Abstract
The Radical SAM (RS) enzyme PqqE catalyzes the first step in the biosynthesis of the bacterial cofactor pyrroloquinoline quinone, forming a new carbon-carbon bond between two sidechains within the ribosomally synthesized peptide substrate PqqA. In addition to the active site RS 4Fe-4S cluster, PqqE is predicted to have two auxiliary Fe-S clusters, like the other members of the SPASM domain family. Here we identify these sites and examine their structure using a combination of X-ray crystallography and Mössbauer and electron paramagnetic resonance (EPR) spectroscopies. X-ray crystallography allows us to identify the ligands to each of the two auxiliary clusters at the C-terminal region of the protein. The auxiliary cluster nearest the RS site (AuxI) is in the form of a 2Fe-2S cluster ligated by four cysteines, an Fe-S center not seen previously in other SPASM domain proteins; this assignment is further supported by Mössbauer and EPR spectroscopies. The second, more remote cluster (AuxII) is a 4Fe-4S center that is ligated by three cysteine and one-aspartate residue. In addition, we examined the roles these ligands play in catalysis by the RS and AuxII clusters using site-directed mutagenesis coupled with EPR spectroscopy. Lastly, we discuss the possible functional consequences that these unique AuxI and AuxII clusters may play in catalysis for PqqE and how these may extend to additional RS enzymes catalyzing the post-translational modification of ribosomally encoded peptides.
Graphical Abstract
INTRODUCTION
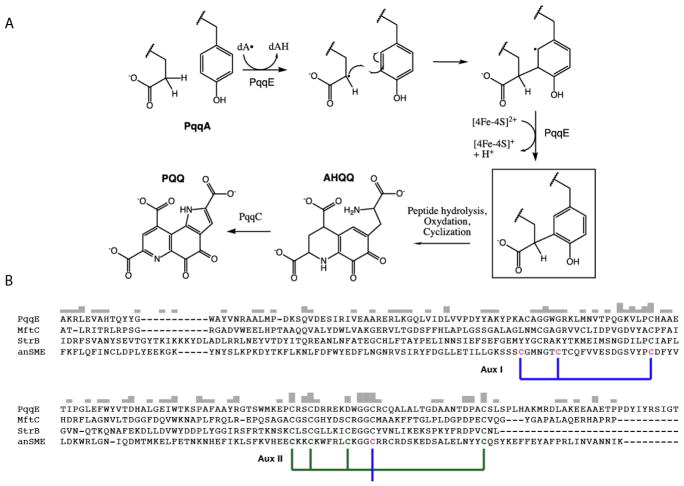
Pyrroloquinoline quinone (PQQ) is a redox active quinocofactor produced by bacteria that is used primarily in the metabolism of simple alcohols and sugars by methanol or glucose dehydrogenases1. The pqq operon encodes the genes required to synthesize PQQ, which contains—at a minimum—pqqA, pqqB, pqqC, pqqD, and pqqE, in a highly conserved order. PqqA is a short peptide of 20–30 amino acids, depending on the species, and is part of a class of peptides known as RiPPs (Ribosomally-produced and Post-translationally modified Peptides) that are translated by the ribosome but later modified with a series of enzymes during their maturation. The biosynthesis of PQQ begins with cyclization of PqqA in a reaction catalyzed by the radical SAM enzyme PqqE2. This reaction takes place with PqqA bound to the peptide binding protein PqqD3, and involves formation of a new carbon bond between glutamate and tyrosine (See Figure 1A). PqqE, a member of the RS superfamily of enzymes, catalyzes the carbon-carbon bond formation. Most members of the RS superfamily of enzymes bind S-adenosyl-L-methionine (SAM) to a [4Fe-4S]+1/+2 cluster (FeS), which upon reduction from the resting +2 state to the active +1 state, generates methionine and a 5′-deoxyadenosyl (5′dA) radical that initiates the chemistry by abstracting a hydrogen atom from the target substrate (Figure 1A).4,5. The RS cluster in PqqE is located at the N-terminus of the protein, while the remaining two clusters are predicted to bind to the C-terminal SPASM domain, performing unknown functions that have, nonetheless, been shown in several cases to be required for activity4. Characterized members of the SPASM family act predominantly via the modification of ribosomally translated peptides, and have been coined “SPASM” for the founding members: AlbA, PqqE, AnSME, and MftC that participate in subtilosin A and PQQ formation and anaerobic sulfatase and mycofactocin maturation, respectively4,5,6. A C-terminal FeS binding motif CX2CX5CX3C is nearly invariant for members of the SPASM subfamily, including AnSME from C. perfringens that acts on a sulfatase protein precursor. A direct role for the auxiliary clusters found in the SPASM domain containing proteins remains to be defined; however, one proposal is that they act as a conduit for the controlled transfer of an unpaired electron during the oxidation of the substrate radical intermediate7, or in the binding and positioning of substrate close to the RS center; as in MoaA8 or AlbA 9.
Figure 1.
A, proposed mechanism for carbon-carbon bond formation in the biosynthesis of PQQ, and its relation to the rest of the biosynthetic pathway. The final product of the PqqE reaction is boxed. B, ClustalW alignment of the C-terminal region of SPASM domain family members. The cysteines ligated to the innermost auxiliary cluster (Aux I) in AnSME are identified by the blue bracket, and the outermost (AuxII) are bracketed in green. In PqqE, D319 is placed exactly where, in the other three examples, there is a conserved cysteine; the latter is seen to ligate to Aux II in the AnSME X-ray structure.
Recent work has suggested, based on mutagenesis and spectroscopic data, that PqqE binds both a [2Fe-2S] cluster and a carboxylate-liganded [4Fe-4S] cluster within its SPASM domain10. In working toward a structural basis that can be linked to such diverse functional roles, we present an X-ray structural model of PqqE from M. extorquens, as well as EPR spectroscopic characterization of the auxiliary FeS centers in PqqE. These data unambiguously identify the ligands to the FeS clusters of PqqE’s SPASM domain and highlight key structural differences between PqqE and other SPASM domain containing enzymes. We also show that these auxiliary clusters are necessary for the formation of the initial carbon-carbon bond in PqqA.
MATERIALS AND METHODS
M. extorquens PqqE was purified and reconstituted as described previously2,3. Samples prepared in this way were used for all of the described biophysical probes with the exception of the X-ray studies. We have found it extremely difficult to obtain diffractable crystals of PqqE. The fortuitous observation of a crystalline form and structure from unreconstituted PqqE, while absent the RS site, has allowed observation of the two Aux centers (Figure 2 below). Site-directed mutagenesis of MexPqqE was accomplished by the following primers and their reverse complements using the QuickChange mutagenesis kit.: D319A (GGCGCGAGAAGGCTTGGGGCGGGTGCC); C323S/C325S (GGGGCGGGTCCCGCTCCCAGGCGC); -RS (CGCGCCCCGCTGCGCGCCCCATACGCCTCGAACC); -AUXII (GGAGCCCGCCCGCTCCGCCGACCGGCGC). The PqqE +RS mutant was made by Genescript, Inc. Kinetics of 5′dA formation were determined using 50 μM PqqE in the presence of 1 mM sodium dithionite and 500 μM SAM. Aliquots were removed at designated time and quenched with 1% formic acid with subsequent analysis by HPLC as described previously11. Iron and sulfide quantification was obtained as previously reported2. Modification of PqqA was detected using LC-MS, according to the published protocol2.
Figure 2.
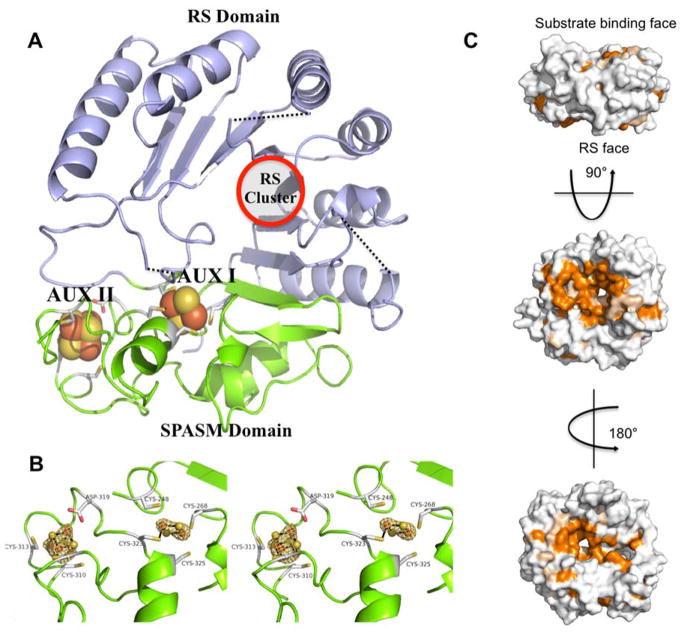
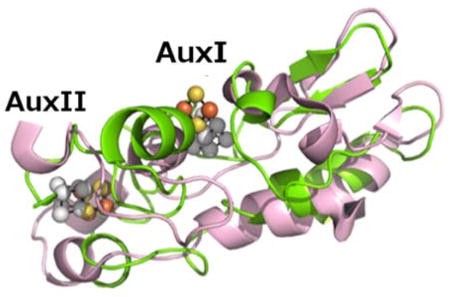
Structure of MePqqE. (A) The Radical SAM (RS) domain (lavender) contains the RS (β/α)6 partial TIM barrel. The RS cluster and canonical motif are missing from the structure, but the location of the cluster has been approximated using a superposition of the AnSMEcpe structure as a model. The SPASM domain (green) harbors two auxiliary clusters. (B) Stereo view of the MePqqE SPASM domain, which contains a [2Fe-2S] cluster ligated by four cysteines and one [4Fe-4S] cluster ligated by three cysteines and one conserved aspartate. The Fe-edge (9.686 keV) anomalous difference electron density map (gold mesh) is contoured at 4.0 RMSD. (C) Surface representation of sequence conservation as calculated by the ConSurf server [http://conseq.bioinfo.tau.ac.il/] between MePqqE and 1061 members of the PqqE family (SFLD, http://sfld.rbvi.ucsf.edu/django/) using pairwise alignment. Strictly conserved residues are shown in orange, neutral substitutions are shown in tan, and variable regions are shown in white. The view is rotated 90° about the horizontal axis (middle) and 180° about the vertical axis (bottom).
EPR Spectroscopy
The X-band (9.43 GHz) continuous-wave (CW) EPR spectra were recorded on a Bruker (Billerica, MA) Biospin EleXsys E500 spectrometer equipped with a super-high Q resonator (ER4122SHQE). Cryogenic temperatures were achieved and controlled using an ESR900 liquid helium cryostat in conjunction with a temperature controller (Oxford Instruments ITC503) and gas flow controller. CW EPR data were collected under slow-passage, non-saturating conditions at all temperatures. The spectrometer settings were as follows: modulation amplitude = 0.2 mT, and modulation frequency = 100 kHz; conversion time = 88 ms. Spectra were simulated using the EasySpin Matlab package.
Mössbauer Spectroscopy
Zero-field 57Fe Mössbauer spectra were recorded in a constant acceleration spectrometer (See Co., Edina, MN) at 4.2 K using a Janis Research Co. cryostat (Willmington, MA). Spectra were analyzed with the WMOSS software package (See Co., Edina, MN). Isomer shifts are reported relative to α-iron (27 μm foil) at room temperature. PqqE samples were prepared by freezing solutions in a Teflon sample holder (thickness 0.2 inch) under an Argon/Nitrogen atmosphere.
Crystallography
Diffraction quality crystals of unreconstituted MePqqE were obtained by sitting-drop vapor diffusion at 20 °C in an anaerobic chamber maintained at < 0.1 ppm oxygen (MBraun). Crystals appeared in 70 days, after combining 0.5 μL protein solution (20 mg mL−1 of MePqqE in 50 mM Tris, 100 mM NaCl, 1 mM TCEP pH 7.4) with 0.5 μL reservoir solution (0.2 M magnesium formate, 20% polyethylene glycol 3,350). Crystals were mounted on nylon loops and flash-cooled in liquid nitrogen inside the anaerobic chamber, without added cryoprotectant, and stored in liquid nitrogen prior to data collection. X-ray diffraction data were collected at 100 K on the beamline X29A (National Synchrotron Light Source, Brookhaven National Laboratory, Upton, NY) at a wavelength of 1.280 Å (9.686 keV), and processed and scaled with HKL300012. Diffraction from these crystals is consistent with the trigonal space group P3121, with unit cell parameters a=b=97.57 Å, c=86.42 Å, with one molecule per asymmetric unit, and solvent content of 57%. Exploiting the intrinsic iron-sulfur cluster, phases were determined by SAD with autoSHARP13, and an initial poly-alanine model was built with ARP/wARP14. Subsequent rounds of automated model building were performed by AutoBuild in PHENIX15, interspersed with manual model building and refinement using Coot16, phenix.refine17, and Refmac518. All figures were generated with PyMOL19. The final model consists of 312 residues, with gaps occurring from residues 1-13, 29-43, 127-140, 202-204, 349-384, and includes 6 iron ions, 6 sulfide ions, and 13 water molecules. Data collection and refinement statistics are shown in Table 1.
Table 1.
Data collection and refinement statistics
| PDB ID 6C8V | MePqqE |
|---|---|
| Data collection statistics | |
| Space group | P3121 |
| Unit-cell parameters (Å) | a = b = 97.57, c = 86.42 |
| Resolution (Å) | 19.75–3.20 (3.46–3.20) |
| Rmerge | 0.227 (0.834) |
| No. of unique reflections | 79485 (16073) |
| Multiplicity | 9.8 (9.7) |
| Completeness (%) | 99.6 (100) |
| Mean I/σ(I) | 9.4 (2.8) |
| CC1/2 | 0.990 (0.741) |
| Wilson B factor (Å2) | 64.03 |
| Refinement statistics | |
| Resolution (Å) | 19.75–3.20 (3.315–3.20) |
| Rwork (%) | 0.1967 (0.2687) |
| Rfree (%) (5% of data) | 0.2514 (0.3115) |
| No. of residues | 312 |
| No. of waters | 13 |
| Total No. of atoms | 2374 |
| B factor (Å2) | |
| Protein | 63.60 |
| Waters | 44.68 |
| Ligand | 50.49 |
| R.m.s.d., bond lengths (Å) | 0.0039 |
| R.m.s.d., angles (°) | 0.894 |
| Ramachandran outliers (%) | 0.0 |
| Side-chain outliers (%) | 1.2 |
Highest-resolution shell data are shown in parentheses
RESULTS
Sequence Comparison of MePqqE to members of the SPASM domain family reveals unique features
Figure 1B shows the multiple sequence alignment of four well-characterized proteins containing SPASM domains. PqqE, AnSME and MftC are founding members of the SPASM domain family, while StrB was recently shown to carry out a reaction similar to PqqE, the formation of a carbon-carbon bond within a small peptide7. Among the defining members of the RS-SPASM family, only a single X-ray crystal structure of AnSME, (PDB 4K37)20 had been available over many years, serving as the structural surrogate for the entire family. Very recently two additional X-ray structures became available: one for the peptide modifying enzyme CteB from Clostridium thermocellum, involved in carbon-sulfur cross-linking21, and another for the enzyme SuiB, a homolog of StrB that forms a carbon-carbon bond between a lysine and tryptophan side chains in the peptide substrate SuiA 22. Significantly, anSME, SuiB, and CteB show highly conserved features at all three iron sulfur sites: cysteine coordination of [4Fe-4S] clusters in the AuxI and AuxII sites, with the major difference being that CteB has an open coordination sphere within its AuxI site21. With the exception of PqqE, all enzymes characterized thus far indicate 5 conserved Cys residues at the C-terminus, four of which are assigned to the AuxII site based on the AnSME structure. Using the side chain numbering for PqqE in this region (310, 313, 319, and 341), PqqE indicates instead an aspartate at position 319, implicating CX2CX5DX3C as the motif for AuxII. Consistent with this feature, PqqE contains only 3 conserved cysteines at its C-terminus. As shown in Figure 1B, the spacing of cysteine ligands for AuxI is much more variable than for AuxII, making it difficult to predict ligands to AuxI based on sequence data alone.
Crystal structure of MePqqE shows the auxiliary clusters and their ligation environment
The final model of MePqqE from Methylobacterium extorquens was refined to 3.20 Å (R=19.67%, Rfree=25.14%) and spanned residues 13 to 348 out of 384 residues. Domain analysis of the structure revealed a partial [β/α6] TIM barrel (amino acids 22-179), followed by the C-terminal SPASM domain (amino acids 268-348, TIGR04085)5. Residues 29 to 43 contain the canonical radical SAM motif (CX3CX2C), however this region was not observed due either to a high degree of structural disorder and/or an incompletely formed RS site.
MePqqE SPASM domain
The two auxiliary clusters of PqqE are easily visualized from the structure (Figure 2A, 2B) as a 2Fe-2S in the AuxI site and a [4Fe-4S] in the II site. The AuxI 2Fe-2S cluster is ligated by residues C248, C268, C323, and C325. The AuxII 4Fe-4S cluster is located ~12 Å from AuxI and is ligated by residues C310, C313, D319, and C341. Notably, the participation of D319 in AuxII ligation—implied from the sequence alignment—(Figure 1) is corroborated by the crystal structure (Figure 2B). This ligand pattern for AuxII in the SPASM domain of MePqqE appears to be highly conserved in all annotated sequences of PqqE: a pair-wise alignment23 using 1061 sequences24 showed D319 to be invariant.
Comparison of MePqqE to AnSME
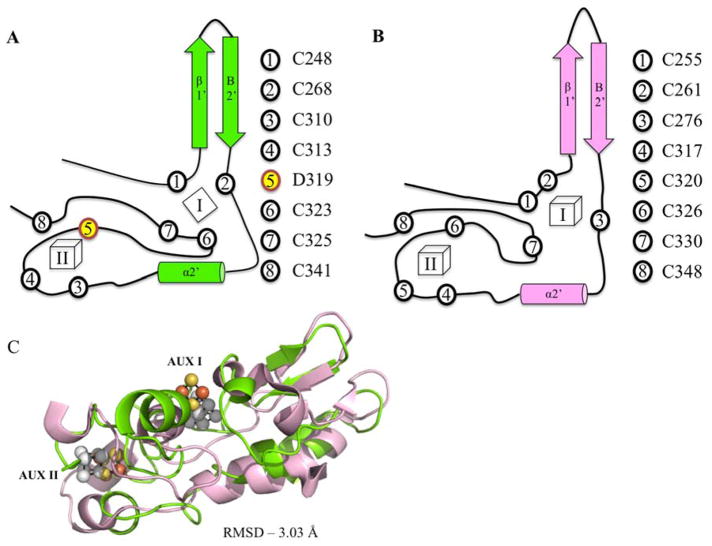
Though the cofactor within the RS-domain of PqqE cannot be visualized, the RS domain of AnSME (amino acids 10-181) aligns well with MePqqE (amino acids 22-179), with an RMSD of 2.69 Å over 217 aligned C-alpha carbons (sequence identity 29%). The C-terminal SPASM domain of MePqqE (amino acids 268-348) diverges more significantly from AnSME (RMSD 3.03 Å, Figure 3) with the aforementioned CX2CX5DX3C motif for PqqE contrasting with the predicted motif of CX2CX5CX3C motif. Furthermore, the last cysteine in this motif, C325, ligates Aux I in MePqqE, whereas the equivalent cysteine (C333) in anSME is unbound. PqqE has a unique ligand set for AuxI, with both C323 and C325 liganded to the same iron of a [2Fe-2S] cluster (Figure 3A, 3B). This is an unusual ligation motif (CXC) found predominantly in certain [2Fe-2S] clusters25,26, and only rarely in [4Fe-4S] clusters27, making it less likely that we are observing a degraded [4Fe-4S] cluster instead of a native [2Fe-2S] cluster. For comparative purposes, a structural overlay of MePqqE and MoaA is given in Fig. S1.
Figure 3.
Structural similarity of MePqqE and AnSMEcpe. Secondary structure and topology of MePqqE (A) and AnSMEcpe (B) show the ligation of auxiliary clusters. AUX I and AUX II are labeled as I and II. Ligating residues are represented by circles and corresponding residue numbers are shown to the right of each topological rendering. Asp 319 is shown by number 5, yellow highlighting. Panel (C) Shows the structural alignment of the SPASM domains of MePqqE in green (Fe is orange and S is yellow) and AnSMEcpe in pink (Fe and S are grey; RMSD 3.03 Å over 78 C-alphas, sequence identity 19.23%).
Impact of conserved residues in PqqE on Fe-S cluster formation and activity of PqqE
In order to determine the effects of removing an Fe-S cluster, we pursued mutagenesis to alter ligating residues within the three unique Fe-S clusters of PqqE. While our goal was to eliminate the RS center, AuxI and AuxII in a step-wise fashion, we were never able to isolate a stable variant that was disrupted solely at AuxI. On the other hand, proteins for which either the RS or AuxII was disrupted were successfully prepared. The iron binding and activity of the resulting variants are summarized in Table 2. Wild type PqqE purifies with ca. 7 irons and sulfides when expressed recombinantly in E. coli. Upon chemical reconstitution up to 13 irons and 12 sulfurs per peptide have been found previously2. The variant in which the RS site was disrupted (C32A, C35A termed –RS cluster) causes a decrease in the iron loading of PqqE, and, as expected, abolishes both the reductive cleavage of SAM and cross-link formation on PqqA (Table 2). The crystal structure shows that C310 and C313 bind to auxiliary AuxII, similar to what is found in anSME, CteB, and SuiB.12–14 The C310A and C313A variant (termed –AuxII protein) likewise causes a decrease in iron content. In this instance, the protein is able to reductively cleave SAM, but is incapable of forming the cross-linked PqqA product. The variant MePqqE that contained only cysteines 28, 32, and 35 (termed +RS protein) was highly unstable and capable of only producing low amounts of 5′dA for each molecule of protein without detectable cross-linking of PqqA. The MePqqE D319A variant led to a decrease in iron loading (~9 Fe per protein) and, importantly, was also unable to cross-link PqqA. This suggests that correct ligation of AuxII is essential for function of PqqE. Several reviewers noted the lack of exact correspondence between the titers for Fe in reconstituted proteins and the Fe-S structures inferred from the combined application of X-ray, EPR and Mossbauer methodologies. It is well known that RS protein preparations can contain significant adventitious Fe that is bound to either contaminating proteins (contributing broad backgrounds to Mossbauer spectra) or possibly to His-tags incorporated to facilitate protein purification (as the case for PqqE). For each of the protein variants described in Table 2, the presented Fe-protein ratios are included as representative of the preparations and cannot be used to draw precise conclusions regarding structures at the individual Fe-S sites28.
Table 2.
Mutants of M. extorquens PqqE studied. Each was reconstituted, assayed for SAM cleavage, Fe content and ability to form cross-linked PqqA. Assays were carried out using established procedures (Materials and Methods).
| Name | Mutations of Cys within RS and 2 Aux Sites | SAM Cleavage | Fe/Protein | Modification of PqqAa |
|---|---|---|---|---|
| WT | None | 0.031/min | 13.0 +/− 0.1 | Yes |
| −RS | C32A,C35A | None Observed | 8.7 +/− 0.8 | No |
| −AuxII | C310A,C313A | 0.0045/min | 7.3 +/− 0.6 | No |
| +RS | All to Ser except C28,32,35 | 0.00043/min | 4.6 +/− 0.7 | No |
| D319A | D319A | 0.0037/min | 8.9 +/− 0.9 | No |
We note the low yields for cross-linked PqqA, optimally 10% for WT enzyme: A failure to observe cross-linked product for the variants described is, thus, “within the limits of detection.”
Distinct Spectroscopic Properties at each of the three Fe-S clusters of PqqE
Low temperature EPR
We first compared the 10 K EPR behavior of the dithionite-reduced forms of WT, -RS and –AuxII PqqE. All samples show a continuous wave (CW) EPR spectrum with multiple components centered near g = 2 (Figure 4). These are distinct from samples not treated with dithionite that typically show signals consistent with a variable amount of [3Fe-4S]+ clusters. By raising the temperature to 60 K, only the signal corresponding to the [2Fe-2S]+ cluster persists, owing to its unique relaxation properties (see below)29,30. At the lower temperature (10 K) the g1-values for two [4Fe-4S]+ signals are also evident (g1 = 2.055 and g1 = 2.035). Examination of a 10 K EPR difference spectrum of the reduced WT PqqE and the –RS PqqE samples indicates loss of the small g = 2.035 feature, which we have assigned to RS cluster cluster (typical RS cluster g =2.037, 1.92, 1.89). The signal at g1 = 2.055 is attributed to an alternate [4Fe-4S]+ center, possibly AuxII. Consistent with the X-ray structure that indicates D319 as the fourth ligand to AuxII, mutation of this residue to Ala leads to a spectrum very similar to the –AuxII variant indicating that D319 is required for proper formation of the 4Fe-4S cluster in AuxII (Figure 4 bottom). The inset in Figure 4, which shows the region surrounding g1 = 2.055, may indicate the persistence of a small signal in both the –AuxII variant and D319A; assignment of this feature will require further study.
Figure 4.
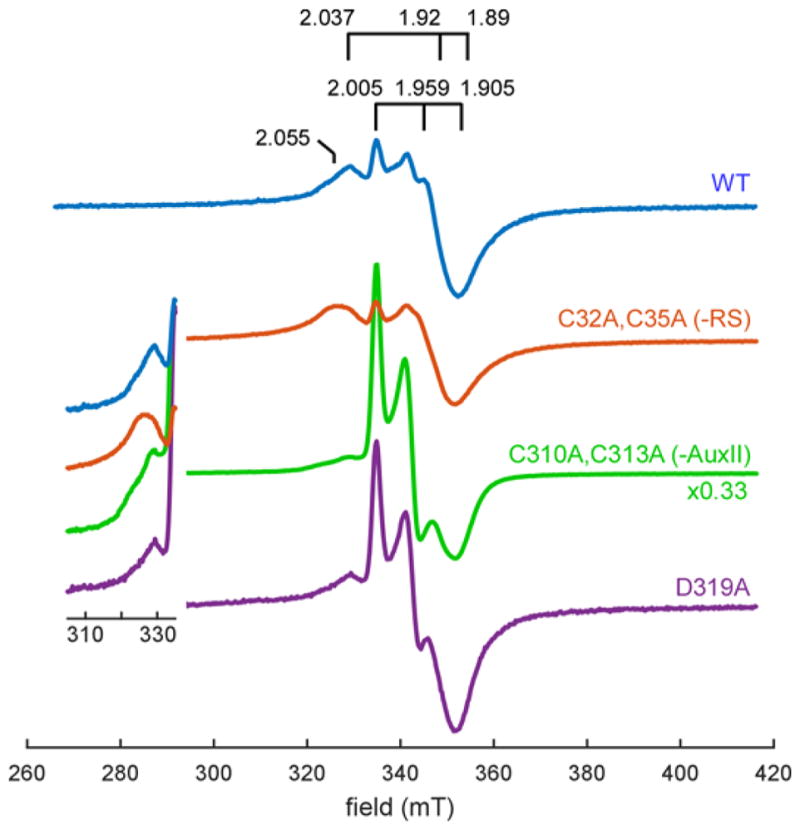
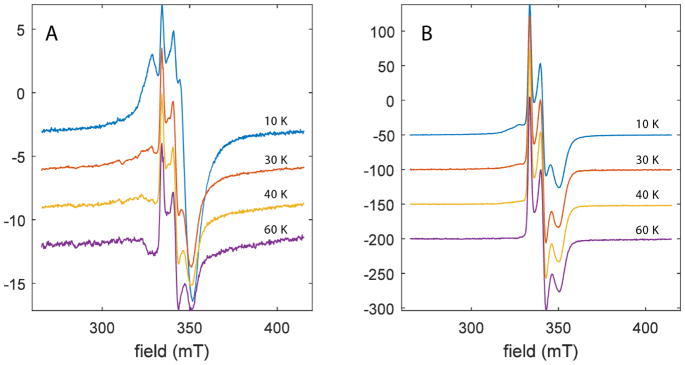
Continuous-wave EPR of dithionite reduced reconstituted wild-type PqqE (100 μM), PqqE -RS (200 μM), PqqE –AuxII (200 μM), and PqqE D319A (95 μM) at 10 K. We see three major EPR signals contributing to the spectrum of reduced WT PqqE: (1) a weak signal with g1 = 2.037 which corresponds to the radical SAM cluster (see simulation in Figure S2), (2) a signal with g1 = 2.055, and (3) a signal with g1 = 2.005, g2 = 1.959, g3 = 1.905, which corresponds to the auxiliary clusters. All spectra have been scaled according to the concentration of PqqE, with the PqqE –AuxII spectrum further divided by a factor of 3 for ease of comparison. Note that disruption of the AuxII cluster induces a dramatic increase in the amount of [2Fe–2S]+ signal relative to the [4Fe–4S]+ cluster signals in most samples. Inset focuses on the g1-region for the [4Fe–4S]+ cluster signals.
Temperature-dependent EPR
The dithionite reduced WT PqqE was further analyzed using CW EPR spectroscopy at varying temperatures. The resulting spectra show a distinctive transition between 10 K and 60 K (Figure 5A) at which only a single paramagnetic species is observed that is characterized by the g-values = [2.005, 1.959, 1.905]. At the elevated temperatures, EPR signals for most [4Fe-4S]+ clusters relax too fast to be observed; by contrast, [2Fe-2S]+ clusters retain strong EPR signals at 60 K. The average g-value of the high temperature signal of WT PqqE (gav = 1.971) is at the high end of the range of values for ferredoxins coordinated by four cysteine sidechains (typically gav =1.96.) (Table S1).
Figure 5.
Temperature dependence of EPR spectra of DTH-reduced wild-type PqqE (A) and –AuxII PqqE (B). As the temperature is elevated above 10 K, the [4Fe-4S]+ signals that are dominated by AuxII (Figure 4) completely disappear, due to fast relaxation that is a signature of [4Fe-4S]+ clusters. At 60 K, all that remains is a signal with g-values = [2.0049, 1.958, 1.906]. The average g-value = 1.9563 for the remaining signal is consistent with that measured for several all cysteinyl-coordinated, ferredoxin-type [2Fe-2S]+ clusters (Table S1).
Of considerable interest, this high temperature feature dominates the spectra for –AuxII PqqE at all temperatures (Figure 5B). This finding argues against the generation of new degradation products in PqqE –AuxII and supports the assignment of the high temperature spectra of WT enzyme to a 2Fe-2S center within the AuxI cluster, because no new spectral features appear in the PqqE –AuxII mutant that do not already exist in the wild-type. For the reduced WT PqqE sample at 10 K, the [2Fe-2S]+ cluster signal can be estimated to account for ca.15% of the total spins detected. We note that this result is variable, with occasional preparations of enzyme failing to show strong signals at the elevated temperatures.
Mössbauer spectroscopy is consistent with the presence of a single 2Fe-2S cluster and two 4Fe-4S clusters
The PqqE variant C32A/C35A (PqqE -RS) was grown in the presence of 57Fe and chemically reconstituted. Figure 6, bottom, shows the zero-field Mössbauer spectrum of PqqE -RS, as compared with the spectrum of WT from ref. (2) (top). Wild-type PqqE shows two doublets: a [4Fe-4S]2+ signal (δ = 0.478, ΔEq = 1.172), comprising 80% of the total iron, and a [2Fe-2S]2+ signal (δ = 0.324, ΔEq = 0.558), comprising 20% of the total iron. In the -RS mutant, the contribution of the 2Fe-2S signal (δ = 0.327, ΔEq = 0.521) increases significantly, accounting for ca 55% of all iron in the sample, whereas the [4Fe-4S] signal (δ = 0.523, ΔEq = 1.264) accounts for the remaining 45%. While we see less 4Fe-4S signal than expected, these data provide strong support for the presence of a [2Fe-2S] cluster within the SPASM domain of PqqE.
Figure 6.
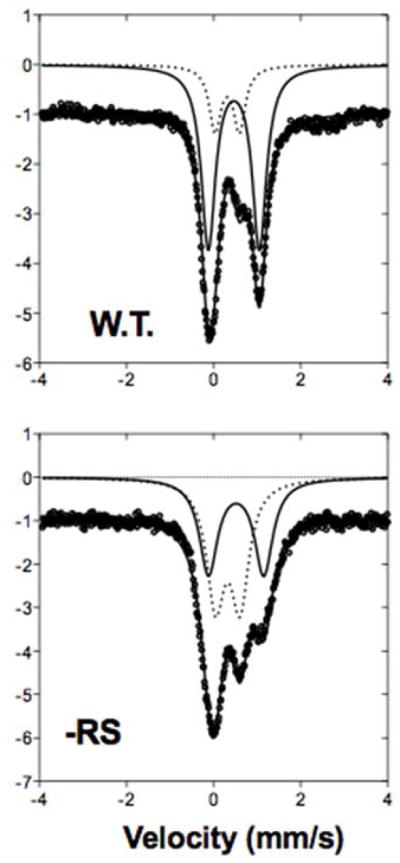
Zero-field, 4 K Mossbauer spectra of reconstituted PqqE WT and PqqE -RS knockout show loss of 4Fe-4S clusters and presence of 2Fe-2S cluster. Data (circles) were fit to two doublets, representing [4Fe-4S] (solid line) and [2Fe-2S] (dotted line). WT spectrum from Barr et al., 2016.
DISCUSSION
From mechanistic considerations alone, it is not clear why an enzyme such as PqqE would require auxiliary clusters in order to function. Many RS family enzymes catalyze complex reactions that require both oxidation and reduction of radical intermediates using only a single RS 4Fe-4S cluster, such as NirJ,31 DesII32, RlmN33, Cfr34, and QueE35: i.e. all RS enzymes that utilized SAM as a cosubstrate rather than a cofactor. For others, the auxiliary clusters function in substrate ligation (MoaA8) or as a source of sulfur or iron (BioB36 and HydG11). Prior to the determination of a structure for the RS SPASM proteins, the most likely assumption was that an auxiliary cluster housed in the SPASM domain would contain at least one open coordination site that enabled the binding of their peptide or protein substrate37. This can no longer be assumed, and focus has shifted to their potential role in electron transfer. What is clear is that in cases where the auxiliary clusters of a SPASM enzyme have either been removed by mutagenesis or incorrectly formed, activity is abolished, as shown in Table 2 and Refs. 11, 18, 19. This remains the most puzzling aspect of the RS SPASM enzymes, and one that experimental results have just begun to address.
Whatever the explanation, an acceptable hypothesis for the unusual Fe-S clusters of PqqE must center on the peculiarities of PqqE’s mechanism, or the biological context in which it operates. The RS enzyme SuiB carries out a reaction very similar to PqqE22,38. In this reaction, a carbon-carbon bond is formed between the β-carbon of a lysine residue and the aromatic carbon-7 of tryptophan in the peptide substrate SuiA, where the reactive side chains are separated by three residues. In PqqE’s reaction, cross-linking occurs between the β-carbon of a glutamic acid and the aromatic carbon-3 of tyrosine, also separated by three residues. SuiB additionally has, like many peptide modifying enzymes, a domain with homology to PqqD, the binding partner of PqqA that also associates with PqqE. While these features alone may have predicted auxillary FeS clusters in PqqE that are highly similar to SuiB, the crystal structure of SuiB shows a SPASM domain with topology much closer to anSME or CteB and neither a 2Fe-2S cluster in AuxI nor an aspartate ligand to AuxII, highlighting how far PqqE has diverged from the other characterized members of the SPASM family.
Asp-ligation and redox tuning
The two notable features of PqqE are the presence of the 2Fe-2S cluster in the Aux I site of the SPASM domain and the participation of aspartate as a ligand for the 4Fe-4S cluster in Aux II. The latter observation may be of functional importance, as the reduction potentials of iron-sulfur clusters are significantly influenced by the nature of the ligating functionalities39–41. Similar to Aux II from MePqqE, Pyrococcus furiosus ferredoxin (PfFd) contains a single 4Fe-4S cluster ligated by 3 Cys residues and 1 Asp residue39,40. The effects of ligand environment of the PfFd were extensively studied by Brereton and co-workers40, who reported that the reduction potential of the wild-type protein was −368 mV, but drops to −427 mV in the Asp to Cys variant. Substituting the Asp ligand to Ser further reduces the redox potential to −505 mV. Because PqqE catalyzes the two-electron oxidation of its substrate, a more positive reduction potential for the unique Asp-ligation in the Aux II cluster may facilitate oxidation of the substrate-derived radical intermediate by providing a thermodynamically favorable electron sink (see Fig. 1A).
Presence of [2Fe-2S] cluster in AuxI site
While the presence of a [2Fe-2S] center in PqqE has been previously proposed,10,42 there is always the potential of degradation within an originally formed 4Fe-4S cluster site. Therefore, characterization of 2Fe-2S clusters in the presence of 4Fe-4S clusters is often difficult. Our inability to produce a soluble AuxI knock-out to directly interrogate the remaining clusters complicated the analysis; the latter would have been expected to show the presence of only 4Fe-4S clusters in the RS and AuxII sites. In the only example of an AuxI variant that could be characterized thus far for PqqE11, the C323S variant has been reported to be capable of binding both 2Fe-2S and 4Fe-4S clusters. Despite some remaining experimental ambiguity, the X-ray, EPR and Mössbauer characterizations presented herein, together with the recent biochemical and Mössbauer studies of PqqE variants11, argue for the presence of a [2Fe-2S] center within AuxI.
Mechanistic Implications
The presence of a [2Fe-2S] cluster in PqqE is unusual as the other known SPASM domain enzymes studied are shown to exclusively bind [4Fe-4S]. The substitution of a four cysteine-liganded [2Fe-2S] cluster in this position in PqqE, where other SPASM enzymes show a [4Fe-4S], further suggests to us that this cluster may be used for electron transfer; electron transfer is one of the demonstrated functions of Fe-S clusters for which [4Fe-4S] and [2Fe-2S] are commonly interchangeable, e.g. in [4Fe-4S] and [2Fe-2S] ferredoxins43. [2Fe-2S] clusters generally have higher redox potentials than [4Fe-4S], commonly in the −400 to −300 mV range. While redox potentials have not yet been measured for any of the clusters of PqqE, this will be an important next step in relating function to structure.
Several reasons for the unusual FeS clusters of PqqE can be devised, and a second plausible explanation centers around the fact that PqqE must operate in the presence of molecular oxygen, which is required for other steps in the biosynthetic pathway 44. As 2Fe-2S clusters have been observed to be less sensitive to oxygen than 4Fe-4S clusters, the 2Fe-2S cluster might represent an evolutionary strategy for a SPASM domain enzyme operating in an oxygen-rich environment. For PqqE this stability has been shown experimentally42 but only for resting, oxidized protein. Another possible effect of having two auxiliary clusters with unusually high redox potentials might be to “dissipate” the energy of the electron being removed from the substrate in the final oxidation step. In the presence of oxygen, an electron exiting the active site by means of a 4Fe-4S cluster with a potential around −500 mV is far more likely to produce a reactive oxygen species than one where the potential has been altered to around −350 mV.
For SPASM-domain containing enzymes, including PqqE, a great deal of work has been done to determine the reaction catalyzed and the number and character of the auxiliary clusters. What is missing is perhaps the most important yet difficult aspect of these enzymes, a mechanistic description of the reaction itself. From the current work, and from previous studies, it is clear that the auxiliary clusters of these enzymes are essential to whatever mechanism is used, and that a complete description of the action of PqqE and its relatives will need to account for the correct structural relationship within a complex containing the peptide substrate, its chaperone (as a separate protein or N-terminally fused domain), SAM, the RS enzyme and a reductase to initiate the reaction. The current work is a necessary first step in that direction, giving us for the first time a three dimensional picture of the SPASM domain of PqqE and the nature of the FeS clusters therein.
Supplementary Material
Acknowledgments
Funding Sources
Financial support was provided by the National Institutes of Health (NIH): GM118117 to J.P.K, GM66569 to C.M.W, P01GM118303-01 and U54GM093342 to J.A.G. and S.C.A., and U54GM094662 to S.C.A., R21 AI133329 to T.L.G and S.C.A.; GM104543 to R.D.B., and the Price Family Foundation to S.C.A.
We thank Prof. Chris Chang of the University of California, Berkeley for the use of his Mössbauer spectrometer, and Teera Chantarojsiri for assistance collecting the spectra in Fig. 6. We acknowledge the use of the Albert Einstein Anaerobic Structural and Functional Genomics Resource (http://www.nysgxrc.org/psi3/anaerobic.html).
ABBREVIATIONS
- PQQ
pyrolloquinoline quinone
- RS
Radical SAM
- SAM
S-adenosyl methionine
- AnSME
anaerobic sulfatase maturing enzyme
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Notes
The authors declare no competing financial interest.
Supporting Figures S1 and S2 and Table S1. The Supporting Information is available free of charge.
References
- 1.Duine JA. The PQQ story. J Biosci Bioeng. 1999;88:231–236. doi: 10.1016/s1389-1723(00)80002-x. [DOI] [PubMed] [Google Scholar]
- 2.Barr I, Latham JA, Iavarone AT, Chantarojsiri T, Hwang JD, Klinman JP. Demonstration That the Radical S-Adenosylmethionine (SAM) Enzyme PqqE Catalyzes de Novo Carbon-Carbon Cross-linking within a Peptide Substrate PqqA in the Presence of the Peptide Chaperone PqqD. J Biol Chem. 2016;291:8877–8884. doi: 10.1074/jbc.C115.699918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Latham JA, Iavarone AT, Barr I, Juthani PV, Klinman JP. PqqD Is a Novel Peptide Chaperone That Forms a Ternary Complex with the Radical S-Adenosylmethionine Protein PqqE in the Pyrroloquinoline Quinone Biosynthetic Pathway. J Biol Chem. 2015;290:12908–12918. doi: 10.1074/jbc.M115.646521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grell TAJ, Goldman PJ, Drennan CL. SPASM and Twitch domains in S-Adenosylmethionine (SAM) radical enzymes. J Biol Chem. 2014;290:3964–3971. doi: 10.1074/jbc.R114.581249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haft DH, Basu MK. Biological systems discovery in silico: radical S-adenosylmethionine protein families and their target peptides for posttranslational modification. J Bacteriol. 2011;193:2745–2755. doi: 10.1128/JB.00040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnison PG, Bibb MJ, Bierbaum G, Bowers Aa, Bugni TS, Bulaj G, Camarero Ja, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach Ma, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Linka J, Liu W, Marahiel Ma, Mitchell Da, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Süssmuth RD, Tagg JR, Tang G-L, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk Wa. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–60. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schramma KR, Bushin LB, Seyedsayamdost MR. Structure and biosynthesis of a macrocyclic peptide containing an unprecedented lysine-to-tryptophan crosslink. Nat Chem. 2015;7:431–437. doi: 10.1038/nchem.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hänzelmann P, Schindelin H. Binding of 5′-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism. Proc Natl Acad Sci U S A. 2006;103:6829–6834. doi: 10.1073/pnas.0510711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benjdia A, Guillot A, Lefranc B, Vaudry H, Leprince J, Berteau O. Thioether bond formation by SPASM domain radical SAM enzymes: C α H-atom abstraction in subtilosin A biosynthesis. Chem Commun. 2016;52:6249–6252. doi: 10.1039/c6cc01317a. [DOI] [PubMed] [Google Scholar]
- 10.Saichana N, Tanizawa K, Ueno H, Pechoušek J, Novák P, Frébortová J. Characterization of auxiliary iron–sulfur clusters in a radical S-adenosylmethionine enzyme PqqE from Methylobacterium extorquens AM1. FEBS Open Bio. 7:1864–1879. doi: 10.1002/2211-5463.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wecksler SR, Stoll S, Tran H, Magnusson OT, Wu SP, King D, Britt RD, Klinman JP. Pyrroloquinoline quinone biogenesis: demonstration that PqqE from Klebsiella pneumoniae is a radical S-adenosyl-L-methionine enzyme. Biochemistry. 2009;48:10151–10161. doi: 10.1021/bi900918b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: The integration of data reduction and structure solution - From diffraction images to an initial model in minutes. Acta Crystallogr Sect D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 13.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated Structure Solution With autoSHARP Macromolecular Crystallography Protocols. Macromolecular Crystallography Protocols, Volume 2 SE - Methods in Molecular Biology. 2007:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 14.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–9. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 16.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 19.DeLano WL. The PyMOL Molecular Graphics System. Schrödinger LLC wwwpymolorg Version 1. 2002 http://www.pymol.org.
- 20.Goldman PJ, Grove TL, Sites LA, McLaughlin MI, Booker SJ, Drennan CL. X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification. Proc Natl Acad Sci U S A. 2013;110:8519–8524. doi: 10.1073/pnas.1302417110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grove TL, Himes PM, Hwang S, Yumerefendi H, Bonanno JB, Kuhlman B, Almo SC, Bowers AA. Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides. J Am Chem Soc. 2017;139:11734–11744. doi: 10.1021/jacs.7b01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis KM, Schramma KR, Hansen WA, Bacik JP, Khare SD, Seyedsayamdost MR, Ando N. Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition. Proc Natl Acad Sci. 2017;114:10420–10425. doi: 10.1073/pnas.1703663114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akiva E, Brown S, Almonacid DE, Barber AE, Custer AF, Hicks MA, Huang CC, Lauck F, Mashiyama ST, Meng EC, Mischel D, Morris JH, Ojha S, Schnoes AM, Stryke D, Yunes JM, Ferrin TE, Holliday GL, Babbitt PC. The Structure-Function Linkage Database. Nucleic Acids Res. 2014;42:D521–D530. doi: 10.1093/nar/gkt1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dailey TA, Dailey HA. Identification of [2Fe-2S] clusters in microbial ferrochelatases. J Bacteriol. 2002;184:2460–2464. doi: 10.1128/JB.184.9.2460-2464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paddock ML, Wiley SE, Axelrod HL, Cohen AE, Roy M, Abresch EC, Capraro D, Murphy AN, Nechushtai R, Dixon JE, Jennings Pa. MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone. Proc Natl Acad Sci U S A. 2007;104:14342–14347. doi: 10.1073/pnas.0707189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner T, Koch J, Ermler U, Shima S. Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction. Science. 2017;357:699–703. doi: 10.1126/science.aan0425. [DOI] [PubMed] [Google Scholar]
- 28.Lanz ND, Grove TL, Gogonea CB, Lee K-H, Krebs C, Booker SJ. RlmN and AtsB as models for the overproduction and characterization of radical SAM proteins. Methods Enzym. 2012;516:125–152. doi: 10.1016/B978-0-12-394291-3.00030-7. [DOI] [PubMed] [Google Scholar]
- 29.Orme-Johnson WH, Orme-Johnson NR. In: Iron-Sulfur Proteins. Spiro TG, editor. John Wiley & Sons; New York: 1982. pp. 67–96. [Google Scholar]
- 30.Rupp H, Rao KK, Hall DO, Cammack R. Electron spin relaxation of iron-sulphur proteins studied by microwave power saturation. BBA - Protein Struct. 1978;537:255–269. doi: 10.1016/0005-2795(78)90509-3. [DOI] [PubMed] [Google Scholar]
- 31.Brindley AA, Zajicek R, Warren MJ, Ferguson SJ, Rigby SEJ. NirJ, a radical SAM family member of the d1 heme biogenesis cluster. FEBS Lett. 2010;584:2461–2466. doi: 10.1016/j.febslet.2010.04.053. [DOI] [PubMed] [Google Scholar]
- 32.Ruszczycky MW, Liu HW. Mechanistic enzymology of the radical SAM enzyme DesII. Isr J Chem. 2015;55:315–324. doi: 10.1002/ijch.201400130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boal AK, Grove TL, McLaughlin MI, Yennawar NH, Booker SJ, Rosenzweig AC. Structural basis for methyl transfer by a radical SAM enzyme. Science. 2011;332:1089–1092. doi: 10.1126/science.1205358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grove TL, Radle MI, Krebs C, Booker SJ. Cfr and RlmN contain a single [4Fe-4S] cluster, which directs two distinct reactivities for s -adenosylmethionine: Methyl transfer by S N2 displacement and radical generation. J Am Chem Soc. 2011;133:19586–19589. doi: 10.1021/ja207327v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarty RM, Krebs C, Bandarian V. Spectroscopic, steady-state kinetic, and mechanistic characterization of the radical SAM enzyme QueE, which catalyzes a complex cyclization reaction in the biosynthesis of 7-deazapurines. Biochemistry. 2013;52:188–98. doi: 10.1021/bi301156w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jarrett JT. The biosynthesis of thiol-and thioether-containing cofactors and secondary metabolites catalyzed by radical S-adenosylmethionine enzymes. J Biol Chem. 2015;290:3972–3979. doi: 10.1074/jbc.R114.599308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Latham JA, Barr I, Klinman JP. At the confluence of ribosomally synthesized peptide modification and radical S-adenosylmethionine (SAM) enzymology. J Biol Chem. 2017;292:16397–16405. doi: 10.1074/jbc.R117.797399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schramma KR, Seyedsayamdost MR. Lysine-Tryptophan-Crosslinked Peptides Produced by Radical SAM Enzymes in Pathogenic Streptococci. ACS Chem Biol. 2017;12:922–927. doi: 10.1021/acschembio.6b01069. [DOI] [PubMed] [Google Scholar]
- 39.Calzolai L, Gorst CM, Bren KL, Zhou ZH, Adams MWW, Lamar GN. Solution NMR study of the electronic structure and magnetic properties of cluster ligation mutants of the four-iron ferredoxin from the hyperthermophilic archaeon Pyrococcus furiosus. J Am Chem Soc. 1997;119:9341–9350. [Google Scholar]
- 40.Brereton PS, Duderstadt RE, Staples CR, Johnson MK, Adams MW. Effect of serinate ligation at each of the iron sites of the [Fe4S4] cluster of Pyrococcus furiosus ferredoxin on the redox, spectroscopic, and biological properties. Biochemistry. 1999;38:10594–10605. doi: 10.1021/bi990671d. [DOI] [PubMed] [Google Scholar]
- 41.Bak DW, Elliott SJ. Alternative Fe-S cluster ligands: Tuning redox potentials and chemistry. Curr Opin Chem Biol. 2014;19:50–58. doi: 10.1016/j.cbpa.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 42.Saichana N, Tanizawa K, Pechoušek J, Novák P, Yakushi T, Toyama H, Frébortová J. PqqE from Methylobacterium extorquens AM1: a radical S-adenosyl-l-methionine enzyme with an unusual tolerance to oxygen. J Biochem. 2015;159:87–99. doi: 10.1093/jb/mvv073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsubara H, Saeki K. Chemistry, R. C. B. T.-A. in I, editor. Structural and Functional Diversity of Ferredoxins and Related Proteins. Academic Press; 1992. pp. 223–280. [Google Scholar]
- 44.Toyama H, Fukumoto H, Saeki M, Matsushita K, Adachi O, Lidstrom ME. PqqC/D, which converts a biosynthetic intermediate to pyrroloquinoline quinone. Biochem Biophys Res Commun. 2002;299:268–272. doi: 10.1016/s0006-291x(02)02603-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.