Abstract
Background
Leptospirosis is an important zoonotic disease that causes considerable morbidity and mortality globally, primarily in residents of urban slums. While contact with contaminated water plays a critical role in the transmission of leptospirosis, little is known about the distribution and abundance of pathogenic Leptospira spp. in soil and the potential contribution of this source to human infection.
Methods/Principal findings
We collected soil samples (n = 70) from three sites within an urban slum community endemic for leptospirosis in Salvador, Brazil. Using qPCR of Leptospira genes lipl32 and 16S rRNA, we quantified the pathogenic Leptospira load in each soil sample. lipl32 qPCR detected pathogenic Leptospira in 22 (31%) of 70 samples, though the median concentration among positive samples was low (median = 6 GEq/g; range: 4–4.31×102 GEq/g). We also observed heterogeneity in the distribution of pathogenic Leptospira at the fine spatial scale. However, when using 16S rRNA qPCR, we detected a higher proportion of Leptospira-positive samples (86%) and higher bacterial concentrations (median: 4.16×102 GEq/g; range: 4–2.58×104 GEq/g). Sequencing of the qPCR amplicons and qPCR analysis with all type Leptospira species revealed that the 16S rRNA qPCR detected not only pathogenic Leptospira but also intermediate species, although both methods excluded saprophytic Leptospira. No significant associations were identified between the presence of pathogenic Leptospira DNA and environmental characteristics (vegetation, rat activity, distance to an open sewer or a house, or soil clay content), though samples with higher soil moisture content showed higher prevalences.
Conclusion/Significance
This is the first study to successfully quantify the burden of pathogenic Leptospira in soil from an endemic region. Our results support the hypothesis that soil may be an under-recognized environmental reservoir contributing to transmission of pathogenic Leptospira in urban slums. Consequently, the role of soil should be considered when planning interventions aimed to reduce the burden of leptospirosis in these communities.
Author summary
Leptospirosis is a globally distributed zoonotic disease that disproportionately affects vulnerable populations in urban slums. The disease is transmitted by direct contact with water, soil, or mud that has been contaminated with infected urine shed from chronically infected animals. Despite the critical role the environment plays in the epidemiology of the disease, the contribution of soil to the transmission cycle remains largely undescribed. Herein, we investigated the distribution of pathogenic Leptospira in soil samples from an endemic urban slum in Brazil. We found pathogenic Leptospira in nearly one-third of the soil samples, predominantly in low concentrations (<5×102 GEq/g). However, we observed considerable variation in the distribution and concentration of the pathogen at the fine spatial scale within the slum. Our results indicate that soil is likely an important additional environmental reservoir of pathogenic Leptospira in urban slums and prevention strategies should consider soil to help prevent the transmission of the disease in similar settings.
Introduction
Leptospirosis is a life-threatening, zoonotic disease of global importance, with more than 1 million cases and approximately 60,000 deaths estimated annually, predominately in developing tropical countries [1]. The disease is caused by spirochetes of the genus Leptospira, which contains 35 species, 13 of which in the pathogenic group [2,3]. Pathogenic Leptospira chronically colonize the renal tubules of animal reservoirs and are excreted with the urine. Humans are incidental hosts, and often acquire the infection after seasonal or intense precipitation events [4–6], when pathogenic Leptospira in contaminated soil, mud, or water penetrate abraded skin or wounds [7,8]. Clinical manifestations of leptospirosis range from mild or asymptomatic to severe illness, such as Weil’s disease [7,9], and pulmonary hemorrhage syndrome [9,10], which cause high case fatality rates.
Urban leptospirosis has emerged as a pandemic which disproportionately affects residents of slum communities around the world [11]. Poor sanitation and an abundance of food sources provide ideal conditions for the maintenance of large rodent populations, specifically the Norway rat (Rattus norvegicus), which are the primary animal reservoirs of pathogenic Leptospira in urban environments [12–16]. Infrastructure deficiencies facilitate the transmission to humans [17–19]: Open sewers and inadequate drainage systems allow contaminated water to pervade surrounding soil and water, and unpaved walkways expose residents to contaminated dirt and mud. Thus, the human-environment interface plays a critical role in the epidemiology and transmission of leptospirosis in urban slums. While previous studies have found Leptospira spp. in puddles, sewers, streams, and soil in endemic regions [20–25], the distribution and dynamics of leptospires in the environment, particularly in soil, are largely unknown.
To date, few publications have reported pathogenic Leptospira in soil, presumably because leptospirosis is generally considered to be a water-borne disease, and thus environmental studies have focused preferentially on aquatic matrices [20,21]. Previous studies that successfully analyzed soil reported very low prevalence among samples (4.9% in China [26]), or the isolation of only a few pathogenic strains in Philippines, Malaysia and New Caledonia [2,23,24,27,28]. These studies, however, were based on culture isolation of leptospires, followed by molecular identification. Culture techniques lack sensitivity because of pathogenic Leptospira spp. being overgrown by the autochthonous soil microbiota or saprophytic and intermediate Leptospira [23,24,29]. Notably, a recent study by Thibeaux et al. [25] reported a 57.7% prevalence in soil samples from a suspected environmental infection site in New Caledonia by using a qPCR targeting lipL32.
In this study, we aimed to quantify the pathogenic Leptospira load in soil samples from an urban slum community in Salvador, Brazil. Previous studies have shown this community has a high burden of disease [6,18,30] and a widespread presence of pathogenic Leptospira in its surface waters [31], making it an excellent location for an environmental survey of the pathogen. Since Norway rats are the predominant pathogenic Leptospira reservoir in this community [14], we hypothesized that the presence of rats would be associated with the occurrence and abundance of the pathogen in soil. As urban slum communities in tropical regions share many socioeconomic, structural, and environmental characteristics [32,33], our study may help inform potential public health interventions in similar epidemiological settings around the world.
Methods
Study site
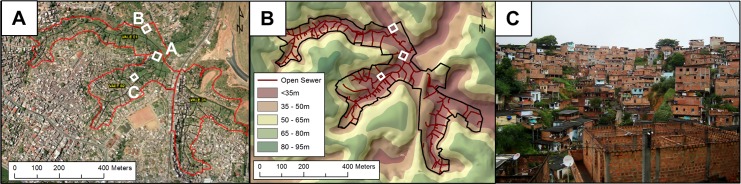
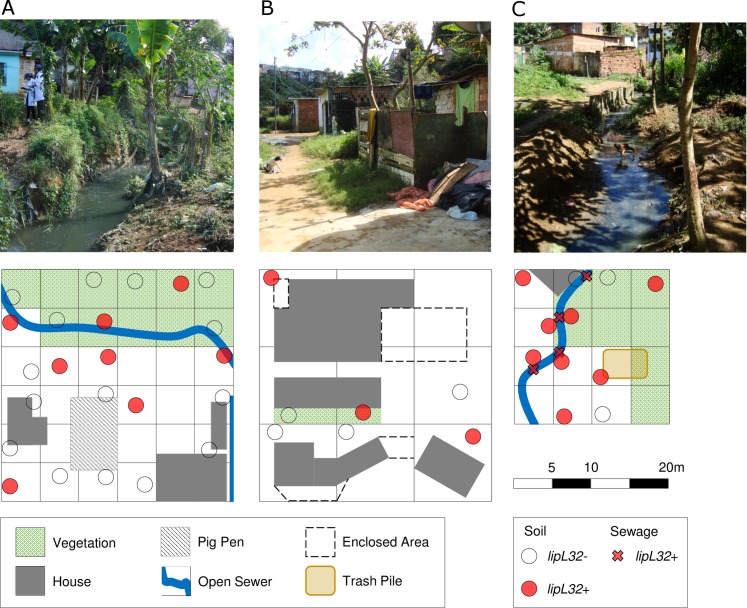
We conducted this study in the community of Pau da Lima, an urban slum located in Salvador, Brazil. This site has been extensively described in previous studies [18,19]. Briefly, the community encompasses four interconnected valleys within 0.46 km2 (Fig 1A), with a population of 14,122 inhabitants residing in 3,689 households, according to a 2003 census [18]. The community lacks adequate sanitary infrastructure, resulting in untreated wastewater and rain runoff flowing through open sewers in the lower parts of the valleys. Leptospirosis is endemic in Pau da Lima, with a mean annual infection incidence of 37.8 per 1,000 inhabitants, and 19.8 severe cases per 100,000 inhabitants [30]. We selected three collection sites to represent a range of microenvironments present within the community (Fig 1B). Site A (900 m2) (12°55'22.2"S, 38°26'04.2"W) was located along an open sewer at the bottom of the valley, and included households, areas of domestic animal raising, and thick vegetation (Fig 2A). The steep, high banks of the sewer served as a barrier to separate households from the sewage, which limited potential flooding. Site B (900 m2) (12°55'17.9"S, 38°26'07.2"W) was situated at a higher elevation next to the valley slope and had closed sewage drains, paved stairs, and patios (Fig 2B). There was limited vegetation, although fences and barriers, coupled with the sheer valley slope, restricted access. Site C (400 m2) (12°55'24.8"S, 38°26'06.3"W) was situated at the bottom of the valley and in proximity to an open sewer with low embankments, allowing frequent flooding of surrounding areas during heavy rainfall periods. The thick vegetation and water-logged soil made part of the site inaccessible. We partitioned collection sites into 5 m x 5 m squares (A and C) or 10 m x 10 m (B) (Fig 2A and 2B). A larger grid was used at Site B, as many areas were impassible due to the decreased number of sampling locations and the challenging terrain (Fig 2B).
Fig 1. The Pau da Lima community site in Salvador, Brazil.
(A) Aerial photograph of the community site (red border), which encompasses three valleys. The locations of the three sites of environmental sampling are indicated by white rectangles. (B) Topographic map that demonstrates differences in elevation at the site. Open sewage draining systems are depicted in dark red. (C) Photograph of the community, depicting a representative valley and squatter households. Panels A and B were created with ArcGIS using copyright-free aerial photographs provided by the Salvador municipality.
Fig 2. Environmental sampling and qPCR detection of Leptospira DNA in the community site.
Sampling of soil and sewage (circles) at three sites (A, B, and C) within the Pau da Lima field site. Positive and negative samples by lipL32 qPCR are depicted as red and open circles, respectively.
Sample collection and rat activity monitoring
We collected soil and sewage samples in the rainy season between July and August 2014 (S1 Fig). A tracking board to monitor rat activity in the collection sites was placed as described previously [34] within all grid squares that were accessible and contained exposed soil. Tracking boards were evaluated daily over the course of three days for evidence of rat activity as ascertained by the identification of footprints, scrapes, and tail slides [34]. After the three days, one or two soil samples of 100–200 g were collected at a depth of 5–10 cm from non-adjacent areas within 30 cm of the edge of each tracking board between 9am and 12.30pm. Grid squares that were inaccessible because of private property, dense vegetation or water-logged soil, or contained no exposed soil were not included in the sampling. In total, we collected soil samples from 23, 7 and 11 grid squares in sites A, B and C, respectively for a total of 35, 14 and 22 soil samples. If a portion of an open sewer was included in the grid square and was accessible, two 50 mL samples for each sampling point were collected. The presence of vegetation and distance to open sewers and households was recorded for each grid square. The soil moisture and clay content were measured for one sample from each grid square. Samples were placed in aseptic containers, transported to the laboratory, and processed within 6 h.
DNA extraction
To maximize the recovery of Leptospira DNA from soil samples, we developed an extraction protocol and determined its efficiency by performing spiking experiments with known concentrations of L. interrogans (S1 Supplementary methods). The final procedure consisted in the following steps: Subsamples of 5 g of soil were mixed with 40 mL of sterile double-distilled water and vortexed at maximum speed for 2 min. Samples were centrifuged at 100 rcf for 5 min. The supernatant was recovered and centrifuged at 12,000 rcf for 20 min at room temperature. The pellets were recovered, resuspended in 1.5 mL of sterile double-distilled water and centrifuged at 12,000 rcf for 20 min. Finally, the samples were decanted and the pellets were frozen at -80°C. Sewage samples (40 mL) were processed as described previously [35]. DNA was extracted from pellets using PowerSoil DNA Isolation Kit (Mo Bio Laboratories). An extraction blank (sterile double-distilled water) was added to each extraction batch to monitor for cross-contamination.
qPCR
We quantified Leptospira spp. loads using two TaqMan assays targeting the lipL32 gene or the 16S rRNA gene as described previously [36,37] with minor modifications. Briefly, all reaction mixtures (25 μL) contained 12.5 μL Platinum qPCR SuperMix (Life Technologies), 0.2 μg/μL of bovine serum albumin (Ambion), and 5 μL of DNA template. The lipL32 reactions included 500 nM each of primers LipL32-45F and LipL32-286R, and 100 nM of LipL32-189P probe (S1 Table). 16S rRNA reactions included 300 nM each of primers Lepto F and Lepto R, and 200 nM of 16S probe. Amplifications were performed using a 7500 Fast Real-Time PCR System (Life Technologies) with the following conditions: an initial step of 2 min at 50°C, followed by 2 min at 95°C, and 40 cycles of amplification (15 s at 95°C and 1 min at 60°C). Genomic DNA obtained from L. interrogans serovar Fiocruz L1-130 [38] was used to construct calibration curves with concentrations ranging from 2 × 102 to 2 × 109 GEq/mL, which we included in each qPCR run. Efficiencies were always higher than 92.5%. All samples were run in duplicate, and included non-template controls in each plate row to detect any contaminating DNA. All negative controls (extraction controls and non-template controls) were negative. All DNA extractions and quantitative PCR (qPCR) analyses were performed according to the minimum information for publication of quantitative real-time PCR experiments (MIQE) guidelines [39].
To determine the specificity of the lipL32 and 16S rRNA qPCRs for pathogenic, intermediate and saprophytic Leptospira, we tested DNA extracts from 21 Leptospira species (S2 Table) as explained above, adjusting the concentration in each well to 107 GEq based on each genome’s size [40].
Sequencing of Leptospira qPCR amplicons
To confirm the specificity of the environmental qPCR reactions, we randomly selected and sequenced (Sanger method) 11/22 (50%) of the soil samples with positive results for lipl32. We also sequenced 27/78 (35%) of samples with a positive result for 16S rRNA qPCR (25 soil and 2 sewage samples). In brief, qPCR products were purified from 2% agarose gels using the QIAquick Gel Extraction Kit (QIAgen) following the manufacturer’s instructions. The products were sequenced using primers LipL32-45F or Lepto-F for lipL32 and 16S rRNA, respectively, corrected using BioEdit 7.2.0 (Ibis Biosciences), and the sequences compared using BLAST to those available in the NCBI. In addition, we performed a phylogenetic analysis for the lipl32 and 16S rRNA amplicons using the Phylogeny.fr platform [41]. Maximum Likelihood trees were inferred by PhyML 3.0 [42] using the Hasegawa-Kishono-Yano (HKY85) substitution model and 1000 bootstrap replicates. We used FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree) to visualize and edit the final trees. The 16S rRNA tree included Leptonema illini DSM 21528 as the outgroup.
Data treatment and statistical analyses
Leptospira concentrations were log10 transformed and analyzed as follows. The theoretical lower limit of quantification (LOQ) of the qPCR assays was calculated using the assumption that 1 copy of the targeted gene was amplified in a reaction (4 GEq/g). We assigned positive samples with concentrations below this threshold a value equal to the LOQ. Fisher’s exact test and the χ2 test were used to compare prevalence among sites and the associations of dichotomized environmental characteristics with the proportion of qPCR-positive samples. t-tests with a Welch’s correction were used to compare soil moisture content and clay component values between positive and negative qPCR samples. The median concentrations between sampling sites were compared using Kruskal-Wallis test with Dunn’s correction for multiple comparisons. Mann-Whitney test was used to compare the concentrations between soil and sewage at site C and the overall concentrations obtained by lipl32 and 16S rRNA qPCRs. Non-parametric tests were chosen due to the non-normal distribution of the data. Although equivalent parametric tests (one-way ANOVA and t-test) were inappropriate, technically, they supported the results of the non-parametric tests in all cases. To analyze the degree of agreement between lipL32 and 16S PCR detection methods, we used Cohen’s kappa coefficient. All statistical analyses were performed using GraphPad Prism 6.05 (GraphPad Software Inc.), and multivariate models were fit using the GENMOD procedure with a GEE model in SAS v9.3 (SAS Institute Inc., Cary, NC) to evaluate the relationship between the measured environmental characteristics and outcome of sample testing.
Results and discussion
Pathogenic Leptospira are present in soil using lipL32 qPCR
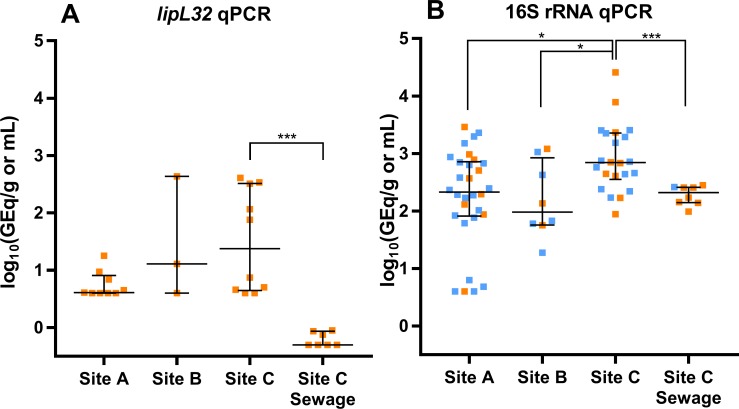
We collected 70 soil samples within the study area (34, 14, and 22 from sites A, B, and C, respectively) (Fig 2). Of those 70 samples, 22 (31%) were positive for pathogenic Leptospira as measured by the lipL32 qPCR (Fig 3A). There were no significant differences in the proportion of positive samples between the three collection sites (26%, 21% and 45%, respectively; p = 0.78) (Table 1), indicating that pathogenic Leptospira were widespread in the study site. These prevalences are lower than those recently reported using the same qPCR method in river bank soils from active leptospirosis transmission sites in New Caledonia (57.7%) [25]. Nevertheless, our results contrast with the very low number of pathogenic Leptospira isolates that are usually obtained from soil in endemic areas using culture-based approaches [23,24,26–28], suggesting that molecular approaches may capture better the presence of the pathogen in soil.
Fig 3. Pathogenic Leptospira concentrations in environmental samples.
The median concentration and interquartile range is shown for each site. (A) Concentrations measured with lipL32 qPCR. (B) Concentrations measured with 16S rRNA qPCR. Samples in orange were positive for both assays, while those in blue were only positive for the 16S rRNA qPCR. The differences between the soil median concentrations in the three sites were analyzed by Kruskal-Wallis test with Dunn’s correction for multiple comparisons. The differences between the median concentrations from soil and sewage at site C were analyzed with the Mann-Whitney test. (*) p≤0.05; (***) p≤0.0001.
Table 1. Detection of pathogenic Leptospira by lipL32 qPCR in soil samples according to environmental characteristics of the sampling sites within the urban slum community.
None of the measured variables showed significant differences between lipL32-positive and lipL32-negative samples in bivariate or multivariate analysis.
| Soil Samples (n = 70) No. (%) or Mean (SD) |
|||
|---|---|---|---|
| Characteristic | lipL32 Positive | lipL32 Negative | |
| Sitea | All | 22 (31%) | 48 |
| A | 9 (26%) | 25 | |
| B | 3 (21%) | 11 | |
| C | 10 (45%) | 12 | |
| Adjacent Vegetation | 14 (64%) | 27 (56%) | |
| Evidence of Rat Activity | 16 (73%) | 28 (58%) | |
| Distance to Open Sewerb | 3.3 (4.6) | 5.3 (5.6) | |
| Distance to Housec | 6.8 (4.5) | 5.9 (5.7) | |
| Soil Moisture Contentd | 28.4 (15.7) | 26.8 (13.7) | |
| Soil Clay Contentd,e | 10.7 (4.2) | 10.8 (5.5) | |
a Percent for the site variables are row percent. The others are column percent.
b n = 56 at sites A and C combined. Site B did not have an open sewer. Measurements are in meters.
c Measured in meters.
d Percent moisture.
e n = 56
The concentration of pathogenic Leptospira among positive samples was predominantly low (median: 6 GEq/g), but varied by two orders of magnitude (range: 4–4.31×102 GEq/g) even within the same collection grid. This indicates a considerable heterogeneity of environmental loads within the slum microenvironment. Furthermore, there were no significant differences in the concentration of the qPCR-positive soil samples among the three collection sites (p = 0.16) (Fig 3A). Among the eight sewage-water samples collected from site C, seven (88%) were positive by the lipL32 qPCR with a median concentration of 0.5 GEq/mL.
Together our results showed that pathogenic Leptospira were present in low concentrations in soils sampled from diverse microenvironments within the urban slum. Contact with mud in the peridomiciliary environment was previously identified as a risk factor for leptospirosis infection in this setting [18], which suggests that soil may serve as an important environmental reservoir of the pathogen. Intense rainfall events during the rainy season would cause mobilization and dispersion of pathogenic Leptospira from soil to run-off as described for other environmental pathogens such as E. coli, Salmonella spp., Cryptosporidium spp. and fecal indicator bacteria [43–48]. Simultaneously, run-off originated in the higher areas of the valley may contaminate soil in the lower areas with pathogenic Leptospira through flooding and sewer overflow. Thus, soil may act as a source and a recipient of the pathogen depending on the specific location and weather conditions. Furthermore, the low concentrations found in soil are in agreement with those found in sewage and standing water in a previous study conducted in the same setting [31], which supports the hypothesis that the environmental load of pathogenic Leptospira is generally low, even in endemic areas. The dynamics and characteristics of water-based mobilization and dispersion of Leptospira to and from the soil reservoir within the slum community, and the role that low environmental concentrations may have on the risk of acquiring leptospirosis, will require detailed studies beyond the scope of our methods.
More soil samples contain Leptospira when using 16S rRNA qPCR
In contrast to the lipL32 qPCR results, the 16S rRNA qPCR detected Leptospira from 60/70 (86%) soil samples, significantly more than detected by the lipL32 qPCR (p<0.0001). Higher prevalences were found at sites A and C (88% and 100%, respectively) than site B (57%) (p<0.0014) (Table 2). Among positive samples, the median concentration was 4.16×102 GEq/g (range: 4–2.58×104 GEq/g), significantly higher than the one detected by lipL32 qPCR (6 GEq/g, p < 0.0001). All eight sewage samples from Site C were also positive using the 16S qPCR, with a mean concentration of 2.09×102 GEq/mL (range: 98–2.81×102), nearly 9-fold higher than that detected by lipL32 qPCR (24 GEq/mL, p = 0.0003). Notably, all soil and sewage samples that were positive using the lipL32 assay were positive with the 16S assay, though there was a poor agreement between the qualitative results obtained by the two methods (Cohen’s Kappa coefficient = 0.14± 0.05) (Table 2)
Table 2. Summary of sampling data and results obtained by lipl32 and 16S rRNA qPCR (presence/absence and concentration) for the three sites investigated.
| lipl32 | 16S rRNA | ||||||
|---|---|---|---|---|---|---|---|
| Site | N of samples | Positive samples | Concentrationa | Positive samples | Concentrationa | Agreementb | |
| Soil | A | 34 | 9 (26%) | 4 (4–17.8) | 30 (88%) | 2.13×102 (4–2.91×103) | 0.09 ± 0.05 Poor |
| B | 14 | 3 (21%) | 13 (4–4.31×102) | 8 (57%) | 96 (19–1.20×103) | 0.34 ± 0.18 Fair | |
| C | 22 | 10 (45%) | 24 (4–4.09×102) | 22 (100%) | 6.95×102 (88.5–2.57×104) | 0.00 ± 0.00 Poor | |
| All | 70 | 22 (31%) | 6 (4–4.31×102) | 60 (86%) | 4.16×102 (4–2.57×104) | 0.14 ± 0.05 Poor | |
| Sewage | C | 8 | 7 (88%) | 0.5 (0.5–0.9) | 8 (100%) | 2.09×102 (97.7–2.81×102) | 0.00 ± 0.00 Poor |
aMedian concentration and range (GEq/g or GEq/mL)
bCohen’s kappa coefficient between qualitative lipl32 and 16S rRNA qPCR results, standard error and strength of agreement.
The 16S rRNA PCR method also detects intermediate group Leptospira
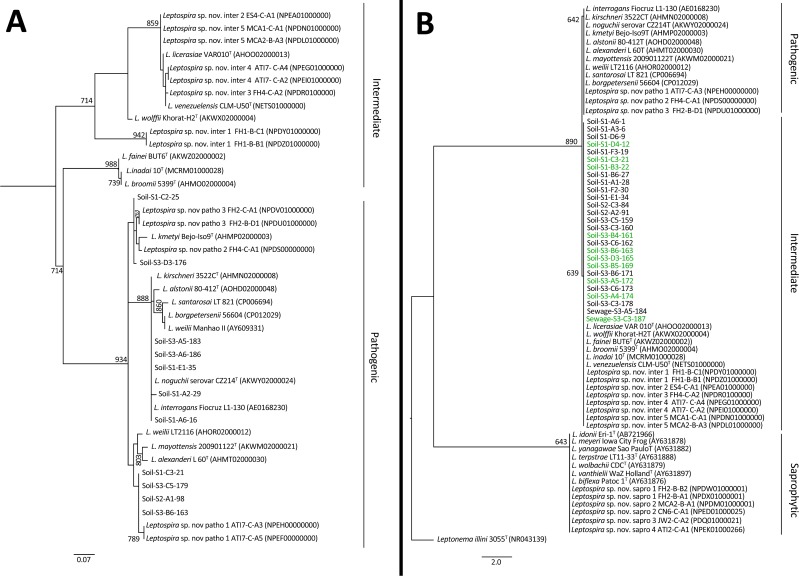
Given the discrepancy of results obtained with the two-qPCR methods (Table 2), we analyzed the specificity for detecting pathogenic Leptospira for each method. We sequenced the lipL32 amplicon from 11/22 (50%) lipL32 qPCR-positive soil samples. In all cases, the sequences presented similarities greater than 92% with lipL32 gene sequences from pathogenic Leptospira deposited in GenBank (S3 Table) and clustered with species from the pathogenic group (Fig 4A). These results confirmed that the lipL32 qPCR was highly specific for pathogenic Leptospira in soil as it was previously shown in sewage [31]. We also sequenced 27/78 (35%) 16S qPCR-positive samples (25 soil and 2 sewage samples). All the sequenced 16S rRNA amplicons clustered with species from the intermediate Leptospira group (Fig 4B). As observed elsewhere [49], a single mismatch in the approximately 50 bp fragment sequenced discriminates between intermediate and pathogenic Leptospira groups. Indeed, the reverse primer used in the 16S qPCR is degenerated at position 14 allowing for the hybridization with T and C bases, and thus, may detect both pathogenic and intermediate species (S1 Table). Of note, 6 sequences that were positive for both lipL32 and 16S presented a double peak in the sequencing chromatogram at the mismatch position. This indicates that both sequences coexisted in the sample [49], although in all cases the highest peak was the one belonging to the intermediate group.
Fig 4.
Phylogenetic trees of (A) lipl32 and (B) 16S rRNA Leptospira amplicons from soil and water samples collected at the community site. The trees were constructed using Maximum Likelihood method with HKY85 substitution model. A bootstrap of 1000 replicates was performed and values above 600 are shown in the nodes. Leptonema illini was used as the outgroup for the 16S rRNA tree, and samples colored in green were positive for both 16S rRNA and lipl32 qPCRs.
To conclusively determine the specificity of the both qPCRs, we tested 21 type strains from the pathogenic, intermediate and saprophytic groups. Lipl32 qPCR showed signal only for pathogenic species, and excluded intermediate and saprophytic ones. In contrast, all pathogenic and intermediate species gave positive results for 16S rRNA qPCR and no signal was observed for the saprophytes (S2 Table). Therefore, 16S rRNA qPCR detects not only pathogenic Leptospira, but also intermediate species. Since the pathogenicity of the intermediate group is not well established, we considered only the results obtained with lipL32 qPCR for subsequent analyses.
The presence of Leptospira is not associated with specific environmental factors except moisture
Bivariate and multivariate analyses identified no significant associations between the presence of pathogenic Leptospira DNA in soil and environmental characteristics such as vegetation, distance to an open sewer or a house, or soil clay content (Table 1). However, we found that 62% of samples with a moisture content higher than 20% were positive, while only 21% were positive when the moisture was lower than 20%, which is consistent with previous observations that higher soil moisture content is associated with increased Leptospira isolation [23,50]. Additionally, previous studies have reported that Leptospira persist for a longer time in moist and super-saturated soils than in drier ones, although the duration of survival is also dependent on the serovar and other characteristics of the soil such as pH [51–53].
Furthermore, against our initial hypothesis, we did not find any significant association between rat activity and the presence of pathogenic Leptospira in soil. A potential explanation is that the direct association with rats is confounded by other animal sources of pathogenic Leptospira both domestic (cows, pigs and dogs) and wildlife (opossums and bats) that coexist in this urban slum [15,34]. Alternatively, as discussed above, some of the pathogenic Leptospira detected may have originated in other parts of the valley and contaminated the soil through run-off or floodwater, making rat activity an unreliable proxy for the presence of the pathogen. Finally, we cannot rule out that the tracking board method was not a sufficient to assess rat activity at the fine scale at which the variation of the presence and concentration of pathogenic Leptospira in the soil seems to occur. Studies with larger sample sizes and an increased diversity of sites sampled are required to track the origin of pathogenic Leptospira in soil and determine relationships with environmental characteristics potentially obscured by our relatively small sample size.
Intermediate Leptospira spp. are common in soil
The concentrations of Leptospira in soil detected using the 16S qPCR were higher than those detected by lipL32 in all samples. Moreover, the difference between both measurements was higher than 0.60 log10 units in all but one sample (mean difference and SD: 2.05±0.89 log10 units). Since 16S qPCR detects pathogenic and intermediate species while lipL32 qPCR detects only pathogenic ones and both methods exclude saprophytic Leptospira (S2 Table), the observed concentration differences suggest that most of the signal detected with 16S qPCR originates from intermediate species. Hence, Leptospira species from the intermediate group may be more ubiquitous and present in significantly higher concentrations in soil from this community relative to pathogenic ones.
Intermediate species such as L. fainei, L. licerasiae, L. wolffii, and L. broomii have been linked to human leptospirosis cases [54–58], although they are not considered as virulent as the species from the pathogenic group, and thus are less relevant from a public health perspective. It is important to note that no cases of leptospirosis caused by an intermediate species has been reported in Pau da Lima during 15 years of active surveillance [6,18,30,59]. Previous studies of pathogenic Leptospira in the environment using 16S qPCR [20,49,60] might have led to a overestimation of the burden of the pathogen. Our results illustrate the importance of using highly specific tests to detect pathogenic bacteria in estimations of disease burden and environmental reservoir load.
Limitations
Inherent limitations of qPCR in environmental samples may influence the accuracy of our estimates and ability to evaluate risk associations. First, qPCR may be detecting DNA from dead or damaged cells that have lost the ability to cause infection and therefore, our results may overestimate the environmental risk. On the other hand, although we optimized sample processing, DNA extraction, and detection methodologies to reduce DNA loss and PCR inhibition, we may have underestimated pathogenic Leptospira loads in the soil if they were below the LOD. Second, our sampling scheme may not have completely captured the heterogeneity in the urban slum environment. While we evaluated three study sites representative of different microenvironments within the slum, there may be additional heterogeneity with respect to soil type, climatic conditions, land use, and rat activity levels, which should be further explored. Finally, the relatively small sample size limited our ability to draw robust conclusions concerning environmental factors contributing to positive and negative soil samples.
Conclusion
To date, most research regarding the environmental reservoirs of pathogenic Leptospira has focused on water matrices such as sewage, puddles, wells, or freshwater. Our results are the first to successfully quantify the burden of pathogenic Leptospira in soil from an endemic region, and indicate that soil is an additional environmental reservoir in the life cycle of pathogenic Leptospira. As with other environmentally transmitted diseases, the mobilization of leptospires from the sub-surface soil, either by heavy rainfall, flooding, or excavation, would contribute to environmental exposures with a sufficient dose to produce infection in humans. Furthermore, understanding the specific role and impact of soil as an environmental reservoir and the relation of low environmental concentrations to the risk of human disease is critical to our knowledge of the leptospirosis transmission cycle. Importantly, our data suggest that efforts to eliminate or reduce access to recognized transmission sources, such as open sewers, may not be sufficiently effective to decrease the risk of infection. Consequently, the role of soil in the transmission dynamics and epidemiology of leptospirosis should be considered when designing public health interventions in endemic areas.
Supporting information
The vertical dashed lines indicate the collection date at each sampling site.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The authors thank the joint collaborative effort of the resident associations, community leaders and residents, which constitute the Urban Health Council of Pau da Lima.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the National Institutes for Health research grants R01 AI052473, U01 AI088752, R25 TW009338, R01 TW009504 and R01 AI121207. AGS was supported by a Downs International Health Student Travel Fellowship and a Field Research Grant from the Tinker Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, et al. (2015) Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Negl Trop Dis 9: e0003898 doi: 10.1371/journal.pntd.0003898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thibeaux R, Iraola G, Ferrés I, Bierque E, Girault D, et al. (2018) Deciphering the unexplored Leptospira diversity from soils uncovers genomic evolution to virulence. Microb Genomics. doi: 10.1099/mgen.0.000144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puche R, Ferrés I, Caraballo L, Rangel Y, Picardeau M, et al. (2018) Leptospira venezuelensis sp. nov., a new member of the intermediate group isolated from rodents, cattle and humans. Int J Syst Evol Microbiol 68: 513–517. doi: 10.1099/ijsem.0.002528 [DOI] [PubMed] [Google Scholar]
- 4.Amilasan AST, Ujiie M, Suzuki M, Salva E, Belo MCP, et al. (2012) Outbreak of leptospirosis after flood, the Philippines, 2009. Emerg Infect Dis 18: 91–94. doi: 10.3201/eid1801.101892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tangkanakul W, Smits HL, Jatanasen S, Ashford DA (2005) Leptospirosis: an emerging health problem in Thailand. Southeast Asian J Trop Med Public Health 36: 281–288. [PubMed] [Google Scholar]
- 6.Ko AI, Reis MG, Dourado CMR, Johnson WD Jr, Riley LW, et al. (1999) Urban epidemic of severe leptospirosis in Brazil. Lancet 354: 820–825. [DOI] [PubMed] [Google Scholar]
- 7.Ko AI, Goarant C, Picardeau M (2009) Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol 7: 736–747. doi: 10.1038/nrmicro2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faine S, Adler B, Bolin C, Perolat P (1994) Leptospira and leptospirosis 2nd ed Faine S, editor Melbourne, Australia: MediSci. [Google Scholar]
- 9.McBride AJA, Athanazio DA, Reis MG, Ko AI (2005) Leptospirosis. Curr Opin Infect Dis 18: 376–386. [DOI] [PubMed] [Google Scholar]
- 10.Gouveia EL, Metcalfe J, Carvalho ALF De, Aires TSF, Caetano Villasboas-Bisneto J, et al. (2008) Leptospirosis- associated Severe Pulmonary Hemorrhagic Syndrome, Salvador, Brazil. Emerg Infect Dis 14: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Himsworth CG. b, Parsons KL., Jardine C., Patrick DM. (2013) Rats, cities, people, and pathogens: A systematic review and narrative synthesis of literature regarding the ecology of rat-associated zoonoses in urban centers. Vector-Borne Zoonotic Dis 13: 349–359. doi: 10.1089/vbz.2012.1195 [DOI] [PubMed] [Google Scholar]
- 12.Easterbrook JD., Kaplan JB., Vanasco NB., Reeves WK., Purcell RH., et al. (2007) A survey of zoonotic pathogens carried by Norway rats in Baltimore, Maryland, USA. Epidemiol Infect 135: 1192–1199. doi: 10.1017/S0950268806007746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa F, Porter FH, Rodrigues G, Farias H, de Faria MT, et al. (2014) Infections by Leptospira interrogans, Seoul virus, and Bartonella spp. among Norway rats (Rattus norvegicus) from the urban slum environment in Brazil. Vector Borne Zoonotic Dis 14: 33–40. doi: 10.1089/vbz.2013.1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa F, Wunder EA, De Oliveira D, Bisht V, Rodrigues G, et al. (2015) Patterns in Leptospira Shedding in Norway Rats (Rattus norvegicus) from Brazilian Slum Communities at High Risk of Disease Transmission. PLoS Negl Trop Dis 9: e0003819 doi: 10.1371/journal.pntd.0003819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faria MT, Calderwood MS, Athanazio DA, McBride AJA, Hartskeerl RA, et al. (2008) Carriage of Leptospira interrogans among domestic rats from an urban setting highly endemic for leptospirosis in Brazil. Acta Trop 108: 1–5. doi: 10.1016/j.actatropica.2008.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villanueva SYAM, Ezoe H, Baterna RA, Yanagihara Y, Muto M, et al. (2010) Serologic and molecular studies of Leptospira and leptospirosis among rats in the Philippines. Am J Trop Med Hyg 82: 889–898. doi: 10.4269/ajtmh.2010.09-0711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarkar U, Nascimento SF, Barbosa R, Martins R, Nuevo H, et al. (2002) Population-based case-control investigation of risk factors for leptospirosis during an urban epidemic. Am J Trop Med Hyg 66: 605–610. [DOI] [PubMed] [Google Scholar]
- 18.Hagan JE, Moraga P, Costa F, Capian N, Ribeiro GS, et al. (2015) Spatiotemporal determinants of urban leptospirosis transmission: Four-year prospective cohort study of slum residents in Brazil. PLoS Negl Trop Dis 10: e0004275 doi: 10.1371/journal.pntd.0004275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reis RB, Ribeiro GS, Felzemburgh RDM, Santana FS, Mohr S, et al. (2008) Impact of environment and social gradient on Leptospira infection in urban slums. PLoS Negl Trop Dis 2: e228 doi: 10.1371/journal.pntd.0000228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganoza CA, Matthias MA, Collins-Richards D, Brouwer K., Cunningham CB, et al. (2006) Determining risk for severe leptospirosis by molecular analysis of environmental surface waters for pathogenic Leptospira. PLoS Med 3: 1329–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muñoz-Zanzi C, Mason MR, Encina C, Astroza A, Romero A (2014) Leptospira contamination in household and environmental water in rural communities in southern Chile. Int J Environ Res Public Health 11: 6666–6680. doi: 10.3390/ijerph110706666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexander AD, Evans LB, Baker MF, Baker HJ, Ellison D, et al. (1975) Pathogenic leptospiras isolated from Malaysian surface waters. Appl Microbiol 29: 30–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito M, Villanueva SYAM, Chakraborty A, Miyahara S, Segawa T, et al. (2013) Comparative analysis of Leptospira strains isolated from environmental soil and water in the Philippines and Japan. Appl Environ Microbiol 79: 601–609. doi: 10.1128/AEM.02728-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saito M, Miyahara S, Villanueva SYAM, Aramaki N, Ikejiri M, et al. (2014) PCR and culture identification of pathogenic Leptospira from coastal soil in Leyte, Philippines after a storm surge during Super Typhoon Haiyan (Yolanda). Appl Environ Microbiol 80: 6926–6932. doi: 10.1128/AEM.02568-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thibeaux R, Geroult S, Benezech C, Chabaud S, Soupé-Gilbert M-E, et al. (2017) Seeking the environmental source of Leptospirosis reveals durable bacterial viability in river soils. PLoS Negl Trop Dis 11: e0005414 doi: 10.1371/journal.pntd.0005414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W, Pang J, Li C (1994) An investigation on the distribution of leptospirae interrogans in water and soil in southwest of Yunnan Province. Zhonghua Liu Xing Bing Xue Za Zhi 15: 289–291. [PubMed] [Google Scholar]
- 27.Azali MA, Yean Yean C, Harun A, Aminuddin Baki NN, Ismail N (2016) Molecular Characterization of Leptospira spp. in Environmental Samples from North-Eastern Malaysia Revealed a Pathogenic Strain, Leptospira alstonii. J Trop Med 2016: 2060241 doi: 10.1155/2016/2060241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slack AT. T, Khairani-Bejo S., Symonds ML. L, Dohnt MF. F, Galloway RL. L, et al. (2009) Leptospira kmetyi sp. nov., isolated from an environmental source in Malaysia. Int J Syst Evol Microbiol 59: 705–708. doi: 10.1099/ijs.0.002766-0 [DOI] [PubMed] [Google Scholar]
- 29.Benacer D, Thong KL, Verasahib KB, Galloway RL, Hartskeerl RA, et al. (2016) Human Leptospirosis in Malaysia: Reviewing the Challenges After 8 Decades (1925–2012). Asia-Pacific J Public Heal. doi: 10.1177/1010539516640350 [DOI] [PubMed] [Google Scholar]
- 30.Felzemburgh RDM, Ribeiro GS, Costa F, Reis RB, Hagan JE, et al. (2014) Prospective study of leptospirosis transmission in an urban slum community: role of poor environment in repeated exposures to the leptospira agent. PLoS Negl Trop Dis 8: e2927 doi: 10.1371/journal.pntd.0002927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casanovas-Massana A, Costa F, Riediger IN, Cunha M, de Oliveira D, et al. (2018) Spatial and temporal dynamics of pathogenic Leptospira in surface waters from the urban slum environment. Water Res 130: 176–184. doi: 10.1016/j.watres.2017.11.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.UN-HABITAT (2003) The Challenge of Slums—Global report on human settlements. [Google Scholar]
- 33.Riley LW, Ko AI, Unger A, Reis MG (2007) Slum health: diseases of neglected populations. BMC Int Health Hum Rights 7. doi: 10.1186/1472-698X-7-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hacker KP, Minter A, Begon M, Diggle PJ, Serrano S, et al. (2016) A comparative assessment of track plates to quantify fine scale variations in the relative abundance of Norway rats in urban slums. Urban Ecosyst 19: 561–575. doi: 10.1007/s11252-015-0519-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riediger IN, Hoffmaster AR, Casanovas-Massana A, Biondo AW, Ko AI, et al. (2016) An Optimized Method for Quantification of Pathogenic Leptospira in Environmental Water Samples. PLoS One 11: e0160523 doi: 10.1371/journal.pone.0160523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoddard RA, Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR (2009) Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn Microbiol Infect Dis 64: 247–255. doi: 10.1016/j.diagmicrobio.2009.03.014 [DOI] [PubMed] [Google Scholar]
- 37.Smythe L, Smith I, Smith G, Dohnt M, Symonds M, et al. (2002) A quantitative PCR (TaqMan) assay for pathogenic Leptospira spp. BMC Infect Dis 2: 13 doi: 10.1186/1471-2334-2-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nascimento ALTO, Ko AI, Martins EAL, Monteiro-Vitorello CB, Ho PL, et al. (2004) Comparative Genomics of Two Leptospira interrogans Serovars Reveals Novel Insights into Physiology and Pathogenesis. J Bacteriol 186: 2164–2172. doi: 10.1128/JB.186.7.2164-2172.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. (2009) The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin Chem 55: 611–622. doi: 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 40.Fouts DE, Matthias MA, Adhikarla H, Adler B, Amorim-Santos L, et al. (2016) What Makes a Bacterial Species Pathogenic?:Comparative Genomic Analysis of the Genus Leptospira. PLoS Negl Trop Dis 10: e0004403 doi: 10.1371/journal.pntd.0004403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–9. doi: 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, et al. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. doi: 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 43.Reischer GH, Haider JM, Sommer R, Stadler H, Keiblinger KM, et al. (2008) Quantitative microbial faecal source tracking with sampling guided by hydrological catchment dynamics. Environ Microbiol 10: 2598–2608. doi: 10.1111/j.1462-2920.2008.01682.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guber AK, Shelton DR, Pachepsky YA, Sadeghi AM, Sikora LJ (2006) Rainfall-induced release of fecal coliforms and other manure constituents: comparison and modeling. Appl Environ Microbiol 72: 7531–7539. doi: 10.1128/AEM.01121-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Semenov A V, van Overbeek L, van Bruggen AHC (2009) Percolation and survival of Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in soil amended with contaminated dairy manure or slurry. Appl Environ Microbiol 75: 3206–3215. doi: 10.1128/AEM.01791-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGechan MB, Vinten AJA (2006) Simulation of transport through soil of E. coli derived from livestock slurry using the macro model. Soil Use Manag 19: 321–330. doi: 10.1111/j.1475-2743.2003.tb00322.x [Google Scholar]
- 47.Ramirez NE, Wang P, Lejeune J, Shipitalo MJ, Ward LA, et al. (2009) Effect of tillage and rainfall on transport of manure-applied Cryptosporidium parvum oocysts through soil. J Environ Qual 38: 2394–2401. doi: 10.2134/jeq2008.0432 [DOI] [PubMed] [Google Scholar]
- 48.García-Aljaro C, Martín-Díaz J, Viñas-Balada E, Calero-Cáceres W, Lucena F, et al. (2017) Mobilisation of microbial indicators, microbial source tracking markers and pathogens after rainfall events. Water Res 112: 248–253. doi: 10.1016/j.watres.2017.02.003 [DOI] [PubMed] [Google Scholar]
- 49.Viau EJ, Boehm AB (2011) Quantitative PCR-based detection of pathogenic Leptospira in Hawai’ian coastal streams. J Water Health 9: 637–646. doi: 10.2166/wh.2011.064 [DOI] [PubMed] [Google Scholar]
- 50.Henry RA, Johnson RC (1978) Distribution of the genus Leptospira in soil and water. Appl Environ Microbiol 35: 492–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khairani-Bejo S, Bahaman AR, Zamri-Saad M, Mutalib AR (2004) The survival of leptospira interrogans in the Malasyan environment. J Anim Vet Adv 3: 123–129. [Google Scholar]
- 52.Hellstrom JS, Marshall RB (1978) Survival of Leptospira interrogans serovar pomona in an acidic soil under simulated New Zealand field conditions. Res Vet Sci 25: 29–33. [PubMed] [Google Scholar]
- 53.Kirschner L, Maguire T (1957) Survival of Leptospira outside their hosts. N Z Med J 56: 385–391. [PubMed] [Google Scholar]
- 54.Arzouni J-P, Parola P, La Scola B, Postic D, Brouqui P, et al. (2002) Human Infection Caused by Leptospira fainei. Emerg Infect Dis 8: 865–868. doi: 10.3201/eid0808.010445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matthias MA, Ricaldi JN, Cespedes M, Diaz MM, Galloway RL, et al. (2008) Human leptospirosis caused by a new, antigenically unique Leptospira associated with a Rattus species reservoir in the Peruvian Amazon. PLoS Negl Trop Dis 2: e213 doi: 10.1371/journal.pntd.0000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krogfelt KA, Schlichting P, Blom J, Petersen AM, Boye K (2001) First isolation of Leptospira fainei serovar Hurstbridge from two human patients with Weil’s syndrome. J Med Microbiol 50: 96–100. doi: 10.1099/0022-1317-50-1-96 [DOI] [PubMed] [Google Scholar]
- 57.Levett PN, Morey RE, Galloway RL, Steigerwalt AG (2006) Leptospira broomii sp. nov., isolated from humans with leptospirosis. Int J Syst Evol Microbiol 56: 671–673. doi: 10.1099/ijs.0.63783-0 [DOI] [PubMed] [Google Scholar]
- 58.Chiriboga J, Barragan V, Arroyo G, Sosa A, Birdsell DN, et al. (2015) High Prevalence of Intermediate Leptospira spp. DNA in Febrile Humans from Urban and Rural Ecuador. Emerg Infect Dis 21: 2141–2147. doi: 10.3201/eid2112.140659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torgerson PR, Hagan JE, Costa F, Calcagno J, Kane M, et al. (2015) Global Burden of Leptospirosis: Estimated in Terms of Disability Adjusted Life Years. PLoS Negl Trop Dis 9: e0004122 doi: 10.1371/journal.pntd.0004122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sumanta H, Wibawa T, Hadisusanto S, Nuryati A, Kusnanto H (2015) Spatial Analysis of Leptospira in Rats, Water and Soil in Bantul District Yogyakarta Indonesia. Open J Epidemiol 5: 22–31. doi: 10.4236/ojepi.2015.51004 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The vertical dashed lines indicate the collection date at each sampling site.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.