Abstract
The TU8 mutant of Arabidopsis previously described to be deficient in glucosinolate metabolism and pathogen-induced auxin accumulation was found to be remarkably less tolerant upon exposure to elevated temperatures than wild-type plants. Although moderately increased temperature only affected shoot growth, exposure to severe heat stress led to a dramatic decay of mutant plants. By contrast, wild-type seedlings showed little or no damage under the same conditions. Analysis of different heat stress proteins (Hsps) in TU8 seedlings revealed that only expression of cytoplasmic Hsp90 was affected in these plants. Although Hsp90 was present under control conditions, its level declined in mutant plants at elevated temperatures. Northern-blot analysis indicated that the decrease in Hsp90 protein was accompanied with a reduction of hsp90 transcript levels. Transient expression of Hsp90 in mutant protoplasts increased their survival rate at higher temperatures to near equivalent that of wild-type protoplasts. These data suggest that the reduced level of Hsp90 in TU8 mutants may be the primary cause for the observed reduction in thermostability.
Pioneer weeds like Arabidopsis are adapted to a broad temperature amplitude, which is a prerequisite for enduring changes in the environment for sessile organisms. Survival above optimal temperature conditions is accompanied by a massive accumulation of heat stress proteins (Hsps). Most of them belong to a group of proteins termed molecular chaperones (Ellis and van der Vries, 1991). Their main task is to assist refolding of partially unfolded or denatured proteins occurring under elevated temperatures (for review, see Forreiter and Nover, 1998). Deficiency in the expression of chaperones often results in increased thermosensitivity or death of the organism even under normal growth conditions (Lindquist, 1986; Nover, 1991). In this article, we describe an Arabidopsis mutant that failed to survive temperatures above 25°C when grown in the greenhouse, whereas wild-type seedlings grown under the same conditions were not affected. This mutant line, designated TU8, has been analyzed for its reduced thermostability and Hsp expression. It is interesting that TU8 showed pleiotrophic effects such as an altered phenotype (shorter stems and reduced apical dominance) and differences in auxin induction upon infection with Plasmodiophora brassicae, the causal agent of clubroot disease of Brassicaceae (Ludwig-Müller et al., 1999). TU8 was first isolated by Haughn et al. (1991) on the basis of its altered glucosinolate patterns. Glucosinolates are mainly synthesized by Brassicaceae but are also present in other plant species (Rodman, 1991). The level of glucosinolates is dependent on tissue type and age (Porter et al., 1991) and increases after wounding and pathogen attack (Doughty et al., 1991). Thus far, three distinct groups have been described (Bennett and Wallsgrove, 1994; Bennett et al., 1995)which contain more than 100 different members (Heaney and Fenwick, 1987; Daxenbichler et al., 1991). It has been discussed that glucosinolates play a role in plant defense against bacterial and fungal pathogens as well as predatory insects, but they may also be involved in host/pathogen recognition (Bennett and Wallsgrove, 1994). Indole glucosinolates are hypothesized to be especially important for symptom development of clubroot disease (Butcher et al., 1974). Altered glucosinolate metabolism of TU8 and its effect upon infection with P. brassicae was already described in detail by Ludwig-Müller et al. (1999). Whereas auxin concentrations in roots of wild-type plants increased markedly after infection, the concentrations in mutant plants remained unaffected. In addition, TU8 mutants showed delayed symptom development accompanied by reduced fungal structures within the root cortex and a slower development of the fungus.
In this study, we analyze the observed reduced thermostability in TU8 mutant plants in more detail and present evidence that the increased sensitivity to high temperatures may be linked to a possible defect in cytoplasmic Hsp90 expression.
RESULTS
TU8 Plants Are Affected in Growth and Survival Rate at Higher Temperatures
Under normal growth conditions (22°C) mature TU8 plants were one-third smaller than the wild type, although shoots revealed more branching (Fig. 1F). When cultivated at higher temperatures in the greenhouse (>25°C), mutant plants showed a decrease in growth development and decayed after 2 weeks, whereas wild-type plants were essentially unaffected. This increased thermosensitivity correlated with the above described TU8 phenotype and segregated in a 3:1 ratio in the F2 generation when TU8 was backcrossed with Columbia (Col) wild-type plants (Table I).
Figure 1.
Phenotypes of Col wild-type (left) and TU8 mutant seedlings (right) at different temperatures. Sixteen-day-old seedlings before exposure to elevated temperatures (A) and after 6 d at 22°C (B), 34°C (C), 37°C (D), and 42°C (E), respectively. Col and TU8 plants after 14 d at 22°C (F), 27°C (G), 32°C (H), and 37°C (J). Bars represent 1 cm.
Table I.
Cross of Col and TU8 Arabidopsis plants
| Parental Plants | Resulting Seedlings | TU8 Phenotype | Col Phenotype | Ratio |
|---|---|---|---|---|
| 1 | 63 | 13 | 50 | 1:3.8 |
| 2 | 57 | 19 | 38 | 1:2 |
| 3 | 52 | 16 | 36 | 1:2.3 |
| 4 | 59 | 16 | 43 | 1:2 |
| 5 | 52 | 9 | 43 | 1:4.8 |
| 6 | 60 | 14 | 46 | 1:3.3 |
| 7 | 57 | 16 | 41 | 1:2.6 |
| 8 | 60 | 18 | 42 | 1:2.3 |
| 9 | 63 | 17 | 46 | 1:2.7 |
| 10 | 57 | 8 | 49 | 1:6.1 |
| 11 | 60 | 13 | 47 | 1:3.6 |
| 640 | 159 | 481 | 1:3 |
F2 was analyzed for segregation of the apparent TU8 phenotype and the results of individual crosses are presented.
Reduced growth of TU8 at elevated temperatures was further investigated in detail by cultivating wild-type and mutant plants for 16 d at 22°C and subsequently transferring them to growth chambers with different temperatures. Wild type and mutant grown at 22°C are displayed in Figure 1A, and Figure 1, B through J, shows plants additionally exposed to the indicated temperatures for 6 or 14 d . After 6 d no major differences were observed in any of the plants below 32°C (Fig. 1B). At 34°C however, TU8 plants showed the first signs of senescence, whereas wild-type seedlings were not affected (Fig. 1C). Higher temperatures caused damage to both types with different severity (Fig. 1, D and E). Both wild-type and TU8 plants were not able to survive long periods at 37°C (Fig. 1J); however, wild-type plants remained alive for an additional 4 and 2 d at 37°C and 42°C, respectively, as compared to TU8 plants (Figs. 1, D, E, H, and J, and 2).
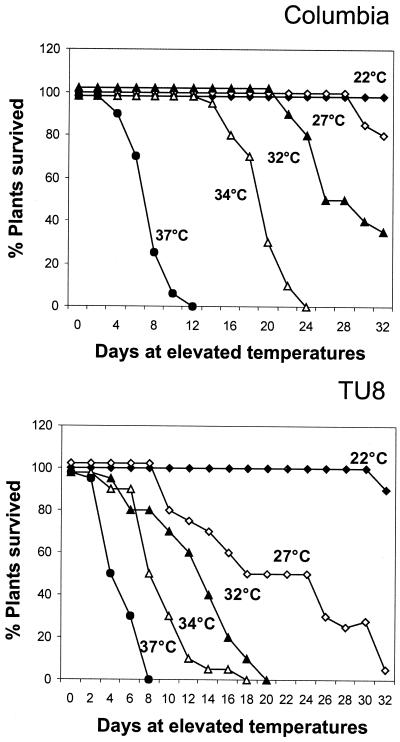
Differences in TU8 and Col decay under elevated temperatures are summarized in Figure 2. At 32°C, no TU8 plant survived after 3 weeks and only 50% of the plants were able to survive at 27°C. At these temperatures, all Col plants were still alive, although they had started to show signs of senescence (yellow leaves, dry flowers, and pod formation was inhibited), which increased as the temperature did. At 34°C, the effect appeared to be more pronounced. At these temperatures, more than 90% of the TU8 plants were dead by d 12. Col plants survived for an additional 10 d. Although other effects like water stress cannot be ruled out, the results indicate that under our growth conditions, the basal thermostability of Arabidopsis wild-type seedlings is approximately 30°C, whereas under the same conditions, TU8 plants were not able to survive more than 22°C. These data were confirmed by T50-values (days after transfer to elevated temperatures with 50% survival rate) at different temperatures. Although 42°C was lethal for Col as well as for TU8, at 27°C a T50-value of 18 d was determined for TU8, while all Col plants were still alive (not shown).
Figure 2.
Survival rate of Col and TU8 plants at different temperatures. Plants were grown at 22°C for 3 weeks and subsequently placed in growth chambers set at the indicated temperatures. Survival rate was estimated every 3rd d.
The previously described data in Figures 1 and 2 were obtained with plants immediately exposed to the indicated temperatures. However, if plants and other organisms are exposed to a short heat pulse followed by a recovery period at normal temperature, prior to a further heat treatment, it leads to an increased synthesis of Hsps, which results in a better survival rate at elevated temperatures. We were interested to know whether a brief non-lethal heat pulse (2 h at 42°C) of Col and TU8 mutant plants after 3 weeks of growth at normal temperature leads to an increased capability to resist higher temperatures during subsequent cultivation at 32°C. After 15 d at 32°C, plants were monitored for their phenotypes and fresh weight.
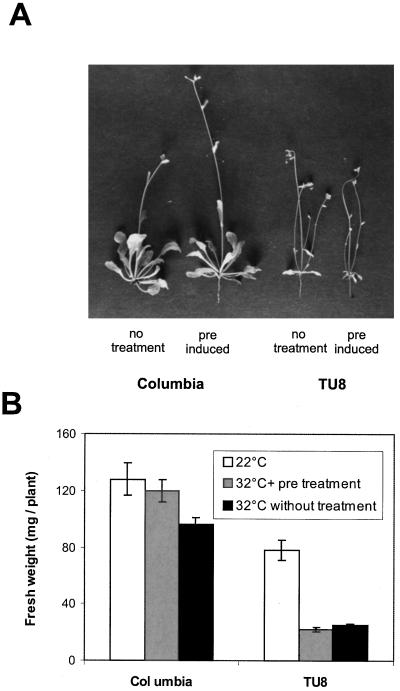
In Figure 3A, representative wild-type and mutant seedlings are shown. Whereas Col plants without pretreatment were slightly reduced in shoot length (Fig. 3A, left side), heat conditioned Col plants were in better shape and showed no differences when cultivated at 32°C, compared to plants grown at 22° (Fig. 1F). On the other hand, TU8 seedlings suffered dramatically at 32°C in general (Fig. 3A, right side), if compared to mutant plants at normal temperatures (Fig. 1). It could be shown that in contrast to Col seedlings, an additional heat pulse of 2 h at 42°C prior to incubation at 32°C did not increase TU8 ability to stand cultivation at 32°C (Fig. 3A, right side). They had strongly reduced and senescing leaf rosettes with multiple tiny shoots and, following an initial heat pulse, TU8 showed slightly more damage when grown at 32°C.
Figure 3.
Comparison of Col and TU8 seedlings after exposure to a brief heat pulse (2 h at 42°C) followed by growth at 32°C for 15 d. A, Col seedlings without heat pretreatment (1); Col with 2 h of heat stress at 42°C prior to incubation at 32°C (2); TU8 without heat pretreatment (3); and TU8 with 2 h of heat stress at 42°C prior to incubation at 32°C (4). B, After 15 d of growth at 32°C, fresh weight of 80 Col and TU8 plants each with or without heat pretreatment (2 h at 42°C) was determined and compared to plants grown at 22°C.
This effect can also be demonstrated when the fresh weight of pretreated and non-treated seedlings after 15 d of incubation at 32°C was determined and compared to plants grown at room temperature (Fig. 3B); only preconditioned Col seedlings had no significant difference in fresh weight compared to control plants kept at room temperature. Non-treated plants showed approximately 25% reduction in fresh weight. In contrast, both non-treated and heat-conditioned TU8 plants had a similar reduction (approximately 60%) in fresh weight compared to mutant plants grown at 22°C after 15 d. This suggests that TU8 plants were not able to benefit from an initial heat pulse and could not acquire increased thermotolerance.
Western-Blot Analysis of Hsp Expression in Wild-Type and TU8 Seedlings
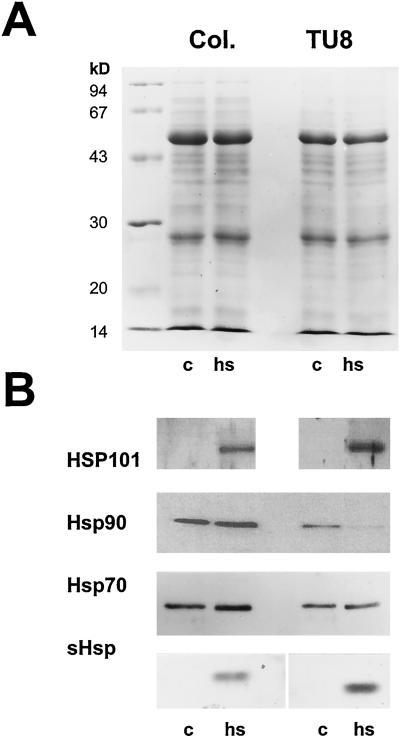
One reason for the observed increased thermosensitivity of TU8 may be reduced synthesis of Hsp proteins. The presence of the four most prominent Hsp representatives, Hsp101, Hsp90, Hsp70, and small Hsps (sHsps) was tested in wild-type and TU8 seedlings kept under either control or heat stress conditions (3 h at 42°C). A Coomassie-stained gel showed no significant change in the overall protein pattern between wild-type and TU8 mutant plants after heat stress (Fig. 4A). Western-blot analysis, however, revealed that Hsp101 and sHsps were strongly induced after exposure to elevated temperatures, whereas Hsp70 was present in both control and heat-treated seedlings, increasing slightly upon heat. The expression pattern between wild-type or TU8 seedlings was identical. Analysis of Hsp90 expression resulted in a different picture. Although Hsp90 was present in both wild-type and mutant plants under control conditions, the signal increased only in wild-type seedlings at elevated temperatures, whereas in mutant plants, the signal dropped below the limit of detection (Fig. 4B).
Figure 4.
Protein pattern of Col and TU8 plants after incubation for 3 h at either 25°C (c) or at 42°C (heat stress). Equal amounts (20 μg) of protein were separated by SDS-PAGE and either stained with Coomassie Blue (A) or transferred onto nitrocellulose and subsequently incubated with antibodies directed against Hsp101, Hsp90, Hsp/Hsc70, or Hsp17.6 (B).
In Situ Localization of Hsps in Wild-Type and TU8 Mesophyll Cells
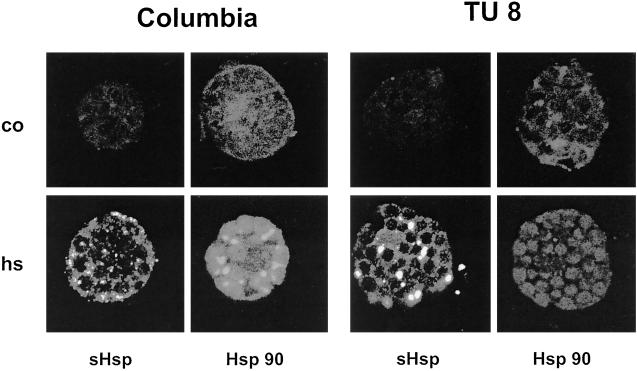
Since Hsp90 homologs are reported to be localized in other cellular compartments like plastids (Schmitz et al., 1996), we were interested to know which Hsp90 was affected in the TU8 mutant. For this reason, we isolated protoplasts from Col and TU8 leaves either kept under control conditions or exposed to a moderate heat stress to induce Hsp synthesis. Protoplasts were incubated either with pre-immunserum, which did not result in a detectable fluorescence signal (not shown), or with sera raised against Hsp90. Expression of Hsp17.6 was monitored for control of the cellular heat stress response. The study of the intracellular Hsp90 distribution revealed two interesting results (Fig. 5): (a) The fluorescence signal in Col and TU8 was spread all over the cell, including plastids, indicating that, in contrast to the sHsp antibody used, Hsp90 antiserum recognized both plastidic and cytoplasmic Hsp90; and (b) compared to wild-type cells, the fluorescence signal in TU8 cells was reduced, even under control conditions. Following heat stress treatment, a strong overall increase was observed in wild-type cells, whereas the signal for Hsp90 disappeared in the cytoplasm of TU8 cells. Still, a signal remained in the plastids, indicating that the organellar Hsp90 in mutant cells was not affected.
Figure 5.
In situ localization of Hsps in wild-type and TU8 mutant cells. Leaf mesophyll cells were treated for 30 min at 38°C, for 30 min at 39°C, and for 2 h at 40°C. After fixation cells were incubated with antibodies against Hsp17.6 or Hsp90. The antibody-protein complex was detected with an FITC-conjugated secondary antibody and analyzed using confocal laserscan microscopy.
As expected, sHsp expression is strictly heat dependent in mesophyll cells and no significant difference could be detected between wild-type and TU8 cells. In contrast to the Hsp90 antiserum, sHsps antibodies recognized only the cytoplasmic protein. As expected from the western analysis, no differences in expression of sHsps were detectable between TU8 and wild-type cells, either in the amount or in their capability of forming heat stress granules. These subcellular particles leading to a spotty appearance of the sHsp fluorescence signal emerge transiently in plant cells under heat stress and contain mostly sHsps (Nover et al., 1983; Stuger et al., 1999).
Transcript Analysis of Hsp90
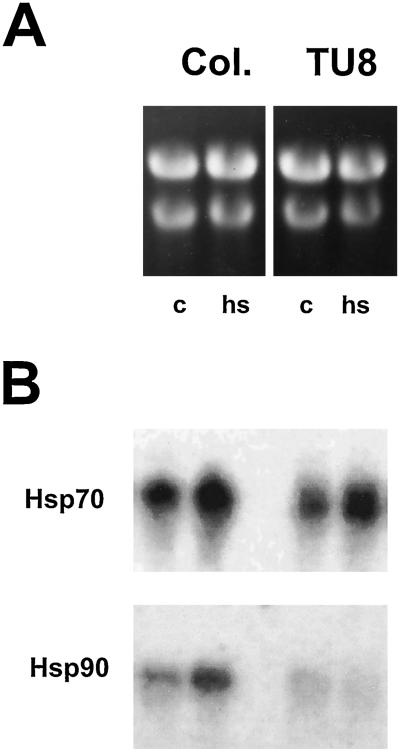
To determine if the reduced levels of Hsp90 protein in TU8 mutants are the result of reduced levels of hsp90 mRNA, total RNA obtained from seedlings grown under normal conditions (25°C) or from seedlings treated for 3 h at 42°C was analyzed for hsp90 transcripts. As a control, hsp70 transcript levels were determined. A strong constitutive signal increasing in intensity at elevated temperatures was detected in both wild-type and mutant plants for hsp70 transcripts (Fig. 6B). hsp90 transcripts, however, showed a different expression pattern in wild-type and mutant seedlings (Fig. 6B). Although an increase in hsp90 transcript levels after heat stress was observed for Col plants, the signal in TU8 plants was weaker and did not increase under heat stress conditions. High homology within the coding region of all currently known genomic Hsp90 members from Arabidopsis makes it difficult to obtain probes specific for individual members of the Arabidopsis Hsp90 family. For this reason, transcript reduction cannot be attributed to an individual member of the Hsp90 family and cross hybridization of the probe with the gene coding for plastid hsp90 cannot be excluded. Together with the data obtained by immunofluorescence, it is likely that the signal obtained after heat stress may arise from hybridization with mRNA encoding organellar hsp90.
Figure 6.
Northern-blot analysis of Hsp transcripts under control (c; 25°C) and heat stress (hs; 42°C) conditions. Total RNA was isolated and separated on denaturing gels (A) or transferred to solid support and hybridized with probes coding for hsp70 and hsp90 (B). Lane 1 represents RNA from seedlings under control conditions; lane 2 RNA from seedlings exposed for 3 h under heat stress conditions.
Transient Transformation with Hsp90 Increases Thermostability in TU8 Mutants
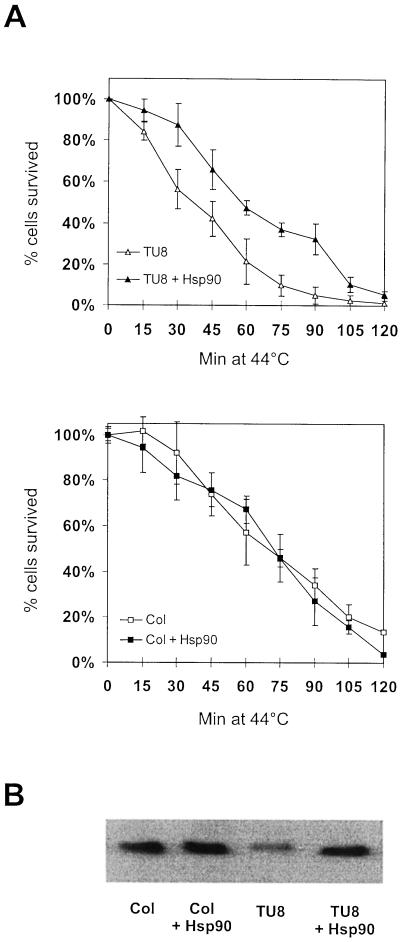
To further investigate if the reduced amount of Hsp90 is responsible for the heat stress sensitivity in TU8 mutant, leaf mesophyll protoplasts were isolated and transiently transformed with a plasmid containing Brassica napus hsp90-1 cDNA under regulation of a double 35S-cauliflower mosaic virus promoter. As controls, wild-type and TU8 cells were mock-treated with empty vector. In addition, Col cells were treated with an Hsp90 coding plasmid to analyze whether additional Hsp90 could further improve their thermostability. After 16 h, cells were analyzed for Hsp90 expression and exposed to a 120-min heat stress at 44°C. Although this treatment killed nearly all protoplasts, TU8 cells died faster than wild-type cells (Fig. 7A). If TU8 cells were substituted with a cDNA coding for cytoplasmic Hsp90, cells declined almost like wild-type protoplasts. This suggests that the reduced thermostability of the TU8 mutant is most likely caused by deficiency in Hsp90 protein levels. Overexpression of Hsp90 in wild-type cells resulted only in slightly increased amounts of Hsp90, which had no apparent effect on the survival rate of these cells. This result may reflect tight control of Hsp90 synthesis in wild-type cells, since overexpression of chaperones, Hsp90 in particular, under normal growth conditions often resulted negative effects, as observed in yeast (Scheibel et al., 1999).
Figure 7.
Transient expression of Hsp90 in TU8 leaf mesophyll protoplasts. TU8 mesophyll protoplasts were transformed either with the hsp90 coding vector or with vector alone. After overnight incubation for protein expression, cells were incubated at 44°C for 120 min. A, Cell survival was estimated by adding Evans blue dye every 15 min. The average value of four independent experiments ± sd was taken. B, Presence of Hsp90 was verified by western-blot analysis of control cells (lane 1), control cells expression construct (lane 2), TU8 cells (lane 3), and TU8 cells transformed with Hsp90, respectively.
DISCUSSION
In the present study we show that the TU8 mutant of Arabidopsis, which is deficient in glucosinolate metabolism and fails to respond with an increased auxin level following infection with P. brassicae (Ludwig-Müller et al., 1999), is considerably less able to survive elevated temperatures than wild-type plants. At 32°C and above TU8 plants decayed progressively. Below this temperature mutant plants developed slightly faster than the wild type, but did not show early senescence. Since TU8 plants started bolting several days before the wild type, it can be argued that the life cycle of these plants is shorter in general. However, careful analysis of differences concerning the survival rate of Col and TU8 at temperatures between 27°C and 34°C suggests that the faster decay of TU8 plants is not in line with this argument.
There are several reasons for reduced thermostability ranging from a deficiency in ribosome assembly to a high temperature-caused block of tubulin assembly. It can also be due to deficiency in expression of Hsps, as has been demonstrated for several yeast mutants (for review, see Lindquist, 1986). Hsps are a group of unrelated proteins involved in (re)folding of partially denatured polypeptides. Referred to as molecular chaperones (Ellis and van der Vries, 1991), Hsps interact with partially folded proteins and keep them in a folding-competent state (for review, see Beissinger and Buchner, 1998; Forreiter and Nover, 1998).
Some Hsp proteins act together as chaperone machines, as described for Hsp70 and Hsp40 or GroEL/GroES proteins (Hartl, 1996; Bukau and Horwich, 1998). These chaperone machines are reported to be linked, causing a cellular network for maintenance of protein folding and refolding, which allows organisms to endure temperatures beyond optimal conditions (Frydman and Höhfeld, 1997; Johnson and Craig, 1997). In this network every component has a special task. Whereas the Hsp70/Hsp40-system (or DnaK/J in prokaryotic organisms) plays the central element in folding of nascent polypeptides or protein transport (Bukau and Horwich, 1998), Hsp100 members are reported to disassemble already aggregated proteins (Schirmer et al., 1996) and small stress proteins provide a non-specific surface for partially unfolded proteins to keep them in a competent state for refolding (Vierling, 1991; Ehrnsperger et al., 1997; Forreiter et al., 1997; Lee et al., 1997).
A special role in this context has been discussed for Hsp90, which is the most abundant constitutively expressed Hsp in eukaryotic cells (Jakob and Buchner, 1994; Buchner, 1996). Although Hsp90 is reported to act as a molecular chaperone (Wiech et al., 1992; Freeman and Morimoto, 1996; Scheibel et al., 1998), the correlation between its abundance and its chaperone activity is not very clear, particularly un-der stress conditions. However, the crucial involvement of Hsp90 in the maturation and folding of several protein kinases and nuclear steroid hormone receptors has been studied extensively (for review, see Kimura et al., 1995; Buchner, 1999), suggesting a regulatory role for Hsp90 in several developmental processes. A hallmark for understanding Hsp90 function in this context was a recent publication by Rutherford and Lindquist (1998) indicating that Hsp90 can serve as capacitor for morphological evolution in fruitfly.
The observed thermosensitivity of TU8 and the reduced amount of cytoplasmic Hsp90 in TU8 plants in the present study point to a more general function of Hsp90 as a molecular chaperone in Arabidopsis. However, the TU8 phenotype is pleiotrophic as outlined above and backcrossing experiments revealed a 3:1 ratio in the F2 generation (data not shown), suggesting that TU8 phenotype is caused by a recessive mutation of a single locus. This suggests that Hsp90 deficiency is also responsible for some, if not all, developmental dysfunctions in TU8 mutant plants.
Since transcript levels of Hsp90 were also reduced in TU8 plants, the main reason for the observed Hsp90 deficiency appears to be the reduced amount of its mRNA under stress. Considering that Arabidopsis contains up to four different members of Hsp90 (Milioni and Hatzopoulos, 1997), we do not know at present which of these members are affected. In situ analysis of Hsp90 in isolated TU8 cells showed a constitutive signal for plastidic Hsp90 signal under stress, whereas the signal for cytoplasmic Hsp90 dramatically decreased. These data are consistent with the results obtained by western-blot analysis. A slightly reduced signal for Hsp90 compared with Col seedlings was detected in mutant plants under control conditions, which declined under heat stress but did not totally disappear under heat stress. It is possible that the remaining signal was due to the plastidic counterpart of cytosolic Hsp90, the levels of which were obviously not affected by heat stress.
Although reduced thermostability of TU8 may be due to other reasons than deficiency in Hsp90 alone, transient substitution by Hsp90 was able to overcome the reduced survival rate of isolated TU8 cells at elevated temperatures, suggesting that Hsp90 plays a major role in this context. To investigate the genetic cause of the pleiotrophic phenotype of TU8, we started to map the locus. In addition, we plan to establish a mutant cell line stably substituted with the B. napus hsp90 to analyze whether stable expression of Hsp90 in TU8 plants will be able to fully restore wild-type thermostability in the mutant. Such a transformed mutant cell line will also be useful in addressing further questions related to influence of Hsp90 on auxin metabolism and glucosinolate synthesis.
MATERIALS AND METHODS
Plant Material
The Arabidopsis mutant line TU8 was derived from Col ecotype by ethylmethylsulfonic acid mutagenesis. The original seeds, described by Haughn et al. (1991) as indole glucosinolate deficient, were a gift from George Haughn (University of British Columbia, Vancouver). Plants were sown on a mixture of compost:peat:sand (3:2:1), vernalized for 24 h at 4°C, and then grown at 22°C in 60% humidity, in a 16-h light, 8-h dark cycle (60 μmol m−2 s−1; Philips fluorescent lights (Phillips, Eindhoven, The Netherlands) TL55 daylight and TL32 Warmton de Luxe). After 18 d of growth (time point just before bolting of TU8 plants), seedlings were placed in growth chambers set at different temperatures (23°C, 27°C, 32°C, 34°C, 37°C, and 42°C) and further cultivated under the light and humidity conditions described above. Plants were inspected and photographed every 3rd d. The number of surviving plants was monitored. For each experiment, 16 plants were used and values represent means of four experiments.
Genetic Analysis
TU8 and Col wild-type plants were backcrossed and the resulting seeds were sown (F1). Seeds of single F1 plants were collected, planted, and analyzed for TU8 phenotype expression (F2). Between d 16 and 21, TU8 mutant seedlings could be easily identified because of their early shoot development. At this stage Col seedlings still remained in the rosette stage. Backcrossing was performed three times and the resulting progeny was used for TU8 analysis.
Protein Analysis
To prepare cytosolic extracts, frozen plant material was ground in liquid nitrogen. Tissue was thawed in electrophoresis buffer (Laemmli, 1970) without β-mercaptoethanol. Protein concentration was determined according to Smith et al. (1985). Twenty micrograms of total protein was boiled in SDS-sample buffer in the presence of β-mercaptoethanol and separated by SDS-PAGE (Laemmli, 1970). After electrophoresis, proteins were blotted onto nitrocellulose (Schleicher & Schüll, Dassel, Germany). Membranes were blocked with 5% (w/v) non-fat dry milk in phosphate-buffered saline and incubated with an antibody against Hsp17.6 class I (obtained from E. Vierling, University of Arizona, Tucson), Hsp70, (obtained from D. Neuman, Institut für Pflanzenbiochemie, Halle, Germany), or Hsp101 (obtained from E. Vierling). Hsp90 was detected by using the R2 antiserum (Krishna et al., 1997). All antibodies were applied in a 1:2,000 dilution. Detection was performed with the ECL-system (NEN Life Science Products, Boston) using an anti-rabbit antibody conjugated to horseradish peroxidase according to the manufacturers' instructions.
Hsp Transcript Analysis
Total RNA was isolated using a modified guanidinium-isothiocyanate method described previously (Forreiter and Apel, 1993). Ten micrograms of total RNA from different tissues was separated on agarose gels containing formaldehyde, transferred to Hybond N+ membrane (Amersham, Buchler, Germany) and fixed by UV-irradiation in a Stratalinker (Stratagene, Amsterdam). Blots were prehybridized in 5× SSC, 50% (v/v) deionized formamide, 0.02% (w/v) SDS, 0.1% (w/v) sodium-lauroylsarcosin, and 2% (v/v) Denhardt's reagent for 5 min. Hybridization was performed at 42°C for 16 h according to standard procedures (Ausubel et al., 1993) with probes coding for hsp90 from Brassica napus and hsp70 from petunia. Unspecific bound probe was removed by washing filters at 55°C in the presence of 0.1% (w/v) SDS and 0.1× SSC.
Indirect Immunofluorescence
Leaves of wild-type and TU8 plants were harvested 14 d after germination. Protoplast preparation, fixation, and antibody treatment were essentially performed according to Lyck et al. (1997). After cell wall digestion, protoplasts were subsequently treated for 30 min at 38°C, for 30 min at 39°C, and for 2 h at 40°C to induce Hsp formation. Antibodies were diluted 1:300 for anti-Hsp17.6 and 1:500 for anti-Hsp90. Fluorescein thiocarbamoyl (FITC)-conjugated antibodies were applied at a 1:200 dilution. Confocal laserscan analysis was performed using a TCS NT confocal microscope and software system (Leica Microsystems, Wetzlar, Germany). Using an argon laser exciting at 488 nm, FITC-fluorescence was monitored at 540 nm.
Construction of an Hsp90 Expression Plasmid
For expression of Hsp90 in Arabidopsis wild-type and mutant protoplasts, the coding region of the Hsp90-1 cDNA from B. napus (Krishna et al., 1995) was placed under regulation of a cauliflower mosaic virus double 35S promoter in a pTRL expression plasmid (details of the construct will be described elsewhere).
Transient Transformation of Leaf Mesophyll Protoplasts
For transient expression of Hsp90 in mesophyll protoplasts, leaves of 14-d-old seedlings were excised and incubated overnight in a solution containing 0.4 m mannitol, 7 mm CaCl2, 3 mm MES [2-(N-morpholino)-ethanesulfonic acid], pH 5.7, 0.2% (w/v) macerocyme, and 0.5% (w/v) cellulase. Resulting protoplasts were adjusted to 500,000 mL−1 and transiently transformed using polyethyleneglycol-mediated plasmid uptake with a vector (10 μg of DNA per 50,000 protoplasts) carrying the coding region of a B. napus hsp90-1 cDNA. For control, TU8 and wild-type protoplasts were mock transformed with empty vector. Cells were incubated overnight to allow Hsp90 expression and then exposed to 44°C for 120 min. Cell vitality was determined every 15 min by adding 1% (w/v) Evans blue dye to the protoplast solution (Gaff and Okong'O-Ogola, 1971). Hsp90 expression was monitored by western-blot analysis using the anti-Hsp90 antibody (see above).
ACKNOWLEDGMENTS
We are gratefully indebted to Sigrid Ranostaj and Kerstin Pieper for skillful technical assistance and to Marc Kirschner for kindly introducing us to confocal laserscan analysis. We would like to thank Jill Winter, Elizabeth Vierling, and Dieter Neumann for providing us with Hsp70, Hsp100, and sHsp antibodies, and Janet P. Slovin, Daniela Löw, Willy Hilgenberg, and Rogier Stuger for carefully reading the manuscript and helpful discussion.
Footnotes
This work was supported by a research grant from the Natural Sciences and Engineering Research Council of Canada and by the Deutsche Forschungsgemeinschaft.
LITERATURE CITED
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1993. [Google Scholar]
- Beissinger M, Buchner J. How chaperones fold proteins. Biol Chem. 1998;379:245–259. [PubMed] [Google Scholar]
- Bennett R, Ludwig-Müller J, Kiddle G, Hilgenberg W, Wallsgrove R. Developmental regulation of aldoxime formation in seedlings and mature plants of Chinese cabbage (Brassica campestris) and oilseed rape (Brassica napus): glucosinolate and IAA biosynthetic enzymes. Planta. 1995;196:239–244. [Google Scholar]
- Bennett R, Wallsgrove R. Secondary metabolites in plant defense mechanisms. New Phytol. 1994;127:617–633. doi: 10.1111/j.1469-8137.1994.tb02968.x. [DOI] [PubMed] [Google Scholar]
- Buchner J. Supervising the fold: functional principles of molecular chaperones. FASEB J. 1996;10:10–19. [PubMed] [Google Scholar]
- Buchner J. Hsp90 & Co.: a holding for folding. Trends Biochem Sci. 1999;24:136–141. doi: 10.1016/s0968-0004(99)01373-0. [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Butcher DN, El-Tigani S, Ingram DS. The role of indole glucosinolates in the clubroot disease of the Cruciferae. Physiol Plant Pathol. 1974;4:127–141. [Google Scholar]
- Daxenbichler ME, Spencer GF, Carlson DG, Rose GB, Brinker AM, Powell RG. Glucosinolate composition of seeds from 297 species of wild plants. Phytochemistry. 1991;30:2623–2638. [Google Scholar]
- Doughty KJ, Porter AJR, Morton AM, Kiddle G, Bock CH, Wallsgrove RM. Variation in the glucosinolate content of oilseed rape leaves: II. Response to infection by Alternaria brassicae (Berk) Sacc. Ann Appl Biol. 1991;118:469–477. [Google Scholar]
- Ehrnsperger M, Gräber S, Gaestel M, Buchner J. Binding of non-native protein to HSP25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 1997;16:221–229. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ, van der Vries SM. Molecular chaperones. Annu Rev Biochem. 1991;60:321–347. doi: 10.1146/annurev.bi.60.070191.001541. [DOI] [PubMed] [Google Scholar]
- Forreiter C, Apel K. Light-independent and light-dependent protochlorophyllide reducing activities and two distinct NADPH-protochlorophyllide oxidoreductase polypeptides in mountain pine (Pinus mugo) Planta. 1993;190:536–545. doi: 10.1007/BF00224793. [DOI] [PubMed] [Google Scholar]
- Forreiter C, Kirschner M, Nover L. Stable transformation of an Arabidopsis cell suspension culture with firefly luciferase providing a cellular system for analysis of chaperone activity in vivo. Plant Cell. 1997;9:2171–2181. doi: 10.1105/tpc.9.12.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forreiter C, Nover L. The heat stress response and the concept of molecular chaperones. J Biosci. 1998;23:287–302. [Google Scholar]
- Freeman BC, Morimoto RI. The human cytosolic molecular chaperones Hsp70 (Hsc70) and Hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J. 1996;15:2969–2979. [PMC free article] [PubMed] [Google Scholar]
- Frydman JE, Höhfeld J. Chaperones get in touch: the Hip-Hop connection. Trends Biochem Sci. 1997;22:87–92. doi: 10.1016/s0968-0004(97)01005-0. [DOI] [PubMed] [Google Scholar]
- Gaff DF, Okong'O-Ogola O. The use of non-permeating pigments for testing the survival of cells. J Exp Bot. 1971;22:756–758. [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Haughn GW, Davin L, Giblin M, Underhill EW. Biochemical genetics of plant secondary metabolites in Arabidopsis thaliana: the glucosinolates. Plant Physiol. 1991;97:217–226. doi: 10.1104/pp.97.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney RK, Fenwick GR. Identifying toxins and their effects. In: Watson DH, editor. Natural Toxicants in Food: Progress and Prospects. Chichester, UK: Ellis Horwood; 1987. pp. 76–109. [Google Scholar]
- Jakob U, Buchner J. Assisting spontaneity: the role of HSP90 and small Hsps as molecular chaperones. Trends Biochem Sci. 1994;19:205–211. doi: 10.1016/0968-0004(94)90023-x. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Craig EA. Protein folding in vivo: unraveling complex pathways. Cell. 1997;90:201–204. doi: 10.1016/s0092-8674(00)80327-x. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Yahara I, Lindquist S. Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- Krishna P, Reddy RK, Sacco M, Frappier JR, Felsheim RF. Analysis of the native forms of the 90-kDa heat shock protein (hsp90) in plant cytosolic extracts. Plant Mol Biol. 1997;33:457–466. doi: 10.1023/a:1005709308096. [DOI] [PubMed] [Google Scholar]
- Krishna P, Sacco M, Cherruti JF, Hill S. Cold-induced accumulation of hsp90 transcripts in Brassica napus. Plant Physiol. 1995;107:915–923. doi: 10.1104/pp.107.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee GH, Roseman AM, Saibil HR, Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. The heat shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J, Pieper K, Ruppel M, Cohen JD, Epstein E, Kiddle G, Bennett R. Indole glucosinolate and auxin biosynthesis in Arabidopsis thaliana L. glucosinolate mutants and the development of the clubroot disease. Planta. 1999;208:409–419. doi: 10.1007/s004250050576. [DOI] [PubMed] [Google Scholar]
- Lyck R, Harmening U, Höhfeld I, Scharf KD, Nover L. Identification of the nuclear localization signal of two tomato heat stress transcription factors. Planta. 1997;202:117–125. doi: 10.1007/s004250050110. [DOI] [PubMed] [Google Scholar]
- Milioni D, Hatzopoulos P. Genomic organization of hsp90 gene family in Arabidopsis. Plant Mol Biol. 1997;35:955–961. doi: 10.1023/a:1005874521528. [DOI] [PubMed] [Google Scholar]
- Nover L. Heat Shock Response. Boca Raton, FL: CRC Press; 1991. [Google Scholar]
- Nover L, Scharf KD, Neumann D. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol Cell Biol. 1983;3:1648–1655. doi: 10.1128/mcb.3.9.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter AJR, Morton AM, Kiddle G, Doughty KJ, Wallsgrove RM. Variations in the glucosinolate content of oilseed rape leaves: I. Effect of leaf age and position. Ann Appl Biol. 1991;118:461–467. [Google Scholar]
- Rodman JE. A taxonomic analysis of glucosinolate-producing plants: Part I. Phenetics Syst Bot. 1991;16:5998–6118. [Google Scholar]
- Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Scheibel T, Weikl T, Buchner J. Two chaperone sites in Hsp90 differing in substrate specificity and ATP dependence. Proc Natl Acad Sci USA. 1998;95:1495–1499. doi: 10.1073/pnas.95.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibel T, Weikl T, Rimmerman R, Smith D, Lindquist S, Buchner J. Contribution of N- and C-terminal domains of the function of Hsp90 in Saccharomyces cerevisiae. Mol Microbiol. 1999;34:701–713. doi: 10.1046/j.1365-2958.1999.01632.x. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Glover JR, Singer MA, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- Schmitz G, Schmidt M, Feierabend J. Characterization of a plastid-specific Hsp90 homologue: identification of a cDNA sequence, phylogenetic descendence and analysis of its mRNA and protein expression. Plant Mol Biol. 1996;30:479–492. doi: 10.1007/BF00049326. [DOI] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Stuger R, Ranostaij S, Materna T, Forreiter C. In vivo and in vitro mRNA binding properties of non-polysomal RNPs from heat stressed tomato cells. Plant Physiol. 1999;120:23–31. doi: 10.1104/pp.120.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierling E. The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:579–620. [Google Scholar]
- Wiech H, Buchner J, Zimmermann R, Jakob U. Hsp90 chaperone protein folding in vitro. Nature. 1992;358:169–170. doi: 10.1038/358169a0. [DOI] [PubMed] [Google Scholar]