Abstract
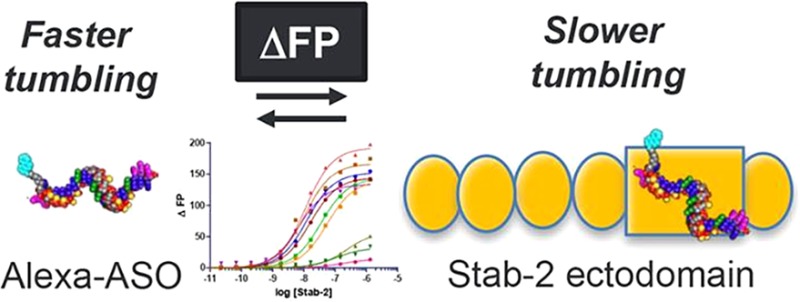
The Stabilin receptors are systemic clearance receptors for some classes of chemically modified nucleic acid therapeutics. In this study, the recombinant human secreted ecto-domain of the small isoform of Stabilin-2 (s190) was purified from cell culture and evaluated for direct binding with a multitude of antisense oligonucleotides (ASOs) using a fluorescence polarization-based assay. The tested ASOs varied in their backbone composition, modification of the ribose 2′ position, overall length of the oligo, and sequence of the nucleotide bases. A fully phosphorothioate (PS) ASO with a 5–10–5 pattern of flanking 2′-O-methoxyethyl modifications was then used to test the effects of pH and salt concentration on receptor binding. These tests concluded that the PS backbone was the primary determinant for ASO binding and that decreasing pH and increasing salt generally increased the rate of ligand dissociation and fit within the biological parameters expected of a constitutive recycling receptor. These results will be useful in the rational design of therapeutic oligonucleotides for enhancing their affinity or avoidance of the Stabilin receptors.
Antisense oligonucleotides (ASOs) are short (14–25) chemically modified nucleic acids that have made rapid progress for the treatment of congenital and acquired metabolic diseases.1 The effectiveness of an ASO relies on several parameters, including biological stability, adherence to cell-surface proteins, internalization within the cells, and escape from endosomes and specificity to the target RNA.2,3 To increase their stability in biological fluids, they are often designed with a phosphorothioate linkage in which the free nonbridging oxygen atom of the phosphodiester backbone is replaced with a sulfur atom, rendering the polymer resistant to nucleases.4 The PS backbone also enhances the avidity of ASO for plasma and cell-surface proteins that promote distribution to tissues and cellular accumulation.5 Gen 2 ASOs typically have the gapmer design in which a central region of DNA nucleotides is flanked by 2′-modified nucleotide analogues that further enhance nuclease stability and RNA binding affinity.6 Commonly used 2′-modified analogues used in gapmers include 2′-methoxyethyl RNA (MOE), constrained ethyl BNA (cEt), and locked nucleic acid (LNA)7 (Figure 1).
Figure 1.
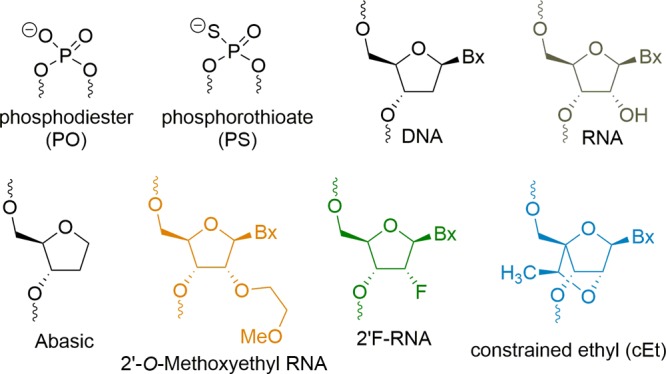
Structures of chemical modifications used in this study.
Our collaborative group discovered that the Stabilin class of receptors, of which there are two members, is responsible for the systemic clearance of phosphorothioate antisense oligonucleotides (PS-ASOs).8 Both human Stabilin-1 and Stabilin-2 are ∼315 kDa type 1 receptors with a single transmembrane domain and a short cytoplasmic tail.9 Stabilin-1 is more widely expressed within endothelial cells and alternatively activated macrophages.10 Stabilin-2 is expressed at a high level in the liver, spleen, bone marrow, and lymph node sinusoidal endothelium and at a lower level in specific tissues within the muscle, brain, and kidney.11−13 Both receptors share the same domain organization in which the extracellular portion consists of seven Fasciclin-1 domains separated by four clusters consisting of four to six EGF/EGF-like domains, and an X-Link domain that binds hyaluronan in Stabilin-2 but is dysfunctional in Stabilin-1.14 Both receptors bind with ligands such as heparin,15 PS-ASOs,8 phosphatidylserine,16,17 and oxidized low-density lipoprotein.18 Each protein can also internalize their own unique ligands such as SPARC19 and placental lactogen20 for Stabilin-1 and hyaluronan21 and chondroitin sulfates A, C, and D for Stabilin-2.22 Sodium dodecyl sulfate–polyacrylamide gel electrophoresis analysis of the receptor demonstrates that Stabilin-1 is expressed as two high-molecular weight proteins (1:1 ratio) that migrate as a tight doublet in contrast to Stabilin-2, which is expressed as 315 and 190 kDa isoforms in an approximately 1:1 ratio in native tissues.23 For the experiments outlined in this report, we utilized the ecto-domain of the recombinant 190 kDa isoform (s190) of Stabilin-2 as it has a high level of expression and/or secretion in cell lines and may be purified to near 100% purity using affinity chromatography. Both isoforms have the same activity against PS-ASOs.8
Previously, we used the recombinant 190 kDa isoform expressed in cell lines and the s190 purified protein to assess PS-ASO binding and internalization. From both enzyme-linked immunosorbent assay (ELISA)-like assays and internalization data with [125I]PS-ASO (5–10–5 oligo), we determined that the binding affinity was ∼140 nM.8 Competition assays were utilized to determine the effect of chemical modifications and oligonucleotide composition on Stab2 binding. The competition assays did not accurately inform the direct binding of the competitors or their lower affinity for the receptor. The objective in this report was to assess direct binding of a variety of ASOs to determine which chemistries (Figure 1) provide the weaker and stronger interaction between the nucleic acid and s190 using a sensitive fluorescence polarization (FP) assay.24,25
A series of experiments were performed using different variants of an ASO targeting phosphatase and tensin homologue (PTEN) mRNA to determine their affinity for s190. The interaction between the protein receptor and PTEN ASO was then challenged by pH and salt dependence.
Table 1 (Figure 2A) provides a summary of results for the initial binding experiments with the PTEN ASOs. It was found that the receptor has a significantly higher affinity for single-stranded ASO (ASO 1) than for double-stranded molecules in which the same ASO was bound with a RNA complement (ASO 8). The phosphorothioate linkage is highly preferred for binding (ASO 1) in contrast to the phosphodiester oligo (ASO 5), and the affinity rapidly drops as the oligo length is reduced to 15 bases (ASO 6) and 10 bases (ASO 7). The five flanking bases with 2′ modifications did not affect the overall affinity, indicating that the PS modification on the oligo backbone is the primary contributor for Stabilin-2 binding (compare ASO 5 with ASO 1, 9, and 10).
Table 1. Binding of PTEN ASOs to s190 (Stab2)a.
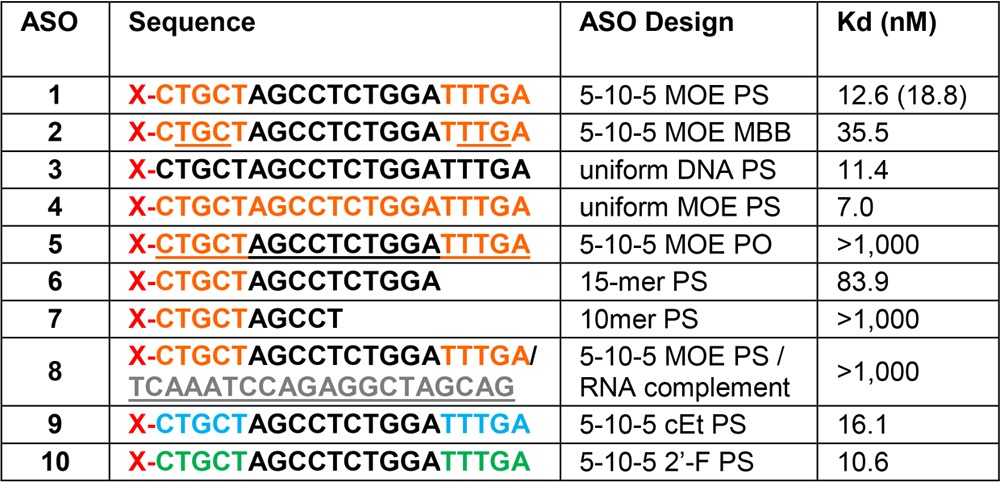
Orange letters indicate MOE, gray letters RNA, black letters DNA, green letters 2′F RNA, and blue letters cEt modifications. All oligonucleotides are PS-modified, except for underlined letters, which are natural phosphodiesters (PO). Oligos with a mixture of PS and PO linkages are mixed backbone (MBB); X = Alexa 647 Fluor. The number in parentheses is a value from duplicate measurements.
Figure 2.
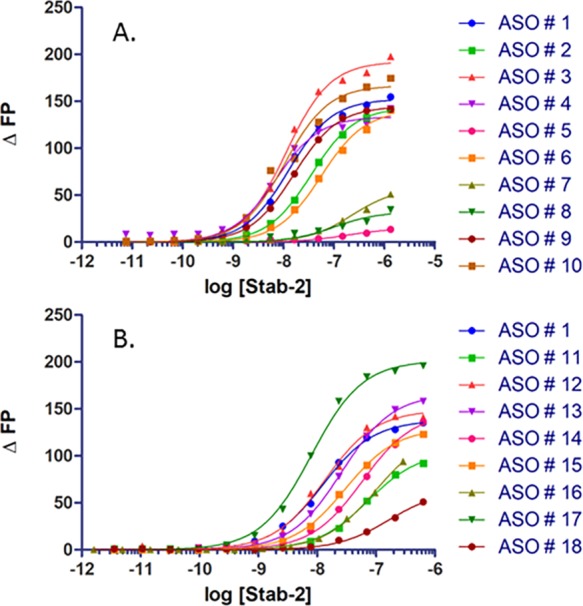
Direct ASO binding to s190 (Stab2). (A) PTEN ASOs with various chemistries. (B) ASOs with differing sequences and designs.
The same experiments were repeated using a “gapmer” designed set of oligos (Figure 2B and Table 2). The control for this group is PTEN ASO 1. All of these PS-based oligos (ASO 11–15) bound with affinities within 1 order of magnitude of each other (15–73 nM). The T20 (ASO 17) and A20 (ASO 18) oligos were also compared against each other, which resulted in A20 having a 20-fold weaker affinity for the receptor. The explanation for this observation is the assumption that because of the helical self-stacking of the purine bases, A20 is more rigid and, therefore, has a lower binding affinity. Rigidity may also be the reason for which the double-stranded PTEN ASO has an affinity lower than that of the single-stranded ASO in Table 1. The tight binding with the A-basic ASO (ASO 16) confirms that the PS modification is the binding motif for Stabilin-2, which is in agreement with the PTEN ASOs in which the PO version of the PTEN ASO does not bind with s190.
Table 2. Effect of Sequence and Design on Binding to s190 (Stab2)a.
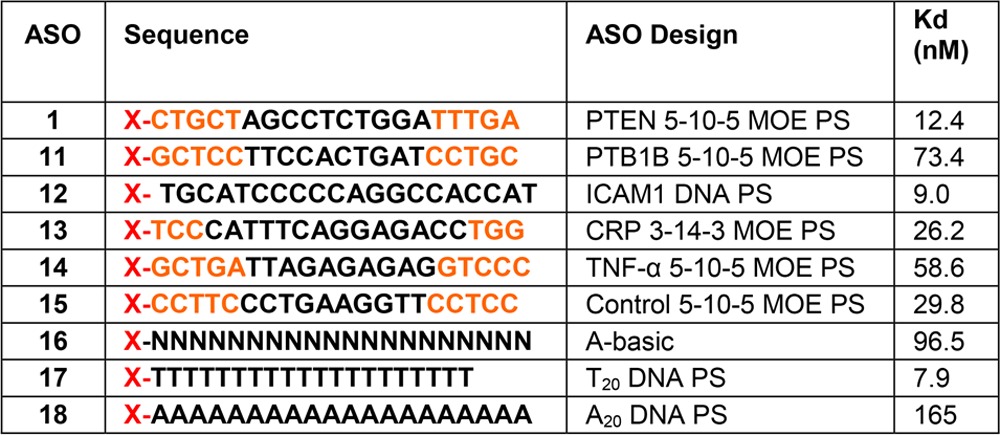
Orange letters indicate MOE and black letters DNA. N indicates a PS-modified abasic oligonucleotide, and X = Alexa 647 Fluor.
The effect of pH on ASO binding is important to examine as once the receptor is internalized in early endosomes, the pH decreases during endosomal maturation. Using a mixture of mono- and divalent 10 mM sodium phosphate buffers containing 150 mM NaCl, the fluorescence-based assay was repeated under four pH conditions (Figure 3A). As the pH decreased from 7.4 to 5.0, the affinity decreased. For most protein–ASO interactions that have been observed thus far, the opposite trend or no change in binding affinity is the typical result. In other proteins and receptors, the higher affinity at lower pH may be the result of a more positive charge that is attractive to the polyanionic PS ASO.26 However, this is clearly not the case with this receptor. Most, if not all, professional endocytic receptors release their cargo under low-pH (<5.5) conditions,27 and this may be the reason for the results observed for Stabilin-2.
Figure 3.
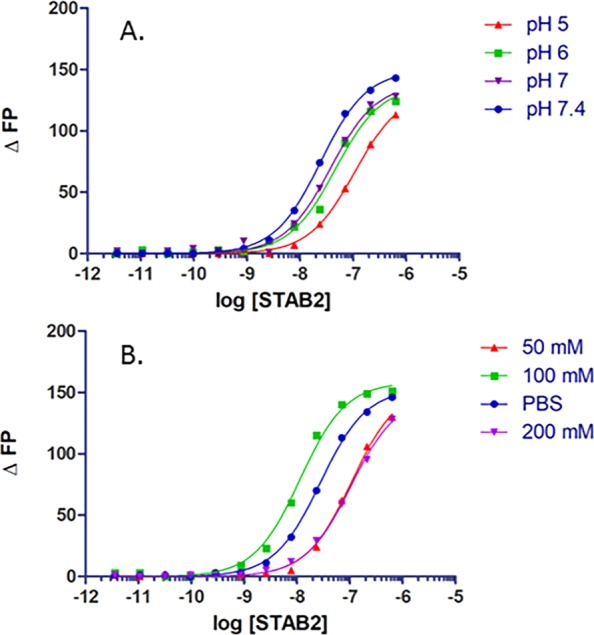
ASO 1 binding to s190 under various (A) pH and (B) salt conditions. The phosphate-buffered saline concentration is 150 mM.
Previous experiments with an ELISA type assay revealed that PS ASO–protein binding is dependent on ionic bonding.8 We repeated this assay with the FP method and found that, as before, the affinity of the ASO for the receptor decreases as the ionic strength increases (Figure 3B). It is somewhat surprising that binding affinity is weakest at the lowest salt concentration and may be a result of an artifact from the experimental method or that low concentrations of salt perturb protein structure enough to decrease the level of ASO binding. Any perturbation to salt concentration may alter the physical and chemical environment of the binding site(s). There are no structural data for this receptor, and the s190 used in these experiments contains 1359 amino acids, including 104 cysteine residues; thus, predicting overall and specific subdomain structures is not possible at this time (Figure S1).
This fluorescence-based assay confirmed the results from our previous report describing the high affinity of the PS-based ASO for the Stabilin receptors.8 This is the first report in which direct binding affinities have been observed with a multitude of different PS and non-PS ASOs that could not have been attained otherwise. With this information, it is clear that the length of the PS backbone and the single-stranded nature of the nucleic acid are the primary determinants for binding to the Stabilin receptors. In addition, the nucleotide sequence does not substantially affect the affinity for the receptor.
The sequence-independent tissue accumulation properties of PS ASOs in the liver have been used advantageously for the clinical development of ASO therapeutics. Our binding data show that PS ASOs can bind the Stabilin receptors, and presumably other cell-surface proteins, in a PS-dependent but sequence-independent manner and provide a rationale for the predictable liver accumulation properties of single-stranded PS ASOs in animals. Our data also emphasize the importance of interactions with cell-surface proteins for the promotion of cellular internalization of nucleic acid-based therapeutics.
Acknowledgments
The authors are grateful to Tracy Reigle (Ionis Pharm.) for her assistance with manuscript preparation.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.8b00126.
Methodology for the purification of s190 and fluorescence polarization assay and the amino acid sequence and domain organization of s190 (Figure S1) (PDF)
The authors are grateful for funding from National Institutes of Health Grant R01HL130864 to E.N.H.
The authors declare no competing financial interest.
Supplementary Material
References
- Shen X.; Corey D. R. (2018) Nucleic Acids Res. 46, 1584–1600. 10.1093/nar/gkx1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P. J.; Anastasopoulos F.; Pouton C. W.; Boyd B. J. (2009) Expert Rev. Mol. Med. 11, e10 10.1017/S1462399409001021. [DOI] [PubMed] [Google Scholar]
- Crooke S. T.; Wang S.; Vickers T. A.; Shen W.; Liang X. H. (2017) Nat. Biotechnol. 35, 230–237. 10.1038/nbt.3779. [DOI] [PubMed] [Google Scholar]
- Eckstein F. (2014) Nucleic Acid Ther. 24, 374–87. 10.1089/nat.2014.0506. [DOI] [PubMed] [Google Scholar]
- Geary R. S.; Norris D.; Yu R.; Bennett C. F. (2015) Adv. Drug Delivery Rev. 87, 46–51. 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- Monia B. P.; Lesnik E. A.; Gonzalez C.; Lima W. F.; McGee D.; Guinosso C. J.; Kawasaki A. M.; Cook P. D.; Freier S. M. (1993) J. Biol. Chem. 268, 14514–22. [PubMed] [Google Scholar]
- Wan W. B.; Seth P. P. (2016) J. Med. Chem. 59, 9645–9667. 10.1021/acs.jmedchem.6b00551. [DOI] [PubMed] [Google Scholar]
- Miller C. M.; Donner A. J.; Blank E. E.; Egger A. W.; Kellar B. M.; Ostergaard M. E.; Seth P. P.; Harris E. N. (2016) Nucleic Acids Res. 44, 2782–94. 10.1093/nar/gkw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz O.; Gratchev A.; McCourt P. A.; Schledzewski K.; Guillot P.; Johansson S.; Svineng G.; Franke P.; Kannicht C.; Kzhyshkowska J.; Longati P.; Velten F. W.; Johansson S.; Goerdt S. (2002) Biochem. J. 362, 155–64. 10.1042/0264-6021:3620155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kzhyshkowska J.; Gratchev A.; Goerdt S. (2006) J. Cell. Mol. Med. 10, 635–49. 10.1111/j.1582-4934.2006.tb00425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel J. A.; Weigel P. H. (2003) J. Biol. Chem. 278, 42802–11. 10.1074/jbc.M307201200. [DOI] [PubMed] [Google Scholar]
- Falkowski M.; Schledzewski K.; Hansen B.; Goerdt S. (2003) Histochem. Cell Biol. 120, 361–9. 10.1007/s00418-003-0585-5. [DOI] [PubMed] [Google Scholar]
- Park S. Y.; Yun Y.; Lim J. S.; Kim M. J.; Kim S. Y.; Kim J. E.; Kim I. S. (2016) Nat. Commun. 7, 10871. 10.1038/ncomms10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyosseva S. V.; Harris E. N.; Weigel P. H. (2008) J. Biol. Chem. 283, 15047–55. 10.1074/jbc.M709921200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pempe E. H.; Xu Y.; Gopalakrishnan S.; Liu J.; Harris E. N. (2012) J. Biol. Chem. 287, 20774–83. 10.1074/jbc.M111.320069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. Y.; Jung M. Y.; Kim H. J.; Lee S. J.; Kim S. Y.; Lee B. H.; Kwon T. H.; Park R. W.; Kim I. S. (2008) Cell Death Differ. 15, 192–201. 10.1038/sj.cdd.4402242. [DOI] [PubMed] [Google Scholar]
- Park S. Y.; Jung M. Y.; Lee S. J.; Kang K. B.; Gratchev A.; Riabov V.; Kzhyshkowska J.; Kim I. S. (2009) J. Cell Sci. 122, 3365–73. 10.1242/jcs.049569. [DOI] [PubMed] [Google Scholar]
- Li R.; Oteiza A.; Sorensen K. K.; McCourt P.; Olsen R.; Smedsrod B.; Svistounov D. (2011) Am. J. Physiol Gastrointest Liver Physiol 300, G71–81. 10.1152/ajpgi.00215.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kzhyshkowska J.; Workman G.; Cardo-Vila M.; Arap W.; Pasqualini R.; Gratchev A.; Krusell L.; Goerdt S.; Sage E. H. (2006) J. Immunol. 176, 5825–32. 10.4049/jimmunol.176.10.5825. [DOI] [PubMed] [Google Scholar]
- Kzhyshkowska J.; Gratchev A.; Schmuttermaier C.; Brundiers H.; Krusell L.; Mamidi S.; Zhang J.; Workman G.; Sage E. H.; Anderle C.; Sedlmayr P.; Goerdt S. (2008) J. Immunol. 180, 3028–37. 10.4049/jimmunol.180.5.3028. [DOI] [PubMed] [Google Scholar]
- Yannariello-Brown J.; Zhou B.; Weigel P. H. (1997) Glycobiology 7, 15–21. 10.1093/glycob/7.1.15. [DOI] [PubMed] [Google Scholar]
- Harris E. N.; Weigel P. H. (2008) Glycobiology 18, 638–48. 10.1093/glycob/cwn045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E. N.; Kyosseva S. V.; Weigel J. A.; Weigel P. H. (2007) J. Biol. Chem. 282, 2785–97. 10.1074/jbc.M607787200. [DOI] [PubMed] [Google Scholar]
- Kornilova A. Y.; Algayer B.; Breslin M.; Uebele V. (2012) Anal. Biochem. 425, 43–6. 10.1016/j.ab.2012.02.024. [DOI] [PubMed] [Google Scholar]
- Schmidt K.; Prakash T. P.; Donner A. J.; Kinberger G. A.; Gaus H. J.; Low A.; Ostergaard M. E.; Bell M.; Swayze E. E.; Seth P. P. (2017) Nucleic Acids Res. 45, 2294–2306. 10.1093/nar/gkx060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers T. A.; Crooke S. T. (2016) PLoS One 11, e0161930 10.1371/journal.pone.0161930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C.; Boutros M. (2010) Sci. Signaling 3, pe26. 10.1126/scisignal.3134pe26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


