Abstract
CRISPR-Cas9 is a genome editing technology with major impact in life sciences. In this system, the endonuclease Cas9 generates double strand breaks in DNA upon RNA-guided recognition of a complementary DNA sequence, which strictly requires the presence of a protospacer adjacent motif (PAM) next to the target site. Although PAM recognition is essential for cleavage, it is unknown whether and how PAM binding activates Cas9 for DNA cleavage at spatially distant sites. Here, we find evidence of a PAM-induced allosteric mechanism revealed by microsecond molecular dynamics simulations. PAM acts as an allosteric effector and triggers the interdependent conformational dynamics of the Cas9 catalytic domains (HNH and RuvC), responsible for concerted cleavage of the two DNA strands. Targeting such an allosteric mechanism should enable control of CRISPR-Cas9 functionality.
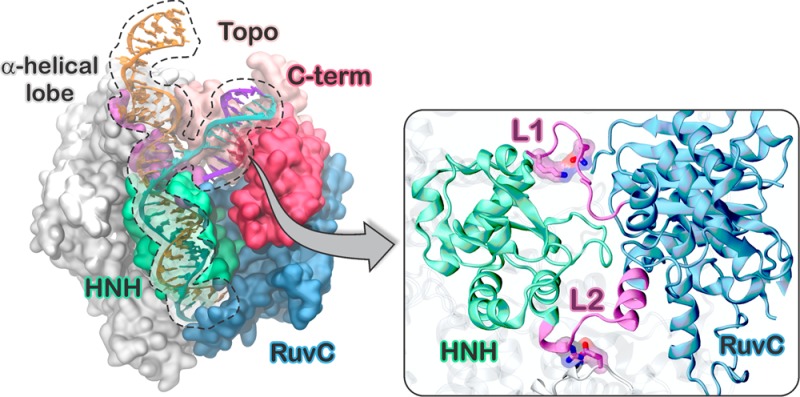
The CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9 system is a bacterial adaptive immune system whose adaptation as a genome editing technology has revolutionized life sciences.1 The major impact of CRISPR-Cas9 in biomedicine and pharmaceutics motivates fundamental studies of the underlying mechanism, where the molecular basis of DNA recognition and cleavage remain unclear. In CRISPR-Cas9, the endonuclease Cas9 associates with guide RNAs and cleaves complementary DNA sequences.2 Site-specific recognition and cleavage requires that a short sequence of 2–5 nucleotides, known as protospacer adjacent motif (PAM), is found immediately adjacent to the complementary DNA sequence, which is otherwise ignored by Cas9.2,3 The X-ray structure of the Streptococcus pyogenes CRISPR-Cas9 complex shows that the DNA binds to Cas9 matching the RNA with the complementary strand (cDNA), whereas the other strand (non complementary, ncDNA) is displaced (Figure 1).4 An α-helical lobe of Cas9 mediates RNA binding via an arginine rich (R-rich) helix, whereas PAM binds in a groove formed by the protein C-terminus (C-term) and a domain structurally similar to type II topoisomerase (Topo).
Figure 1.
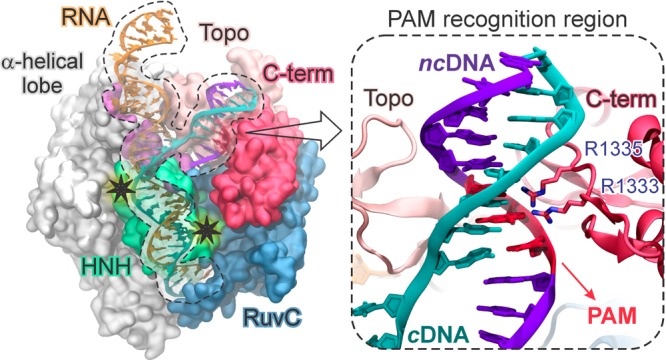
Architecture of the S. pyogenes CRISPR-Cas9 bound to a guide RNA and a target DNA (4UN3.pdb),5 with close-up view of the PAM recognition region. Two stars highlight the cleavage sites.
The 5′-TGG-3′ PAM sequence initiates DNA association and cleavage by binding to the C-term amino acid residues R1333 and R1335.3,5 Two spatially distant domains, HNH and RuvC, cleave the cDNA and ncDNA, respectively, via a concerted mechanism that relies on interdependent conformational dynamics.6 It is clear that PAM recognition is essential for triggering Cas9 nuclease activity at distant catalytic sites.3 However, the nature of the allosteric mechanism communicating PAM-Cas9 interactions with the HNH and RuvC domains remains unknown. Elucidating that mechanism is essential for understanding the system’s activation and for rational engineering of controlled functionality. Here, microsecond time scale molecular dynamics (MD) simulations reveal that PAM is an allosteric effector that triggers interdependent conformational dynamics of the catalytic domains, responsible for concerted cleavage of the two DNA strands.6
PAM is an allosteric effector. MD simulations, including configurational sampling over ∼13 μs, have been performed on the complex of Cas9 with guide RNA and its matching DNA containing a 5′-TGG-3′ PAM sequence (referred as Cas9 with PAM: Cas9-wPAM),5 and on its analogue, crystallized without the PAM segment (Cas9-w/oPAM),7 employing simulation conditions tailored for DNA/RNA endonucleases.8
MD simulations reveal that PAM induces a conformational transition of Cas9, as shown by the conformational space explored along its principal modes of motions (i.e., principal components PC1 and PC2) (Figure 2A and SI (paragraph S2.1 and Figures S1, S3)). The distinct conformations adopted by Cas9 when interacting with DNA containing PAM (PC1 < 0) or without PAM (PC1 > 0), reveal an “open-to-close” conformational transition (Figure 2B, Movie S1), in agreement with the conformational change required for nucleic acid association.9,10 The generalized correlation matrix (GCij), which captures both linear and nonlinear correlated motions of amino acids based on the Shannon entropy,11 reveals that PAM binding significantly strengthens the correlations between motions of Cas9, including the interdomain correlations of the RuvC and HNH catalytic domains (Figure 2C). The net effect is clearly shown by the distributions of GCij, centered at ∼0.4 (w/oPAM) and ∼0.5 (wPAM, Figure 2D). As known, the binding of an allosteric effector reshapes the protein conformational landscape, resulting in a shift of the conformational ensemble.12 This is associated with an overall increase of coupled motions, which mediate the communication between distant sites.12,13 Accordingly, PAM induces a shift in the conformational dynamics of CRISPR-Cas9, functioning as an allosteric effector that enhances correlations between distant domains. Notably, the PAM-induced correlated motions involve the RuvC and HNH catalytic domains and are associated with the observed “open-to-close” conformational transition.
Figure 2.
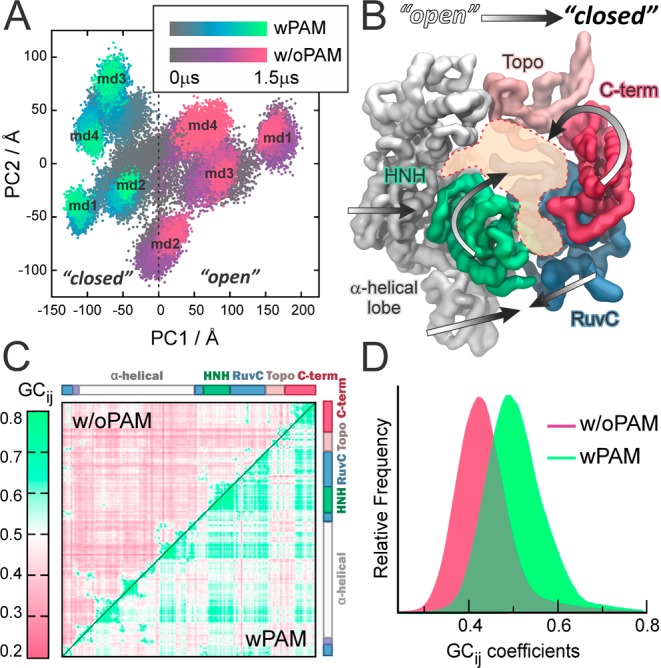
(A) Distribution of conformations in eight independent MD runs (each of ∼1.5 μs) for Cas9-wPAM (gray-to-green) and Cas9-w/oPAM (gray-to-violet), along the principal components of motion (PC1 vs PC2, see also Figure S3). (B) “Open-to-close” conformational transition of Cas9 induced by PAM. (C) GCij matrix for Cas9-wPAM (lower triangle) and w/oPAM (upper triangle). GCij are color-coded green (correlated) to violet (not correlated). (D) Probability distribution of the GCij coefficients wPAM (green) and w/oPAM (violet).
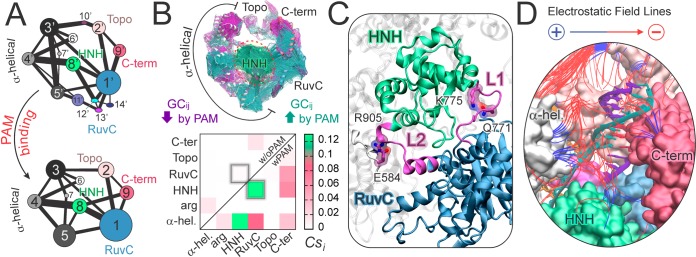
PAM induced reorganization and signaling. Community network analysis (CNA) provides valuable insights on the structure of correlations, including motions that are essential for conformational reorganization and allosteric signaling.12,14 CNA identifies groups (i.e., “communities”) of closely correlated residues and the strength of correlation between them, as described by a set of nodes and edges weighted according to GCij (details in SI). When comparing the structure of the communities in Cas9 with and without PAM, CNA shows that PAM reduces the number of communities (Figure 3A), enhancing the allosteric signaling.12,15 In the absence of PAM, the communities are much more fragmented, weakening correlations that are essential for allosteric signaling. In addition, PAM strengthens the correlation between communities #1 and #8, composed of the RuvC and HNH domains, respectively, as represented in Figure 3A by a thicker bond between those communities. The connection between communities #1 and #8 is weaker in Cas9-w/oPAM due to fragmentation of communities. Thus, it is clear that PAM induces a stronger communication channel between HNH and RuvC.6
Figure 3.
(A) CNA of Cas9–w/oPAM (top) and wPAM (bottom), shown in a 2D representation of the community network. Bonds connect communities and measure their intercommunication strength. (B) GCij increased (green) and decreased (magenta) by PAM are plotted on the 3D structure (top), indicating an increase of intercorrelations involving the HNH domain. A per-domain correlation score (Csi) matrix (bottom) measures the interdomain correlations wPAM (lower triangle) and w/oPAM (upper triangle). Two boxes highlight the HNH–RuvC intercorrelation. (C) Interactions between essential edges of the allosteric pathway (Q771–K775 and E584–R905), involving the L1 and L2 loops (magenta), which interconnect HNH (green) and RuvC (blue). (D) Electrostatic properties of Cas9–wPAM. Field lines indicate the direction of the electrostatic field from positive (blue) to negative (red) regions. Field lines are filtered according to a gradient magnitude >4.9 kT/e/Å and length >13.5 Å.
Mechanism of allosteric signalling. Here, we analyze the effector-induced (i.e., PAM-induced) changes in the dynamic correlations to elucidate the underlying allosteric mechanism.
By plotting the difference in correlations (ΔGCij) between the systems wPAM and w/oPAM on the 3D structure (Figure 3B), we identify correlated motions that are decreased (ΔGCij < −0.3) or increased (ΔGCij > 0.3) by the presence of PAM. We find that PAM increases the interdomain connectivity, with the majority of the correlations involving the central HNH domain. A measure of the interdependent domain motions is given by a per-domain correlation score (Csi) matrix, which accumulates interdomain correlations (Figure 3B, details in SI).16 In Cas9-wPAM, the HNH and RuvC domains are highly intercorrelated, as opposite to Cas9-w/oPAM. This indicates that PAM is key in triggering the interdependent conformational dynamics of HNH and RuvC, which is required for concerted cleavage of the DNA strands.6 In the presence of PAM, HNH also correlates strongly with the α-helical domain and results the most correlated domain (see SI). These findings are consistent with the experimental evidence that the HNH dynamics exert a conformational control over Cas9 nuclease activity,6 and shows that this depends on the presence of PAM.
Electrostatic interactions play important roles in allosteric signaling,16 because charged amino acid residues are key nodes of the dynamic network in Cas9-wPAM (Figure S8). Interestingly, Q771 and E584 engage in interactions with K775 and R905, respectively, forming essential edges of the allosteric pathway (Figure 3C, details in SI). These interactions connect HNH to RuvC and the α-helical lobe via the L1 (residues 765–780) and L2 (residues 906–918) loops, which effectively function as “allosteric transducers”.6,17 By computing the optimal pathways (i.e., “shortest pathways”) for information transfer between pairs of catalytic residues of the RuvC and HNH domains, we find that the information flows through the L1 loop, via key residues in the path (K772, T770, Figure S9). As such, PAM triggers signal transduction through L1 and L2, suggesting that modulatory strategies might arise from mutagenesis of the residues in the path (e.g., K772, T770) and of the nodes in the dynamic network (e.g., E584, R905, Q771 K775).15,16 Consistently, Cas9 engineering studies have shown that mutation of charged residues leads to sensible changes in the specificity of the enzyme.18 In particular, the K775A mutation (identified as a key node residue) has shown a decrease in the off-target cleavage of non-fully complementary DNAs. Therefore, the mutation of the above-mentioned residues that have not been tested yet, and the modulation of the HNH-L1/L2-RuvC allosteric pathways elucidated here, could lead to the design of CRISPR-Cas9 systems with controllable activity, opening new routes for CRISPR-Cas9 rational design. This allosteric mechanism also establishes the basis for investigating the selectivity of Cas9 toward alternative PAM sequences,19 currently ongoing in our laboratories with the aim of expanding the DNA targeting capability of Cas9.
Electrostatic analysis also provides fundamental insights on the origin of interactions responsible for the “open-to-close” transition, observed in the presence of PAM (Figure 3D).20 Upon site-specific binding to PAM, positively charged groups of the α-helical, C-term and HNH domains are attracted by the negatively charged DNA backbone. The resulting rearrangements lead to the “open-to-close” conformational transition, further stabilized by H-bonds and ionic interactions (detailed in SI).
In summary, multi-microsecond time scale molecular simulations (∼13 μs) disclose here an allosteric mechanism in CRISPR-Cas9, which is induced by the presence of PAM. We reveal that PAM functions as an allosteric effector that triggers the interdependent conformational dynamics of the HNH and RuvC catalytic domains, required for concerted cleavage of the DNA strands.6 These findings provide fundamental understanding of the CRISPR-Cas9 mechanistic action that should be valuable for rational engineering and modulation of the CRISPR-Cas9 allosteric communication.
Acknowledgments
G.P. thanks the Swiss National Science Foundation for her postdoctoral grant P300PA_164698. J.A.M. thanks NIH, NSF, HHMI, NBCR and SDSC grants. V.S.B. thanks NIH grant 1R01GM106121-01A1 and computational time from NERSC. I.R. thanks ENS-Lyon (grants: 900/S81/BS81-FR14, MI-LOURD-FR15) and PSMN. Computing time was provided by the XSEDE grant MCA93S013 to J.A.M.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b05313.
The authors declare no competing financial interest.
Supplementary Material
References
- Doudna J. A.; Charpentier E. Science 2014, 346, 1258096. 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Jinek M.; Chylinski K.; Fonfara I.; Hauer M.; Doudna J. A.; Charpentier E. Science 2012, 337, 816. 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg S. H.; Redding S.; Jinek M.; Greene E. C.; Doudna J. A. Nature 2014, 507, 62. 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F.; Doudna J. A. Annu. Rev. Biophys. 2017, 46, 505. 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- Anders C.; Niewoehner O.; Duerst A.; Jinek M. Nature 2014, 513, 569. 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg S. H.; LaFrance B.; Kaplan M.; Doudna J. A. Nature 2015, 527, 110. 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H.; Ran F. A.; Hsu P. D.; Konermann S.; Shehata S. I.; Dohmae N.; Ishitani R.; Zhang F.; Nureki O. Cell 2014, 156, 935. 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo G.; Cavalli A.; Klein M. L.; Alfonso-Prieto M.; Dal Peraro M.; De Vivo M. Acc. Chem. Res. 2015, 48, 220. 10.1021/ar500314j. [DOI] [PubMed] [Google Scholar]
- Palermo G.; Miao Y.; Walker R. C.; Jinek M.; McCammon J. A. Proc. Natl. Acad. Sci. USA 2017, 114, 7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo G.; Miao Y.; Walker R. C.; Jinek M.; McCammon J. A. ACS Cent. Sci. 2016, 2, 756–763. 10.1021/acscentsci.6b00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange O. F.; Grubmuller H. Proteins: Struct., Funct., Genet. 2006, 62, 1053. 10.1002/prot.20784. [DOI] [PubMed] [Google Scholar]
- Guo J.; Zhou H. X. Chem. Rev. 2016, 116, 6503. 10.1021/acs.chemrev.5b00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhireksan Z.; Palermo G.; Riedel T.; Ma Z.; Muhammad R.; Rothlisberger U.; Dyson P. J.; Davey C. A. Nat. Commun. 2017, 8, 14860. 10.1038/ncomms14860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi A.; Eargle J.; Black A. A.; Luthey-Schulten Z. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 6620. 10.1073/pnas.0810961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivalta I.; Sultan M. M.; Lee N. S.; Manley G. A.; Loria J. P.; Batista V. S. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, E1428. 10.1073/pnas.1120536109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci C. G.; Silveira R. L.; Rivalta I.; Batista V. S.; Skaf M. S.. Sci. Rep. 2016, 6, doi: 10.1038/srep19940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F. G.; Taylor D. W.; Chen J. S.; Kornfeld J. E.; Zhou K. H.; Thompson A. J.; Nogales E.; Doudna J. A. Science 2016, 351, 867. 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker I. M.; Gao L.; Zetsche B.; Scott D. A.; Yan W. X.; Zhang F. Science 2016, 351, 84. 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B. P.; Prew M. S.; Tsai S. Q.; Topkar V. V.; Nguyen N. T.; Zheng Z. L.; Gonzales A. P. W.; Li Z. Y.; Peterson R. T.; Yeh J. R. J.; Aryee M. J.; Joung J. K. Nature 2015, 523, 481. 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N. A.; Sept D.; Joseph S.; Holst M. J.; McCammon J. A. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 10037. 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.