Abstract
Chondrogenic cell differentiation constitutes a multistep program that is spatially and temporally modulated by combinations of bioactive factors that drives the establishment of specific cellular phenotypes. This sequence of events results in the fabrication of a distinctive structural and functional extracellular matrix which determines the quality of the cartilaginous tissue and, thus, its potential in vivo implantability as a tissue-engineered implant. Current assessments of engineered cartilage rely on destructive methodologies typically applied at the end of the fabrication period that make it difficult to predict failures early in the process. The high inherent variability of engineered tissues raises questions regarding reproducibility and the validity of using such end-stage representative samples to characterize an entire batch of engineered tissues. Therefore, the development of dynamic, multimodal, nondestructive, and noninvasive technology toolsets to monitor cell differentiation (and secondarily tissue phenotypes) in real time is of paramount importance. In this study, we report the creation of cell-based probes to directly interrogate cell differentiation events during in vitro chondrogenesis and in vivo osteogenesis. For that, native promoters of well-established chondrogenic (Sex Determining Region Y-Box 9 [Sox9] and Aggrecan [AGG]) and osteogenic (Osteocalcin [OC]) differentiation biomarkers were used to create independent probes incorporating a traceable signal (Luciferase) and transduced into human bone marrow-derived mesenchymal stem cells. The probes were used to monitor the progression throughout in vitro chondrogenic differentiation program in aggregate (pellet) cultures and in vivo osteogenic differentiation in heterotopic ossicles. These tissue differentiation constructs were positively tested in conditions known to modulate the differentiation program at various phases that confirmed their sensitivity and reproducibility. This technology toolset allows a nondestructive and noninvasive, imaging-based longitudinal reconstruction of the in vitro chondrogenic differentiation program, while providing an analytical assessment of phenotypic changes of engineered cartilage in real time.
Keywords: : MSCs, chondrogenesis, differentiation, cell-based probes, bioluminescence
Introduction
Tissue-engineered (TE) cartilage manufactured from mesenchymal stem cells (MSCs) is a promising strategy for cartilage repair.1–3 However, to date, the tissue produced has inferior functional properties compared with native articular cartilage and, thus, fails when implanted.4,5 There is a need to modify preimplantation fabrication procedures to improve the biological quality and functional performance of engineered cartilage products derived from MSCs. In this regard, a critical component is to develop technologies that will allow predicting outcomes early in the fabrication process thereby limiting the number of constructs that end in poorly performing implants. To avoid implantation of suboptimal tissues, assessment of molecular predictors of failure and the acquisition of desirable features (related with differentiation events) during the fabrication period becomes essential. We report an approach which allows spatiotemporal monitoring and reconstruction of the chondrogenic cell differentiation program and the resulting tissue phenotypic changes. These molecular-based differentiation examinations are driven at a high temporal resolution and are useful to both predict success and failure, to monitor interventions in real time aimed at modifying the outcomes and to define and test release criteria for implantation.
Chondrogenic differentiation from mesenchymal precursors (i.e., MSCs) constitutes a multistep program mainly controlled by the transcription factor Sex Determining Region Y-Box 9 (Sox9).6,7 This cell differentiation process drives the fabrication of a specific extracellular matrix (ECM) with distinctive structural and functional properties defined by the assembly of multiple components, including proteoglycans (PGs) such as Aggrecan (AGG) and collagen fibers such as type 2 collagen (Col2), with specific three-dimensional directionality.8 The retention of specific glycosaminoglycans controls the tissue's mechanical properties in compression as the ECM structures the water content, while collagen fibers offer tensile resistance.9 These features are difficult to reproduce in vitro as well as the multistep/multimolecular stimulation of chondrogenic precursors that control their cellular differentiation into chondrocytes manufacturing anatomic site-specific hyaline articular cartilage.
The above-mentioned “technical” limitations are further complicated by the intrinsic endochondral bone formation program that is observed during MSCs in vitro chondrogenesis that ultimately develops into a transient type of cartilaginous ECM.10–12 This ECM is reminiscent of both embryonic skeletal formation and that observed during fracture healing in the adult, in which chondrocytes undergo terminal hypertrophic differentiation.13 This transient ECM is different from the permanent hyaline articular cartilage and constitutes a “placeholder” matrix contributing to posterior bone formation as well as a component of osteoarthritic changes.14 Various markers of hypertrophic chondrocytes (e.g., type 10 collagen, Runx2, and PTHR1) as well as osteoblastic differentiation (e.g., Osteocalcin [OC] and type 1 collagen) can be used to establish the presence of these undesired cartilaginous traits.
The process of chondrogenesis during the fabrication of an engineered implant is influenced by several factors, including the cell type, the stimulatory factor(s) used to induce or maintain their differentiation, the scaffolds used to physically support the growth of the tissue, and the presence of an inductive environment to precondition the construct before implantation.5,15,16 The evaluation of TE cartilage has traditionally relied on the presence of a small set of specific markers evaluated at arbitrarily time points, averaged from multiple samples and acquired through destructive methods. The validity of using only a few representative samples to characterize an entire batch is questionable in the face of the high variability inherent of engineered tissues, which raises questions of reproducibility across implants.17 Reported here is the generation and validation of molecular probes based on functional promoters of known biomarkers of chondrogenic and osteogenic differentiation. The resulting library allows the nondestructive and noninvasive examination and tracking of differentiation events of cells bearing the reporter probes inside the growing chondrogenic structure through the acquisition of bioluminescence imaging (BLI) signals secondary to activation of the promoters. This technology then helps to perfect Tissue Engineering-based approaches at the preimplantation phase.
Materials and Methods
Cell cultures
Cultures of human bone marrow (BM)-derived MSCs (hBM-MSCs) from three healthy deidentified adult volunteer donors were established as previously described.18 The BM was collected using a procedure reviewed and approved by the University Hospitals of Cleveland Institutional Review Board. Informed consent was obtained from all deidentified donors. Cells were further expanded in Dulbecco's modified Eagle's medium low glucose (DMEM-LG) supplemented with 10% fetal bovine serum (FBS) that had been screened to support hMSC culture and used in first passage for the experiments.19 Cells were grown without (C) or in the presence of fibroblast growth factor-2 (FGF2; 10 ng/mL of) for 14 days.20
Chondrocyte isolation
Human chondrocytes were isolated from articular cartilage harvested from discarded femoral head samples collected by the Tissue Procurement Facility at Case Western Reserve University. Cartilage tissue was cut in ∼2 × 2 pieces and rinsed with phosphate-buffered saline and subjected to sequential enzymatic digestion with 0.1% trypsin during 30 min, then with 0.1% hyaluronidase for 60 min, and with 0.1% collagenase type II overnight (37°C). The enzymatic digestion is stopped with the addition of FBS and the cell suspension is filtered through a sterile cell strainer (pore size 70 μM; Falcon). After centrifugation, the resulting pellet is washed twice and resuspended in DMEM-LG supplemented with 10% FBS. Cells are seeded in 100-mm culture dishes at a density of 200,000 cells per dish, expanded at high density, and used in first passage.
Fibroblasts isolation
Human fibroblasts were isolated from human skin dermis of paid volunteers under an approved IRB protocol of the Skin Study Center, Skin Diseases Research Center, Case Western Reserve University School of Medicine, as described previously.21
Chondrogenic differentiation
hMSCs were trypsinized and then resuspended in a chondrogenic differentiation medium consisting of DMEM-high glucose supplemented with 1% antibiotic–antimycotic solution, 1% sodium pyruvate, 1% nonessential amino acids (Invitrogen), 1% ITS+ (BD Bioproducts), 10−7 M dexamethasone (Sigma-Aldrich, St Louis, MO), and 80 μM ascorbic acid-2 phosphate (Wako Chemicals) and transforming growth factor β-1 (10 ng/mL). Two hundred microliters of this cell suspension containing 250,000 cells was added per well of a 96-well polypropylene V-bottom, multiwell dish (Phenix Research). The multiwell plates were centrifuged at 500 g for 5 min and then incubated at 37°C. The differentiation medium was changed every other day.22 Chondrogenic pellets were harvested after 21 days for histological analysis.
In vitro osteogenic differentiation
hMSCs were seeded at high density (1 × 105 cells/cm2) and cultured in DMEM-LG supplemented with 10−7 M dexamethasone and 120 μM ascorbic acid-2-phosphate. After 10 days, the medium was augmented with 2 mM β-glycerophosphate and maintained in this medium for another 11 days.23
Construction of promoter reporters
Lentiviral promoter reporter constructs for Sox9 and AGG were custom-ordered from GeneCopoeia (HIV-based lentiviral third generation self-inactivating expression system) containing a 1.0–1.3 kb insert 1.3 kb upstream and up to 200 bp downstream of the transcription initiation site. This insert was placed upstream of the secreted Gaussia luciferase (GLuc) reporter gene. The reporter construct for OC was custom ordered from VectorBuilder (Cyagen) with an insert upstream of the firefly luciferase gene.
Production of the lentiviral vectors
To produce the third-generation replication-incompetent lentivirus, near confluent 293Ta cells (GeneCopoeia Cat. No. Clv-PK-01) were transfected with the promoter reporter lentiviral vectors and the Lenti-Pac HIV mix (GeneCopoeia) in Opti-MEM culture medium (Gibco) and EndoFectin (GeneCopoeia). Opti-MEM was replaced with fresh hMSC medium after 12 h, and viral supernatant was collected after 36 h and again after another 24 h at half the volume.
Transduction of hBM-MSCs
hBM-MSCs were transduced as described previously.24 Cells were seeded at a density of 1 × 104 cells/cm2. Transduction was performed with a multiplicity of infection of 5 in the presence of 100 μg/mL protamine sulfate (Cat. No. P4020-1G; Sigma-Aldrich). Cells were transduced for 24 h.
Luciferase reporter assays
For secreted luciferase (Gaussia), 50 μL of conditioned medium from chondrogenic aggregates at different time points were transferred to a well of a black 96-well plate. Coelenterazine substrate (20 μM) was added to the wells and the GLuc activity measured using Xenogen IVIS 200 series system (PerkinElmer). To quantify the BLI signal in terms of photon flux (photons/s/cm2/steradian) and the area covered by signal (cm2/e), a predefined geometrical shape was used. For nonsecreted luciferase (firefly), luciferin substrate (150 μg/mL) was added directly to the chondrogenic aggregate and imaged as described above.
Humanized heterotopic bone formation assay
A total of 5 × 106 nontransduced (NT) hMSC and hMSC expressing the promoter reporter constructs were vacuum-loaded into sterile porous ceramic cube carriers (hydroxyapatite/tricalcium phosphate 40/60; Zimmer, Warsaw, IN) precoated with a 100 μg/mL solution of fibronectin.25 The cell-loaded cubes were subcutaneously implanted into immunocompromised mice (CB17-Prkdc SCID) for 6 weeks to form extraskeletal bone structures (ossicles).26–28 Every animal (n = 8) received four cubes. All animal procedures were approved by the Case Western Reserve University Institutional Animal Care and Use Committee. After 6 weeks, ossicles were harvested and imaged individually before processed for histological analysis.
In vivo imaging
BLI was performed after subcutaneous injection of 200 mL of 12.5 mg/mL of luciferin substrate (NanoLight) using a Xenogen IVIS 200 series system. To quantify the BLI signal in terms of photon flux (photons/s/cm2/steradian) and the area covered by signal (cm2/e), a predefined geometrical shape was used.
Hybrid pellets
Hybrid pellets were created with the same cell type (homogeneous), in which a fraction of the cells is engineered, and with different cell types (heterogeneous), in which one of them is engineered. Hybrid pellets were made using different ratios of hBM-MSC and chondrocytes or fibroblasts (3:1, 1:1, and 1:3).
Immunohistochemistry
Chondrogenic pellets were fixed with 10% phosphate-buffered formalin and paraffin-embedded. Samples were cut into sections 5 μm thick, which were deposited onto glass slides and then deparaffinized and stained with Safranin O to evaluate PG content.
Statistical analysis
Statistical analysis was conducted using Prism 7 for MAC (GraphPad, Inc., San Diego, CA). Visual analysis of the data shows no skewedness and Grubb's test excluded the presence of outliers within samples. Parametric tests were used to calculate statistical significance, taking into account that with small samples the nonparametric tests have little power to find significant differences. Student's paired two-tailed t-test was used to compare two groups. Values are represented as means ± standard deviations and significance assigned at p < 0.05.
Results
Early outcome correlation using a Sox9 promoter reporter
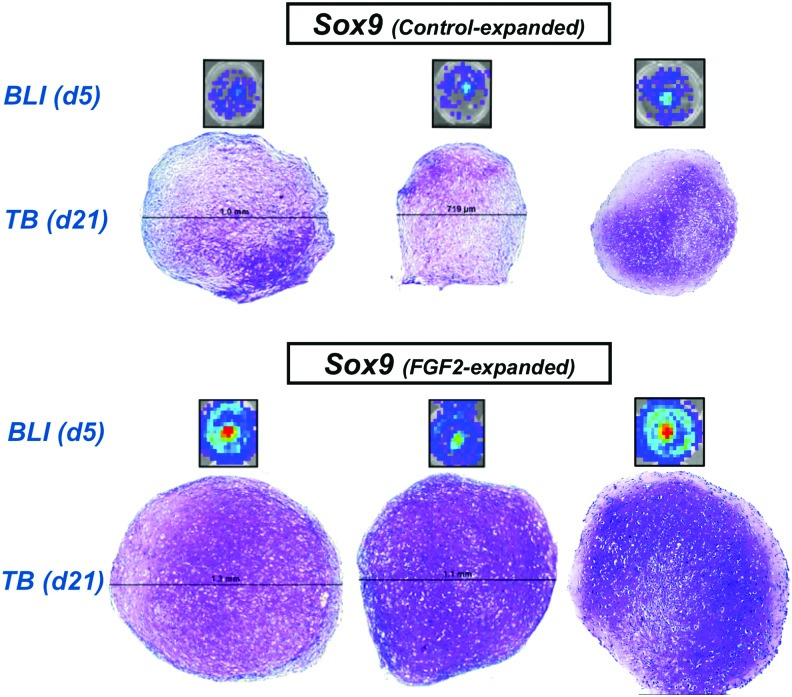
Our group has previously described the prochondrogenic effect of expanding hMSCs in the presence of FGF2.20,29 We have reported a mechanistic early activation of the Sox9-dependent molecular machinery with this exposure evidenced through destructive methods involving immunolocalization and gene expression analysis of the constitutive cells.30 We tested whether a nondestructive approach through an imaging-based Sox9-probe transiently present in hMSCs undergoing chondrogenesis could be used to detect those same differences. We observed that this particular probe was suitable to detect early changes in Sox9 promoter activity (evaluated with BLI at day 5) which were correlated with the final outcomes of hMSCs-derived chondrogenesis (histology at day 21) (Fig. 1). In that respect, a qualitative correlation was observed between the BLI signal intensity at early time points and the pellet development as seen at the end of the differentiation program.
FIG. 1.
Transient transfection of hMSCs with a Sox9 promoter reporter. FGF2-expanded hMSC transiently transfected with a Sox9-luc reporter show an increased signal at day 5 compared with control-expanded cells, later manifested in an accelerated cell differentiation and ECM deposition at day 21, as previously reported.20,29 n = 5 (different donors), three representative samples shown. ECM, extracellular matrix; FGF2, fibroblast growth factor-2; hMSCs, human mesenchymal stem cells; Sox9, Sex Determining Region Y-Box 9. Color images available online at www.liebertpub.com/tea
Generation of stable cell-based probes
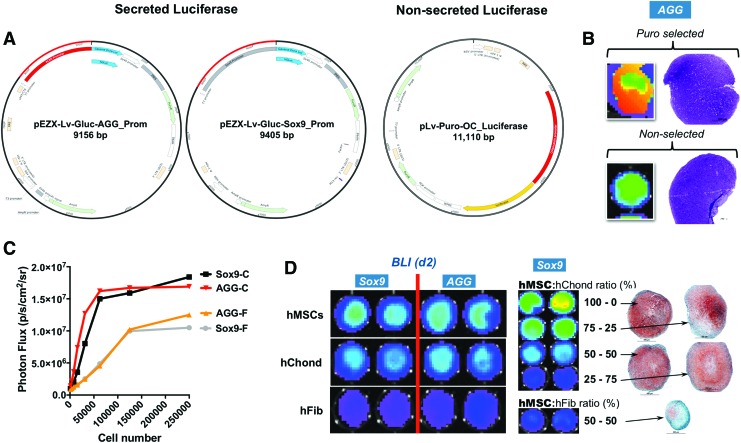
Transient probes have obvious time-dependent constraints, especially in long-lasting programs such as chondrogenesis. Thus, we shifted to a viral-mediated, permanent cell transduction to insure the steady expression of the transgene beyond 5–7 days. We have previously reported modifying hMSCs with traceable viral constructs (including luciferase) to capture MSC fate after infusion via different routes.31 We also have reported that the viral transduction adjuvant hexadimethrine bromide (Polybrene®) compromises hMSCs proliferative and differentiation potential after transduction.32 Consequently, we developed a method that uses protamine sulfate instead of polybrene, resulting in transduction rates in hMSCs greater than 50% without compromising cell proliferation, differentiation, and cell survival,24 all of which are critical for this technology. Lentiviral probes have been generated, which can be traced by BLI using two approaches to detect luciferase activity, thus avoiding signal overlap. In addition to using different substrates, helping in their signal discrimination, one relies on the enzyme activity within the growing structure (i.e., nonsecreted), while the other on the secreted enzyme (GLuc) to the surrounding medium, which can be collected and measured (Fig. 2A).
FIG. 2.
Lentiviral transduction of hMSCs with various promoter reporters. (A) Vector maps for the secreted and nonsecreted luciferase probes showing their structures. (B) BLI and histology (day 21) analysis evidencing that selecting transduced hMSC with puromycin does not affect their chondrogenic potential. FGF2-expanded cells are shown here (similar results obtained with control-expanded cells). (C) Dilution curve showing an increased transduction efficiency in control-expanded (C) versus FGF2-expanded (F) hMSC for both Sox9 and AGG probes. A plateau effect is also seen earlier in control-expanded than in FGF2-expanded cells. (D) Day 2 BLI showing a baseline, high transgene activity for both Sox9 and AGG probes only in chondrogenic cells (hMSCs and human chondrocytes [hChond]) (left panel), which is sensitive to the number of transduced cells used (right panel). In cocultures, hMSC (in bold) were the only cells virally transduced. Early transgene activation is present only in chondrogenic cells as hybrid cultures with human fibroblasts (hFib) exhibited a marginal signal compared with cultures with the same cell ratio made with hChond (50:50 in this case). n = 3 (different donors), one representative set of data shown. BLI, bioluminescence imaging. Color images available online at www.liebertpub.com/tea
Transduced cells were selected with puromycin to generate a pure population of reporter cells as evidenced by a substantial increase on the signal compared with nonselected cells; importantly, their chondrogenic differentiation was not affected by these procedures as evidenced by a comparable histology at day 21 (Fig. 2B). For the signal quantification, all data points were normalized using the control-expanded cells at day 2 due to the following: First, a marked difference in the transduction efficiency between control and FGF2-expanded cells in all donors as evidenced in the dilution curve (Fig. 2C); Second, an early high signal at day 2 in all probes which we would argue constitutes a transduction artifact. In that regard, cell–virus interactions have been shown to produce epigenetic plasticity that may allow a temporary promoter dysregulation effect.33 Of the cells tested, this was evident only in cells with chondrogenic potential especially hMSCs; this effect was reduced when the pellets were made with heterogeneous cells at varying proportions and when using human fibroblasts (Fig. 2D).
Sox9 and AGG probes can detect early prochondrogenic changes in FGF2-expanded hMSC
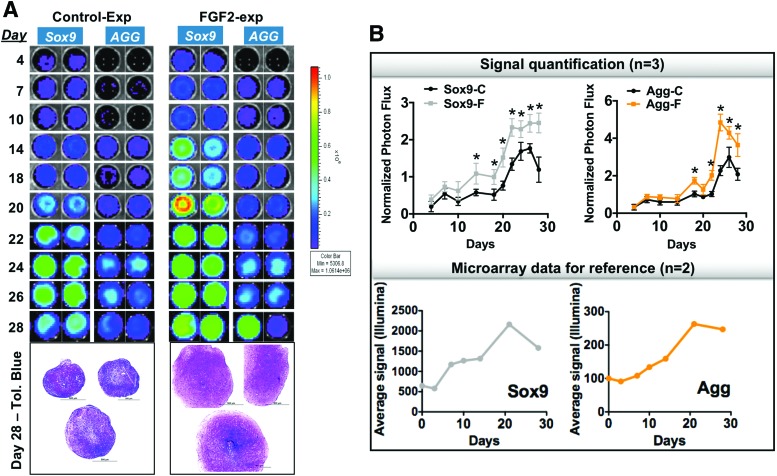
Sox9 and AGG lentivirus-based probes were tested in their ability to detect differences in the molecular machinery controlling chondrogenesis. Unlike the transiently present probe reported in Figure 1, these viral-transduced probes allow a complete reconstruction of the chondrogenic program in pellets formed from both control and FGF2-expanded cells. These probes were able to detect the expected early presence in both Sox9 and AGG in FGF2-expanded cell pellets with high specificity (Fig. 3A, B). The signal profile obtained with the imaging probes correlates with the expression profile of both markers analyzed with a high-throughput microarray (complete dataset submitted for publication) performed at similar time points under the same conditions and also with multiple donors (Fig. 3B).
FIG. 3.
Sox9 and AGG probes early predict the ultimate prochondrogenic effect of FGF2 exposure during hMSC expansion. (A) Secreted luciferase analyzed by BLI throughout the chondrogenic program shows an early Sox9 and AGG presence in FGF2-expanded hMSCs compared with control-expanded cells, evidenced by expected histological differences at the end of differentiation. (B) Quantification of the BLI signal, showing a similar pattern of expression with data obtained in a microarray analysis at similar time points and with multiple donors. n = 3; BLI run in duplicate for each donor; one representative donor shown; pellets correspond to one of the replicates of each donor; BLI quantification with all three donors incorporated; microarray data from two independent donors (used for reference only). *Statistical significance when p < 0.05. Color images available online at www.liebertpub.com/tea
AGG probe confirms the negative effect of FGF9 when present from day 0 of the chondrogenic program
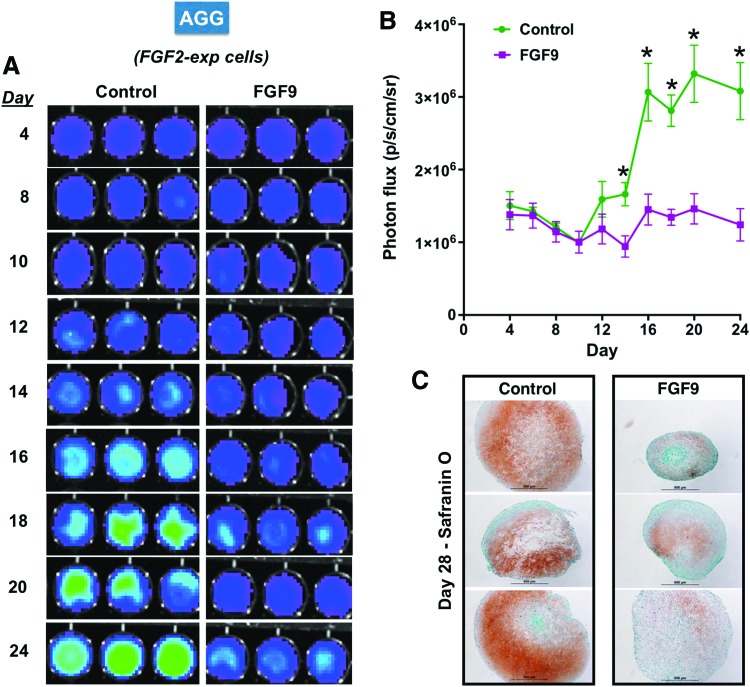
The exposure of FGF9 to pellet cultures distinctively affects chondrogenic differentiation of hMSCs depending on the time of initiation.30 In contrast to the anabolic effect when started at day 14, FGF9 effects on FGF2-expanded hMSCs starting at day 0 was observed to have a negative impact on the initiation and progression of the chondrogenic differentiation program. The AGG probe detected this negative effect and predicted the impaired activation of the chondrogenic program as early as day 10–12, at which time AGG is starting to be expressed in those hMSCs (Fig. 4A, B); this exposure of FGF9 results in underdeveloped pellets (Fig. 4C) similar to previous results. These data further confirm that such early signal detection by the appropriate probe can be used to predict ultimate failures in the differentiation program.
FIG. 4.
AGG probe confirms the negative effect of FGF9 when present from day 0 in hMSC chondrogenesis. (A) Secreted luciferase analyzed by BLI throughout the chondrogenic program shows that AGG is not expressed at all in FGF2-expanded hMSC, subsequently exposed to FGF9 from day 0 of chondrogenesis, evidencing its negative effect on the differentiation program as reported in Correa et al.30 (B) Quantification of the BLI signal and (C) Histology analysis of the resulting pellets (independent donors), confirming the negative effect of FGF9 when present throughout the differentiation program. n = 3; BLI run in triplicate for each donor; one representative donor shown; pellets correspond to one of the replicates of each donor; BLI quantification with all three donors incorporated; *statistical significance when p < 0.05. Color images available online at www.liebertpub.com/tea
The OC probe can discriminate osteogenesis and chondrogenesis in vitro and in vivo
The utilization of the OC probe can be useful for displaying a potential osteogenic shift in differentiation as well as late stages of chondrocyte hypertrophic differentiation and possibly osteoarthritis-like phenotypes as part of in vitro models of inflammation in osteoarthritis. A basal OC expression was observed in hMSCs in expansion and early in pellet culture chondrogenesis up to day 3–5 (Fig. 5A). These observations are independently confirmed by microarray analysis of hMSCs subjected to chondrogenic conditions (submitted article). This OC expression observed on day 3–5 was followed by a significant reduction until the beginning of the fourth week when it starts to rise again, which we interpret as terminal hypertrophic differentiation. As a negative control of the probe, transduced human chondrocytes did not exhibit OC expression throughout the examination period. Conversely, and as positive control, hMSCs subjected to in vitro osteogenic conditions exhibited an increased OC signal with time (Fig. 5B).
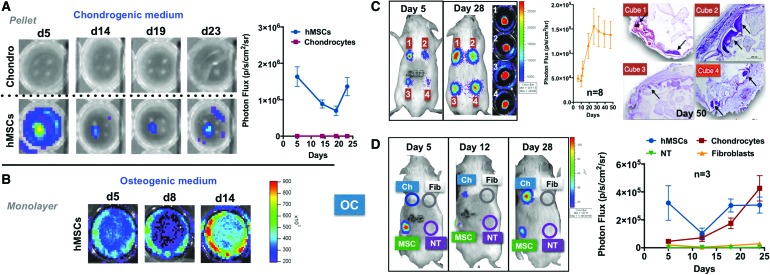
FIG. 5.
The OC probe discriminates chondrogenesis from osteogenesis in vitro and in vivo. (A) Transduced hMSCs exhibit a basal OC expression, which decreases with time until the beginning of the fourth week when terminal differentiation is characterized by its presence. Control human chondrocytes are negative throughout the same time frame, while same hMSCs subject to osteogenic conditions (B) show the expected activation of the probe. n = 3, with each experiment run in triplicate; one representative experiment shown. (C) Transduced hMSCs implanted in vivo (immunocompromised mice) in a porous hydroxyapatite ceramic matrix (n = 8, made from three donors; four shown; all eight with signal quantified) exhibit a characteristic increasing OC expression (inside the back of the mice and after harvesting) with a signal peak at day 28 and associated full osteoblastic differentiation evidenced histologically at day 50 (black arrows). (D) Separate experiment in which hMSCs (n = 3, from independent donors) exhibited a similar ultimate osteoblastic differentiation, while human chondrocytes (n = 3) show a progressive increase in the signal, suggesting hypertrophic differentiation within an endochondral bone formation environment. Human fibroblasts (n = 3) and NT hMSCs (n = 3, from independent donors) were negative throughout the time frame as expected. NT, nontransduced; OC, osteocalcin. Color images available online at www.liebertpub.com/tea
The probes were also validated in vivo using the heterotopic bone formation assay, in which porous calcium phosphate ceramic cubes were loaded with OC-transduced hMSCs and implanted subcutaneously into immunocompromised mice (four cubes per mouse).25–28 These implanted specimens exhibited an initial (day 5) signal (as in vitro) followed by an increased signal with time as evidence of an osteoblastic differentiation responsible for the deposition of bone matrix verified histologically at the end of the program (day 50) (Fig. 5C).25 In a separate experiment, human chondrocytes, fibroblasts, and NT hMSCs were used as negative controls (Fig. 5D): human chondrocytes showed a progressive increase in the signal presumably due to terminal differentiation within an endochondral program supported by in vivo conditions within this extraskeletal experimental construct. Cubes seeded with either fibroblasts or NT-hMSCs were observed to be negative. These observations support the use of the probes in in vivo settings since the signal is visible with the reporter cells present in subcutaneous tissue sites.
Discussion
The progression of cell differentiation during the fabrication of engineered cartilage constructs determines the resulting tissue structure and function, and hence, its analysis is clinically useful. This progression is influenced by several factors,5,15,16 including the cells used (e.g., progenitors vs. mature cells); the use of a single uniform cell type or combinations of different cells (e.g., MSCs with chondrocytes)34–36; the exogenous factor(s) used to induce and maintain differentiation30,37–40; the use of three-dimensional scaffolds to physically support the growth of the tissue41; and the presence of a mechanical environment that mimics the conditions the tissue will experience upon implantation.42 All these interventions have direct effects on the differentiation status of the cells that manifest in quantity and type of ECM secreted and eventually the clinical utility of the engineered tissue.
The evaluation of engineered cartilage is usually performed at the end of the fabrication period at arbitrarily defined intermediate stages as determined by the presence of specific biomarkers that should predict the functional properties of the tissue. These destructive evaluations require the analysis of the changing tissue to assess its mechanical properties and gene expression markers and render the specimen unusable for further implantation. Approaches have been developed to nondestructively determine key structural parameters (i.e., ECM components) based on imaging technologies such as MRI43 and fluorescence spectroscopy incorporated with ultrasound backscattered microscopy.44,45 Although they hold great promise for an end-stage structural characterization, the dynamic molecular changes bringing the tissue to this end-stage cannot be assessed. This is especially problematic at the critical early stages of the fabrication program when the molecular signature assessment is of greater importance compared with the evaluation of the primitive ECM. The key to the successful fabrication process is to quantify, in real time, the effects of outcome-modifying interventions needed to bring about an appropriate end-stage cartilage.
The development and validation of nondestructive molecular imaging-based tools to analytically monitor the dynamic cell differentiation events and the resulting forming tissue in real time is essential for obtaining clinically relevant tissues for future implantation. This objective can be realized by interrogating the fabricating cells as they transit throughout their appropriate and tissue unique chondrogenic lineage. Functional promoters of established, stage-dependent chondrogenic differentiation markers can be used to develop molecular probes by incorporating a traceable signal (luciferase) that can be detected by nondestructive BLI. A library of probes tuned to the dynamically changing differentiation of hMSCs allows an imaging-based time-lapse approach to acquire a longitudinal differentiation assessment of the cells within, for example, the chondrogenic pellet model culture system.
To test this longitudinal logic, three markers were tested: Sox9, AGG, and OC which span the entire chondrogenic program, including commitment (Sox9), ECM assembly (AGG), and terminal hypertrophic differentiation (OC). The latter probe can also be used to access the potential deviation toward osteogenic differentiation.46 Additional markers (e.g., Col2, other Sox family members, and type 10 collagen) and their probes can be generated to address specific mechanistic questions. The incorporation of the probes into nonviral constructs for transient transfection was used as a proof-of-concept understanding their limitations in terms of illuminating of the entire fabrication sequence. The lentivirus-based probes provided adequate cell transduction efficiencies, transgene expression, and BLI signals throughout the entire differentiation process. Importantly, the possibility of using either secreted or nonsecreted luciferase gives the probes additional experimental versatility for data acquisition avoiding signal overlap when two probes are used simultaneously. Fluorescence imaging-based probes can be generated in parallel to guarantee a multiprobe assessment with more than two markers.
One of the main goals of this technology is to establish predictive indicators to reflect the end-stage tissue properties based on early observations (nondestructive snapshots) within the differentiation program. A good theory47 must accurately describe a large class of observations in a model that contains few arbitrary elements and make definitive predictions about the results of future observations. A useful technology toolset in turn must be capable of theoretically achieving these goals. The probes were validated by using them in an established in vitro model of chondrogenesis (aggregate cultures) in an effort to assess their predictive potential based on quantifiable observations. The probes accurately detected early changes in the chondrogenic differentiation molecular program, which were reflected later in the end-stage tissue. Specifically, the anabolic effect of expanding hMSCs with FGF220,29 as evidenced as an early signal from Sox9 and AGG; the negative effect of FGF9 on chondrogenesis when applied at the beginning of the program30 as evidenced with an absent AGG; and the prediction of chondrogenesis and osteogenesis from specific progenitors both in vitro and in vivo using the OC probe.48,49
The information gathered from the probes permits adjustments to interventions to the cultures in real time and the quantification of their short-term effects on the fabrication dynamics. Signaling pathways driving cell differentiation can be described by a molecular cascade of events started at discrete time points typically by master regulators (e.g., Sox9).6,7 These initial control elements are followed by other molecular signaling effectors that culminate in the expression of a particular arrays of target genes (e.g., AGG and Col2).7 An evaluation carried out at a particular arbitrary time point during this ever-changing program only captures a limited state of the cell/tissue at that particular moment. Such static snapshots disregard the dynamic molecular events “in progress” at the evaluation time and, thus, this view is incomplete and represents “past” molecular events. These gaps and a potential cause–effect relationship between interventions and outcomes can be overcome with a complete reconstruction of the entire program using continuous signals in real time such as the ones that are potentially captured with the probes such as those used in this study.
In conclusion, the availability of molecular probes that nondestructively and noninvasively report the progression of the differentiation program, while documenting transient phenotypic changes, offer significant advantages to attempt to improve the fabrication of site-specific engineered cartilage. Such advantages include the following: (i) avoiding end-point and destructive assessments to determine success or failures, thus establishing ways to predict failure early in the process of fabrication; (ii) making spatiotemporal correlations between specific interventions and resulting phenotypes; (iii) the ability to make adaptations and tune ups “on the go”; and (iv) the possibility of defining release criteria for implantation based on established features and events monitored in real time throughout the fabrication period. These advantages positively impact both the design strategies and the testing approaches of engineered cartilage by improving the quality of the implantable constructs.
Acknowledgments
We thank Amad Awadallah for the histological processing of the samples, Margie Harris for hMSC preparations, and NIH/NIBIB grants 1R01EB020367, 1P41EB021911 (Case Center for Multimodal Evaluation of Engineered Cartilage), and the Virginia and David Baldwin fund for support.
Disclosure Statement
No competing financial interests exist.
References
- 1.Caplan A.I. Mesenchymal stem cells. J Orthop Res 9, 641, 1991 [DOI] [PubMed] [Google Scholar]
- 2.Caplan A.I., and Goldberg V.M. Principles of tissue engineered regeneration of skeletal tissues. Clin Orthop Relat Res 367 Suppl, S12, 1999 [DOI] [PubMed] [Google Scholar]
- 3.Vinatier C., Bouffi C., Merceron C., Gordeladze J., Brondello J.-M., Jorgensen C., Weiss P., Guicheux J., and Noël D. Cartilage tissue engineering: towards a biomaterial-assisted mesenchymal stem cell therapy. Curr Stem Cell Res Ther 4, 318, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L., Hu J., and Athanasiou K. A. The role of tissue engineering in articular cartilage repair and regeneration. Crit Rev Biomed Eng 37, 1, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Correa D., and Lietman S.A. Articular cartilage repair: current needs, methods and research directions. Semin Cell Dev Biol 62, 67, 2016 [DOI] [PubMed] [Google Scholar]
- 6.Lefebvre V., Huang W., Harley V.R., Goodfellow P.N., and de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol 17, 2336, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Crombrugghe B., Lefebvre V., Behringer R.R., Bi W., Murakami S., and Huang W. Transcriptional mechanisms of chondrocyte differentiation. Matrix Biol 19, 389, 2000 [DOI] [PubMed] [Google Scholar]
- 8.Buckwalter J.A., and Mankin H.J. Articular cartilage: tissue design and chondrocyte-matrix interactions. Instr Course Lect 47, 477, 1998 [PubMed] [Google Scholar]
- 9.Han L., Grodzinsky A.J., and Ortiz C. Nanomechanics of the cartilage extracellular matrix. Annu Rev Mater Res 41, 133, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mueller M.B., and Tuan R.S. Functional characterization of hypertrophy in chondrogenesis of human mesenchymal stem cells. Arthritis Rheum 58, 1377, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mueller M.B., Fischer M., Zellner J., Berner A., Dienstknecht T., Prantl L., Kujat R., Nerlich M., Tuan R.S., and Angele P. Hypertrophy in mesenchymal stem cell chondrogenesis: effect of TGF-β isoforms and chondrogenic conditioning. Cells Tissues Organs 192, 158, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelttari K., Winter A., Steck E., Goetzke K., Hennig T., Ochs B.G., Aigner T., and Richter W. Premature induction of hypertrophy during in vitro chondrogenesis of human mesenchymal stem cells correlates with calcification and vascular invasion after ectopic transplantation in SCID mice. Arthritis Rheum 54, 3254, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Gerstenfeld L.C., Cullinane D.M., Barnes G.L., Graves D.T., and Einhorn T.A. Fracture healing as a post-natal developmental process: molecular, spatial, and temporal aspects of its regulation. J Cell Biochem 88, 873, 2003 [DOI] [PubMed] [Google Scholar]
- 14.van der Kraan P.M., and van den Berg W.B. Chondrocyte hypertrophy and osteoarthritis: role in initiation and progression of cartilage degeneration? Osteoarthritis Cartilage 20, 223, 2012 [DOI] [PubMed] [Google Scholar]
- 15.Huang B.J., Hu J.C., and Athanasiou K.A. Cell-based tissue engineering strategies used in the clinical repair of articular cartilage. Biomaterials 98, 1, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kock L., van Donkelaar C.C., and Ito K. Tissue engineering of functional articular cartilage: the current status. Cell Tissue Res 347, 613, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marin A.C., and Lacroix D. The inter-sample structural variability of regular tissue-engineered scaffolds significantly affects the micromechanical local cell environment. Interface Focus 5, 20140097, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lennon D.P., and Caplan A.I. Isolation of human marrow-derived mesenchymal stem cells. Exp Hematol 34, 1604, 2006 [DOI] [PubMed] [Google Scholar]
- 19.Lennon D.P., Haynesworth S.E., Bruder S.P., Jaiswal N., and Caplan A.I. Human and animal mesenchymal progenitor cells from bone marrow: identification of serum for optimal selection and proliferation. In Vitro Cell Dev Biol Anim 32, 602, 1996 [Google Scholar]
- 20.Solchaga L.A., Penick K., Goldberg V.M., Caplan A.I., and Welter J.F. Fibroblast growth factor-2 enhances proliferation and delays loss of chondrogenic potential in human adult bone-marrow-derived mesenchymal stem cells. Tissue Eng Part A 16, 1009, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sorrell J.M., Baber M.A., and Caplan A.I. Clonal characterization of fibroblasts in the superficial layer of the adult human dermis. Cell Tissue Res 327, 499, 2007 [DOI] [PubMed] [Google Scholar]
- 22.Yoo J.U., Barthel T.S., Nishimura K., Solchaga L., Caplan A.I., Goldberg V.M., and Johnstone B. The chondrogenic potential of human bone-marrow-derived mesenchymal progenitor cells. J Bone Joint Surg Am 80, 1745, 1998 [DOI] [PubMed] [Google Scholar]
- 23.Jaiswal N., Haynesworth S.E., Caplan A.I., and Bruder S.P. Osteogenic differentiation of purified, culture-expanded human mesenchymal stem cells in vitro. J Cell Biochem 64, 295, 1997 [PubMed] [Google Scholar]
- 24.Lin P., Lin Y., Lennon D.P., Correa D., Schluchter M., and Caplan A.I. Efficient lentiviral transduction of human mesenchymal stem cells that preserves proliferation and differentiation capabilities. Stem Cells Transl Med 1, 886, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dennis J.E., Konstantakos E.K., Arm D., and Caplan A.I. In vivo osteogenesis assay: a rapid method for quantitative analysis. Biomaterials 19, 1323, 1998 [DOI] [PubMed] [Google Scholar]
- 26.Ohgushi H., Goldberg V.M., and Caplan A.I. Heterotopic osteogenesis in porous ceramics induced by marrow cells. J Orthop Res 7, 568, 1989 [DOI] [PubMed] [Google Scholar]
- 27.Ohgushi H., Okumura M., Tamai S., Shors E.C., and Caplan A.I. Marrow cell induced osteogenesis in porous hydroxyapatite and tricalcium phosphate: a comparative histomorphometric study of ectopic bone formation. J Biomed Mater Res 24, 1563, 1990 [DOI] [PubMed] [Google Scholar]
- 28.Ohgushi H., and Caplan A.I. Stem cell technology and bioceramics: from cell to gene engineering. J Biomed Mater Res 48, 913, 1999 [DOI] [PubMed] [Google Scholar]
- 29.Solchaga L.A., Penick K., Porter J.D., Goldberg V.M., Caplan A.I., and Welter J.F. FGF-2 enhances the mitotic and chondrogenic potentials of human adult bone marrow-derived mesenchymal stem cells. J Cell Physiol 203, 398, 2005 [DOI] [PubMed] [Google Scholar]
- 30.Correa D., Somoza R.A., Lin P., Greenberg S., Rom E., Duesler L., Welter J.F., Yayon A., and Caplan A.I. Sequential exposure to fibroblast growth factors (FGF) 2, 9 and 18 enhances hMSC chondrogenic differentiation. Osteoarthritis Cartilage 23, 443, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin P., Correa D., Kean T.J., Awadallah A., Dennis J.E., and Caplan A.I. Serial transplantation and long-term engraftment of intra-arterially delivered clonally derived mesenchymal stem cells to injured bone marrow. Mol Ther 22, 160, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin P., Correa D., Lin Y., and Caplan A.I. Polybrene inhibits human mesenchymal stem cell proliferation during lentiviral transduction. PLoS One 6, e23891, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sayed N., Wong W.T., Ospino F., Meng S., Lee J., Jha A., Dexheimer P., Aronow B.J., and Cooke J.P. Transdifferentiation of human fibroblasts to endothelial cells: role of innate immunity. Circulation 131, 300, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Acharya C., Adesida A., Zajac P., Mumme M., Riesle J., Martin I., and Barbero A. Enhanced chondrocyte proliferation and mesenchymal stromal cells chondrogenesis in coculture pellets mediate improved cartilage formation. J Cell Physiol 227, 88, 2012 [DOI] [PubMed] [Google Scholar]
- 35.Bian L., Zhai D.Y., Mauck R.L., and Burdick J.A. Coculture of human mesenchymal stem cells and articular chondrocytes reduces hypertrophy and enhances functional properties of engineered cartilage. Tissue Eng Part A 17, 1137, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qing C., Wei-Ding C., and Wei-Min F. Co-culture of chondrocytes and bone marrow mesenchymal stem cells in vitro enhances the expression of cartilaginous extracellular matrix components. Braz J Med Biol Res 44, 303, 2011 [DOI] [PubMed] [Google Scholar]
- 37.Johnstone B., Hering T.M., Caplan A.I., Goldberg V.M., and Yoo J.U. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res 238, 265, 1998 [DOI] [PubMed] [Google Scholar]
- 38.Pittenger M.F., Mackay A.M., Beck S.C., Jaiswal R.K., Douglas R., Mosca J.D., Moorman M.A., Simonetti D.W., Craig S., and Marshak D.R. Multilineage potential of adult human mesenchymal stem cells. Science 284, 143, 1999 [DOI] [PubMed] [Google Scholar]
- 39.Mackay A.M., Beck S.C., Murphy J.M., Barry F.P., Chichester C.O., and Pittenger M.F. Chondrogenic differentiation of cultured human mesenchymal stem cells from marrow. Tissue Eng 4, 415, 1998 [DOI] [PubMed] [Google Scholar]
- 40.Barry F., Boynton R.E., Liu B., and Murphy J.M. Chondrogenic differentiation of mesenchymal stem cells from bone marrow: differentiation-dependent gene expression of matrix components. Exp Cell Res 268, 189, 2001 [DOI] [PubMed] [Google Scholar]
- 41.Kon E., Roffi A., Filardo G., Tesei G., and Marcacci M. Scaffold-based cartilage treatments: with or without cells? A systematic review of preclinical and clinical evidence. Arthroscopy 31, 767, 2015 [DOI] [PubMed] [Google Scholar]
- 42.Khozoee B., Mafi P., Mafi R., and Khan W. Mechanical stimulation protocols of human derived cells in articular cartilage tissue engineering—a systematic review. Curr Stem Cell Res Ther 12, 260, 2017 [DOI] [PubMed] [Google Scholar]
- 43.Ramaswamy S., Uluer M.C., Leen S., Bajaj P., Fishbein K.W., and Spencer R.G. Noninvasive assessment of glycosaminoglycan production in injectable tissue-engineered cartilage constructs using magnetic resonance imaging. Tissue Eng Part C Methods 14, 243, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fite B.Z., Decaris M., Sun Y., Sun Y., Lam A., Ho C.K.L., Leach J.K., and Marcu L. Noninvasive multimodal evaluation of bioengineered cartilage constructs combining time-resolved fluorescence and ultrasound imaging. Tissue Eng Part C Methods 17, 495, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Y., Responte D., Xie H., Liu J., Fatakdawala H., Hu J., Athanasiou K.A., and Marcu L. Nondestructive evaluation of tissue engineered articular cartilage using time-resolved fluorescence spectroscopy and ultrasound backscatter microscopy. Tissue Eng Part C Methods 18, 215, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura A., Dohi Y., Akahane M., Ohgushi H., Nakajima H., Funaoka H. and Takakura, Y. Osteocalcin secretion as an early marker of in vitro osteogenic differentiation of rat mesenchymal stem cells. Tissue Eng Part C Methods 15, 169, 2009 [DOI] [PubMed] [Google Scholar]
- 47.Hawking H. A Brief History of Time. New York, NY: Bantam Book, 1988 [Google Scholar]
- 48.Chen W.-H., Lai M.-T., Wu A.T.H., Wu C.-C., Gelovani J.G., Lin C.-T., Hung S.-C., Chiu W.-T., and Deng W.-P. In vitro stage-specific chondrogenesis of mesenchymal stem cells committed to chondrocytes. Arthritis Rheum 60, 450, 2009 [DOI] [PubMed] [Google Scholar]
- 49.Song I.-H., and Dennis J.E. Simple evaluation method for osteoinductive capacity of cells or scaffolds using ceramic cubes. Tissue Cell 46, 372, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]