SUMMARY
Treatment of human pancreatic non-endocrine tissue with Bone Morphogenetic Protein 7 (BMP-7) leads to the formation of glucose-responsive β-like cells. Here, we show that BMP-7 acts on extrainsular cells expressing PDX1 and the BMP receptor activin-like kinase 3 (ALK3/BMPR1A). In vitro lineage tracing indicates that ALK3+ cell populations are multipotent. PDX1+/ALK3+ cells are absent from islets but prominently represented in the major pancreatic ducts and pancreatic duct glands. We identified the puri-nergic receptor P2Y1 (P2RY1) as a surrogate surface marker for PDX1. Sorted P2RY1+/ALK3bright+ cells form BMP-7-expandable colonies characterized by NKX6.1 and PDX1 expression. Unlike the negative fraction controls, these colonies can be differentiated into multiple pancreatic lineages upon BMP-7 withdrawal. RNA-seq further corroborates the progenitor-like nature of P2RY1+/ALK3bright+ cells and their multilineage differentiation potential. Our studies confirm the existence of progenitor cells in the adult human pancreas and suggest a specific anatomical location within the ductal and glandular networks.
In Brief
Qadir et al. describe and characterize a population of multipotent, BMP-7-responsive progenitor-like cells within the human exocrine pancreas. These cells are characterized by the expression of PDX1 and ALK3, a canonical BMP receptor. Their findings shed new light on potential regenerative pathways in the human pancreas.
INTRODUCTION
The existence of progenitor-like cells within the adult human pancreas has been hypothesized for decades (Bonner-Weir et al., 2008; Wang et al., 2013), but their characterization has proven elusive. The study of their nature and potency may help us tap into an endogenous cell repository for pancreatic β cell regeneration, which could lead to therapeutic applications for type 1 and type 2 diabetes. We have previously shown that bone morphogenetic protein 7 (BMP-7), a transforming growth factor β (TGF-β) family member with dual BMP activation and TGF-β inhibition potential, stimulates progenitor-like cells within cultured human non-endocrine pancreatic tissues (hNEPTs) (Klein et al., 2015). Our studies suggested that BMP-7-responsive cells express both pancreatic duodenal homeobox 1 (PDX1) and the BMP receptor 1A (BMPR1A, also known as activin-like receptor 3, ALK3), whose engagement has been associated with regeneration in multiple tissues (Sugimoto et al., 2012; Yasmin et al., 2013; Zhang et al., 2015). These cells were also negative for insulin and the hitherto-considered pan-ductal marker carbonic anhydrase II (CAII). Here, we present additional evidence that genetically tagged ALK3+ cells within hNEPT have multilineage differentiation potential. Progenitor-like cells can be sorted using ALK3 and the purinergic receptor P2Y1 (P2RY1), which we have validated as a surrogate surface marker for PDX1-expressing cells. P2RY1+/ALK3bright+ cells can be cultured in defined conditions, respond to BMP-7 by expanding, and then differentiate into multiple pancreatic cell types upon BMP-7 withdrawal, including C-peptide/ NKX6.1/PDX1-expressing β-like cells. qRT-PCR and RNA sequencing (RNA-seq) analyses further confirm the BMP-7-induced transcriptional activation of inhibitor of binding/differentiation (ID) genes associated with progenitor cell proliferation, as well as the upregulation of differentiation markers of all pancreatic lineages following BMP-7 withdrawal. We further show the anatomic location of PDX1+/ALK3bright+ cells in the human pancreas, mostly within the major pancreatic ducts (MPDs) and associated pancreatic duct glands (PDGs). Our studies shed new light on the nature and niche of pancreatic progenitor cells and suggest potential interventions to induce β cell regeneration in situ.
RESULTS
In Vitro Lineage Tracing Supports ALK3+ Origin of BMP-7-Stimulated C-Peptide-Expressing Cells and Suggests Multilineage Differentiation Potential
Previous in vitro lineage-tracing experiments suggested that, while BMP-7-responsive cells within hNEPT are largely negative for CAII and elastase 3a (Elas3a, acinar marker), they were positive for PDX1 (Klein et al., 2015). Tagged residual β cells (which are also PDX1+) had a lower contribution to the resulting C-peptide+ cells, with additional evidence ruling out that they were responsible for the reported BMP-7-mediated effects. Further assays also determined that ALK3 is the most likely BMP receptor mediating the effect of BMP-7 in our system (Klein et al., 2015). To confirm that ALK3-expressing cells exhibit multilineage differentiation potential upon BMP-7 stimulation, similar to that previously reported for PDX1-expressing cells, we performed further lineage tracing. The strategy entails transducing fresh hNEPT with a lentiviral reporter (CMV-LoxP-dsRED-STOP-LoxP-EGFP) for expression of a dsRed fluorescent marker flanked by loxP sites. Expression of a second adenoviral construct, in which Cre is driven by the ALK3 promoter (Calva-Cerqueira et al., 2010), results in the excision of the dsRed/STOP sequence and expression of EGFP (Figure 1A). As a result, cells with active ALK3 expression at the time of transduction, as well as their progeny, are labeled in green.
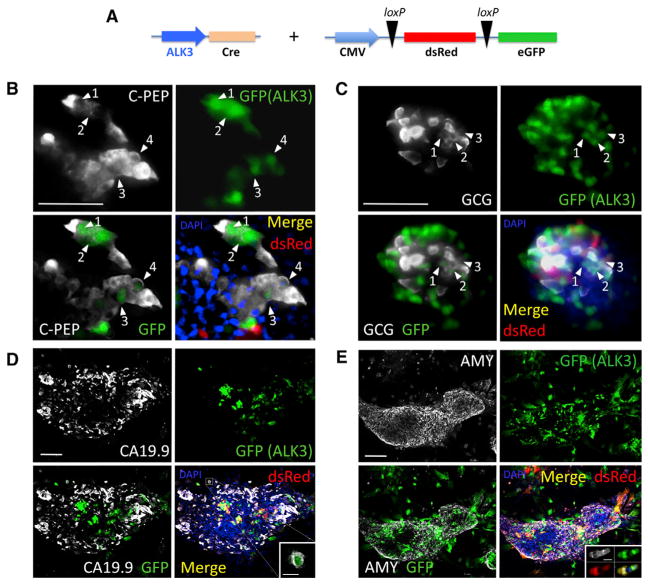
Figure 1. Lineage-Tracing Studies.
(A) Experimental design. The ALK3 promoter was used to drive Cre expression. The reporter expresses dsRed (red) or EGFP (green) upon Cre-mediated loxP excision. Both constructs were simultaneously transduced into hNEPT cells. Upon BMP-7 treatment, we set out to determine whether EGFP was present in cells expressing C-peptide (C-PEP), glucagon (GCG), CA19.9, or amylase (AMY). These markers are shown in white in (B) through (E), respectively. EGFP (green) and dsRed (red) + channel merge (DAPI, blue) are shown for all experiments. Each panel also shows an EGFP (green) + mature marker (white) merge to facilitate the identification of cells that co-express both markers. Confocal microscopy was used in the acquisition of all these images.
(B) ALK3-Cre + reporter + C-peptide IF. Abundant C-peptide+ cells expressed EGFP (white arrows), suggesting a significant participation of ALK3+ cells in BMP-7-induced C-peptide+ cells.
(C) ALK3-Cre + reporter + GCG IF. As previously observed for C-PEP, a significant percentage of EGFP-tagged cells were also GCG+ (white arrows).
(D) ALK3-Cre + reporter + CA19.9 IF. In contrast with C-PEP and GCG, very few CA19.9 cells expressed the EGFP tag. A representative microphotograph shows co-localization in only one cell of a large cluster (magnified in the inset).
(E) ALK3-Cre + reporter + amylase IF. As with CA19.9, less than 1% of amylase-expressing cells were EGFP-tagged (representative microphotograph showing no co-localization). Inset: example of a rare small cluster of Amy+/EGFP+ cells elsewhere. Numbered white arrows denote incidences of co-localization between the indicated marker and GFP.
4 hNEPT preparations were used for this purpose. hNEPT cultures were transduced as described previously (Klein et al., 2015), i.e., following a regimen in which cells are exposed to BMP-7 for 8–10 days, and then BMP-7 is withdrawn to allow for differentiation. We typically observe 7% and 20% efficiency of transduction for lentivirus and adenovirus, respectively. In our experience, neither exhibits preferential transduction for any particular cell type in hNEPT. We then determined whether there was co-localization of EGFP+ signal (labeling ALK3+ cells) with C-peptide (Figure 1B), glucagon (Figure 1C), CA19.9 (Figure 1D), and amylase (Figure 1E). Similar to our previous findings with labeled PDX1+ cells (Klein et al., 2015), by tagging ALK3+ cells, we also detected co-localization of EGFP/C-peptide (59.2 ± 4.0% of the EGFP-tagged cells were also C-peptide+) and, to a lesser extent, of EGFP/glucagon (19.6 ± 5.5%). However, the degree of co-localization of EGFP with either CA19.9 or amylase (mature markers of ductal and acinar tissues, respectively) was almost negligible (<1%). These values provide only an orientation of differentiation trends, as limitations of in vitro lineage tracing make accurate quantification impossible. While we cannot rule out the possibility that ALK3+ cells may give rise to ductal and acinar cells under different culture conditions, our results suggest that the potential of ALK3+ progenitor-like cells in hNEPTs is chiefly restricted to endocrine fates. Coupled with our earlier findings (Klein et al., 2015), these data further suggest that the cells within hNEPT that convert to C-peptide+ cells upon BMP-7 stimulation are characterized by PDX1 and ALK3 expression.
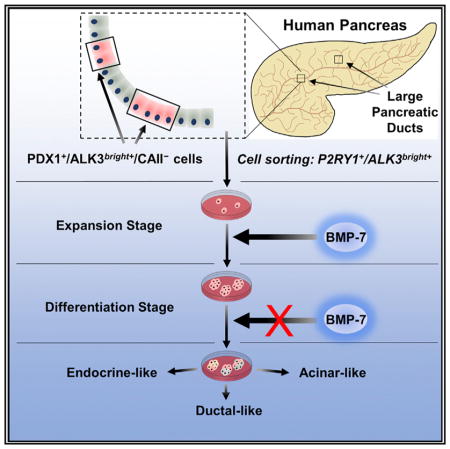
PDX1+/ALK3+ Cells Are Found throughout the Non-endocrine Compartment of the Human Pancreas, but Not in Islets
Based on the aforementioned data, we sought to identify cells with PDX1+ and ALK3+ co-expression in human pancreatic tissue sections (n = 6 donors). In addition to the PDX1+/hormone+ cells within the islet, PDX1+/hormone− cells are abundant in the exocrine compartment of human pancreatic tissues (Figures 2A and S1A). Two populations of non-endocrine PDX1+ cells could be identified: one in ducts of all types, including large/major (main pancreatic duct and interlobular ducts, characterized by columnar epithelium and fibromuscular stroma) as well as small/minor (terminal ductules, squamous intercalated ducts, intralobular ducts, and interlobular ducts of small diameter, featuring flattened cuboidal epithelium and little or no surrounding stroma); and the second population in PDGs (pockets of epithelial cells embedded into the stroma of the MPDs) (Figures S1A and S1B). MUCIN 6 (MUC6), a glycoprotein previously associated with exocrine PDX1+ cells by our team (Wang et al., 2013), is highly expressed in PDGs and can thus be used to distinguish the two populations (Figures S1B and S1C). To further characterize extrainsular PDX1+ populations, we also examined SOX9 expression. This is a transcription factor expressed by multipotent PDX1+ pancreatic progenitors and by ductal cells in the mouse (Furuyama et al., 2011). In human pancreatic samples, nuclear SOX9 was detected in most ductal cells (Figures 2B and 2C) and some PDG cells (Figure 2H). However, while nuclear SOX9 and PDX1 co-localized regularly in the cells of small ducts (Figures 2B and 2C), their expression was often observed at a very low level for one if the other was present both in PDGs (Figure 2H) and MPDs (Figure S1D).
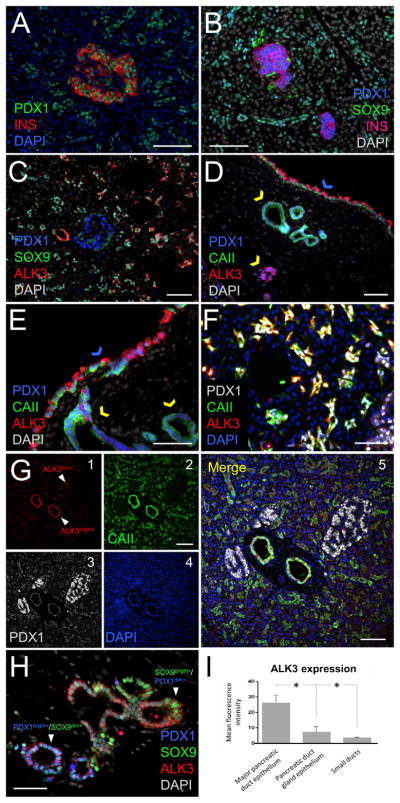
Figure 2. PDX1 and ALK3 Patterns of Expression in the Human Pancreas.
(A) PDX1 (green) is expressed in insulin-producing β cells (red) and in exocrine cells.
(B) Extrainsular PDX1+ cells are found in ductal structures. In most, PDX1 (blue) co-localizes with the ductal marker SOX9 (green), yielding a cyan color.
(C) Many extrainsular PDX1+ cells co-express SOX9 (green) and ALK3 (red). Islets (blue PDX1+ nuclei) are invariably ALK3−.
(D) The epithelium of the MPDs (blue arrow) has the largest population of PDX1+/ ALK3bright+/CAII− cells across the organ. PDGs within the stroma of the MPDs (yellow arrows) contain comparatively less ALK3bright+ and more CAII+ cells.
(E) Higher magnification of the pattern observed in (D).
(F) Nearly 100% extrainsular PDX1+/ALK3+ cells within small ducts co-express CAII, unlike in PDGs and MPDs, where CAII− are also found.
(G) A microphotograph showing the two distinct subpopulations of ALK3+ cells in the human pancreas. 1–4: ALK3 (red), CAII (green), PDX1 (gray), and DAPI (blue), respectively. 5: channel merge. Cells in major ducts are ALK3bright+, whereas those in minor ducts are ALK3dim+.
(H) PDX1/SOX9 staining in ALK3+ epithelial cells within PDGs is heterogeneous, in contrast with the homogeneous pattern observed in small ducts shown in (C). There were PDX1bright+/SOX9dim+ and PDX1dim+/SOX9bright+ cells, as indicated by arrows.
(I) Quantification of ALK3 expression in the epithelium of the MPDs, PDGs, and intercalated ducts. The mean fluorescence intensity (MFI) is reported as mean ± SD (n = 3 samples). Differences were significant among all groups (p < 0.05). Cells in the epithelial lining of the MPDs express ALK3 at the highest level.
Nuclear counterstaining for all panels: DAPI, indicated in blue in (A), (F), and (G) and indicated in white in (B)–(E) and (H). Scale bars: 100 μm in (A)–(D) and (G); and 50 μm in (E), (F), and (H). n = 6 biological replicates. In (A)–(E) and (H), standard fluorescence microscopy was used; in (F) and (G), confocal microscopy was used. *p < 0.05.
We found ALK3 to co-localize with many extrainsular PDX1+ cells (Figures 2C–2G, S1, and S2). In contrast, PDX1-expressing cells within the islets were ALK3− (Figures 2C and 2G). Fluorescence-activated cell sorting (FACS) analysis of human pancreatic samples further confirmed that ALK3+ and insulin+ cells are two independent populations (Figure S3).
Extrainsular PDX1+/ALK3+ cells were abundant both in ducts and in PDGs. We found two distinct subpopulations of PDX1+/ ALK3+ cells: PDX1+/ALK3dim+ cells were widely distributed throughout all the minor ducts, where they almost invariably (>98%) co-localized with the panductal marker CAII (Inada et al., 2006) (Figures 2F, 2G, and S2). PDX1+/ALK3bright+ cells, in contrast, were relatively more abundant in the MPDs and, to a lesser extent, in PDGs. These cells were often negative for CAII (Figures 2D, 2E, 2I, and S2). PDX1+/ALK3bright+/CAII− cells were also generally positive for the ductal markers CK19 (Figure S1E) and SOX9, although in the latter case, we often found SOX9bright+ and SOX9dim+ cells (Figure S1F).
In summary, the histological analysis of the cells co-expressing PDX1 and ALK3 indicates that they reside in ducts and PDGs, but not in islets. Importantly, not all ductal cells expressed CAII. We found populations of PDX1+/ALK3+ cells in the epithelium of the MPDs (as well as in some PDGs and, more rarely, in smaller ducts) in which CAII and ALK3 are mutually exclusive. PDX1+/ALK3+/CAII− cells within the MPD are also characterized by the strongest ALK3 signal (ALK3bright+) detected across the organ (Figure 2I). These observations are aligned with our published lineage-tracing data (Klein et al., 2015), which exclude CAII-expressing cells as a major source of β-like cells upon BMP-7 stimulation.
PDX1+/ALK3bright+ Cells Can Be Sorted and Cultured in Defined Conditions and Show BMP-7 Responsiveness
The sorting and culture of PDX1+/ALK3+ cells would afford us the possibility of studying them without the confounding effects of all other cell types present in hNEPT. For instance, the observation of AMY+ or CA19.9+ clusters that lack the EGFP tag when ALK3 is lineage traced (Figures 1D and 1E) would suggest that these cells are carried over in hNEPT and not arising as a result of BMP-7 stimulation of progenitors.
ALK3 is a surface receptor and, as such, may be used for sorting. However, PDX1 is typically nuclear. In order to define surrogate surface markers for PDX1, we interrogated a published dataset with the transcriptome of the major human pancreatic populations (Dorrell et al., 2011). Gene set enrichment analysis was applied on a matrix of expression data obtained from the datasets E-MTAB-463 and −465 (Dorrell et al., 2011). The PDX1 expression profile was used as continuous phenotype labels, and the Pearson’s correlation was used as the metric to select which genes showed concordant or opposite expression patterns with PDX1. PDX1 was found to be strongly associated with P2RY1, a purinergic receptor. Indeed, P2RY1 was found to be co-expressed with PDX1 (both in ducts and islets) in the human pancreas (Figure 3A).
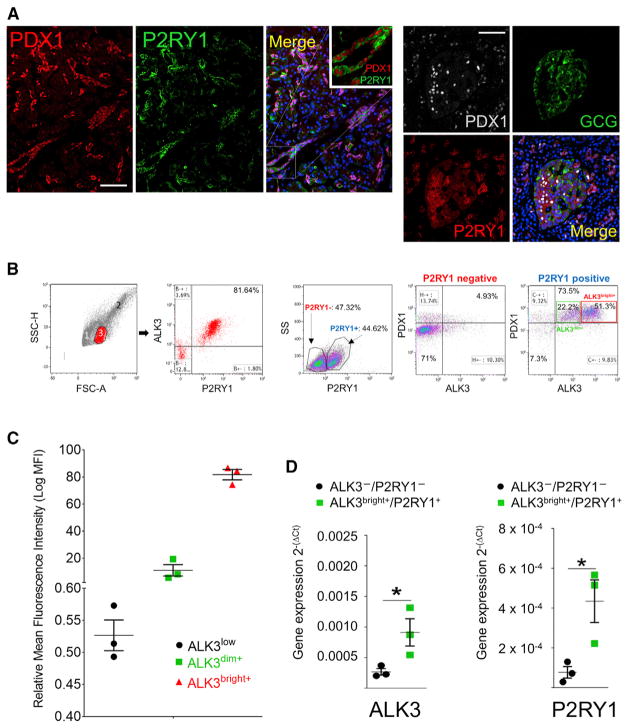
Figure 3. P2RY1 Is a Surrogate Surface Marker for PDX1.
Expression of the P2RY1 surface marker was predicted by bioinformatics to co-localize with that of PDX1 in human pancreatic samples.
(A) Left: from left to right, PDX1 (red), P2RY1 (green), and merged (DAPI, blue). A magnified detail is shown in the inset. Scale bar: 50 μm. Right: P2RY1 stains both PDX1+ ducts and islet β cells, but not α cells. From top left, clockwise: PDX1 (gray), glucagon (green), P2RY1 (red), and merged channels. Scale bar: 50 μm.
(B) ALK3/P2RY1 FACS analysis of hNEPT. A forward/side scatter of hNEPT shows three distinct subpopulations identified in a forward/side scatter (1–3). Population no. 3 corresponds to ~21% of the total population and is 81% enriched in ALK3+/ P2RY1+ cells. P2RY1 sorting alone results in a significant enrichment (>73%) in cells that are both ALK3+ and PDX1+. Insets in the last panel show the percentage of cells with ALK3bright+ (51.3%, red) and ALK3dim+ (22.2%, green) phenotypes.
(C) Mean fluorescence intensity (MFI) of the two populations (ALK3dim+ and ALK3bright+) detected by FACS analysis of hNEPT preparations. A third fraction (ALK3low) is usually considered negative, owing to its very low detection threshold. MFI of each fraction is shown in the y axis (error bars indicate SE).
(D) ALK3bright+/P2RY1+ cells are enriched in their corresponding mRNAs after sorting when compared to the ALK3−/P2RY1− fraction, as determined by qRT-PCR. Gene expression (y axis) is measured as 2−(ΔCt). *p < 0.05.
In (A): n = 4 biological replicates; in (B)–(C): n = 3 biological replicates; in (D): n = 3 biological replicates. Confocal microscopy was used in the acquisition of all these images.
Individually, neither P2RY1 nor ALK3 is a selective marker for BMP-7-responsive cells. However, we hypothesized that double sorting with the two markers would enrich for progenitor-like cells while excluding mature β cells. We tested this hypothesis on freshly isolated hNEPT (n = 3 preparations). Representative results are shown in Figure 3B. One of the three cell subpopulations identified in a forward/side scatter (corresponding to ~21% of the population according to the gating performed) is 81% enriched in ALK3+/P2RY1+ cells (Figure 3B, left). Moreover, in additional experiments, we found that P2RY1 sorting alone results in a significant enrichment (>73%) in cells that are both ALK3+ and PDX1+ (Figure 3B, right). Therefore, sorting by ALK3+/P2YR1+ ensures the isolation of most ALK3+/PDX1+ cells (Figures S4A–S4D). It is important to reiterate that ALK3 sorting alone specifically excludes insulin+ cells, as shown in Figure S3. Therefore, no residual β cells are present in the sorted population. Of note, as no surrogate surface markers for CAII have been identified yet, we could not sort live P2RY1+/ALK3+/CAII− cells for further study. However, we circumvented this limitation by gating on the ALK3bright+ fraction. Once the ALK3+ population is gated away from the isotype control, two discrete populations (ALK3dim+ and ALK3bright+) can be observed (Figures 3B and 3C), with a 7.4-fold difference in mean fluorescence intensity (MFI) between them. As shown in Figure 3D, ALK3bright+/ P2RY1+ cells are enriched in their corresponding mRNAs after sorting when compared to the ALK3−/P2RY1− fraction. Gating on the ALK3bright+ cells (Figure 3B) excludes more than 90% of CAII+ small ductal cells (ALK3dim+) (Figures 2, S2, and S4G). Moreover, following treatment with BMP-7, the ALK3bright+/CAII− fraction exhibited a ~2-fold (when gating on PDX1+) or ~4-fold (when gating on P2RY1+) increase in SMAD1/5/9 phosphorylation (Figures S4J and S4L). In contrast, the ALK3bright+/CAII+ fraction had no meaningful SMAD1/5/9 phosphorylation response to BMP-7, whether gating for PDX1 (Figure S4K) or P2RY1 (Figure S4M). In summary: (1) P2RY1 is an effective surrogate surface marker for PDX1; (2) only CAII− cells are responsive to BMP-7; and (3) sorting for ALK3bright+ cells effectively excludes most CAII+ cells. Based on the aforementioned data, we hypothesized that BMP-7-responsive progenitor-like cells within the human pancreas have a P2RY1+/ALK3bright+ phenotype.
FACS analysis of n = 3 hNEPTs shows that 9.9 ± 0.2% of the cells have a PDX1+/P2RY1+/ALK3bright+ phenotype. However, it is important to consider that the rigors of isolation already enrich for ductal cells at the expense of acinar cells. We have reported that only ~35% of the cells in hNEPT are acinar, when this percentage is much higher (>80%) in the native pancreas (Klein et al., 2015). Therefore, the aforementioned percentage is probably higher than that in the native organ.
We next sorted P2RY1+/ALK3bright+ cells for further study. The culture and differentiation strategy is outlined in Figure 4A. Since the viability of cells sorted from fresh hNEPT is very low (1%–5%; data not shown), cells were allowed to attach for 48 hr in starting medium and then were placed in STEMPRO medium for an additional 72 hr prior to sorting. FACS analysis of hNEPT plated for 48 hr indicates that the relative percentage of PDX1+/P2RY1+/ ALK3bright+ remained unchanged versus that determined for fresh tissue (10.2 ± 0.4%, n = 3 preparations). 72 hr later, at sorting, 4.6 ± 5.2% of the live cells were PDX1+/P2RY1+/ALK3bright+ (n = 12 hNEPT preparations; Figure S5F).
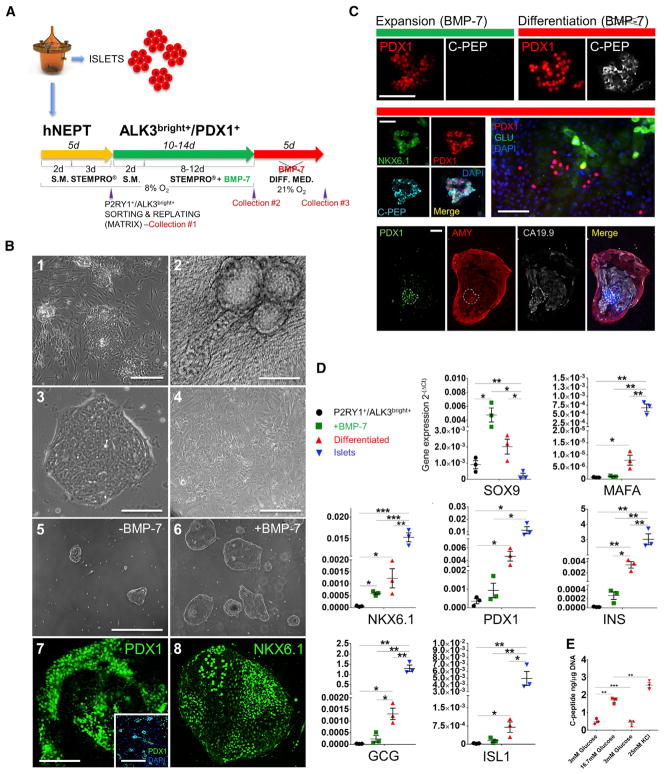
Figure 4. Culture, Expansion and Differentiation of P2RY1+/ALK3bright+ Cells.
(A) hNEPT are obtained as the by-product of islet isolation. After 5 days in culture in low-oxygen conditions (48 hr in starting medium, S.M., and 72 hr in STEMPRO), P2RY1/ALK3 (bright fraction) live-cell sorting is performed on the attached cells (collection point 1). Sorted cells are then re-plated in low oxygen (48 hr in S.M. and then STEMPRO for the remainder of this stage) and cultured with BMP-7 for 10–14 days, a period during which colonies form and expand. Collection point 2 is at the end of this stage. In a third phase, BMP-7 is removed, and cells are cultured in differentiation medium for an additional 5 days at 21% oxygen. Collection point 3 is at the end of this stage.
(B) P2RY1+/ALK3bright+ cells grow both atop (1) and embedded in (2) fibroblast feeder layers. In the latter case, three-dimensional ductal-like structures are often observed. Using a 1:1 collagen I/fibronectin matrix and low-oxygen concentration, P2RY1+/ALK3bright+ cells form colonies of small cells with well-defined edges (3). In contrast, the negative fraction (P2RY1−/ALK3− cells) grows as a mesenchymal-like monolayer (4). In the absence of BMP-7, colonies from P2RY1+/ ALK3bright cells form but grow slowly (5). BMP-7 administration, however, induces sustained colony growth. Panels 5 and 6 show cultures of P2RY1+/ALK3bright+ cell-derived colonies without or with BMP-7, respectively, at the same time point (day 10 after sorting). Regardless of whether BMP-7 is supplemented, P2RY1+/ ALK3bright cell-derived colonies exhibit an IF signal for PDX1 (7) and NKX6.1 (8). Shown in the pictures are BMP-7-treated colonies at day 10 after sorting. Inset in (7) shows PDX1 staining (green) and DAPI (blue) IF of P2RY1+/ALK3bright cell-derived colonies at day 2 after sorting. Scale bars: 100 μM for 1, 5, 6, and 7-inset; and 50 μM for 2, 3, 4, 7, and 8.
(C) During the expansion (top left, green bar), colonies exhibit PDX1 (red) but not C-peptide (white) IF signal. In contrast, after BMP-7 withdrawal and differentiation (top right, red bar), C-peptide signal is detected in many PDX1+ structures. The middle left panel shows a representative C-peptide+ colony with nuclear PDX1 and NKX6.1 signal. The middle right panel shows glucagon+ cells, and the bottom panel the different channels of a colony displaying ductal (CA19.9 signal, light gray) and acinar (amylase, red) cells with a PDX1+ core that remains largely undifferentiated (white dotted lines). Scale bars: 50 μm.
(D) qRT-PCR of representative pancreatic endocrine markers at sorting (black data points), at the end of the BMP-7-mediated expansion phase (green data points) and following differentiation (red data points). y axis: 2−(ΔCt) values against 18S. Error bars indicate SD. *p < 0.05; **p < 0.01; ***p < 0.005.
(E) Glucose-stimulated C-peptide release of differentiated cells (n = 3 preparations) at 3 mM (first low), 16.7 mM (high), 3 mM (second low), and 25 mM KCl-induced depolarization. y axis: nanograms of C-peptide/micrograms of DNA. Error bars indicate SE. *p < 0.05; **p < 0.01; ***p < 0.005.
In (B): n = 5 biological replicates; in (C): n = 4 biological replicates; in (D) and (E): n = 3 biological replicates. In (B): 1–6, brightfield microscopy was used; in 7 and 8, confocal microscopy was used. In (C), standard fluorescence microscopy was used.
Sorted primary pancreatic cells are difficult to culture, but plating P2RY1+/ALK3bright+ cells either atop or embedded in mitomycin C-inactivated human fibroblasts results in the formation of flat colonies or three-dimensional duct-like structures, respectively (Figure 4B, 1 and 2). To perform a thorough characterization of these cells in the absence of other confounding populations, we conducted a three-pronged optimization of culture conditions (n = 11 preparations). First, we screened 36 extracellular matrix combinations for optimal sorted P2RY1+/ALK3bright+ cell attachment. As shown in Figure S5, a 1:1 mix of collagen I and fibronectin resulted in maximal attachment. Second, we screened defined medium formulations specifically designed for stem cells, among which STEMPRO hESC-SFM (Thermo Fisher Scientific, Waltham, MA, USA) turned out to be the most effective at sustaining the attachment and growth of P2RY1+/ ALK3bright+ cells. Finally, based on earlier reports (Prasad et al., 2009) and our own experience on the influence of oxygen partial pressure (pO2) in pancreatic progenitor cell proliferation and differentiation (Cechin et al., 2014; Fraker et al., 2007, 2009), we used low pO2 (8% O2 concentration) to favor their expansion in an undifferentiated state (Figure S5D).
Using a combination of the aforementioned conditions (n = 5 preparations), colonies from sorted P2RY1+/ALK3bright+ cells could be readily grown, even in the absence of BMP-7 (Figure 4B, 3). The P2RY1−/ALK3− fraction, in contrast, grew as a mesenchymal-like monolayer (Figure 4B, 4). When exposed to BMP-7 (100 ng/mL), colonies from P2RY1+/ALK3bright+ sorted cells expanded in size versus non-treated colonies (Figure 4B, 5 and 6; Figure S5G). A 2.4-fold increase in the mitotic index of BMP-7-treated cells was observed versus that of untreated cultures (Figure S5G). Regardless of BMP-7 exposure, colonies almost uniformly expressed PDX1 (Figure 4B, 7) and NKX6.1 (Figure 4B, 8), widely accepted markers of pancreatic progenitors (Rezania et al., 2013). Neither C-peptide (Figure 4C) nor glucagon (data not shown) were readily detected during this BMP-7-induced expansion phase, although some CA19.9+ ductal cells could be observed in the periphery (data not shown). Following a 10- to 14-day period of exposure to BMP-7 (Figure 4A), cultures were placed in differentiation medium without BMP-7 (Cechin et al., 2014; Russ et al., 2015). We have previously reported that BMP-7 withdrawal induces maturation of BMP-7-induced progenitor-like cells within whole hNEPT into endocrine cells (Klein et al., 2015). BMP-7-treated P2RY1+/ ALK3bright+ cells differentiated into C-peptide-expressing cells in the same manner, as shown by immunofluorescence analysis 5 days after BMP-7 withdrawal (Figure 4C). We could also detect the expression of glucagon and, in some colonies, the ductal and acinar markers CA19.9 and amylase (Figure 4C). Imaging quantification (n = 4 preparations) shows that 12.5 ± 4.6% of the cells were PDX1+/NKX6.1+/C-peptide+ (β-like cells). An additional 7.2 ± 4.5% were PDX1+/NKX6.1+ but negative for C-peptide (likely progenitor cells). Finally, 12.4 ± 3.5% were PDX1+ but negative for both NKX6.1 and C-peptide (possibly mature ductal cells). Out of all PDX1+ cells, 39.5 ± 14.1% were also NKX6.1+/ C-peptide+ (β-like cells), 22.3 ± 14% were also NKX6.1+ but C-peptide− (likely progenitors), and 38.1 ± 3.3% were negative for both NKX6.1 and C-peptide (probably ductal).
As previously reported on whole hNEPT (Klein et al., 2015), colonies often exhibit a PDX1+ core surrounded by differentiated cell types (Figure 4C), suggesting an active process of multilineage differentiation from PDX1-expressing progenitor cells.
qRT-PCR analysis confirmed that BMP-7 exposure of P2RY1+/ALK3bright+ cells (Figure 4D, green data points) results in an upregulation of pancreatic progenitor genes versus sorted/untreated cells, whereas its withdrawal (Figure 4D, red data points) leads to an increase in the expression of endocrine differentiation markers. In contrast, the P2RY1−/ALK3− fraction does not respond to BMP-7. Similarly, when separating ALK3+ and ALK3− cells using ALK3-conjugated magnetic beads, only the former show BMP-7 responsiveness and C-peptide production (data not shown). An analysis of n = 3 additional preparations indicates that differentiated cells respond to glucose stimulation in a physiological manner (average stimulation index = 3.4 ± 0.4), with an average maximum C-peptide secretion of 1.7 ± 0.1 ng/μg of DNA at 16.7 mM of glucose (Figure 4E).
Taken together, our data suggest that sorted P2RY1+/ ALK3bright+ cells have pancreatic progenitor-like characteristics and respond to sequential BMP-7 addition and withdrawal by proliferating and differentiating, respectively.
RNA-Seq Analysis Suggests Upregulation and Maintenance of Progenitor/Cell-Cycling Genes after BMP-7 Exposure and a Transition toward a Differentiated State upon Its Withdrawal
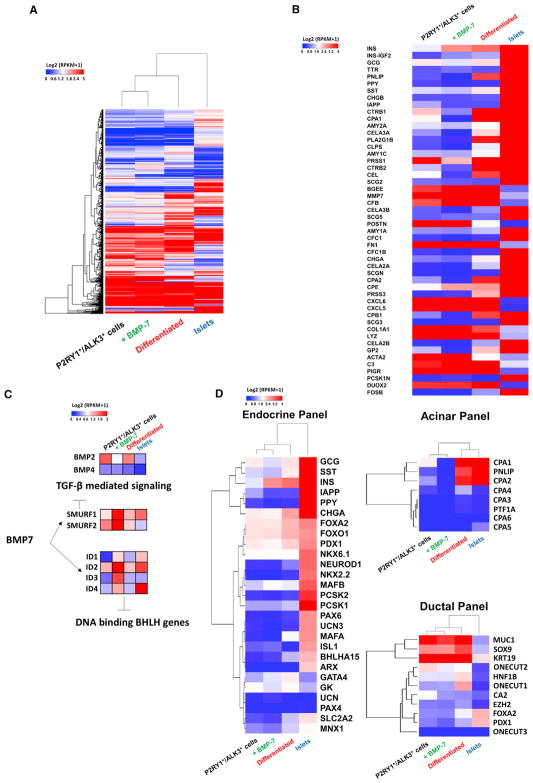
Next, we set out to explore the cellular transcriptome of sorted P2RY1+/ALK3bright+ cells (the sorted progenitor cell group), as well as P2RY1+/ALK3bright+ cells after 10–14 days of BMP-7 treatment (the BMP-7-expanded group), and P2RY1+/ALK3bright+ cells after BMP-7 withdrawal and induction of differentiation (the differentiated group) (n = 3 preparations for each). Human isolated islets (n = 3 donor preparations) were also analyzed as a positive control for all pancreatic differentiated lineages. This is because islet preparations are typically ~60% pure and nearly always include a substantial percentage of acinar and ductal cells (Balamurugan et al., 2014). We conducted RNA-seq of the 4 populations as detailed in the Experimental Procedures. Figure 5 shows a heatmap of the top 10,000 most abundant homologous coding genes. Hierarchical clustering of the 4 sub-populations indicates that the differentiated cells are transcriptionally closer to islets than sorted progenitors and BMP-7-expanded cells (Figure 5A). The top 50 differentially expressed genes across all 4 populations are shown in Figure 5B.
Figure 5. RNA-Seq of P2RY1+/ALK3bright+ Cells at Sorting and after Treatment with and Subsequent Removal of BMP-7.
(A) Hierarchical clustering for the top 10,000 most abundant genes ranked based on highest SD across all 4 cell types (sorted P2RY1+/ALK3bright, BMP-7-treated P2RY1+/ALK3bright, differentiated P2RY1+/ALK3bright cells, and human isolated islets). Heatmap colors correspond to standardized log2 expression values.
(B) Top 50 differentially expressed genes across all cell populations.
(C) Addition of BMP-7 induces downregulation of the TGF-β pathway through SMURF1 and SMURF2, suppresses BMP-2 and BMP-4, and elevates the expression of ID proteins 1–4.
(D) Analysis of gene expression of endocrine, acinar, and ductal panels. Ward linkages above each panel show closer proximity of differentiated cells to islet controls for all three lineages. n = 3 biological replicates.
Since BMP-7 is known to inhibit the TGF-β signaling cascade (Zeisberg et al., 2003), it was not unexpected to see that expression of the TGF-β inhibitors SMURF1 and SMURF2 was high in the sorted progenitors, and even more elevated after BMP-7 treatment, but that it dropped in differentiated cells and islets (Figure 5C). BMP-7 also upregulated all the inhibitor of DNA binding/differentiation (ID) proteins (Figure 5C). High levels of ID1–ID4 are found in progenitor cells, where they sequester basic helix-loop-helix (bHLH) transcription factors, thus leading to the inhibition of lineage-specific and cell-cycle-inhibitory genes (Lasorella et al., 2014). In fact, at least one BMP family member (BMP-4) regulates pancreatic progenitor cell expansion in mice through ID2 (Hua et al., 2006). All IDs are downregulated after BMP-7 withdrawal and differentiation (Figure 5C). Interestingly, both BMP-4 and BMP-2, unlike BMP-7, are expressed in sorted P2RY1+/ALK3bright+ progenitors. This suggests that these two BMPs may be involved in the ID-mediated maintenance of progenitor identity through an autocrine process, as reported in numerous biological examples (Martínez et al., 2014; Yokoyama et al., 2017). However, upon BMP-7 addition, both BMP-2 and −4 are downregulated (Figure 5C) as BMP-7 takes over (and increases) ID upregulation. One potential candidate to explain this downregulation is Gremlin-1 (Grem1), a TGF-β inhibitor known to predominantly antagonize BMP-2 and BMP-4 (Church et al., 2015). Grem1 is among the top 100 differentially expressed genes throughout the 4 populations (high in progenitors, low in differentiated cells/islets) and is strongly upregulated in PANC-1 cells 1 hr after addition of an ALK3 agonist as detected by auto-western capillary immunoblotting (data not shown).
The differentiated cells had also higher overall level of expression of endocrine, ductal, and acinar markers than the BMP-7-expanded ones (Figure 5D), providing additional confirmation of the multilineage differentiation potential. Differentiated cells also clustered closer to islets than both P2RY1+/ALK3bright+ and BMP-7-treated P2RY1+/ALK3bright+ cells for endocrine, acinar, and ductal (Figure 5D) genes. As previously observed by qRT-PCR (Figure 4D), the BMP-7-mediated expansion phase was characterized by an overall downregulation of differentiation markers.
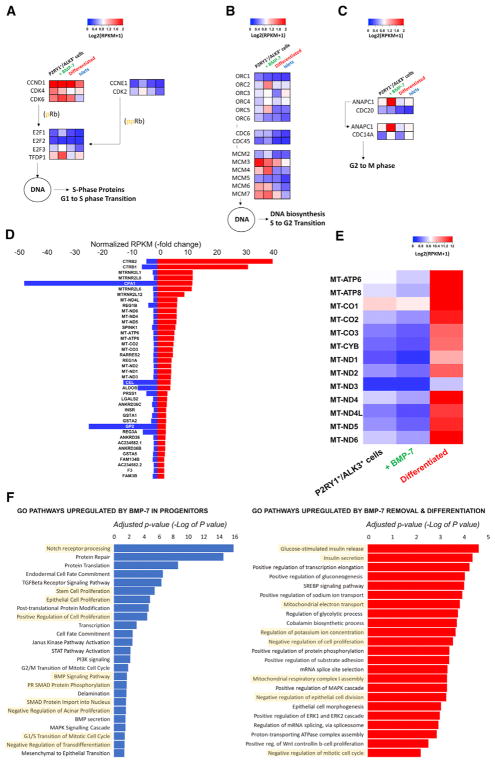
BMPs have a well-documented role in proliferation, as we have reported. To further investigate it, we analyzed 95 genes involved in cell-cycle progression (KEGG: hsa04110). As shown in Figure S6, we found similar expression signatures for P2RY1+/ ALK3bright+ cells and BMP-7-treated P2RY1+/ALK3bright+ cells, suggesting that the former are proliferation ready and that BMP-7 “locks in” that program. However, both islets and P2RY1+/ALK3bright+ cells that were differentiated following BMP-7 withdrawal cluster together at the other end of the spectrum, showing a globally lower degree of expression of genes implicated in cell division. This was further confirmed when we specifically investigated the expression profile of genes involved in G1-to-S phase transition (Figure 6A) as well as those either implicated in S-to-G2 phase transition (Figure 6B) or relevant to the spindle assembly checkpoint transition (Figure 6C). In all these cases, P2RY1+/ALK3bright+ cells and BMP-7-treated P2RY1+/ALK3bright+ cells had generally higher expression levels of cell-cycle progression genes than both the differentiated group and islets.
Figure 6. Analysis of BMP-7-Elicited Cell Replication and Differentiation Induced following BMP-7 Withdrawal.
(A–C) Key regulators of cell-cycle progression shown with their standardized log2 expression values across all four cell types. BMP-7 induces an upregulation of replication genes at all stages of cell division. In contrast, BMP-7 withdrawal (differentiation) leads to the overall downregulation of the replicative machinery.
(A) High cyclin D, CDK4, and CDK6 transcription is needed during the G1-to-S transition.
(B) Genes involved in the pre-replication Cdc45-Mcm2–7.CDC6 complex are recruited to the hetero-hexameric origin recognition complex (ORC2–6) and function as a replicative helicase unwinding pre-replicative DNA.
(C) The spindle assembly checkpoint is suppressed by the ANAPC1-CDC20 (APC/C) complex, allowing the G2-M transition (anaphase). Genes were chosen based on the KEGG Orthology reference hierarchy (KEGG: ko04110) and KEGG pathway entry (KEGG: map04110).
(D) BMP7-mediated gene expression down-regulation is inverted in differentiated cells. Fold changes in RPKM are shown for BMP7 treated versus P2RY1+/ALK3bright+ cells (blue) and differentiated versus P2RY1+/ALK3bright+ (red). The chart shows the top 40 coding genes with a fold change > 1 for the first comparison (blue bars) and >1.2 for the second (red bars). Only genes with a false discovery rate (FDR) <10% were considered.
(E) Heatmap showing gene expression across the three tissue types for mitochondrial protein-coding genes. (n = 3 biological replicates).
(F) Gene ontology (GO) analysis of P2RY1+/ ALK3bright cells treated with BMP-7 versus sorted P2RY1+/ALK3bright cells (left). GO analysis of differentiated cells versus BMP-7-treated (expanded progenitors) P2RY1+/ALK3bright cells (right).
n = 3 biological replicates.
We then focused on P2RY1+/ALK3bright+ cells following the addition and subsequent withdrawal of BMP-7. We interrogated our RNA sequence dataset for genes whose expression may counter proliferation (i.e., decrease with BMP-7) but induce differentiation (i.e., increase after BMP-7 withdrawal). Figure 6D shows the top 40 genes that follow this pattern while exhibiting an RPKM fold change of <1 for BMP-7-treated (expanded) versus untreated (non-stimulated P2RY1+/ALK3bright+ progenitor) cells and >1 for differentiated versus untreated (non-stimulated progenitor) cells. Interestingly, 11 of the 13 protein-coding mitochondrial genes (and all 13 using less restrictive criteria; see Figure 6E) are in this 40-gene list. Since all mitochondrial protein-encoding genes are involved in oxidative phosphorylation (Taanman, 1999), our observation is consistent with the well-described notion that there is a switch from glycolysis to oxidative phosphorylation in the transit from progenitor cells to mature pancreatic (and especially β; Conget et al., 1997) cell types (Xu et al., 2013). Also on that list are CPA1 (carboxypepti-dase-1, a marker of both acinar cells and multipotent pancreatic progenitors) (Pagliuca and Melton, 2013), several regenerating proteins (Reg1a, Reg1b, and Reg3a, linked to islet regeneration) (Parikh et al., 2012), and GSTA1–2 (MAFA-regulated genes involved in reducing β cell stress) (Matsuoka et al., 2015). Gene Ontology (GO) analysis (Figure 6F; Table S1) further confirms that, compared to unstimulated cells, BMP-7-treated P2RY1+/ ALK3bright+ cells (expanded) exhibit an overall enrichment of gene expression in pathways such as Notch receptor processing (associated with the maintenance of cycling status in pancreatic progenitors) (Apelqvist et al., 1999), stem cell proliferation, positive regulation of cell division, and BMP signaling (Figure 6F, left). Conversely, P2RY1+/ALK3bright+ cells differentiated upon BMP-7 withdrawal show enrichment in β cell function pathways (including insulin secretion) and mitochondrial oxidative phosphorylation, as well as a global negative regulation of proliferation (Figure 6F, right).
Taken together, our data further confirm the notion that BMP-7 engages the cell division-ready phenotype of P2RY1+/ALK3bright+ cells and that BMP-7 withdrawal induces cell-cycle arrest and multilineage differentiation.
DISCUSSION
We present an in-depth characterization of progenitor-like cells previously identified by our team within the human exocrine pancreas (Klein et al., 2015). Their responsiveness to BMP-7 led us to the identification of ALK3 as a key mediator of the activation of the canonical BMP pathway. The validation of P2RY1 as a surrogate surface marker for PDX1 has allowed us to use antibodies against these two markers to sort progenitor cells with a high degree of accuracy. Indeed, we show that only P2RY1+/ALK3bright+ populations respond to the administration and subsequent withdrawal of BMP-7 by proliferating first and then differentiating. The negative fractions are unresponsive.
While ductal cells have long been associated with pancreatic regeneration, the use of mature ductal markers for the isolation of duct-residing progenitor cells may have confounded the interpretation of earlier data. Such is the case of CAII, a protein used by ductal cells to produce bicarbonate and, thus, inactivate digestive enzymes until they complete their transit to the duodenum (Hegyi et al., 2011). Despite its terminal differentiation marker credentials, CAII has been extensively used to isolate putative progenitors (Bonner-Weir et al., 2008; Inada et al., 2008). Several lines of evidence align with our conclusion that true progenitors may be CAII−: (1) first, that these cells exist in the first place. To our knowledge, cells that are negative for CAII (a marker widely considered to be pan-ductal; Inada et al., 2006) have not been described in the ductal tree. The notion that ductal progenitors are CAII− would be consistent with the general principle that progenitor cells rarely express terminal differentiation markers; (2) the concurrent observation that a large proportion of such CAII− cells also happen to have the highest expression of ALK3 throughout the pancreas; (3) their residence in MPDs and PDGs, locations long suspected to harbor progenitor cells, as found in studies published by our team (Carpino et al., 2016; Wang et al., 2013) and others (Yamaguchi et al., 2015); and (4) our earlier lineage-tracing results excluding CAII+ cells as the origin of new β cells following BMP-7 stimulation (Klein et al., 2015). Our immunofluorescence (IF) data strongly suggests that sorting for ALK3bright+ cells (the only fraction with progenitor-like characteristics) excludes most CAII+ cells, which are typically ALK3dim+ and found in smaller ducts.
It is important to stress that ALK3 is not expressed in β cells in any meaningful manner. While low expression of ALK3 transcripts has been reported in α and β cells (Dorrell et al., 2011), this study used microarray hybridization, and ALK3 expression was not confirmed by RT-PCR. Likewise, in a recently published study using RNA-seq single-cell transcriptomics, ALK3 transcripts were detected only at very low levels (Li et al., 2016) (nearly 3 orders of magnitude less than insulin; Segerstolpe et al., 2016). Such difference was 20,000-fold in another report (Nica et al., 2013). There are no studies showing ALK3 expression at the protein level, be it by western blot, proteomics, or immunostaining. In fact, ALK3 has not been detected in proteomic analysis of either human (Zhang et al., 2017) or murine (Waanders et al., 2009) isolated islets. By sorting ALK3bright+ cells (the only fraction that shows responsiveness to BMP-7), we selectively exclude any residual β cells that may have been carried over in the hNEPT. Combined with our IF data that ALK3+ cells are never detected in islets, these observations make us confident that the progenitor-like cells described herein are extrainsular.
Earlier work on hNEPT suggested the multilineage differentiation potential of progenitor cells residing therein (Klein et al., 2015). IF analysis of sorted and cultured P2RY1+/ALK3bright+ cells shows that these cells differentiated along at least two of the major endocrine lineages (β and α), as well as ductal (CA19.9+) and acinar (amylase+) cells. We could not detect either somatostatin or pancreatic polypeptide (PP)-expressing cells, but their differentiation cannot be ruled out. RNA-seq data also suggest that the differentiated populations share gene expression profiles with both isolated islets as well as exocrine cells. Interestingly, while lineage tracing shows some contribution of ALK3-tagged cells to both acinar and ductal cells, it was almost negligible compared to that observed for C-peptide and glucagon. This may be due to the culture setting, where confounding cell types in hNEPT (but not in sorted P2RY1+/ ALK3bright+ colonies) may partially block/promote specific differentiation outcomes. Although lineage tracing is useful to provide general, non-quantitative indications about the likely origin of specific cell types, its limitations when used in vitro may also contribute to explain the aforementioned findings. For instance, it is conceivable that the bulk of acinar and ductal differentiation may have already taken place at the onset of ALK3-Cre expression. This would also be consistent with our IF observation that ductal/acinar differentiation preceded that of endocrine fates and could be observed even during the BMP-7-mediated expansion.
Of note, we could not detect the pro-endocrine factor NGN3 (Domínguez-Bendala et al., 2005) either in sorted P2RY1+/ ALK3bright+ cells (expanded or differentiated) or in human islet preparations (which also contain a high percentage of acinar and ductal cells). However, our knowledge of NGN3 function in the pancreas is limited to embryonic development, and mostly in the mouse. The notion that regenerative processes in the adult pancreas must necessarily mimic those involved in its development is not supported yet by experimental evidence and remains only speculation.
Previous analyses on hNEPT showed that BMP-7 induced the formation of cells with islet-like insulin content and glucose responsiveness both in vitro and in vivo (Klein et al., 2015). Despite the relatively low percentage of β-like cells (~12%) in differentiated P2RY1+/ALK3bright+ colonies, we determined that these cultures also respond to glucose stimulation. Given the observation that all mitochondrial oxidative phosphorylation genes are elevated upon differentiation, it would be interesting to conduct follow-up studies on their oxygen consumption rates.
When studying hNEPT-sorted P2RY1+/ALK3bright+ cells, we observed great variability between preparations in their differentiation output, with varying degrees of bias toward endocrine or exocrine fates. Examples of the latter were the preparations used for RNA-seq, where we observed a strong tendency toward acinar differentiation (e.g., 82-fold in pancreatic lipase versus BMP-7-expanded progenitors, 70-fold in carboxypeptidase A1, 96-fold in Elas3a, etc.) but more modest increases in endocrine marker expression. We acknowledge that the β-like cells generated in our study are not as competent as native β cells. This observation was also probably compounded by the fact that effective differentiation of pancreatic progenitors into mature β cells remains a bottleneck in the field (Domínguez-Bendala et al., 2016) and the lack of optimization in this particular setting. It is important to emphasize that our study was aimed at establishing proof of principle and not the generation of fully functional, terminally differentiated cells.
The inconsistency in the ability of individual hNEPT preparations to yield colonies after P2RY1+/ALK3bright+ sorting, as well as the variable differentiation profiles, may be due to multiple reasons. While our methods adhere to strict operating procedures, factors such as donor age, sex, weight, and ischemia time, as well as tissue digestion, may affect reproducibility. The latter is of especial importance, since islet isolation requires the pancreas to be cannulated through the main pancreatic duct. The MPDs and adjacent structures, where we have found a higher relative abundance of PDX1+/ALK3bright+ cells, thus bear the brunt of the enzymes used to dissociate the organ. Even small variations in the process result in drastic differences in yield and potency.
RNA-seq profiles show BMP-7-dependent engagement of the canonical BMP pathway, ID protein upregulation, activation of cell division, and inhibition of TGF-β—all hallmarks of BMP-mediated progenitor cell stimulation. The profile of the top 50 differentially expressed genes between all four populations also shows very low levels of expression of pancreatic differentiation markers during the BMP-7 exposure (proliferation) phase, followed by an upregulation upon BMP-7 withdrawal and differentiation. Upregulated markers within the top 50 differentially expressed genes include insulin, somatostatin, pancreatic polypeptide (PPY), islet amyloid polypeptide, and chromogranins A and B for the endocrine lineage, as well as an assortment of acinar markers (amylases, Elas3a, phospholipase, colipase, cholesterol esterase, etc.). As mentioned before, such acinar markers are also highly expressed in the islet control (Balamurugan et al., 2014). GO pathway analysis further confirms the proliferative/progenitor nature of the P2RY1+/ALK3bright+ cells stimulated by BMP-7, as well as the differentiation induced upon its removal. RNA-seq analyses are always somewhat speculative, but all the observed trends are consistent with our hypothesis.
While it may seem counterintuitive that P2RY1+/ALK3bright+ cells exhibit a “proliferation-ready” phenotype even before BMP-7 is added, these cells have been growing in vitro for 5 days prior to RNA collection. The reason for this pre-culture stage is that P2RY1+/ALK3bright+ cells sorted from fresh tissue have poor viability, which is inconsistent with the procurement of good quality RNA. This necessary pre-culture step does not change the “stemness” of the progenitors. Low oxygen, which improves the attachment and yield of colonies in this pre-culture stage, is known to induce proliferation (Ezashi et al., 2005; Prasad et al., 2009), which may explain the observed proliferative signature. At any rate, we cannot discard the hypothesis that some of these cells may be dividing in the native organ, perhaps in the context of normal tissue turnover. This has not been studied yet.
Our studies cannot rule out the possibility that these cells may have de-differentiated from ducts. However, if BMP-7 induced the progenitor-like proliferation of de-differentiated ductal cells, and these differentiated into multiple lineages after its removal, the overall conclusions of our research would remain unchanged.
Our findings go well beyond the comprehensive characterization of this progenitor cell population. The identification of their anatomical niche will allow us to screen for their existence in the exocrine pancreas of both type 2 and type 1 diabetes patients. Provided that these cells remain intact after the autoimmune destruction of β cells in type 1 diabetes, their responsiveness to BMP-7 (a Food and Drug Administration [FDA]-approved agent) could be potentially used to restore β cell mass in situ in the context of concomitant interventions to quell autoimmunity.
EXPERIMENTAL PROCEDURES
Culture of hNEPT and P2RY1+/ALK3bright+ Cells
hNEPT was obtained as a by-product of human islet isolation. This program has an IRB (institutional review board) exemption from the University of Miami. Organs were obtained with appropriate consent. See Table S2 for demographic information of all donors used in this study. Details on the culture are described in the Supplemental Experimental Procedures.
Lineage Tracing, Sorting of P2RY1+/ALK3bright+ Cells, and Flow Cytometry of hNEPT
hNEPT cells were plated with starting medium. Transduction was done with equal amounts of lentiviral reporter and adenoviral ALK3-Cre constructs. Additional details on this method, as well as on the sorting of P2RY1+/ ALK3bright+cells and flow cytometry of hNEPT, are provided in the Supplemental Experimental Procedures.
RNA-Seq
Specific details on RNA isolation and quality control, library preparation, and sequencing are provided in the Supplemental Experimental Procedures.
IF
Methods for IF are detailed in the Supplemental Experimental Procedures.
Glucose-Stimulated C-Peptide Release
These assays were performed as previously reported (Fraker et al., 2013), with some modifications (see Supplemental Experimental Procedures).
Real-Time qPCR
Real-time qPCR was conducted as in Nieto et al. (2012).
Statistics
GraphPad Prism v5 was used for statistical analysis. Following the Shapiro-Wilk normality test, statistical differences between groups were calculated by two-tailed paired t test or Wilcoxon signed-rank test, with 95% confidence intervals (*p < 0.05; **p < 0.01; ***p < 0.001). Results are expressed as mean ± SD. See the Supplemental Experimental Procedures for a statistical description of specific experiments.
Supplementary Material
Highlights.
Multipotent cells expressing PDX1 and ALK3 reside in the human exocrine pancreas
PDX1+/ALK3bright+ cells are found mostly in major pancreatic ducts and duct glands
Surface markers ALK3 and P2RY1 can be used to sort and culture progenitor cells
BMP-7 stimulates pancreatic progenitor cell proliferation by binding to ALK3
Acknowledgments
We thank D. Sant, M. Bellio, J. van Dijk, K. Johnson, M. Boulina, A. Méndez, and the staff of the Translational Models Core and the Clinical Chemistry, Biomarkers and Immunoassay Laboratory (all at the Diabetes Research Institute) for their technical contribution and helpful discussions. Similarly, we are grateful for the contribution of Rocío Muñiz (VUMC School of Medical Sciences, Amsterdam, the Netherlands) while in training at the corresponding authors’ laboratories. We also thank the Network of Pancreatic Donors with Diabetes (nPOD) for control samples. This work was funded by the Diabetes Research Institute Foundation (DRIF), the Inserra family, the Fred and Mabel R. Parks Foundation, the Foundation for Diabetes Research, the Tonkinson Foundation, the Michael J. and Katherine E. Franco Foundation, the Frank Strick Foundation, Mildred Graff, and NIH grants 1R43DK105655-01 and 2R44DK105655-02. M.M.F.Q. is funded by an International Fulbright pre-doctoral fellowship/grant administered by the Foreign Fulbright Scholarship Board and the International Institute of Education. R.L.P. and J.D.-B. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity and accuracy of the analysis.
Footnotes
DATA AND SOFTWARE AVAILABILITY
The accession number for the raw sequencing data and normalized data reported in this paper is GEO: GSE104120.
Supplemental Information includes Supplemental Experimental Procedures, six figures, and three tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.02.006.
AUTHOR CONTRIBUTIONS
M.M.F.Q., S.A.-C., D.K., G.L., and C.G.-S. designed and performed experiments along with analysis and interpretation of data. A.M. and F.P.-U. performed experiments and analyzed data. E.M.C.M. performed bioinformatics analysis. C.R. and L.A.I. contributed discussion and advice on experimental design. R.L.P. and J.D.-B. conceived and designed the study and compiled the manuscript. M.M.F.Q., S.A.-C., D.K., G.L., R.L.P., and J.D.-B. revised the manuscript.
DECLARATION OF INTERESTS
C.G.S. was an employee of Ophysio, Inc., at the time of data generation but does not hold stock in Ophysio, Inc. The University of Miami, C.R., L.I., and J.D.-B., are stockholders in Ophysio, Inc., licensee of the intellectual property used in this study, but do not receive royalties. C.R., L.I., J.D.-B., and R.L.P. are named inventors in a patent application on intellectual property presented in the manuscript, but the patent has not been awarded yet.
References
- Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U, Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Balamurugan AN, Naziruddin B, Lockridge A, Tiwari M, Loganathan G, Takita M, Matsumoto S, Papas K, Trieger M, Rainis H, et al. Islet product characteristics and factors related to successful human islet transplantation from the Collaborative Islet Transplant Registry (CITR) 1999–2010. Am J Transplant. 2014;14:2595–2606. doi: 10.1111/ajt.12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner-Weir S, Inada A, Yatoh S, Li WC, Aye T, Toschi E, Sharma A. Transdifferentiation of pancreatic ductal cells to endocrine beta-cells. Biochem Soc Trans. 2008;36:353–356. doi: 10.1042/BST0360353. [DOI] [PubMed] [Google Scholar]
- Calva-Cerqueira D, Dahdaleh FS, Woodfield G, Chinnathambi S, Nagy PL, Larsen-Haidle J, Weigel RJ, Howe JR. Discovery of the BMPR1A promoter and germline mutations that cause juvenile polyposis. Hum Mol Genet. 2010;19:4654–4662. doi: 10.1093/hmg/ddq396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpino G, Renzi A, Cardinale V, Franchitto A, Onori P, Overi D, Rossi M, Berloco PB, Alvaro D, Reid LM, Gaudio E. Progenitor cell niches in the human pancreatic duct system and associated pancreatic duct glands: an anatomical and immunophenotyping study. J Anat. 2016;228:474–486. doi: 10.1111/joa.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cechin S, Alvarez-Cubela S, Giraldo JA, Molano RD, Villate S, Ricordi C, Pileggi A, Inverardi L, Fraker CA, Domínguez-Bendala J. Influence of in vitro and in vivo oxygen modulation on β cell differentiation from human embryonic stem cells. Stem Cells Transl Med. 2014;3:277–289. doi: 10.5966/sctm.2013-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church RH, Krishnakumar A, Urbanek A, Geschwindner S, Meneely J, Bianchi A, Basta B, Monaghan S, Elliot C, Strömstedt M, et al. Gremlin1 preferentially binds to bone morphogenetic protein-2 (BMP-2) and BMP-4 over BMP-7. Biochem J. 2015;466:55–68. doi: 10.1042/BJ20140771. [DOI] [PubMed] [Google Scholar]
- Conget I, Barrientos A, Manzanares JM, Casademont J, Viñas O, Barceló J, Nunes V, Gomis R, Cardellach F. Respiratory chain activity and mitochondrial DNA content of nonpurified and purified pancreatic islet cells. Metabolism. 1997;46:984–987. doi: 10.1016/s0026-0495(97)90265-1. [DOI] [PubMed] [Google Scholar]
- Domínguez-Bendala J, Klein D, Ribeiro M, Ricordi C, Inverardi L, Pastori R, Edlund H. TAT-mediated neurogenin 3 protein transduction stimulates pancreatic endocrine differentiation in vitro. Diabetes. 2005;54:720–726. doi: 10.2337/diabetes.54.3.720. [DOI] [PubMed] [Google Scholar]
- Domínguez-Bendala J, Lanzoni G, Klein D, Álvarez-Cubela S, Pastori RL. The human endocrine pancreas: new insights on replacement and regeneration. Trends Endocrinol Metab. 2016;27:153–162. doi: 10.1016/j.tem.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Dorrell C, Schug J, Lin CF, Canaday PS, Fox AJ, Smirnova O, Bonnah R, Streeter PR, Stoeckert CJ, Jr, Kaestner KH, Grompe M. Transcriptomes of the major human pancreatic cell types. Diabetologia. 2011;54:2832–2844. doi: 10.1007/s00125-011-2283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezashi T, Das P, Roberts RM. Low O2 tensions and the prevention of differentiation of hES cells. Proc Natl Acad Sci USA. 2005;102:4783–4788. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraker CA, Alvarez S, Papadopoulos P, Giraldo J, Gu W, Ricordi C, Inverardi L, Domínguez-Bendala J. Enhanced oxygenation promotes beta-cell differentiation in vitro. Stem Cells. 2007;25:3155–3164. doi: 10.1634/stemcells.2007-0445. [DOI] [PubMed] [Google Scholar]
- Fraker CA, Ricordi C, Inverardi L, Domínguez-Bendala J. Oxygen: a master regulator of pancreatic development? Biol Cell. 2009;101:431–440. doi: 10.1042/BC20080178. [DOI] [PubMed] [Google Scholar]
- Fraker CA, Cechin S, Álvarez-Cubela S, Echeverri F, Bernal A, Poo R, Ricordi C, Inverardi L, Domínguez-Bendala J. A physiological pattern of oxygenation using perfluorocarbon-based culture devices maximizes pancreatic islet viability and enhances β-cell function. Cell Transplant. 2013;22:1723–1733. doi: 10.3727/096368912X657873. [DOI] [PubMed] [Google Scholar]
- Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- Hegyi P, Maléth J, Venglovecz V, Rakonczay Z., Jr Pancreatic ductal bicarbonate secretion: challenge of the acinar acid load. Front Physiol. 2011;2:36. doi: 10.3389/fphys.2011.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua H, Zhang YQ, Dabernat S, Kritzik M, Dietz D, Sterling L, Sarvetnick N. BMP4 regulates pancreatic progenitor cell expansion through Id2. J Biol Chem. 2006;281:13574–13580. doi: 10.1074/jbc.M600526200. [DOI] [PubMed] [Google Scholar]
- Inada A, Nienaber C, Fonseca S, Bonner-Weir S. Timing and expression pattern of carbonic anhydrase II in pancreas. Dev Dyn. 2006;235:1571–1577. doi: 10.1002/dvdy.20754. [DOI] [PubMed] [Google Scholar]
- Inada A, Nienaber C, Katsuta H, Fujitani Y, Levine J, Morita R, Sharma A, Bonner-Weir S. Carbonic anhydrase II-positive pancreatic cells are progenitors for both endocrine and exocrine pancreas after birth. Proc Natl Acad Sci USA. 2008;105:19915–19919. doi: 10.1073/pnas.0805803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D, Álvarez-Cubela S, Lanzoni G, Vargas N, Prabakar KR, Boulina M, Ricordi C, Inverardi L, Pastori RL, Domínguez-Bendala J. BMP-7 induces adult human pancreatic exocrine-to-endocrine conversion. Diabetes. 2015;64:4123–4134. doi: 10.2337/db15-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasorella A, Benezra R, Iavarone A. The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer. 2014;14:77–91. doi: 10.1038/nrc3638. [DOI] [PubMed] [Google Scholar]
- Li J, Klughammer J, Farlik M, Penz T, Spittler A, Barbieux C, Berishvili E, Bock C, Kubicek S. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. 2016;17:178–187. doi: 10.15252/embr.201540946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez VG, Hidalgo L, Valencia J, Hernández-López C, Entrena A, del Amo BG, Zapata AG, Vicente A, Sacedón R, Varas A. Autocrine activation of canonical BMP signaling regulates PD-L1 and PD-L2 expression in human dendritic cells. Eur J Immunol. 2014;44:1031–1038. doi: 10.1002/eji.201343693. [DOI] [PubMed] [Google Scholar]
- Matsuoka TA, Kaneto H, Kawashima S, Miyatsuka T, Tochino Y, Yoshikawa A, Imagawa A, Miyazaki J, Gannon M, Stein R, Shimomura I. Preserving Mafa expression in diabetic islet β-cells improves glycemic control in vivo. J Biol Chem. 2015;290:7647–7657. doi: 10.1074/jbc.M114.595579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nica AC, Ongen H, Irminger JC, Bosco D, Berney T, Antonarakis SE, Halban PA, Dermitzakis ET. Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Res. 2013;23:1554–1562. doi: 10.1101/gr.150706.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M, Hevia P, Garcia E, Klein D, Alvarez-Cubela S, Bravo-Egana V, Rosero S, Molano RD, Vargas N, Ricordi C, et al. Antisense miR-7 impairs insulin expression in developing pancreas and in cultured pancreatic buds. Cell Transplant. 2012;21:1761–1774. doi: 10.3727/096368911X612521. [DOI] [PubMed] [Google Scholar]
- Pagliuca FW, Melton DA. How to make a functional β-cell. Development. 2013;140:2472–2483. doi: 10.1242/dev.093187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh A, Stephan AF, Tzanakakis ES. Regenerating proteins and their expression, regulation and signaling. Biomol Concepts. 2012;3:57–70. doi: 10.1515/bmc.2011.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad SM, Czepiel M, Cetinkaya C, Smigielska K, Weli SC, Lysdahl H, Gabrielsen A, Petersen K, Ehlers N, Fink T, et al. Continuous hypoxic culturing maintains activation of Notch and allows long-term propagation of human embryonic stem cells without spontaneous differentiation. Cell Prolif. 2009;42:63–74. doi: 10.1111/j.1365-2184.2008.00571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezania A, Bruin JE, Xu J, Narayan K, Fox JK, O’Neil JJ, Kieffer TJ. Enrichment of human embryonic stem cell-derived NKX6.1-expressing pancreatic progenitor cells accelerates the maturation of insulin-secreting cells in vivo. Stem Cells. 2013;31:2432–2442. doi: 10.1002/stem.1489. [DOI] [PubMed] [Google Scholar]
- Russ HA, Parent AV, Ringler JJ, Hennings TG, Nair GG, Shveygert M, Guo T, Puri S, Haataja L, Cirulli V, et al. Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. EMBO J. 2015;34:1759–1772. doi: 10.15252/embj.201591058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segerstolpe A, Palasantza A, Eliasson P, Andersson EM, Andreasson AC, Sun X, Picelli S, Sabirsh A, Clausen M, Bjursell MK, et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 2016 doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto H, LeBleu VS, Bosukonda D, Keck P, Taduri G, Bechtel W, Okada H, Carlson W, Jr, Bey P, Rusckowski M, et al. Activin-like kinase 3 is important for kidney regeneration and reversal of fibrosis. Nat Med. 2012;18:396–404. doi: 10.1038/nm.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1999;1410:103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- Waanders LF, Chwalek K, Monetti M, Kumar C, Lammert E, Mann M. Quantitative proteomic analysis of single pancreatic islets. Proc Natl Acad Sci USA. 2009;106:18902–18907. doi: 10.1073/pnas.0908351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lanzoni G, Carpino G, Cui CB, Dominguez-Bendala J, Wauthier E, Cardinale V, Oikawa T, Pileggi A, Gerber D, et al. Biliary tree stem cells, precursors to pancreatic committed progenitors: evidence for possible life-long pancreatic organogenesis. Stem Cells. 2013;31:1966–1979. doi: 10.1002/stem.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Duan S, Yi F, Ocampo A, Liu GH, Izpisua Belmonte JC. Mitochondrial regulation in pluripotent stem cells. Cell Metab. 2013;18:325–332. doi: 10.1016/j.cmet.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Yamaguchi J, Liss AS, Sontheimer A, Mino-Kenudson M, Castillo CF, Warshaw AL, Thayer SP. Pancreatic duct glands (PDGs) are a progenitor compartment responsible for pancreatic ductal epithelial repair. Stem Cell Res (Amst) 2015;15:190–202. doi: 10.1016/j.scr.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasmin N, Bauer T, Modak M, Wagner K, Schuster C, Köffel R, Seyerl M, Stöckl J, Elbe-Bürger A, Graf D, Strobl H. Identification of bone morphogenetic protein 7 (BMP7) as an instructive factor for human epidermal Langerhans cell differentiation. J Exp Med. 2013;210:2597–2610. doi: 10.1084/jem.20130275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama Y, Watanabe T, Tamura Y, Hashizume Y, Miyazono K, Ehata S. Autocrine BMP-4 signaling is a therapeutic target in colorectal cancer. Cancer Res. 2017;77:4026–4038. doi: 10.1158/0008-5472.CAN-17-0112. [DOI] [PubMed] [Google Scholar]
- Zeisberg M, Hanai J, Sugimoto H, Mammoto T, Charytan D, Strutz F, Kalluri R. BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med. 2003;9:964–968. doi: 10.1038/nm888. [DOI] [PubMed] [Google Scholar]
- Zhang H, Klausen C, Zhu H, Chang HM, Leung PC. BMP4 and BMP7 suppress StAR and progesterone production via ALK3 and SMAD1/5/8-SMAD4 in human granulosa-lutein cells. Endocrinology. 2015;156:4269–4280. doi: 10.1210/en.2015-1494. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lanzoni G, Battarra M, Inverardi L, Zhang Q. Proteomic profiling of human islets collected from frozen pancreata using laser capture microdissection. J Proteomics. 2017;150:149–159. doi: 10.1016/j.jprot.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.