Genome-wide association studies have identified thousands of genetic variants that are associated with disease.1 Most of these variants have small effect sizes, but their downstream expression effects, so-called expression quantitative trait loci (eQTLs), are often large2 and cell type-specific3–5. To identify these cell type-specific eQTLs using an unbiased approach, we used single-cell RNA sequencing (scRNA-seq) to generate expression profiles of ~25,000 peripheral blood mononuclear cells (PBMCs) from 45 donors. We identified previously reported cis-eQTLs, but also identified new cell type-specific cis-eQTLs. Finally, we generated personalized co-expression networks, and identified genetic variants that significantly alter co-expression relationships (which we termed ‘co-expression QTLs’). Single-cell eQTL analysis thus allows for the identification of genetic variants that impact regulatory networks.
Previously, purified cell types4,6–8 or deconvolution methods9,10 have been used to identify cell type-specific eQTLs. However, these methods are biased towards specific cell types, or are of limited use for less abundant cell types and dependent on accurately defined marker genes.11 In contrast, scRNA-seq can be used to investigate rare cell types12, and thus, enables identification of cell type-specific eQTLs using an unbiased approach. Indeed, proof of concept was previously shown in a study on 15 individuals, where 92 genes were studied in 1,440 cells.13
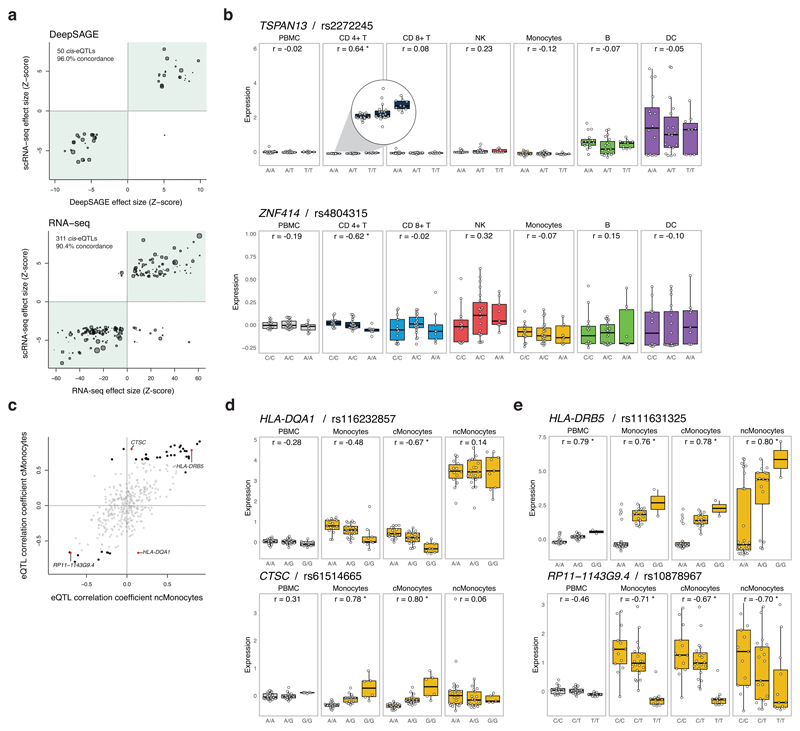
Here, we studied cell type-specific effects of genetic variation on genome-wide gene expression by generating scRNA-seq data of ~25,000 PBMCs from 45 donors of the population-based cohort study Lifelines Deep14. After quality control (Online Methods, Suppl. Fig. 1), we first assessed to which extent previously reported cis-eQTLs from bulk whole blood, using either 94 DeepSAGE samples 15 (a 3’-end oriented RNA-sequencing strategy similar to our scRNA-seq approach) or 2,116 RNA-seq11 samples, also show significant effects in the scRNA-seq dataset. For this analysis, we treated the scRNA-seq data as being bulk PBMCs (by averaging expression levels of all cells per gene per sample, referred to as ‘bulk-like PBMCs’). We detected 50 and 311 significant cis-eQTLs (gene-level false-discovery rate (FDR) of 0.05) that were previously reported in the DeepSAGE15 and RNA-seq11 study, respectively (Fig. 1a, Suppl. Table 1). Although only a small proportion (8% and 1%) of previously reported cis-eQTLs were significant in our scRNA-seq analysis, 96% and 90.4% had identical allelic directions as in the DeepSAGE15 and RNA-seq11 study, respectively, indicating that these cis-eQTLs reflect similar regulatory effects. The few discordant eQTLs may reflect the slightly different sample composition of both datasets (PBMCs versus whole blood) and the relatively few sequence reads targeting the 3’-end of genes in the bulk RNA-seq dataset.
Figure 1. Cis-eQTL analysis in single-cell RNA-seq data.
(a) Effect size of the cis-eQTLs detected in the bulk-like PBMC scRNA-seq sample in which the analysis was confined to previously reported cis-eQTLs in (top) whole blood DeepSAGE or (bottom) bulk RNA-seq data. The number and percentage represent, respectively, the detected cis-eQTLs and their concordance (i.e. same allelic direction – green quadrants) between the bulk-like PBMC population scRNA-seq eQTLs and (top) whole blood DeepSAGE or (bottom) bulk RNA-seq data. The size of each dot represents the mean expression of the cis-regulated gene in the total scRNA-seq dataset. (b) Examples of undetectable cis-eQTLs in the bulk-like PBMC population caused by (top) masking of the cis-eQTL present in CD4+ T cells but absent in DCs with comparatively high expression of the cis-regulated gene or (bottom) opposite allelic effects in CD4+ T and NK cells. (c) Spearman’s rank correlation coefficient for the cMonocytes against the ncMonocytes of all top eQTLs that were identified in the total dataset or at least one (sub)cell cluster (see Suppl. Table 2). Significant correlations are shown in black (four red highlighted examples are shown in d and e), the non-significant in gray. (d) Cis-eQTLs specifically affecting expression in the cMonocytes, and not the ncMonocytes. (e) Cis-eQTLs significantly affecting the expression in both the cMonocytes and ncMonocytes. Each dot represents the mean expression of the eQTL gene in a donor. Box plots show the median, the first and third quartiles, and 1.5 times the interquartile range. r, Spearman’s rank correlation coefficient; *FDR≤0.05.
We subsequently performed a genome-wide cis-eQTL discovery analysis on the bulk-like PBMCs. Separate cis-eQTL analyses were conducted on each of the identified major cell types (cell type classification was performed using Seurat16, Suppl. Fig. 2a, 2b) by averaging the normalized gene expression of all cells per cell type, gene and donor. In total, 379 unique top cis-eQTLs were identified, reflecting 287 unique eQTL genes (gene-level FDR of 0.05) (Table 1), as sometimes in different cell types different SNPs showed the most significant association for an eQTL gene. While 331 (reflecting 249 unique cis-eQTL genes) of these 379 cis-eQTLs were significant in the bulk-like PBMC eQTL analysis, 48 cis-eQTLs (reflecting 38 unique cis-eQTL genes) were only detected in specific cell types (i.e. ‘cell type-dependent’ eQTLs, Suppl. Table 2).
Table 1. Cis-eQTL genes identified per cell type.
| Cell type | Median number of cells/donor | Unique genes with significant cis-eQTL effect | |
|---|---|---|---|
| PBMC | 507 | 249 | |
| CD4+ T | 282 | 145 | |
| CD8+ T | 74 | 21 | |
| NK | 59 | 14 | |
| Monocyte | 44 | 23 | |
| B | 18 | 6 | |
| DC | 11 | 9 | |
| Total (unique) | 287 | ||
The median number of cells per donor (column 2) correlates fairly well with the number of detected cis-eQTL genes (column 3). In total, 379 unique top cis-eQTL effects, reflecting 287 unique eQTL genes, have been identified in the total dataset. Within each cell type, the number of unique cis-eQTL genes that we identified was equal to the number of unique, top cis-eQTL effects.
We subsequently attempted to replicate these eQTLs. For the 249 eQTL genes found in the bulk-like PBMC analysis, 233 cis-eQTLs were testable and 181 (78%) were associated with the same SNP (90.1% shared allelic direction, Suppl. Table 2) in the whole blood RNA-seq eQTL data set11. For the 48 cell type-dependent cis-eQTLs, 29 (60%) replicated in the RNA-seq dataset11. This lower percentage suggests that in bulk RNA-seq datasets cell type-dependent eQTLs might become too diluted, resulting in low statistical power to recover these. While this most likely happens for rare cell types, we also observed this in common cell types. For instance, in the most abundant cell type (CD4+ T cells), rs2272245 significantly affects the expression of the TSPAN13 gene in cis (P = 2.21x10-6). However, this effect was not significant in the bulk-like PBMCs (P = 0.88), because TSPAN13 is lowly expressed in CD4+ T-cells, whereas it is highly expressed in dendritic cells (DCs) where it did not show a cis-eQTL effect (Fig. 1b). Cis-eQTLs might also be missed in bulk data, because they might show opposite allelic effects across different cell types. We could not study this in detail, due to lack of power given the sample size and limited number of cells for rare cell types (Suppl. Fig. 2c). Nevertheless, in CD4+ T cells, the A allele of rs4804315 significantly decreased expression of ZNF414 in cis (P = 6.09x10-6), whereas in natural killer (NK) cells this allele increased expression of ZNF414 at nominal significance (P = 0.0339) (Fig. 1b). However, it cannot be excluded that specifically in NK cells, the effect of rs4804315 on ZNF414 expression is the result of a residual effect on ZNF414 expression of a second, independent variant.
Since some cis-eQTLs did not replicate in whole blood bulk RNA-seq data, we subsequently investigated eQTL datasets of purified cell types. Indeed, 3 out of 19 remaining cell type-dependent cis-eQTLs were detected (each with consistent allelic direction) in purified eQTL datasets of the Blueprint consortium (naïve CD4+ T cells and CD14+ monocytes)17 or Kasela et al. (CD4+ and CD8+ T cells)6 (Suppl. Table 3). Hence, only 16 cell type-dependent cis-eQTLs were not identified before using bulk eQTL datasets of blood or purified immune cells. Although some cis-eQTLs were only significant in specific cell types, this does not prove cell type-specificity; particularly in less abundant cell types power is lacking to detect many cis-eQTLs. Ways to partially overcome this, would be to use methods that consider multiple eQTL datasets together, such as eQTL-BMA18 or Meta-Tissue19. However, these methods are currently computationally too demanding for large scRNA-seq data or do not define the cell type in which the eQTL effect occurs.19,20
A major advantage of using scRNA-seq data is the flexibility by which any cell population of interest can be selected for eQTL analysis. In contrast, when using RNA-seq data of purified cell types, one cannot retrieve data from subcell types anymore. Moreover, while finer differences between subcell types may be detectable using gene expression profiles, it is not always recapitulated by different cell membrane markers, complicating cell sorting. Here, we show the added value of performing eQTL analysis on subcell types using two monocyte subsets: classical (cMonocytes) and non-classical monocytes (ncMonocytes). When plotting Spearman’s rank correlation of each top eQTL for the cMonocytes against the ncMonocytes, several examples were revealed that pinpointed the eQTL effect specifically to cMonocytes (Fig. 1c). Two such examples, which were previously identified in RNA-seq data of purified CD14+ monocytes17, are shown in Figure 1d. The scRNA-seq data now allowed us to specifically assign these effects to cMonocytes (Fig. 1d). Despite having lower power for detecting eQTLs in ncMonocytes due to an almost five times lower abundance compared to cMonocytes (Suppl. Fig. 2b), power in the ncMonocytes remains sufficiently high to detect several other significant ncMonocyte cis-eQTLs (Fig. 1e, Suppl. Table 2).
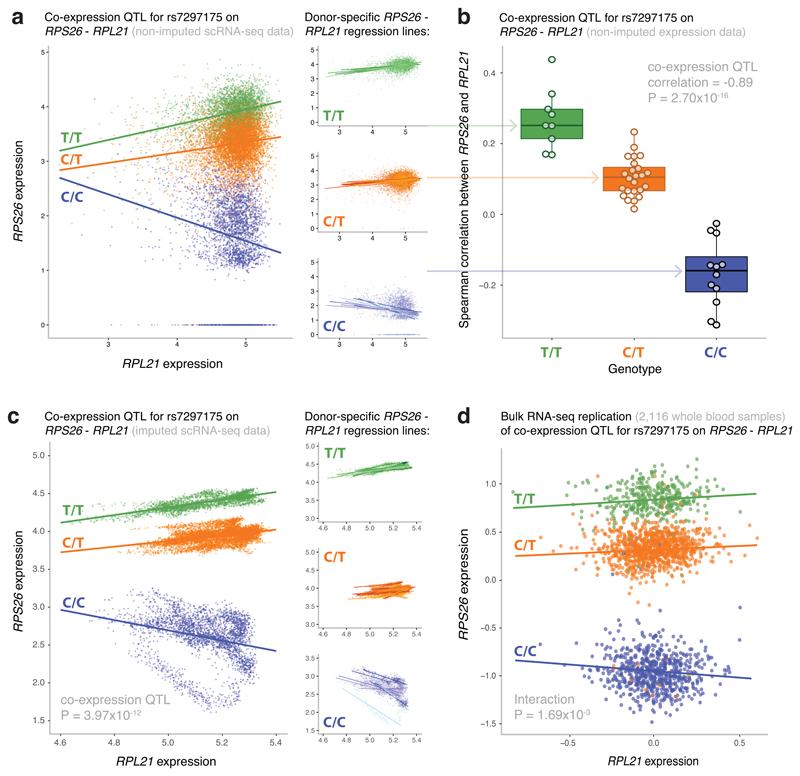
Another opportunity of scRNA-seq data is to use it for determining whether genetic variants can alter gene co-expression. Although recently genes and environmental factors altering the effect size of eQTLs (‘context-specific eQTLs’) have been identified in bulk RNA-seq eQTL datasets11,21, a large sample size was required to ensure sufficient power. In contrast, scRNA-seq data enables generation of co-expression networks on an individual donor basis, which vastly reduces the number of samples required to identify SNPs altering co-expression relationships. This enabled us to study whether SNPs showing cis-eQTL effects also affect the co-expression relationship of the cis-eQTL genes with other genes, which we further define as ‘co-expression QTLs’. We confined our analysis to the most abundant cell type (CD4+ T cells), and calculated the co-expression between individual pairs of genes using Spearman’s rank correlation. We restricted the analysis to the 145 cis-eQTL genes identified in CD4+ T cells (Table 1), thereby increasing the likelihood of finding co-expressed genes that are modulated by the same genetic variant. Out of these, 102 genes showed variance in gene expression within each of the 45 donors and were investigated. For two of these genes we identified significant co-expression QTLs: 93 co-expression QTLs were detected for RPS26 and one for HLA-B (P-value ≤ 1.27x10-7, corresponding to an eQTL-gene level FDR of 0.05). The most significant interaction was found for rs7297175 affecting the co-expression between RPS26 and RPL21 (P ≤ 2.70x10-16) (Fig. 2a, 2b). When using a more liberal FDR of 0.10 (P-value = 4.72x10-7), we identified significant co-expression QTLs for three eQTL genes (Suppl. Table 4): 13 additional co-expression QTLs were found for RPS26 and one for SMDT1. As a result of co-expression between genes, we cannot rule out that the 106 co-expression QTLs identified for RPS26 are actually representing just one effect.
Figure 2. Most significant co-expression QTL in the CD4+ T cells.
(a) The non-imputed expression of RPS26 and RPL21 of all individual CD4+ T cells colored by genotype (left panel) and stratified per SNP rs7297175 genotype (right panels). Genotype- and donor-specific regression lines are shown in the left and right panel, respectively. Each data point represents a single cell. The nominal P-value is given for the co-expression QTL. (b) The Spearman’s rank correlation coefficient (r) between RPS26 and RPL21 expression stratified by SNP rs7297175 genotype in the CD4+ T cells per donor. Each data point represents a single donor. Box plots show the median, the first and third quartiles, and 1.5 times the interquartile range. The nominal P-value is given for the co-expression QTL. (c) The imputed expression of RPS26 and RPL21 of all individual CD4+ T cells colored by genotype (left panel) and stratified per SNP rs7297175 genotype (right panels). Genotype- and donor-specific regression lines are shown in the left and right panel, respectively. Each data point represents a single cell. (d) The expression of RPS26 and RPL21 of whole blood bulk RNA-seq samples colored by SNP rs7297175 genotype. Genotype-specific regression lines are shown. Each data point represents a single bulk RNA-seq sample. The nominal P-value is given for the interaction effect.
To assess the robustness of the identified co-expression QTLs, we tested whether they remained significant after gene expression imputation, which was used to overcome the problem that in scRNA-seq data usually many genes are undetected despite being expressed (i.e. zero-inflated expression). Several computational strategies have been developed to do this.22–24 However, most current methods are either computationally too demanding for large datasets like ours23, or cannot sufficiently impute the 94.1% zero values present in our dataset24. To overcome this, we used MAGIC22, a method that imputes gene expression levels for nearly every gene. To prevent that imputation removes effects of genetic differences between donors or cell types, we performed imputation for each donor separately and again only for CD4+ T cells (see Data availability). In general, imputation worked well, but in some circumstances artifacts were introduced (Suppl. Fig. 3). Therefore, we only used the imputed gene expression data to determine whether the co-expression QTLs, identified prior to imputation, remained significant after imputation (Suppl. Table 4). For the three eQTL genes that were involved in a co-expression QTL, two out of three top co-expression QTLs (rs7297175 affecting the co-expression between RPS26 and RPL21, P = 3.97x10-12 (Fig. 2c) and rs4147641 affecting the co-expression between SMDT1 and RPS3A, P = 2.57x10-4) remained after imputation (Suppl. Table 4). Subsequently, we were able to replicate both effects in a whole blood bulk RNA-seq eQTL dataset11 (P = 1.69x10-3 for RPS26-RPL21 (Fig. 2d), P = 1.59x10-4 for SMDT1-RPS3A) (Suppl. Table 4). Interestingly, SNP rs7297175, affecting the co-expression between RPS26 and 106 other genes, is in near perfect linkage disequilibrium with the type I diabetes (T1D) SNP rs1117173925 (r2 = 0.98). Therefore, the numerous co-expression QTLs for RPS26 may shed new light on RPS26 and its link with T1D. This interaction effect was also observed in other cell types (Suppl. Fig. 4), indicating it is not cell type-specific. In addition, various analyses were performed to rule out potential technical confounders (see Online Methods).
The co-expression QTL analysis as outlined above highlights another advantage of scRNA-seq data; with PBMCs from only 45 donors, we could identify effects that would otherwise only become apparent in large-scale (2,116 samples) bulk RNA-seq eQTL datasets11. Due to Simpson’s paradox26, it may occur that when looking at all individuals together, the interaction between two genes does not show a correlation, while each of the individuals separately do show a correlation. So, even though the effect may be observed in bulk RNA-seq data, the true correlation will only be revealed using scRNA-seq data.
The eQTL and co-expression QTL analyses performed in this study show the benefit of scRNA-seq data for linking genetic variation to gene expression regulation. In addition to these analyses, we expect scRNA-seq data to offer many other opportunities for selecting cells of interest for eQTL and co-expression QTL analysis. For example, one could use the intercellular variation within scRNA-seq data to group cells along the cell cycle13, along a differentiation path27 or along a response to an environmental stimulus28. By doing so, one might identify eQTLs or co-expression QTLs that are influenced by cell cycle phase, differentiation or environmental status.
In conclusion, this proof of concept study shows the feasibility of using scRNA-seq data for eQTL and gene-gene interaction analysis. The identified eQTLs and co-expression QTLs replicated well with earlier reported whole blood RNA-seq data. Moreover, we extended the list of genes known to be under genetic control or specified the cell type in which the effect is most prominent. Finally, several SNPs were linked to modulation of gene co-expression, implying that gene regulatory networks can be highly personal. We expect that larger single-cell eQTL datasets will enable the identification of many cell type-specific eQTLs and genetic variants that affect regulatory network relationships.
Online methods
Isolation and preparation of PBMCs
Whole blood of 47 donors from the general population Lifelines Deep (LLD) cohort14 was drawn into EDTA-vacutainers (BD). Within 2h, peripheral blood mononuclear cells (PBMCs) were isolated using Cell Preparation Tubes with sodium heparin (BD). For all procedures, PBMCs were kept in RPMI1640 supplemented with 50 µg/mL gentamicin, 2 mM L-glutamine and 1 mM pyruvate. Isolated PBMCs were cryopreserved in RPMI1640 containing 40% FCS and 10% DMSO. Within one month, PBMCs were further processed for scRNA-seq. First, cells were thawed in a 37°C water bath until almost completely thawed, after which the cells were slowly washed in warm medium. After washing, cells were resuspended in medium and incubated for 1h in a 5° slant rack at 37°C in a 5% CO2 incubator. After this 1h resting period, cells were washed twice in medium supplemented with 0.04% bovine serum albumin. Cells were counted using a haemocytometer and cell viability was assessed by Trypan Blue. Eight, sex-balanced sample pools were prepared each containing 1750 cells/donor from 6 (or 5) donors (10,500 cells).
Single-cell library preparation and sequencing
Single cells were captured using the 10X Chromium controller (10X Genomics) according to the manufacturer’s instructions (document CG00026), and as previously described.29 Each sample pool was loaded into a different lane of a 10X chip (Single cell chip kit, 120236). cDNA libraries were generated using the Single Cell 3’ Library & Gel Bead kit version 2 (120237) and i7 Multiplex kit (120262) in line with the company’s guidelines. These libraries were sequenced using a custom program (27-9-0-138) on 8 lanes of an Illumina HiSeq4000 using a 75bp paired-end kit, per GenomeScan (Leiden, the Netherlands) sequencing guidelines. In total, 28.855 cells were captured and sequenced to an average depth of 74k.
Alignment and initial processing of sequence-data
CellRanger v1.3 software with default settings was used to demultiplex the sequencing data, generate FASTQ files, align the sequencing reads to the hg19 reference genome, filtering of cell and UMI (unique molecular identifier) barcodes, and counting gene expression per cell (see Data availability).
Demuxlet algorithm: demultiplexing samples per lane and doublet detection
Genotypes of the LLD-samples were previously generated14 and were phased using Eagle v2.330 and imputed with the HRC-reference panel31 using the Michigan Imputation Server32. As genotype data of each donor (except 2) was available, we could use the Demuxlet method33 that uses variable SNPs between the pooled individuals to determine which cell belongs to which individual and to identify doublets (two cells encapsulated in a single droplet by the 10X Chromium controller).
To determine how well every genotype matches each cell, a likelihood score was calculated by the formula: Here, c is the cell, s is the individual, v are the unique genetic variants (SNPs) found on the reads of the cell, dcv the number of unique reads overlapping with the vth variant from the cth cell. bcvi is the variant-overlapping base call from the ith read, representing reference (R), alternate (A), and other (O) alleles respectively. ecvi is a latent variable indicating whether the base call is correct (0) or not (1) and finally g is the true genotype. This likelihood score was calculated by taking into account the genotype probabilities of a sample at all known SNPs, the variant-overlapping base calls with base quality (Phred quality score) > 15, and a probability that the base was not called correctly, which is fixed at 0.001. In this way, for each pool of cells, the genotype within this pool with the highest likelihood was assigned as the most likely person the cell belonged to.
To identify doublets, likelihoods for a 50/50 ratio of all possible combinations of two genotypes were calculated, similarly as for singlets but now considering two genotypes at the same time. To consider a mix of genotypes from two individuals, the following formula was used: Here, s1 and s2 are the two individuals, g1 and g2 the corresponding true genotypes and α is the expected proportion of the SNPs in every cell for each of the individuals. An α of 0.5 was consistently used, assuming a 50/50 ratio. The maximum likelihood in the mixed-genotype case was divided by the maximum likelihood in the singlet case to obtain a likelihood ratio. If this ratio was less than 1/t for some number t, the cell was assigned to be a singlet of the sample corresponding to the maximum singlet likelihood. If the ratio was greater than t, the cell was assigned to be a doublet. When the ratio was in between 1/t and t, the cell was called inconclusive: no confident call could be made from which sample(s) the cell originated. The decision boundary factor t was fixed at 2. In theory, if there are n samples in a lane, (n – 1)/n doublets can be identified using the Demuxlet algorithm, because doublets from the same individual (1/n) cannot be identified. Further details of the algorithm can be found in Kang et al.33
Using the Demuxlet algorithm, we could confidently assign the majority (99.8%) of cells to one of the individual donors (singlets) or to two different donors (doublets) (Suppl. Fig. 1a, Suppl. Table 5). Remarkably, in two out of eight sample pools, no cells were assigned to one of the six donors within the pool. Moreover, the detected doublet rate in those sample pools was abnormally high (17.5% and 21.1%, while 3-4% was expected) (Suppl. Table 5). This is most probably due to a sample mix-up in the lab which resulted in an artificially high doublet rate. Since the genotypes of these two mixed-up samples were not available, those samples were excluded from the analysis (marked as “doublet”).
Two additional tests were performed to confirm the correct assignment of cells using Demuxlet. First, we determined what would happen if the cells did not match with their genotypes by taking six random genotypes not present in the sample pool itself. This resulted in 0.02% of the cells being a singlet, 0.03% being inconclusive and 99.95% being a doublet. Second, the number of reads mapping to the Y-chromosome was determined for the singlets of each donor. Cells belonging to a female donor showed (almost) no Y-reads (mismapping reads34 may explain the few sporadic Y-reads), whereas the majority of cells from male donors did (Suppl. Fig. 1b). So, the correct gender for each of the donors could be confirmed by looking at the number of Y-reads. These tests indicated that the Demuxlet method is correctly assigning cells to their respective donor and is suitable for detecting sample swaps.
Cell type classification
Version 1.4 of the R package Seurat16 was used to determine the cell types using the raw UMI counts from CellRanger. First, all genes that were not detected in ≥3 cells were removed. Cells in which >5% of the UMIs mapped to the mitochondrial-encoded genes were discarded as this can be a marker of bad quality cells; broken cells will leak cytoplasmic RNA, while the mitochondrial RNA content is retained inside the mitochondria.35 Also, cells expressing >3,500 genes were considered outliers and discarded (Suppl. Fig. 1c, Suppl. Table 6). Finally, all cells that were marked as doublet or inconclusive by the Demuxlet method were discarded. Supplementary figure 1d shows a t-Distributed Stochastic Neighbor Embedding (t-SNE) plot36 in which all cells failing the above QCs are visualized. Library size normalization was performed on the UMI-collapsed gene expression for each barcode by scaling the total number of transcripts per cell to 10,000. The data was then log-2 transformed. In total, 25,291 cells and 19,723 genes (average of 1147 detected genes/cell) (see Data availability) were used in the cell type determination.
Linear regression was used to regress out the total number of UMIs and the fraction of mitochondrial transcript content per cell. The variable genes were identified using Seurat’s MeanVarPlot function which places all genes in 20 bins based on their average expression (the mean of non-zero values) and calculates the dispersion (standard deviation of all values) within each bin. Standard parameters were used except the bottom gene expression cut-off (x.low.cutoff) was set to 0 and the bottom dispersion cut-off (y.cutoff) was set to 1.0, resulting in the identification of 1,090 genes. These 1,090 variable genes were used in the principle component analysis (PCA). The first 16 principal components were used for cell clustering using Seurat’s FindCluster function (default parameters, resolution 1.2) and a t-SNE plot was used to visualize this. Based on known marker genes and differentially expressed genes per cluster (found using Seurat’s FindMarkers function), we could assign 11 cell types to the clusters, including some smaller subcell types (Suppl. Fig. 2a, 2b, Suppl. Table 7). The smallest cluster we could detect consisted of plasma cells, making up 0.3% of the total PBMC population.
eQTL analysis
To find the association between genotype and expression per cell type, genome-wide cis-eQTL analysis for 18,264 genes (only autosomal genes, gene expressed in at least 3 cells within the total dataset and in at least 1 cell within the cell type queried, within 100 kb distance of the SNP and the gene midpoint, MAF>0.1, call rate >0.95, a Hardy-Weinberg equilibrium P value of >0.001) was performed using our previously described eQTL pipeline, version 1.2.4F (Suppl. Table 2, see Data availability).11 To assure sufficient power, cell types were merged to a more general classification: CD4+ T cells, CD8+ T cells, NK cells (CD56dim CD16+ and CD56bright CD16+/-), monocytes (CD14bright CD16- classical, cMonocyte, and CD14dim CD16+ non-classical, ncMonocyte), B cells and DCs (CD1C+ myeloid, mDC and plasmacytoid, pDC). The mean expression per gene per cell type per donor was calculated on the normalized (Z-score transformed) expression and used as input for the eQTL analysis. eQTLs were mapped using Spearman’s rank correlation coefficient on imputed genotype dosages. eQTLs were considered significant at a gene-level FDR of 0.05. To control the FDR at 0.05 we used the permutation method described previously by us.2 Here we permute the link between the genotypes and expression data and create an overall null distribution using all genes. We performed in total 10 permutations and use for each gene the total null distribution of all genes to determine a gene-level FDR: during FDR estimation only the most significant SNP per gene is used, both for the real analysis and for each of the permutations.
Concordance and detection
Concordance with previously found independent top eQTLs from a whole blood DeepSAGE (3’-end transcriptomics)15 and RNA-seq study11 were computed. For this, the mean expression per gene per individual of all cells was calculated and the cis-eQTL mapping was confined to the independent top eQTLs found in the DeepSAGE15 or RNA-seq study11. Subsequently, detection of the same SNP-gene combination and concordance (with same allelic direction) were assessed between the significant top effects (Suppl. Table 1). Vice-versa, we also determined how many of the 379 top eQTLs in our scRNA-seq dataset could be detected and with which allelic direction within the whole blood RNA-seq study11. Similarly, we assessed detection rate and concordance with two studies containing RNA-seq data of purified cell types: Kasela et al. performed eQTL analysis on purified CD4+ and CD8+ T cells6, whereas the data from the Blueprint consortium contains purified CD14+ monocytes and naïve CD4+ T cells17 (Suppl. Table 2, 3). Moreover, for the eQTLs that were specifically detected in the cMonocytes and not the ncMonocytes (Fig. 2d), detection rate and concordance were determined using the RNA-seq data of the purified CD14+ monocytes from the Blueprint consortium17.
Single-cell gene expression imputation
To overcome the zero-inflated expression, the computational method MAGIC22 was used to impute practically all values of genes with at least some expression. MAGIC imputation (using the following parameters: 20 PCs, t=4, k=9, ka=3, ε=1) was performed per donor separately and only in the CD4+ T cells (see Data availability). The effect of MAGIC imputation was validated by comparing the co-expression of typical cell type-specific marker genes (Suppl. Fig. 3).
Co-expression QTL analysis
For every individual, a Spearman’s rank correlation coefficient was calculated between the expression of the cis-eQTL gene and all other genes. Given the large zero-inflation of scRNA-seq data, we only tested those 7,975 genes that showed variance in expression for each of the 45 samples. As a consequence we could study 102 eQTL genes out of the 145 unique genes that showed a significant cis-eQTL effect in CD4+ T-cells. For each of these combinations, a weighted linear model was used (co-expression ~ genotype, where weight is in which the explained variable is a Spearman correlation coefficient that describes the co-expression between the two genes and the genotype is the predictor and the weights are the square root of the number of CD4+ T cells within the given sample (Suppl. Fig. 5).
In order to determine for how many cis-eQTL genes we had identified a significant co-expression QTL we performed 100 permutations (see Data availability). For the real analysis we denoted for each of the tested 102 eQTL genes what was the most significant co-expression QTL P-Value (Suppl. Table 4). For each permutation we shuffled the genotype identifiers and reran the above analysis and also determined for each of the 102 eQTL genes what was the most significant co-expression QTL P-Value (see Data availability). This subsequently enabled us to calculate an eQTL-gene level FDR2 (using exactly the same multiple testing correction procedures as we employ for the detection of cis-eQTLs, see paragraph “eQTL analysis”). An eQTL gene-level FDR of 0.05 was considered significant, i.e. the p-value threshold of the most significant co-expression QTL p-values at which 5% of the co-expression QTLs are significant in the permuted compared to the real data.
All significant co-expression QTLs were discovered using non-imputed gene expression data. We then assessed whether these co-expression QTLs were also significant when using the MAGIC-imputed gene expression data. Subsequently, we tested whether these co-expression QTLs replicated using a large whole blood bulk RNA-seq dataset11 (Suppl. Table 4). We finally attempted to falsify the observed co-expression QTL for rs7297175 on the co-expression between RPS26 and RPL21, by checking the following potential confounders:
-
-
Potential sequence homology: no evidence was found for sequence homology between RPS26 and RPL21.
-
-
Genotype-dependent mapping problems of RNA sequence reads: no evidence was found that the RPS26 cis-eQTL SNP rs7297175 has any SNP proxies (r2 > 0.8) that are coding and that map within RPS26. As such this suggests that potential genotype-dependent mapping biases of sequence-reads are unlikely.
-
-
Multi-mapping of RNA sequence reads: no differences were found between individuals with regards to the amount of sequence reads that were discarded due to multi-mapping of sequence reads to RPS26.
-
-
Unexpected trans-eQTL on RPL21: no evidence was found that the RPS26 cis-eQTL SNP rs7297175 is affecting the expression of RPL21 in trans.
-
-
Genotype-dependent subcell-type composition effects: the RPS26-RPL21 co-expression QTL is unlikely the result of a subcell-type within the CD4+ T cell population, as this co-expression QTL effect is also significant within CD8+ T cells, within monocytes and within NK cells (Suppl. Figure 4).
Data availability
Raw gene expression counts, MAGIC imputed CD4+ T cell gene expression, and eQTL and co-expression QTL summary statistics can be found under “Supplementary Data” at the website accompanying this paper (https://molgenis58.target.rug.nl/scrna-seq/).
Processed (deanonimyzed) single-cell RNA-seq data, including a text file that links each cell barcode to its respective donor, has been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001002560. Gene expression and genotype data can be obtained and requested by filling in a single and short web form at https://molgenis58.target.rug.nl/scrna-seq/. This form is subsequently reviewed by a single Data Access Committee, who will be able to approve access to both the raw gene expression and genotype data within 5 working days (during the holiday season there might be a slight delay). Once the proposed research is approved, access to the relevant gene expression or genotyped data will be free of charge. Access to the genotype and gene expression data is facilitated via the Lifelines workspace and the EGA, respectively. Sample metadata (age, gender, processing batch) is presented in Suppl. Table 8.
Code availability
The original R code for Seurat16 (https://github.com/satijalab/seurat), Demuxlet33 (https://github.com/statgen/demuxlet), MAGIC22 (https://github.com/KrishnaswamyLab/magic) and our in-house eQTL pipeline2 (https://github.com/molgenis/systemsgenetics/tree/master/eqtl-mapping-pipeline) can be found at Github. All custom-made code is made available via GitHub (https://github.com/molgenis/scRNA-seq).
Supplementary Material
Acknowledgements
We are very grateful to all the volunteers who participated in this study. Moreover, we thank J. Dekens for arranging informed consent and contact with LifeLines. We thank A. Maatman and M. Platteel for their assistance in the lab. M.S and L.F. are supported by grants from the Dutch Research Council (ZonMW-VIDI 917.164.455 to M.S. and ZonMW-VIDI 917.14.374 to L.F.) and L.F. is supported by an ERC Starting Grant, grant agreement 637640 (ImmRisk). The Biobank-Based Integrative Omics Studies (BIOS) Consortium is funded by BBMRI-NL, a research infrastructure financed by the Dutch government (NWO 184.021.007).
Footnotes
Author contribution
MW generated the scRNA-seq data. MW, HB and DV performed bioinformatics and statistical analyses. PD and BIOS consortium performed replication of co-expression QTLs. MW and LF designed the study and wrote the manuscript. MS and the LLD consortium provided biomaterials, genotype data and computational resources. All authors discussed the results and commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Ethics approval and consent to participate
The LifeLines DEEP study was approved by the ethics committee of the University Medical Centre Groningen, document number METC UMCG LLDEEP: M12.113965. All participants signed an informed consent form prior to study enrollment. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
References
- 1.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Westra HJ, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown CD, Mangravite LM, Engelhardt BE. Integrative modeling of eQTLs and cis-regulatory elements suggests mechanisms underlying cell type specificity of eQTLs. PLoS Genet. 2013;9:e1003649. doi: 10.1371/journal.pgen.1003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fairfax BP, et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012;44:502–510. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu J, et al. Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS Genet. 2012;8:e1002431. doi: 10.1371/journal.pgen.1002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasela S, et al. Pathogenic implications for autoimmune mechanisms derived by comparative eQTL analysis of CD4+ versus CD8+ T cells. PLOS Genetics. 2017;13:e1006643. doi: 10.1371/journal.pgen.1006643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naranbhai V, et al. Genomic modulators of gene expression in human neutrophils. Nat Commun. 2015;6:7545. doi: 10.1038/ncomms8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishigaki K, et al. Polygenic burdens on cell-specific pathways underlie the risk of rheumatoid arthritis. Nat Genet. 2017 doi: 10.1038/ng.3885. [DOI] [PubMed] [Google Scholar]
- 9.Westra H, et al. Cell Specific eQTL Analysis without Sorting Cells. PLOS Genetics. 2015;11:e1005223. doi: 10.1371/journal.pgen.1005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venet D, Pecasse F, Maenhaut C, Bersini H. Separation of samples into their constituents using gene expression data. Bioinformatics. 2001;17(Suppl 1):S279–87. doi: 10.1093/bioinformatics/17.suppl_1.s279. [DOI] [PubMed] [Google Scholar]
- 11.Zhernakova DV, et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nat Genet. 2017;49:139–145. doi: 10.1038/ng.3737. [DOI] [PubMed] [Google Scholar]
- 12.Villani AC, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356 doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wills QF, et al. Single-cell gene expression analysis reveals genetic associations masked in whole-tissue experiments. Nat Biotechnol. 2013;31:748–752. doi: 10.1038/nbt.2642. [DOI] [PubMed] [Google Scholar]
- 14.Tigchelaar EF, et al. Cohort profile: LifeLines DEEP, a prospective, general population cohort study in the northern Netherlands: study design and baseline characteristics. BMJ Open. 2015;5 doi: 10.1136/bmjopen-2014-006772. e006772-2014-006772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhernakova DV, et al. DeepSAGE reveals genetic variants associated with alternative polyadenylation and expression of coding and non-coding transcripts. PLoS Genet. 2013;9:e1003594. doi: 10.1371/journal.pgen.1003594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, et al. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell. 2016;167:1398–1414.e24. doi: 10.1016/j.cell.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flutre T, Wen X, Pritchard J, Stephens M. A Statistical Framework for Joint eQTL Analysis in Multiple Tissues. PLOS Genetics. 2013;9:e1003486. doi: 10.1371/journal.pgen.1003486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sul JH, Han B, Ye C, Choi T, Eskin E. Effectively identifying eQTLs from multiple tissues by combining mixed model and meta-analytic approaches. PLoS Genet. 2013;9:e1003491. doi: 10.1371/journal.pgen.1003491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duong D, et al. Applying meta-analysis to genotype-tissue expression data from multiple tissues to identify eQTLs and increase the number of eGenes. Bioinformatics. 2017;33:i67–i74. doi: 10.1093/bioinformatics/btx227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knowles DA, et al. Allele-specific expression reveals interactions between genetic variation and environment. Nat Methods. 2017;14:699–702. doi: 10.1038/nmeth.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Dijk D, et al. MAGIC: A diffusion-based imputation method reveals gene-gene interactions in single-cell RNA-sequencing data. bioRxiv. 2017 [Google Scholar]
- 23.Huang M, et al. Gene Expression Recovery For Single Cell RNA Sequencing. bioRxiv. 2017 doi: 10.1038/s41592-018-0033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li WV, Li JJ. An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat Commun. 2018;9:997. doi: 10.1038/s41467-018-03405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson EH. The Interpretation of Interaction in Contingency Tables. Journal of the Royal Statistical Society.Series B (Methodological) 1951;13:238–241. [Google Scholar]
- 27.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shalek AK, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng GXY, et al. Massively parallel digital transcriptional profiling of single cells. Nature Communications. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loh PR, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang HM, et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol. 2017 doi: 10.1038/nbt.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosser ZH, Balaresque P, Jobling MA. Gene conversion between the X chromosome and the male-specific region of the Y chromosome at a translocation hotspot. Am J Hum Genet. 2009;85:130–134. doi: 10.1016/j.ajhg.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ilicic T, et al. Classification of low quality cells from single-cell RNA-seq data. Genome Biol. 2016;17 doi: 10.1186/s13059-016-0888-1. 29-016-0888-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van de Maaten L, Hinton G. Visualizing data using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw gene expression counts, MAGIC imputed CD4+ T cell gene expression, and eQTL and co-expression QTL summary statistics can be found under “Supplementary Data” at the website accompanying this paper (https://molgenis58.target.rug.nl/scrna-seq/).
Processed (deanonimyzed) single-cell RNA-seq data, including a text file that links each cell barcode to its respective donor, has been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001002560. Gene expression and genotype data can be obtained and requested by filling in a single and short web form at https://molgenis58.target.rug.nl/scrna-seq/. This form is subsequently reviewed by a single Data Access Committee, who will be able to approve access to both the raw gene expression and genotype data within 5 working days (during the holiday season there might be a slight delay). Once the proposed research is approved, access to the relevant gene expression or genotyped data will be free of charge. Access to the genotype and gene expression data is facilitated via the Lifelines workspace and the EGA, respectively. Sample metadata (age, gender, processing batch) is presented in Suppl. Table 8.