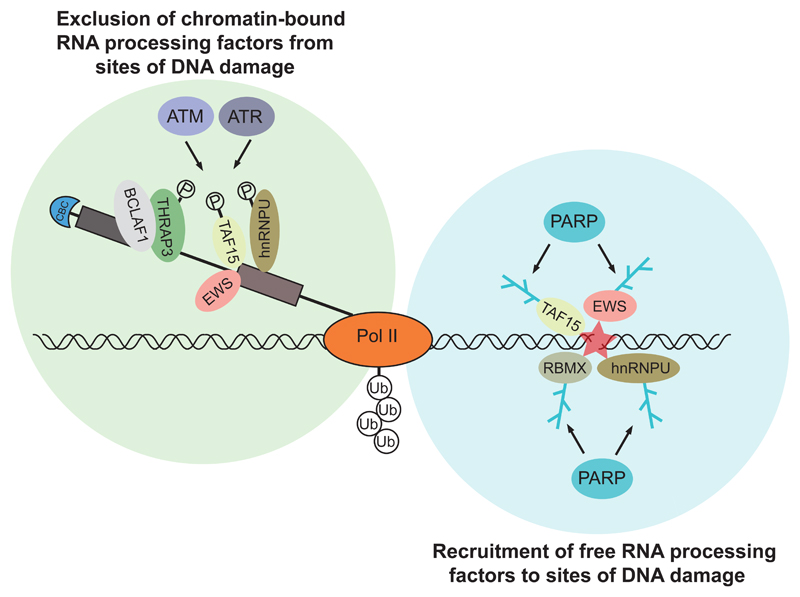
How the DNA damage signal is transduced to RNA processing is not yet clear. RNA processing factors themselves may directly participate in protein complexes that assemble at sites of DNA damage (Figures 2 and 3) (Adamson et al., 2012; Marechal et al., 2014; Polo et al., 2012; Wang et al., 2013), as well as in downstream steps that affect the DNA damage response (Decorsiere et al., 2011; Moumen et al., 2005; Tresini et al., 2015). Interestingly, some RNA processing factors such as TAF15 and hnRNPU display a bimodal dynamic in their localization following DNA damage: an initial recruitment to damage sites, which is often PARP dependent, followed by exclusion (Adamson et al., 2012; Britton et al., 2014; Izhar et al., 2015; Polo et al., 2012). Their exclusion seems to predominantly depend on the activity of phosphatidylinositol 3-kinase-related kinases (PIKKs) ATM, ATR and DNA-PK and whether they are actively involved in RNA processing at sites of DNA damage (Beli et al., 2012) (Figure 2). It is interesting to speculate that recruitment may reflect a direct role of these factors in the DDR, whereas exclusion may be a consequence of the general repression of transcription and processing after DNA damage.
Figure 2. Recruitment and exclusion of RNA processing factors at sites of DNA damage.
The left panel represents exclusion of chromatin-bound RNA processing factors actively participating in RNA processing at or near sites of DNA damage, which is dependent on ATM and ATR activity. The right panel represents recruitment of free RNA processing factors to sites of DNA damage, which is dependent on PARP activity.
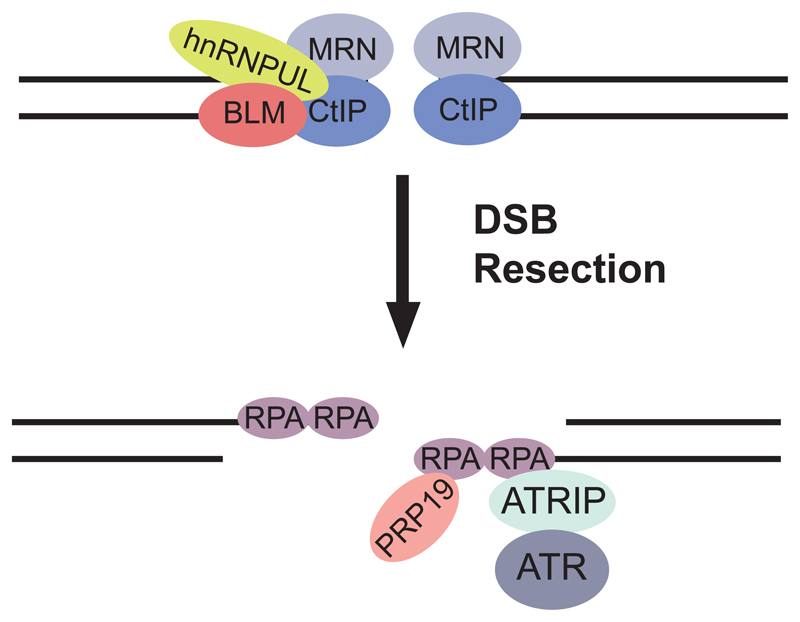
Figure 3. Direct roles for RNA processing factors in the response to DNA damage.
hnRNPUL1 and 2 proteins are recruited to DNA double strand breaks (DSBs) following binding to the MRN (Mre11-Rad50-Nbs1) complex and CtIP and contribute to DSB resection by stimulating recruitment of the BLM helicase implicated in Bloom’s syndrome. Following resection, RPA is recruited to (ss)DNA, where PRP19 can interact directly with RPA and single stranded (ss)DNA, promoting ubiquitylation of RPA and recruitment of ATRIP, the regulatory partner of the ATR kinase, as well as promoting ATR activation.
Conversely, genotoxic stress may also directly influence the activity of the RNA splicing and processing machineries. UV induced pausing of RNA pol II at DNA lesions triggers chromatin displacement of late-stage spliceosomes and initiates a positive feedback loop that activates ATM (Tresini et al., 2015). Spliceosome remodelling also activates ATM signalling, and ATM modulates the DDR by influencing pre-mRNA processing, forming a feedback loop. Interestingly, ATM activation alone (via IR) is not sufficient to influence spliceosome mobility (Tresini et al., 2015). However, transcriptional inhibition and IR have a combinatorial effect on spliceosome mobility and intron retention, suggesting that ATM amplifies a mobilization signal imposed by transcriptional arrest (Tresini et al., 2015). Importantly, other studies have shown that ATM mediates transcriptional silencing in cis to DSBs (Shanbhag et al., 2010) and, as mentioned above, exclusion of RNA processing factors from sites of DNA damage (Beli et al., 2012), suggesting that ATM signalling may be activated in this context in response to multiple forms of DNA damage, not just those that physically block the transcription machinery. It is noteworthy that DNA repair factors identified in mass spectrometric studies of RNA processing complexes were previously considered to be contaminants (Shi et al., 2009); increasing evidence now suggests direct functional connections.
PRPF19 is an ubiquitin ligase that forms a complex that remodels the U4/U6.U5 tri-snRNP complex during spliceosome activation (Figure 3). PRPF19 can interact directly with RPA and single stranded ssDNA at sites of DNA damage, promoting ubiquitylation of RPA and recruitment of ATRIP, the regulatory partner of the ATR kinase, as well as promoting ATR activation (Marechal et al., 2014). As RPA is preferentially localized to transcribed genes in response to DNA damage (Jiang and Sancar, 2006), there is the possibility that PRPF19 may act as a sensor of genomic instability during transcription.
Similarly, distinct from their well-characterized role in processing heterogeneous nuclear (hn)RNA into mature mRNA, a number of hnRNPs also function in various aspects of the DDR. hnRNPU-like proteins are recruited to DSBs following binding to the MRN (Mre11-Rad50-Nbs1) complex and CtIP and contribute to DSB resection by stimulating recruitment of the BLM helicase implicated in Bloom’s syndrome (Polo et al., 2012) (Figure 3). hnRNP FUS participates in both HR and NHEJ repair and is rapidly recruited to sites of DNA damage in an ATM-dependent manner in neurons (Gardiner et al., 2008; Wang et al., 2013). hnRNP RBMX accumulates at sites of DNA damage in a PARP-dependent manner and may regulate the expression of BRCA2 (Adamson et al., 2012). Apart from their direct role in the DDR, hnRNPs can also participate in selective transcription, 3’-end processing and translation of DNA damage response genes. For example, ATM dependent phosphorylation of hnRNPK promotes transcriptional activation of p53 in response to DNA damage (Moumen et al., 2013; Moumen et al., 2005). Furthermore, hnRNP H/F promotes 3’-end processing of p53 pre-mRNA in DNA damaged cells through its interaction with a G-quadruplex RNA structure at the p53 polyA signal (Decorsiere et al., 2011).
RNA processing factors can also relocalise from the nucleus to the cytoplasm in response to DNA damage, to regulate translation of specific genes required for the DDR. Thus, translation of p53 mRNA is promoted by the cytoplasmic relocalisation of RNA binding protein HuR (Mazan-Mamczarz et al., 2003) and polypyrimidine tract binding protein PTB (Grover et al., 2008), whereas hnRNPA1 limits translation of pro-apoptotic Apaf-1 mRNA (Cammas et al., 2007). Translation of GADD45α mRNA in response to DNA damage is also regulated by the cytoplasmic relocalisation of the p38/MK2 complex, where MK2 phosphorylates hnRNPA0, to stabilize Gadd45α mRNA, promoting its translation (Reinhardt et al., 2010). In contrast, re-localisation of spliceosome associated factor YB-1 from the cytoplasm to the nucleus in response to DNA damage is expected to impact translation of specific mRNAs (Cohen et al., 2010; Dutertre et al., 2014; Sorokin et al., 2005).
RNA processing mechanisms, R-loops and genome instability
We have thus far discussed the role of RNA processing mechanisms in regulating the expression of DDR genes both constitutively and after DNA damage. In this section, we examine a different role – how RNA processing mechanisms regulate the formation of nucleic acid intermediates, like DNA/RNA hybrid R-loops, which also affect genome integrity. Work in yeast and mammals revealed that RNA packaging is pivotal to prevent formation of R-loops during transcription, which may contribute to chromosome rearrangements/loss, recombination and mutation (Huertas and Aguilera, 2003; Li and Manley, 2005) (Figure 4). After the RNA is transcribed by RNA pol II, it has the opportunity to anneal to the transcribed strand, forming a stable DNA-RNA hybrid, thus leaving the non-transcribed strand as an exposed single strand of DNA that is vulnerable to DNA damage (reviewed in Aguilera and Garcia-Muse, 2012; Kim and Jinks-Robertson, 2012; Skourti-Stathaki and Proudfoot, 2014). This three-stranded nucleic acid structure is known as an R-loop, which may be deleterious to cells, and RNA processing mechanisms have evolved to address this problem both through local and distant activities. For example, the RNase H family of enzymes can specifically degrade the RNA component of the RNA/DNA hybrids, generating a 3’ end that can be extended by the DNA replication machinery (Itoh and Tomizawa, 1980). It is unclear whether the physiological role of RNase H is to degrade aberrantly forming or intentionally formed R-loops. RNA/DNA hybrids at G-rich sequences that pause transcription can be unwound by RNA-DNA helicases such as Senataxin (Sen1 in yeast) (Kim et al., 1999; Mischo et al., 2011), releasing RNA for degradation by the Xrn2 exonuclease prior to termination (Skourti-Stathaki et al., 2011). R-loop formation at highly transcribed regions can also be prevented by Topoisomerase 1 (Top1) (El Hage et al., 2010; Tuduri et al., 2009), which relaxes co-transcriptionally generated negative supercoiling of DNA behind elongating RNA polymerases (Pommier, 2006). If unresolved, this may lead to local unwinding of DNA strands, increasing the probability that RNA hybridizes to the DNA. Recent findings suggest positive functions for scheduled R-loops in regulating distinct biological processes. For example, R-loops formed over the 5’ GC-rich regions downstream from CpG promoters may promote gene activation (Ginno et al., 2013; Ginno et al., 2012), whilst their formation at the 3’ ends of genes may effect efficient transcriptional termination of a subset of human genes, provided that they do not accumulate excessively (Skourti-Stathaki et al., 2011). Thus, programmed R-loops may have a role in regulating gene expression, but the balance between their formation and resolution is critical to limit the potentially deleterious effects of unscheduled or excessive accumulation (e.g. Aguilera and Garcia-Muse, 2012; Hamperl and Cimprich, 2014; Skourti-Stathaki and Proudfoot, 2014).
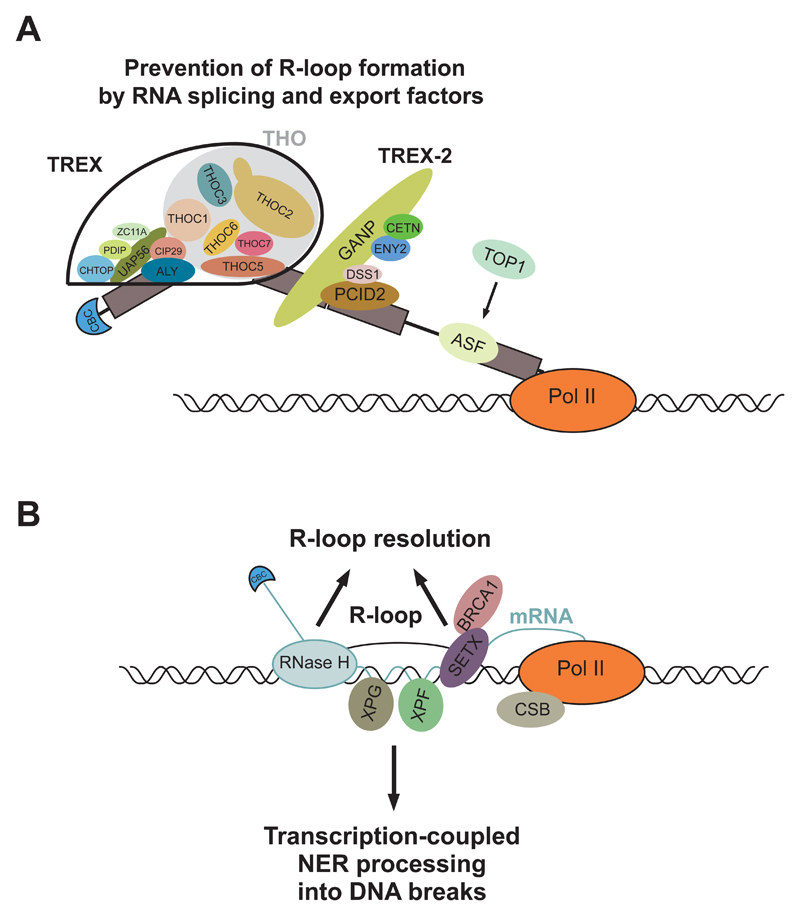
Figure 4. R-loop prevention and resolution.
A, Prevention of R-loop formation by RNA splicing and export factors. The TREX and TREX-2 mRNA export complexes coat newly transcribed and processed mRNA to form export-competent mRNPs, thus minimizing the time for the nascent mRNA to rehybridize with the transcribed DNA strand. The ASF/SF2 SR protein pre-mRNA splicing factor performs a similar function during RNA splicing, in a Top1-dependent manner. B, R-loop resolution is promoted by RNase H1 and H2, which specifically degrade the RNA component of the RNA/DNA hybrids, generating a 3’ end that can be extended by the DNA replication machinery. A BRCA1-Senataxin complex can resolve R-loops at termination elements, releasing RNA for degradation by Xrn2 prior to termination. In contrast, R-loops are actively processed into DSBs by the nucleotide excision repair (NER) endonucleases XPF and XPG.
The role of RBPs in R-loop stability
Depletion or mutation of RBPs that normally coat RNA species to form mRNA complexes during processing and export can increase the probability of R-loop formation (Bhatia et al., 2014; Dominguez-Sanchez et al., 2011; Gonzalez-Aguilera et al., 2008; Huertas and Aguilera, 2003; Mischo et al., 2011; Wahba et al., 2011). Both TREX (TRanscription-EXport) and TREX-2 mRNA export complexes have been implicated in R-loop prevention (Bhatia et al., 2014; Dominguez-Sanchez et al., 2011; Gomez-Gonzalez et al., 2009; Gonzalez-Aguilera et al., 2008). This is most likely through their function in proper packaging of mRNA into export-competent mRNPs (Strasser et al., 2002), which occurs co-transcriptionally, thus minimizing the time for the nascent mRNA to rehybridize with the transcribed DNA strand (Figure 4A). If this process is inefficient, naked stretches of the nascent RNA may pair with the transcribed strand in cis or away from the transcription start point in trans, as was recently demonstrated in yeast (Wahba et al., 2013). These considerations raise the possibility that defects in RNA processing mechanisms, whether constitutive or damage-induced, may promote unscheduled R-loop formation by interfering with mRNP assembly.
Processing of unscheduled R-loops into breaks
Unscheduled R-loops can be processed into both single and double strand DNA breaks. ssDNA breaks may form through processing of unstable structures on exposed ssDNA by various DNA repair enzymes (reviewed in Hamperl and Cimprich, 2014). They include the formation of a covalent Top1 - DNA complex that is trapped during its cleavage-ligation cycle, whose removal generates 2-3bp deletions (Lippert et al., 2011; Takahashi et al., 2011). Another mechanism involves the activity of activation-induced cytidine deaminase (AID) to convert cytidine to uracil residues on ssDNA, which may then be recognised by the mismatch or base excision repair machineries for eventual processing into ssDNA breaks (Chaudhuri et al., 2003; Hamperl and Cimprich, 2014). The resolution of hairpin structures or G-quadruplexes that form on the displaced ssDNA may also result in DNA breaks ((Hamperl and Cimprich, 2014; Kim and Jinks-Robertson, 2012; Lopes et al., 2011; Sun et al., 2001).
DSBs can be generated from a collision between a replication fork and an R-loop, stalling the fork which can then be cleaved to generate a DSB (Gomez-Gonzalez et al., 2009; Helmrich et al., 2011; Tuduri et al., 2009). In addition, R-loops induced by the absence of RNA processing factors such as Senataxin, Aquarius and ASF are actively processed into DSBs by NER (transcription-coupled nucleotide excision repair) endonucleases XPF and XPG (Sollier et al., 2014) (Figure 4B). This is consistent with a primary role of the NER machinery being to rapidly remove transcription-impeding DNA lesions to prevent the prolonged arrest of RNA polymerases, which may occur as a consequence of R-loop formation. This raises the intriguing possibility that the NER machinery may remove unscheduled R-loops in order to restore gene expression from these sites in a timely manner (Lin and Pasero, 2014).
Finally, recent work in yeast and mammalian cells implicates components of the homologous recombination machinery in R-loop formation and genome instability, although it remains unclear whether their role in R-loop accumulation is connected with homologous recombination. In yeast, the RAD51 recombinase promotes R-loop formation in vivo. Interestingly, RAD51 can promote R-loop formation in cis at the site of transcription, or away from the site of transcription in trans (Wahba et al., 2013). This suggests that R-loops may not exclusively form co-transcriptionally, raising the possibility that R-loops may be a larger threat to genome integrity than previously thought. The breast cancer suppressor and RAD51 interacting protein, BRCA2, has also recently been implicated in processing R-loops, but how this may be achieved remains puzzlingly obscure. BRCA2 is reported to interact with the PCID2 component of the TREX-2 mRNA export complex (Jani et al., 2012), as detected indirectly by a proximity ligation assay (Bhatia et al., 2014). PCID2 depletion using RNA interference increased DNA breakage marked by γH2AX formation, but without measurably enhancing R-loop formation. In contrast, however, BRCA2 depletion increases R-loop formation at several actively transcribed genes. The nature of the interaction between BRCA2 and PCID2 remains undefined, leaving open the possibility that it may be indirect, and mediated via proteins like BRCA1, with which BRCA2 interacts on chromatin (Chen et al., 1998; Sy et al., 2009; Zhang et al., 2009). These issues leave unclear if or how BRCA2 may work directly with PCID2 or other TREX-2 components to assist in R-loop processing.
The related breast cancer suppressor protein, BRCA1, has been implicated in damage-induced RNA processing at multiple levels. Apart from its roles in RNA pol II ubiquitylation and in a DNA-damage induced mRNA splicing complex that facilitates splicing of genome maintenance factors described above, BRCA1 forms a complex with Senataxin, which is recruited to R-loop rich termination regions in a subset of actively transcribed genes. Disruption of the Senataxin-BRCA1 complex causes ssDNA breakage at these loci (Hatchi et al., 2015; Hill et al., 2014), suggesting a potentially important role for BRCA1 in the resolution of R-loop structures during transcription. Molecular analyses suggest that BRCA1 is recruited to R-loops at transcription termination sites in certain loci via its interaction with components of paused RNA pol II complexes (Hatchi et al., 2015; Skourti-Stathaki et al., 2011). How recruitment is selectively directed to a subset of actively transcribed genes remains unclear – but this is important because mutations affecting some of these genes are found in BRCA1-mutant breast cancer samples (Hatchi et al., 2015). These recent results thoroughly define a novel function for BRCA1 that affects the pattern of genomic mutations associated with carcinogenesis in mutation carriers.
Implications
This field is now reaching a new understanding of the complex connections between RNA processing and DNA damage and repair. An added layer of complexity not discussed here is brought by different species of non-coding RNA molecules that also regulate the DDR. These include miRNAs, long non-coding RNAs (lncRNAs), and novel non-coding transcripts (reviewed in (Chowdhury et al., 2013; d'Adda di Fagagna, 2014; Sharma and Misteli, 2013; Wouters et al., 2011).
There is also growing evidence that the connections between mRNA processing and DNA damage responses may be disrupted in human diseases, potentially contributing to genome instability and carcinogenesis. For example, as noted above, the tumor suppressor proteins BRCA1 and BRCA2 have recently been implicated in pathways that that prevent the accumulation of R-loops (Bhatia et al., 2014; Hatchi et al., 2015; Hill et al., 2014; Savage et al., 2014). The case is particularly strong for BRCA1, as described in the preceding section (Hatchi et al., 2015).
Armed with knowledge of the mechanisms described in this review, the field can now turn to address how disrupting these processes could promote carcinogenesis. For example, do RNA processing mechanisms contribute to the DNA damage responses proposed to be triggered by oncogenic signals early during carcinogenesis? Germline mutations affecting certain hereditary tumour suppressors like BRCA1 act via RNA processing to prevent DNA damage and genome instability: do somatic mutations affecting similar pathways occur commonly during carcinogenesis? Since any disruption of these pathways may not target all loci equally, might they dictate the genome-wide patterns of mutation found in advanced cancer? If the findings that implicate R-loop formation in trans in yeast are extended to humans, might such a process underlie cancer-associated instability in distal regions of the human genome? The field has a long way to go in connecting the underlying mechanisms to disease states; however, the studies discussed in this review present a solid foundation on which to begin the journey.
References
- Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nature cell biology. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Molecular cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Albrecht H, Durbin-Johnson B, Yunis R, Kalanetra KM, Wu S, Chen R, Stevenson TR, Rocke DM. Transcriptional response of ex vivo human skin to ionizing radiation: comparison between low- and high-dose effects. Radiation research. 2012;177:69–83. doi: 10.1667/rr2524.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantha RW, Alcivar AL, Ma J, Cai H, Simhadri S, Ule J, Konig J, Xia B. Requirement of heterogeneous nuclear ribonucleoprotein C for BRCA gene expression and homologous recombination. PLoS One. 2013;8:e61368. doi: 10.1371/journal.pone.0061368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anindya R, Aygun O, Svejstrup JQ. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol Cell. 2007;28:386–397. doi: 10.1016/j.molcel.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Beli P, Lukashchuk N, Wagner SA, Weinert BT, Olsen JV, Baskcomb L, Mann M, Jackson SP, Choudhary C. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Molecular cell. 2012;46:212–225. doi: 10.1016/j.molcel.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia V, Barroso SI, Garcia-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014 doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- Blackinton JG, Keene JD. Post-transcriptional RNA regulons affecting cell cycle and proliferation. Semin Cell Dev Biol. 2014 doi: 10.1016/j.semcdb.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius M, Wagner SA, Choudhary C, Bartek J, Jackson SP. A quantitative 14-3-3 interaction screen connects the nuclear exosome targeting complex to the DNA damage response. Genes Dev. 2014;28:1977–1982. doi: 10.1101/gad.246272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek D, Kohoutek J, Bartholomeeusen K, Johansen E, Hulinkova P, Luo Z, Cimermancic P, Ule J, Peterlin BM. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes & development. 2011;25:2158–2172. doi: 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton S, Dernoncourt E, Delteil C, Froment C, Schiltz O, Salles B, Frit P, Calsou P. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res. 2014;42:9047–9062. doi: 10.1093/nar/gku601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammas A, Pileur F, Bonnal S, Lewis SM, Leveque N, Holcik M, Vagner S. Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Mol Biol Cell. 2007;18:5048–5059. doi: 10.1091/mbc.E07-06-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM, et al. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell. 1998;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Choi YE, Brault ME. Charity begins at home: non-coding RNA functions in DNA repair. Nat Rev Mol Cell Biol. 2013;14:181–189. doi: 10.1038/nrm3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SB, Ma W, Valova VA, Algie M, Harfoot R, Woolley AG, Robinson PJ, Braithwaite AW. Genotoxic stress-induced nuclear localization of oncoprotein YB-1 in the absence of proteolytic processing. Oncogene. 2010;29:403–410. doi: 10.1038/onc.2009.321. [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F. A direct role for small non-coding RNAs in DNA damage response. Trends Cell Biol. 2014;24:171–178. doi: 10.1016/j.tcb.2013.09.008. [DOI] [PubMed] [Google Scholar]
- Decorsiere A, Cayrel A, Vagner S, Millevoi S. Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3'-end processing and function during DNA damage. Genes & development. 2011;25:220–225. doi: 10.1101/gad.607011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Harding HP, Raught B, Gingras AC, Berlanga JJ, Scheuner D, Kaufman RJ, Ron D, Sonenberg N. Activation of GCN2 in UV-irradiated cells inhibits translation. Curr Biol. 2002;12:1279–1286. doi: 10.1016/s0960-9822(02)01037-0. [DOI] [PubMed] [Google Scholar]
- Dominguez-Sanchez MS, Barroso S, Gomez-Gonzalez B, Luna R, Aguilera A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet. 2011;7:e1002386. doi: 10.1371/journal.pgen.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre M, Lambert S, Carreira A, Amor-Gueret M, Vagner S. DNA damage: RNA-binding proteins protect from near and far. Trends Biochem Sci. 2014;39:141–149. doi: 10.1016/j.tibs.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Dutertre M, Sanchez G, De Cian MC, Barbier J, Dardenne E, Gratadou L, Dujardin G, Le Jossic-Corcos C, Corcos L, Auboeuf D. Cotranscriptional exon skipping in the genotoxic stress response. Nature structural & molecular biology. 2010;17:1358–1366. doi: 10.1038/nsmb.1912. [DOI] [PubMed] [Google Scholar]
- El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YW, Cattoglio C, Tjian R. The intertwined roles of transcription and repair proteins. Mol Cell. 2013;52:291–302. doi: 10.1016/j.molcel.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC. Relationships between DNA repair and transcription. Annu Rev Biochem. 1996;65:15–42. doi: 10.1146/annurev.bi.65.070196.000311. [DOI] [PubMed] [Google Scholar]
- Fry RC, Begley TJ, Samson LD. Genome-wide responses to DNA-damaging agents. Annual review of microbiology. 2005;59:357–377. doi: 10.1146/annurev.micro.59.031805.133658. [DOI] [PubMed] [Google Scholar]
- Gardiner M, Toth R, Vandermoere F, Morrice NA, Rouse J. Identification and characterization of FUS/TLS as a new target of ATM. The Biochemical journal. 2008;415:297–307. doi: 10.1042/BJ20081135. [DOI] [PubMed] [Google Scholar]
- Ginno PA, Lim YW, Lott PL, Korf I, Chedin F. GC skew at the 5' and 3' ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013;23:1590–1600. doi: 10.1101/gr.158436.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, Felipe-Abrio I, Aguilera A. The S-phase checkpoint is required to respond to R-loops accumulated in THO mutants. Mol Cell Biol. 2009;29:5203–5213. doi: 10.1128/MCB.00402-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Aguilera C, Tous C, Gomez-Gonzalez B, Huertas P, Luna R, Aguilera A. The THP1-SAC3-SUS1-CDC31 complex works in transcription elongation-mRNA export preventing RNA-mediated genome instability. Mol Biol Cell. 2008;19:4310–4318. doi: 10.1091/mbc.E08-04-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover R, Ray PS, Das S. Polypyrimidine tract binding protein regulates IRES-mediated translation of p53 isoforms. Cell Cycle. 2008;7:2189–2198. doi: 10.4161/cc.7.14.6271. [DOI] [PubMed] [Google Scholar]
- Hamperl S, Cimprich KA. The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair (Amst) 2014;19:84–94. doi: 10.1016/j.dnarep.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- Harreman M, Taschner M, Sigurdsson S, Anindya R, Reid J, Somesh B, Kong SE, Banks CA, Conaway RC, Conaway JW, et al. Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc Natl Acad Sci U S A. 2009;106:20705–20710. doi: 10.1073/pnas.0907052106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, Dimitrov S, Pathania S, McKinney KM, Eaton ML, et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell. 2015;57:636–647. doi: 10.1016/j.molcel.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Hill SJ, Rolland T, Adelmant G, Xia X, Owen MS, Dricot A, Zack TI, Sahni N, Jacob Y, Hao T, et al. Systematic screening reveals a role for BRCA1 in the response to transcription-associated DNA damage. Genes Dev. 2014;28:1957–1975. doi: 10.1101/gad.241620.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Molecular cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Itoh T, Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A. 1980;77:2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhar L, Adamson B, Ciccia A, Lewis J, Pontano-Vaites L, Leng Y, Liang AC, Westbrook TF, Harper JW, Elledge SJ. A Systematic Analysis of Factors Localized to Damaged Chromatin Reveals PARP-Dependent Recruitment of Transcription Factors. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani D, Lutz S, Hurt E, Laskey RA, Stewart M, Wickramasinghe VO. Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export. Nucleic Acids Res. 2012;40:4562–4573. doi: 10.1093/nar/gks059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G, Sancar A. Recruitment of DNA damage checkpoint proteins to damage in transcribed and nontranscribed sequences. Molecular and cellular biology. 2006;26:39–49. doi: 10.1128/MCB.26.1.39-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Kim HD, Choe J, Seo YS. The sen1(+) gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry. 1999;38:14697–14710. doi: 10.1021/bi991470c. [DOI] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S. Transcription as a source of genome instability. Nat Rev Genet. 2012;13:204–214. doi: 10.1038/nrg3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman FE, Manley JL. The BARD1-CstF-50 interaction links mRNA 3' end formation to DNA damage and tumor suppression. Cell. 2001;104:743–753. doi: 10.1016/s0092-8674(01)00270-7. [DOI] [PubMed] [Google Scholar]
- Kleiman FE, Wu-Baer F, Fonseca D, Kaneko S, Baer R, Manley JL. BRCA1/BARD1 inhibition of mRNA 3' processing involves targeted degradation of RNA polymerase II. Genes Dev. 2005;19:1227–1237. doi: 10.1101/gad.1309505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le May N, Mota-Fernandes D, Velez-Cruz R, Iltis I, Biard D, Egly JM. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol Cell. 2010;38:54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Lin YL, Pasero P. Caught in the Act: R-loops are cleaved by structure-specific endonucleases to generate DSBs. Mol Cell. 2014;56:721–722. doi: 10.1016/j.molcel.2014.12.011. [DOI] [PubMed] [Google Scholar]
- Lippert MJ, Kim N, Cho JE, Larson RP, Schoenly NE, O'Shea SH, Jinks-Robertson S. Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proc Natl Acad Sci U S A. 2011;108:698–703. doi: 10.1073/pnas.1012363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes J, Piazza A, Bermejo R, Kriegsman B, Colosio A, Teulade-Fichou MP, Foiani M, Nicolas A. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30:4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal A, Li JM, Ji XY, Wu CS, Yazinski SA, Nguyen HD, Liu S, Jimenez AE, Jin J, Zou L. PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Molecular cell. 2014;53:235–246. doi: 10.1016/j.molcel.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Mayne LV, Lehmann AR. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne's syndrome and xeroderma pigmentosum. Cancer Res. 1982;42:1473–1478. [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci U S A. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Hagner PR, Zhang Y, Dai B, Lehrmann E, Becker KG, Keene JD, Gorospe M, Liu Z, Gartenhaus RB. ATM regulates a DNA damage response posttranscriptional RNA operon in lymphocytes. Blood. 2011;117:2441–2450. doi: 10.1182/blood-2010-09-310987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, Beech J, Kulshrestha R, Abdelmohsen K, Weinstock DM, et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Molecular cell. 2011;41:210–220. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moumen A, Magill C, Dry KL, Jackson SP. ATM-dependent phosphorylation of heterogeneous nuclear ribonucleoprotein K promotes p53 transcriptional activation in response to DNA damage. Cell Cycle. 2013;12:698–704. doi: 10.4161/cc.23592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moumen A, Masterson P, O'Connor MJ, Jackson SP. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–1078. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- Muller-McNicoll M, Neugebauer KM. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet. 2013;14:275–287. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- Park E, Kim H, Kim JM, Primack B, Vidal-Cardenas S, Xu Y, Price BD, Mills AA, D'Andrea AD. FANCD2 activates transcription of TAp63 and suppresses tumorigenesis. Mol Cell. 2013;50:908–918. doi: 10.1016/j.molcel.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto MP, Minana B, Valcarcel J. The Ewing sarcoma protein regulates DNA damage-induced alternative splicing. Molecular cell. 2011;43:353–368. doi: 10.1016/j.molcel.2011.05.035. [DOI] [PubMed] [Google Scholar]
- Polo SE, Blackford AN, Chapman JR, Baskcomb L, Gravel S, Rusch A, Thomas A, Blundred R, Smith P, Kzhyshkowska J, et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Molecular cell. 2012;45:505–516. doi: 10.1016/j.molcel.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nature reviews. Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- Powley IR, Kondrashov A, Young LA, Dobbyn HC, Hill K, Cannell IG, Stoneley M, Kong YW, Cotes JA, Smith GC, et al. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev. 2009;23:1207–1220. doi: 10.1101/gad.516509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashi-Elkeles S, Warnatz HJ, Elkon R, Kupershtein A, Chobod Y, Paz A, Amstislavskiy V, Sultan M, Safer H, Nietfeld W, et al. Parallel profiling of the transcriptome, cistrome, and epigenome in the cellular response to ionizing radiation. Sci Signal. 2014;7:rs3. doi: 10.1126/scisignal.2005032. [DOI] [PubMed] [Google Scholar]
- Reinhardt HC, Hasskamp P, Schmedding I, Morandell S, van Vugt MA, Wang X, Linding R, Ong SE, Weaver D, Carr SA, et al. DNA damage activates a spatially distinct late cytoplasmic cell-cycle checkpoint network controlled by MK2-mediated RNA stabilization. Molecular cell. 2010;40:34–49. doi: 10.1016/j.molcel.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger KE, Chu G. Portrait of transcriptional responses to ultraviolet and ionizing radiation in human cells. Nucleic Acids Res. 2004;32:4786–4803. doi: 10.1093/nar/gkh783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Savage KI, Gorski JJ, Barros EM, Irwin GW, Manti L, Powell AJ, Pellagatti A, Lukashchuk N, McCance DJ, McCluggage WG, et al. Identification of a BRCA1-mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Molecular cell. 2014;54:445–459. doi: 10.1016/j.molcel.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Misteli T. Non-coding RNAs in DNA damage and repair. FEBS Lett. 2013;587:1832–1839. doi: 10.1016/j.febslet.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, 3rd, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3' processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28:1384–1396. doi: 10.1101/gad.242990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J, Stork CT, Garcia-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA. Transcription-Coupled Nucleotide Excision Repair Factors Promote R-Loop-Induced Genome Instability. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin AV, Selyutina AA, Skabkin MA, Guryanov SG, Nazimov IV, Richard C, Th'ng J, Yau J, Sorensen PH, Ovchinnikov LP, et al. Proteasome-mediated cleavage of the Y-box-binding protein 1 is linked to DNA-damage stress response. EMBO J. 2005;24:3602–3612. doi: 10.1038/sj.emboj.7600830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Sun H, Yabuki A, Maizels N. A human nuclease specific for G4 DNA. Proc Natl Acad Sci U S A. 2001;98:12444–12449. doi: 10.1073/pnas.231479198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy S, Vazquez-Novelle MD, Lekomtsev S, Howell M, Petronczki M. Functional genomics identifies a requirement of pre-mRNA splicing factors for sister chromatid cohesion. EMBO J. 2014;33:2623–2642. doi: 10.15252/embj.201488244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U S A. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Burguiere-Slezak G, Van der Kemp PA, Boiteux S. Topoisomerase 1 provokes the formation of short deletions in repeated sequences upon high transcription in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2011;108:692–697. doi: 10.1073/pnas.1012582108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresini M, Warmerdam DO, Kolovos P, Snijder L, Vrouwe MG, Demmers JA, van IWF, Grosveld FG, Medema RH, Hoeijmakers JH, et al. The core spliceosome as target and effector of non-canonical ATM signalling. Nature. 2015;523:53–58. doi: 10.1038/nature14512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lelij P, Stocsits RR, Ladurner R, Petzold G, Kreidl E, Koch B, Schmitz J, Neumann B, Ellenberg J, Peters JM. SNW1 enables sister chromatid cohesion by mediating the splicing of sororin and APC2 pre-mRNAs. EMBO J. 2014;33:2643–2658. doi: 10.15252/embj.201488202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaraman AR. Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science. 2014;343:1470–1475. doi: 10.1126/science.1252230. [DOI] [PubMed] [Google Scholar]
- Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Molecular cell. 2011;44:978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Gore SK, Koshland D. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. eLife. 2013;2:e00505. doi: 10.7554/eLife.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WY, Pan L, Su SC, Quinn EJ, Sasaki M, Jimenez JC, Mackenzie IR, Huang EJ, Tsai LH. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nature neuroscience. 2013;16:1383–1391. doi: 10.1038/nn.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe VO, Gonzalez-Porta M, Perera D, Bartolozzi AR, Sibley CR, Hallegger M, Ule J, Marioni JC, Venkitaraman AR. Regulation of constitutive and alternative mRNA splicing across the human transcriptome by PRPF8 is determined by 5’ splice site strength. Genome Biol. 2015;16:201. doi: 10.1186/s13059-015-0749-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe VO, Laskey RA. Control of mammalian gene expression by selective mRNA export. Nat Rev Mol Cell Biol. 2015;16:431–442. doi: 10.1038/nrm4010. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe VO, Savill JM, Chavali S, Jonsdottir AB, Rajendra E, Gruner T, Laskey RA, Babu MM, Venkitaraman AR. Human inositol polyphosphate multikinase regulates transcript-selective nuclear mRNA export to preserve genome integrity. Molecular cell. 2013;51:737–750. doi: 10.1016/j.molcel.2013.08.031. [DOI] [PubMed] [Google Scholar]
- Workman CT, Mak HC, McCuine S, Tagne JB, Agarwal M, Ozier O, Begley TJ, Samson LD, Ideker T. A systems approach to mapping DNA damage response pathways. Science. 2006;312:1054–1059. doi: 10.1126/science.1122088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters MD, van Gent DC, Hoeijmakers JH, Pothof J. MicroRNAs, the DNA damage response and cancer. Mutation research. 2011;717:54–66. doi: 10.1016/j.mrfmmm.2011.03.012. [DOI] [PubMed] [Google Scholar]
- Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, Yu X. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19:524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]