Summary
Prior crystal structures of the vault have provided clues of its structural variability but are non-conclusive due to crystal packing. Here, we obtained vaults by engineering at the N-terminus of rat major vault protein (MVP) an HIV-1 Gag protein segment and determined their near-atomic resolution (~4.8 Å) structures in a solution/non-crystalline environment. The barrel-shaped vaults in solution adopt two conformations, 1 and 2, both with D39 symmetry. From N- to C-terminus, each MVP monomer has three regions: body, shoulder and cap. While conformation 1 is identical to one of the crystal structures, the shoulder in conformation 2 is translocated longitudinally up to 10 Å, resulting in an outward-projected cap. Our structures clarify the structural discrepancies in the body region in the prior crystallography models. The vault’s drug-delivery potential is highlighted by the internal disposition and structural flexibility of its Gag-loaded N-terminal extension at the barrel waist of the engineered vault.
In brief
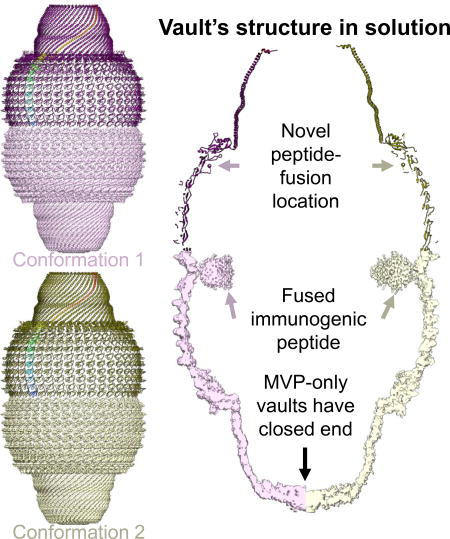
Ding et al. analyzed structures of vaults, large barrel-shaped ribonucleoprotein complexes found in most eukaryotic cells. Vaults adopt multiple conformations in solution. Comparison with crystallography results shows a major flexible region at the shoulder, suggesting that loops near this region can be utilized as peptide fusion sites for engineering purposes.
Introduction
First discovered in the mid-1980’s (Kedersha and Rome, 1986), vaults are large barrel-shaped ribonucleoprotein complexes existing in most eukaryotic cells. The vault’s function remains unknown. Each native vault has a mass of 13 MDa and is composed of multiple copies of at least three different proteins—the major vault protein (MVP, 100 kDa), vault poly(ADP-ribose) polymerase (VPARP), and telomerase-associated protein 1 (TEP1)—and several copies of small untranslated vault-associated RNA (vRNAs). Recombinantly expressed MVP assembles into vault-like nano-particles that can package small molecules and protein antigens, thus can be engineered to deliver drugs and vaccines, respectively. Its expression alone results in an ordered assembly of hollow barrel-shaped structures (Mrazek et al., 2014; Stephen et al., 2001), whose morphology is identical to natural vaults when examined by electron microscopy. Peptides genetically fused to the N-terminus of MVP were found inside the vault at the waist of the barrel (Mikyas et al., 2004), while those fused to the C-terminus were outside the vault at its two poles of the barrel (Kickhoefer et al., 2009). Recombinant vaults have been engineered to enable cell targeting (Kickhoefer et al., 2009), to trigger specific immune responses (Champion et al., 2009), and to deliver drugs (Buehler et al., 2011), demonstrating great potential for biomedical applications (Berger et al., 2009; Qi et al., 2012). However, the detailed structures of these recombinant vaults are not known, hampering efforts to engineer MVP for such applications.
Early structural characterization of the vault was accomplished by cryo electron microscopy (cryoEM) and single particle analysis with resolution limited to ~31 Å, mainly due to the low image contrast and the featureless nature of the vault (Kong et al., 1999). D48 symmetry was applied to this early cryoEM structure (Kong et al., 1999) and an X-ray crystal structure at 9 Å resolution (Anderson et al., 2007). Subsequently, another crystal structure of the rat native vault was solved with D39 symmetry at 3.5 Å resolution (PDB 4V60) (Tanaka et al., 2009). Of the 861-amino-acid (aa) long MVP, PDB 4V60 contains full atom models for aa. 1-427, 449-607,621-813, and Cα-only model for the C-terminal segment (aa 814-845), with a prominent gap from aa 428-448. From the N- to C-terminus, each MVP monomer consists of a body region containing 9 repeats (domains R1-R9) of an antiparallel β-sheet fold, followed by a shoulder region containing a single domain with 4 α-helices and a 4-stranded β-sheet, and a cap region containing a 155-amino-acid-long cap-helix domain and a cap-ring domain. In this crystal structure, the Cα-only model for the C-terminal segment is encapsulated inside the vault, rather than being exposed outside the vault (Kickhoefer et al., 2009). Meanwhile, a crystal structure of a truncated MVP monomer (PDB 3GF5, which contains only the first, N-terminal 387 aa residues) was solved to 2.1 Å resolution (Querol-Audi et al., 2009). These two crystal structures (PDB 4V60 for the vault and PDB 3GF5 for the N terminal segment) differ in the main chain tracing near the N-terminus (R1 and R2 domain). Further model refinement based on the electron density map of PDB 4V60 yielded a new model (PDB 4HL8) (Casanas et al., 2013). This refined, new model is basically a montage of PDB 3GF5 and 4V60: with its N-terminal domains (R1 and R2) similar to PDB 3GF5 and the following domains similar to those in PDB 4V60. Because PDB 3DF5 was obtained from a crystal containing segmented MVP, which lacked constraints from neighboring MVP subunits as those in the assembled vault, N-terminus domains in PDB 3GF5 are less curved than those in PDB 4HL8. These authors indicated that the backbone-tracing error near the N-terminus in the previous, PDB 4V60 model was a result of weak electron densities at the waist region and suggested that the N-terminal region is the major flexible region of MVP and may undergo large conformational change during assembly (Casanas et al., 2013). Such conformational changes might result in a leaky vault in solution and thus have bio-engineering significance, when vaults are engineered to package therapeutic compounds, such as hydrophobic all-trans retinoic acid (Buehler et al., 2011). Questions remain whether vaults in solution undergo conformational changes.
Here we have performed cryoEM and single particle analysis on a recombinant vault engineered at MVP N-terminus with a portion of HIV-1 Gag (amino acids 148-214) and obtained structures at near-atomic resolution (~4.8 Å). This highly conserved HIV-1 Gag segment has been shown to trigger immune response in peripheral blood mononuclear cells (Yang et al., 2015). Our results clarify previous symmetry and structure discrepancies. Further analysis explains the vault’s intrinsic structural flexibility and suggests optimization strategies to engineer MVP-only vaults for vaccine delivery applications.
Results
The vault has multiple conformations in solution
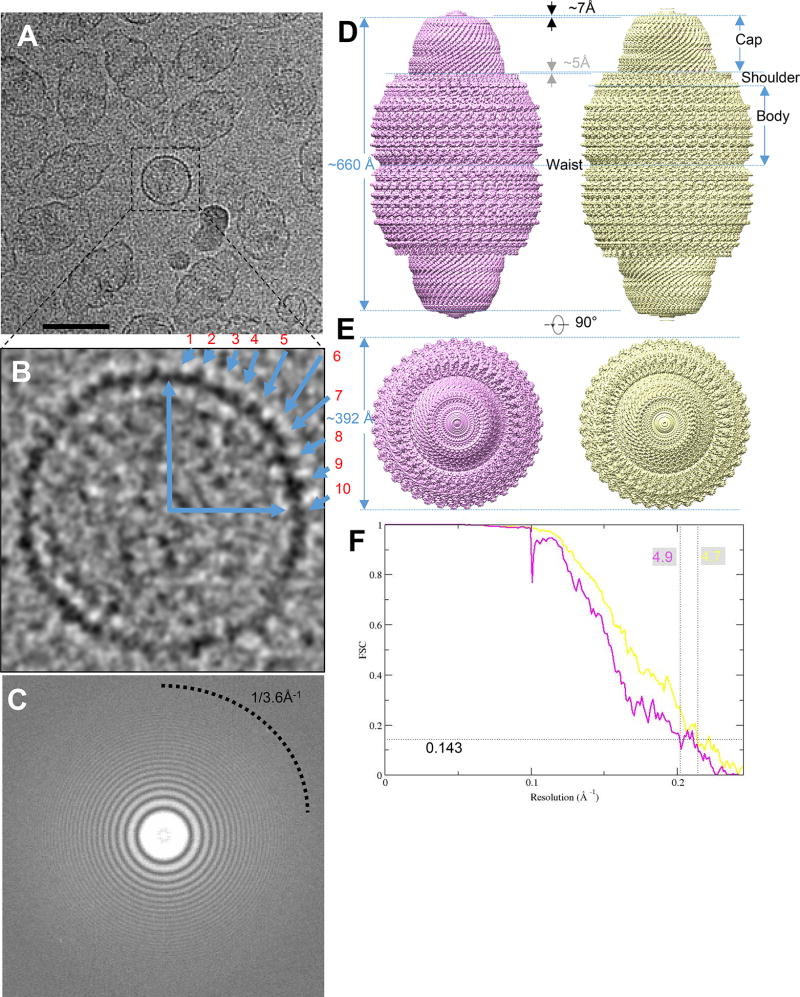
To enhance image contrast and to help clarify the number of subunits/vault, we recorded movies of recombinant rat vaults embedded in vitreous ice in a Titan Krios 300 kV electron microscope equipped with a K2 Summit direct electron-counting detector. Though vaults appeared mostly in their side views in our movies, occasionally top views of the vault can be spotted, showing features that indicate the separation of individual subunits (i.e., one MVP monomer on the top half of the vault closer to the viewer and the other on the bottom half) of MVP subunits lining along the direction of the view (Figure 1A). The number of MVP pairs within one of the four quadrants of the top view (Figure 1B) is between 9 and 10, consistent with 39 MVP pairs (i.e., 78 MVP subunits/vault, as in PDB 4HL8 and 4V60) (Casanas et al., 2013; Tanaka et al., 2009), but different from those used in other studies, such as PDB 2QZV (96 MVP subunits/vault) (Anderson et al., 2007).
Figure 1. CryoEM single particle analysis result on engineered MVP-only vault.
(A) Aligned sum of rat vault raw image stack, showing this dataset has nice orientation distribution. Typical top views are boxed in black square. The scale bar is 50nm. (B) Magnified raw image of top view to show there are ~10 copies in a quadrant of circle, implying close to 40-fold related symmetry. (C) Fourier transform of a sum micrograph. Thon rings can reach to water signal at close to 3.6Å−1. (D) Density map of two vault conformations refined from a single dataset. Conformation 1 (displayed at 4.4 σ) is in pink and conformation 2 (displayed at 4.5 σ) is in yellow. They are all in D39 symmetry. (E) top view of vault density in (D). No diameter change can be observed. (F) FSC curve showing that the resolution (FSC 0.143) of the two conformations are 4.9 Å and 4.7 Å, respectively.
The power spectrum of drift-corrected images shows that the Thon rings extend to 1/3.6 Å−1 (Figure 1C and Figure S1), indicating that our images have structural information beyond 3.6 Å resolution. However, the best resolution we achieved after exhaustive attempts to carry out single-particle reconstruction by Frealign (Lyumkis et al., 2013) was only 13.5 Å. This structure did not resolve individual MVP subunits at the cap region, hence the handedness of the reconstruction could not be established. This observation suggests existence of multiple conformations in the sample. To sort out multiple conformations, we subsequently carried out three-dimensional (3D) classification with Relion (Scheres, 2012) following the scheme illustrated in Figure S2 (see Method Details). Reconstructions with D38 or D40 symmetry did not converge to structures resolving any detailed features to establish handedness in the cap, even after exhaustive 3D classification and refinement. By contrast, reconstruction with D39 symmetry yielded two structure classes (i.e., conformations) that both converged to near atomic resolution [4.9 Å for conformation 1 and 4.7 Å for conformation 2] (Figures 1D, 1E, and 1F). Both conformations reveal extensive secondary structures and some bulky side chains of amino acid residues (Figures 2A, 2B, and 2E).
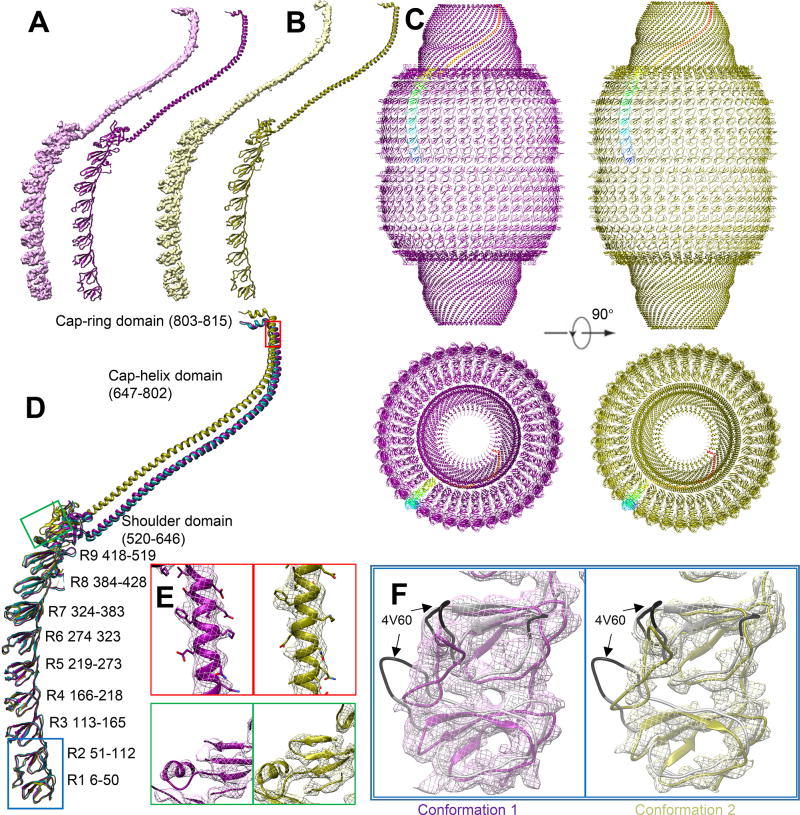
Figure 2. Structural comparison.
(A) Conformation 1 and corresponding model. (B) Conformation 2 and corresponding model. (C) Model comparison between conformation 1 (purple) and conformation 2 (olive). One copy of major vault protein (MVP) is colored in rainbow. The back half is hidden for clarity. (D) Overlapped model comparison. R1-R7 has no major conformational change. PDB 4HL8 is colored in aqua to show similarities between conformation 1 model and PDB 4HL8. (E) Near-atomic resolution feature at shoulder and cap-helix domain in both conformations, including an α-helix (red) and β-sheet (green). Large side chains can be identified and is consistent with current resolution estimation. Position is labeled in (D). Contour displayed at 6.7 σ. (F) Magnified view at R1 and R2 domain of two conformations. 4V60 model (grey) are displayed. The mismatch region in 4V60 is colored with black. No significant flexible region can be found at R1 and R2 domain. The major conformational change of cryoEM vault structure is not at waist region. Mesh contour is displayed at 5 σ for conformation 1 (pink) and 6.4 σ for conformation 2 (yellow)
In these two conformations, the two halves of the barrel-shaped vault are joined at the waist. Extending away from the waist to the distal end of the vault are the body, the shoulder and the cap regions of each half (Figure 1D). Similar to previously reported vault X-ray structures (Casanas et al., 2013; Tanaka et al., 2009), 39 MVP subunits line in parallel to form half of the vault. The top center of cap region is closed with no discernable features in our D39-symmetry-imposed maps, indicating that the D39 symmetry is not maintained in this location (Figure 1E).
While conformation 1 and conformation 2 have the same waist radius, conformation 2 is 14 Å longer than conformation 1 along the D39 symmetry axis direction (Figure 1D). This observation suggests that the conformational changes are real and not an artifact of display differences or magnification variation in the microscope. This is direct evidence that multiple vault conformations exist in solution.
Engineered MVP-only vaults can adopt the structure of naturally-occurring vaults
The availability of the crystal structure of naturally-occurring vaults (Casanas et al., 2013) (PDB 4HL8) allowed us to interpret our cryoEM structures of conformation 1 and conformation 2 at moderate resolutions of 4.9 and 4.7 Å, respectively. Consistent with crystal structure PDB 4HL8, the cryoEM densities of vault monomers in both conformation 1 and conformation 2 reveal the characteristic 9 repeats (R1 through R9) of β-sheet domains, followed by a shoulder domain with 4 α-helices and a 4-stranded β-sheet, and a long (~230 Å) cap helix (Figures 2A and 2B). PDB 4HL8 can be fitted only into conformation 1 density as a rigid body, indicating that conformation 1 is very close to crystal structure PDB 4HL8.
Therefore, we chose the PDB 4HL8 crystal structure as the starting model for real-space refinement against conformation 1 monomer. The refinement result shows that it is nearly identical to PDB 4HL8 with a root-mean-square deviation (RMSD) value of 1.3 Å. The traceable region covers most of the vault body and cap side walls, including R1-R9, shoulder, cap-helix and cap-ring domains (Figure 2A and 2D). The traceable sequence ends at P815 in the cap-ring domain. The rest of the density at the cap region is insufficiently resolved for reliable tracing of the C-terminal segment, from aa 816 to 861 (Figure 1D and 2C). Unlike naturally-occurring vaults, our recombinant vaults do not contain TEP1, VPARP or vRNA, yet the density at the center of the cap top is still solid, suggesting that the density observed at the center of the cap top in the two cryoEM conformations do not exclusively correspond to TEP1, VPARP, or vRNA as previously suggested (Kong et al., 1999; Tanaka et al., 2009; Tanaka and Tsukihara, 2012).
At the waist region inside the vault is the Gag 148-214 peptide fused to N-terminus of MVP (Figures 4A and 4C). The sectional view shows that the fused Gag peptide is fully encapsulated inside the vault, thus not exposed to the external environment (Figures 4A, C). The thickness of the MVP shell varies: close to 20 Å from R1 through R7 domains and thicker than 25 Å from R8, R9 and the shoulder domains (Figure 4E). R1 through R7 domains are also less structurally complex than R8, R9 and the shoulder domains. For example, in addition to R8 and R9’s the antiparallel β-sheet fold sub-domain that resembles the antiparallel β-sheet fold in each of R1 through R7 domains, R8 and R9 domains both contain an inner attachment sub-domain: an α-helix in R8 (R8-helix) and a hairpin in R9 (R9-hairpin) atop R8-helix (Figure 3B). The shoulder domain has folding motif different from the antiparallel β-sheet fold of R1 through R9 and can be roughly divided into two sub-domains: a shoulder-helix sub-domain next to R9’s antiparallel β-sheet fold and a shoulder-hybrid sub-domain (featuring a combination of α-helix and β-sheet) next to the inward-projecting R9-hairpin (Figure 3B). It is the presence of the inner-layer sub-domains in R8, R9 and shoulder domains that contributes to extra thickness of these domains as compared to R1 through R7 domains. The segment from N428 to S449 that connects R8 with R9 missing in X-ray crystal structure (Casanas et al., 2013; Tanaka et al., 2009) remained unresolved in conformation 1 cryoEM structure (Figure 3B).
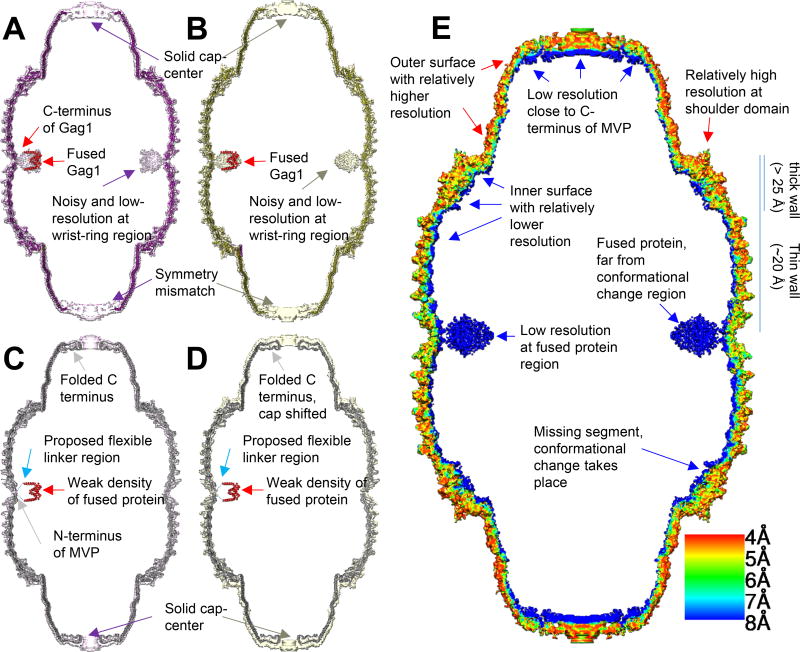
Figure 4. Model and density comparison among models and densities via longitudinal section.
(A) Conformation 1 model (purple), conformation 1 density (pink, displayed at 3 σ) and docked segmented Gag dimer (PDB 1AFV, red). (B) Conformation 2 model (olive), conformation 2 density (yellow, displayed at 3.7 σ) and docked Gag dimer (red). (C, D) utilizing similar color code with (A) and (B), respectively, with higher visualization threshold (displayed 6.2 σ and 5.1 σ for conformation 1 and 2, respectively). PDB 4V60 (grey) is docked into conformation 1 and conformation 2 density. C-terminus of Gag and N-terminus of MVP is connected by flexible linker, shown as dashed line. (E) Local resolution estimation of conformation 2 density map calculated by Resmap (Kucukelbir et al., 2014). The flexible region is of low resolution and appear blue (fused protein at waist, C terminus near cap and inner surface). The major conformational change takes place at shoulder domain but the resolution is relatively high.
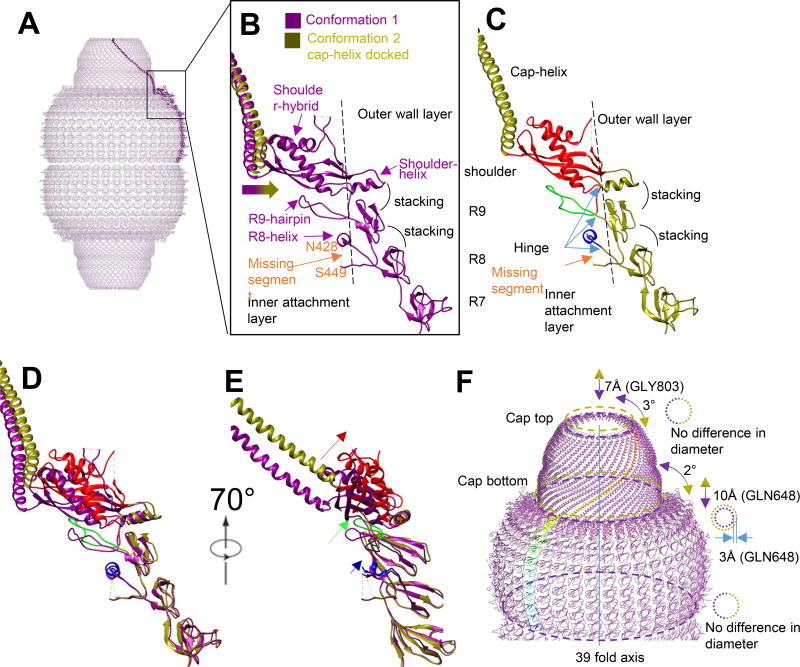
Figure 3. Conformational change diagram.
(A) Conformation 1 monomer model as a side view in vault over all model. (B) Magnified view of R7 to cap-helix domain. The missing segment between N428 to S449 locates inside vault. The docking of helix-cap domain of conformation 2 into conformation 1 density shows that cap-helix domain in conformation 2 bends outwards comparing with conformation 1. R8 to shoulder domain can all be roughly divided into two parts, separated by the dashed line. The attachment layer locates inside and the wall layer locates outside. (C) R7 to cap-helix domain of conformation 2. Like conformation 1, the R8 to shoulder domain is double layered. The inner-layer is colored from blue to red, from N-terminus to C-terminus. The outer layer is colored in original olive color. (D, E) Direct overlapping of conformation 1 and conformation 2 model. Position shift is magnified from R8 to shoulder domain. The relative movement of attachment layer is labeled with corresponding color and the movement is larger from R8 to shoulder domain. (F) A diagram to show the cap movement and conformational change between conformation 1 (purple) and conformation 2 (olive). In the refinement result applied D39 symmetry, the relative movement freedom of cap is limited to axial (up and down) and rotational (rotation around 39-fold axis). The movement from conformation 1 to conformation 2 of cap region can be described as “being rotated clockwise by 2 degrees and lifted by 10Å”. There is minor morphing of cap between the two conformations, when conformation 2 is relatively shorter and more twisted at cap region based on shorter translocation distance along axis and more angular rotation at cap-ring region.
Multiple conformations of the vault in solution
The local resolutions of most regions in the density map of conformation 2 are between 4 Å to 6 Å (Figure 4E), with the best resolution in the R1-R9 and the shoulder and the lowest resolution in the folded C-terminal region at the cap and the fused protein (Gag 148-214) region at the waist. The resolution of the entire inner surface of the vault is lower than that of the outer surface. The elongation of MVP monomer towards vault’s pole in conformation 2 prevented satisfactory fitting of PDB 4HL8 as a rigid body into the density. R1 through R7 domains in PDB 4HL8 fit well with conformation 2 density, however R8, R9, shoulder, cap-helix and cap-ring domains do not. Thus, the atomic model for R1 through R7 domains of PDB 4HL8 was fitted into the conformation 2 map. To address the previous discrepancies at the waist region, PDB 4V60 was also docked into the density of conformation 2 to see if this earlier X-ray model could represent conformation 2 of the vault in solution. This docking revealed that R1 and R2 domains in PDB 4V60 do not match the cryoEM density of conformation 2 (Figure 2F). Recall that conformation 1 of our cryoEM structures adopts the structure of PDB 4HL8, which differs from PDB 4V60 at R1 and R2 domains. Taken together, we conclude that PDB 4V60 does not represent vault’s conformation in solution.
The atomic model of individual α-helixes and β-sheets in R8, R9, shoulder domain, cap-helix domain and cap-ring domain of PDB 4HL8 were fitted into their corresponding secondary structure elements visible in the cryoEM density map as rigid bodies. These fitted secondary structures were connected with linkers derived from the MVP sequence to create a full trial atomic model, which was subjected to model refinement against the cryoEM map. The resulting model (Figure 2B) has a Cα RMSD value of 5.95 Å when compared with PDB 4HL8 (see Table S1). The conformational changes in the refined model of conformation 2 take place at the R8 domain (P420) and become larger as one moves toward the C terminus (Figure 2D). R8 and R9 domains undergo minor conformational changes by slightly bending outwards, pivoting around their respective N-termini. Like that in conformation 1, we were unable to model the segment from N428 to S449 in conformation 2. This flexible segment likely occupies some space near the inner surface of R8 and R9 domains (Figure 3C, labeled “missing segment”). In general, while the secondary structure elements in R8 and R9 domains are conserved between conformation 1 and conformation 2, the relative positions of these secondary structure elements and their connecting linkers are not. Compared that in conformation 1, the shoulder domain in conformation 2 is twisted further outward, along with the attached cap-helix domain (Figure 2D and Figure 3D).
Because of the linear arrangement of domains from N- to C-terminus, the large conformational change at the shoulder domain introduces a large translocation of the cap-helix and cap-ring domains (Figure 2D). In both conformations, α-helix and β-strands are resolved in the shoulder and cap-helix domains, allowing us to perform detailed comparisons of these high-resolution features in the two conformations (Figure 2E). When viewed along the symmetry axis, the cap of conformation 2 is lifted by 10 Å and rotated by 2° clockwise from the bottom (Figure 3F and Movie S1, measurement based on Cα of GLN648) as compared to conformation 1. Further alignment shows that the cap top of conformation 2 was further rotated by 1° and shortened along the 39-fold axis by about 3 Å (measurement based on Cα of GLY803). The diameter of the cap top (i.e., the distance from Cα of GLY803 the symmetry axis) is the same in both conformations, while the cap bottom diameter of conformation 2 is 6 Å (i.e., 3%) larger than that of conformation 1. The 4.7 Å resolution of the cryoEM density lends support for these model-based measurements.
Position and structure of the engineered HIV-1 Gag 148-214 peptide inside the vault
Weak densities are observed in both conformations at the waist region close to the N-terminus of MVP. We interpret these densities as the fused HIV-1 Gag 148-214 peptide because of its connection to the N-terminus of MVP and its size matching that expected for the engineered Gag segment (Figure 4A, B). The density of the fused peptide is weaker than that of MVP (Figures 4C, D) and the boundary of fused protein at the waist region is at lower resolution. A dimer of Gag 148-214 (PDB 1AFV) (Momany et al., 1996) can be docked into the donut shaped density seen inside the waist region. However, the docking was non-unique and no secondary structure can be identified in the waist-ring density, suggesting that the fusion peptide is flexible inside the vault with only limited interactions with MVP. This result is consistent with previous observations that peptides fused to N-terminus of MVP tend to extend towards the center of a vault particle (Mikyas et al., 2004).
The total number of amino acids for the fused Gag 148-214 and the GFLGL linker is 73. Applying the free chain model assuming a persistence length of 5 amino acids, the engineered peptide would give rise to a maximal end-to-end length of 73 Å (i.e., ). This maximal length of the engineered segment is much shorter than the 140 Å axial linear distance between the R8 domain and the waist. Therefore, the observed conformational change starting at the R8 domain is unlikely caused by interactions with the fused Gag 148-214 peptide.
Discussion
Our near-atomic cryoEM structures demonstrate the co-existence of two conformations of the vault in solution. The pole regions of the vault contain solid densities in both conformations. Previous studies proposed that MVP C-terminus (P815-K861) folded back inside the vault, leaving an opening at both poles of the vault (Tanaka et al., 2009). Our MVP-only vault structure suggests that at least some of the C-terminal segment must extend all the way towards the symmetry axis to seal the vault particle, thus different from that as described before (Figure 4C, D). This result is consistent with a previous low-resolution cryoEM study showing that an antibody could bind to its antigen fused at MVP’s C-terminus (Kickhoefer et al., 2009). The relative lower resolution of this region of our vault reconstruction suggests that the symmetry at the pole region might deviate from D39.
The comparison between the two conformations suggests varying levels of structural flexibility for different domains of the vault. Targeting flexible region of the vault for engineering applications is desirable in order to reduce possible steric hindrance introduced by peptide insertion. Domains R1 through R7 on MVP contains only antiparallel β-sheet motifs and exhibit no structural changes between the two conformations. Four hydrogen bonds and eight hydrophobic interactions (Tanaka et al., 2009) exist between neighboring MVP monomers at the R1-R7 domains, which are believed to play a key role in assembling 78 MVP monomers to an intact vault (Casanas et al., 2013). The major difference between the outer shell of our recombinant vault and the naturally-occurring vault is the 73 aa Gag and linker peptides fused to the N-terminus of MVP. The folded peptide appears to only directly interact with the R1 domain through a flexible linker. The structures of R1-R7 domains in our cryoEM structures of both conformations in solution are identical to those in the crystal structure PDB 4HL8, indicating that the fused peptide does not change the conformation of these domains near the site of the engineered peptide and that the cryoEM structures of our recombinant vault can inform about structural dynamics of naturally-occurring vaults in solution.
It is interesting to note that conformation 1 is similar to PDB 4HL8, which was determined from crystals where crystal packing might have constrained the flexible elements of the vault (Tanaka et al., 2009). The cryoEM structures reported here were determined from vaults distributed on carbon film to overcome the preferred orientation problem. Whether this support film influenced the observed structural variability is hard to establish as many cryoEM structures have been determined to near-atomic resolution using similar strategies. R1 through R7 act like rigid plates laced together akin to the arrangement of ancient lamellar armor. Each domain contains antiparallel β-strands and corresponds to a solid plate of the armor. The 12 interactions between these domains (Tanaka et al., 2009) are similar to the lace that holds lamellae together. We suggest a “wall-and-attachment” mechanism for the structural dynamics of the vault, whereby the laced R1-R7 plates, the identical antiparallel-β-sheet motif sub-domains in R8-R9, and the shoulder-helix sub-domain together form a rigid wall to which flexible regions (R8-helix, R9-hairpin and the shoulder-hybrid sub-domains) attach. These flexible regions constitute an “attachment” layer (Figure 3B) that undergo conformational changes while holding to a relatively rigid wall (olive in Figure 3C). Interestingly, the conformational changes increase from R8 domain to the shoulder domain (Figure 3D) and all these domains are more structurally complex than R1 through R7 domains. The protein sequences for the attachment layer are conserved between vaults isolated from different species (Kedersha et al., 1990), suggesting its observed structural dynamics is likely an intrinsic property of vaults. Although the vault is closed in both conformations presented here, our observed dynamics may be the structural basis for vault breathing and opening as suggested previously (Casanas et al., 2013).
Engineered vaults have been proposed as bio-compatible vehicles for vaccine delivery (Champion et al., 2009). In both conformations, the largest opening on the side wall of the vault is estimated to be of 5 Å by 5 Å. Previous study (Poderycki et al., 2006) shows that vault associate protein (TEP1) can enter assembled MVP-only vault, but truncated TEP1 cannot. Our cryoEM observation is consistent with this specific-access model. This feature reduces the chance of cargo contamination and leakage. Our results can also inform how to engineer vaults for carrying peptides and small proteins for therapeutic applications. It is known that the high solubility of vaults under physiological conditions enables packaging of highly potent but often insoluble drugs to improve efficacy and reduce toxicity. Our results show that MVP alone can form a completely sealed enclosure that would protect packaged materials and that the fused Gag 148-214 peptide is located adjacent to the vault waist with size and shape expected for properly folded Gag. This is consistent with previous observation that GFP fused at N-terminal of MVP functioned properly (Slesina et al., 2006). Therefore, N-terminal fusion is a viable strategy for vaccine delivery though a more complete understanding of the rules governing the position and structure of fused target antigens is needed. To the contrary, antigens fused at the MVP C-terminus might not work because they may fold outwards and become exposed.
One remaining caveat of the N-terminal fusion strategy is potential crowdedness that might interfere with dimerization of the two halves of the vault during assembly (Mrazek et al., 2014). The strong interactions between neighboring MVP monomers in their regions spanning R1-R7 domains leave little room to insert peptide between those repeats. Our structure results reveal other locations inside the vault that can be targeted for peptide fusion to overcome this caveat. For example, the missing segment (N428 to S449) is located inside the vault and is flexible. Fusion at this location would divide the payload into two parts of 39 copies each to be packaged inside the top and bottom halves away from the waist. Proteins fused near flexible region (R8, R9 and shoulder domain) can also potentially reduce impact on the assembly of MVP.
STAR ⋆ Methods
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to, and will be fulfilled by the Lead Author, Dr. Z. Hong Zhou (Hong.Zhou@UCLA.edu).
Method Details
Recombinant vault sample preparation
The HIV Gag148-214-GFLGL fragment was PCR amplified and cloned into pFastBac 1 containing rat MVP. The NcoI cloning site at the MVP 5’ end was used as a site of insertion by employing In-Fusion HD cloning kit (cat # 638910) and by strictly following the In-Fusion® HD Cloning kit manual from Takara/Clontech Inc. The resulting pFastBac1-Gag1-M1-GFLGL-rMVP construct was recombined with Bacmid DNA in MAX Efficiency® DH10Bac™ Competent Cells from Invitrogen (cat # 10361012), see manufacturers protocol. The Bacmid containing the Gag 148-214-GFLGL-MVP recombinant DNA was purified following Bac-to-Bac® Baculovirus Expression System manual from Invitrogen (cat # 10359-016). Generation of Baculovirus expressing the recombinant MVP was accomplished by transfecting sf9 cells in Gibco® Sf-900™ II SFM (Cat# 11496015) with Bacmid DNA, using Cellfectin™ II Reagent (Cat# 10362100) and following the user guide (Pub. No. MAN0007821 Rev. 2.0). To produce the recombinant vaults, 1 × 108 sf9 cells were infected with the recombinant Baculovirus in 50 ml Sf-900™ II SFM (cat # 10902096). The infected cells were shaken for 3 to 4 days at 28 °C , then harvested by centrifuging at 500×g for 5 min at room temperature. Cell pellet was stored at −80°C or used directly for vault purification.
For Sf9 cell lysis, buffer A (50 mM Tris-Cl buffer, 75 mM NaCl, 0.5 mM MgCl2) containing 2% Triton-X-100, 2% PI (Protease Inhibitor; Sigma-Aldrich P8849-5ML) and 1 mM PMSF (Phenylmethylsulfonyl fluoride) was prepared. 1 mg of RNase A was added to 1 g of Sf9 cells expressing Gag1-M1-GFLGL-rMVP, then 5 ml of lysis buffer was added and incubated on ice for 15 min. 2 mM DTT was then added and the cell lysate was further incubated on ice for additional 5 min. The lysate was centrifuged at 20,000×g at 4 °C for 20 min. The supernatant was collected and centrifuged at 40K in Ti 70.1 rotor for 1 h at 4 °C. The pellet was resuspended in 1 ml buffer A supplemented with 2% PI, 2 mM PMSF, and 2 mM DTT. 1 ml Ficoll-sucrose was added and the mixture was further vortexed and centrifuged at 25K in Ti 70.1 rotor at 4 °C for 10 minutes. The supernatant was diluted with 5.5 ml buffer A supplemented with 1% PI, 1 mM PMSF and 1 mM DTT, which was then centrifuged at 40 K in Ti 70.1 rotor for 1h, 30 minutes at 4°C. The pellet was resuspended in 1 ml buffer A containing 1% PI, 1 mM PMSF and 1 mM DTT. 5 μg streptomycin sulfate was added; the mixture was tumbled at room temperature for 30 minutes, then centrifuged 16,100 × g at room temperature for 10 minutes. Clarified supernatant was overlaid on a stepwise sucrose gradient (20%, 30%, 40%, 45%, 50%, 60% sucrose, 1.5 ml each) and then centrifuged at 25K in sw41 rotor at 4 °C for 16 hours. The 40% and 45% sucrose fractions were collected. The fractions were diluted in 4.5 ml PBS, then centrifuged at 40K in Ti 70.1 rotor at 4°C for 2 hours. The pellet was resuspended in 210 μl PBS to serve as cryoEM grid ready sample.
Electron microscopy and movie processing
For cryoEM, an aliquot of 2.5 μl of recombinant vault sample was applied to each EM grid with Lacey carbon films. The grid was blotted with Vitrobot in 100% humidity for 10s and then plunged into liquid ethane to vitrify the sample. Movies were obtained in Titan Krios 300kV electron microscope with Gatan K2 direct election detection camera in super-resolution mode with Leginon (Mindell and Grigorieff, 2003) at ×49000. The pixel size was measured to be 1.036 Å on the specimen scale. We used an electron dose rate of 8 electrons/pixel/second and each movie contains 20 frames recorded in 5 seconds. Image stacks in each movie were aligned with UCSF MotionCorr (Li et al., 2013). The first 16 frames in each stack were averaged to obtain an image sum of 32 e−/Å2. The whole dataset has 1218 movies.
Data processing and 3D reconstruction
Micrographs after alignment were used for contrast transfer function (CTF) determination in CTFFIND3 (Mindell and Grigorieff, 2003), with defocus values ranging from −1.7 μm to −4.2 μm. A total of 63751 particles were manually picked with 900 × 900 box size in pixel. Particles were first directly refined with Frealign (Lyumkis et al., 2013) and reported resolution was 13.5 Å with little features showing handedness. Then, all particles were subjected RELION 1.2 (Scheres, 2012) for two-dimensional classifications (Class2D). Top views were intentionally excluded from further classification to limit sampling space and to accelerate refinement process. Also, classes with no interpretable features were discarded. 32702 particles were selected for further three-dimensional classifications (Class3D). Particles are classified based on D38, D39 and D40 symmetry in different runs. Classification result with D38 and D40 symmetry also showed little feature with handedness. The following class3D are all conducted applying D39 symmetry with finer searching grid (Figure S2). The initial model for Class3D was generated from previously published atomic model (PDB 4HL8) of rat vault to 50 Å resolution to eliminate the potential risk of model bias. Class3D analysis was conducted with D39 fold symmetry applied and 2 distinguish classes with relatively good resolution (~9 Å) were found. These two classes were further refined separately with RELION 1.2 with D39 symmetry. To further enhance signal, mask is generated from cryoEM data to focus the refinement on MVP region. Following ‘gold standard’ refinement protocol, the two conformations were refined both to near-atomic resolution after RELION post-processing and automatic soft masking (Scheres, 2012). The resolution was determined based on a ‘gold standard’ Fourier shell correlation (FSC) coefficient of 0.143.
Atomic model building, refinement, and visualization
The atomic model of engineered vault was derived from crystal structure PDB 4HL8. By calibrating pixel size from 1.036 Å to 1.000 Å, we achieved an optimal docking of PDB 4HL8 into conformation 1 density. The fitted PDB 4HL8 was subjected to real-space refinement in Phenix (Adams et al., 2010) using the MVP monomer as density map input. Ramachandran and rotamer outliers were manually corrected with Coot (Emsley et al., 2010) for this conformation 1 model.
The pixel size of conformation 2 density was also adjusted to 1.000 Å accordingly. R1-R7 domains in PDB 4HL8 was first fitted into conformation 2 density. In R8 to cap-helix domains of PDB 4HL8, individual secondary structures were fitted into corresponding densities in conformation 2 map. Those secondary structures were further connected with linker accordingly to create a “morphed” model. Following the same protocol as refining model of conformation 1, this “morphed” PDB 4HL8 was subjected to real-space refinement with segmented density of conformation 2 in Phenix. Ramachandran and rotamer outliers were also manually corrected with Coot (Emsley et al., 2010) for conformation 2.
Visualization and map segment were achieved with UCSF Chimera (Pettersen et al., 2004). Local resolution was calculated by Resmap (Kucukelbir et al., 2014).
Data Availability
3D cryoEM density maps have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-7126 (conformation 1), EMD-7125 (conformation 2) and their corresponding coordinates of atomic models have been deposited in the Protein Data Bank under the accession number 6BP8 and 6BP7, respectively. The data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Movie S1. Conformation change of vaults in solution, Related to Figure 3
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant protein: HIV Gag1-M1-GFLGL (MTPRTLNAWVKVVEEKAFSPEVIPMFTALSEGATPSD LNTMLNTIGGHQAAMQMLKDTINEEAAEWDRGFLGL) | Genbank | MH020171 |
| Gibco® Sf-900™ II SFM | Gibco | 10902096 |
| Critical Commercial Assays | ||
| In-Fusion® HD Cloning Kit | Takara Bio | 638910 |
| Bac-to-Bac® Baculovirus Expression System | Invitrogen | 10359-016 |
| Deposited Data | ||
| Vault conformation 1 model | This paper | PDB-6BP8 |
| Vault conformation 2 model | This paper | PDB-6BP7 |
| Vault conformation 1 cryoEM map | This paper | EMDB-7126 |
| Vault conformation 2 cryoEM map | This paper | EMDB-7125 |
| Experimental Models: Cell Lines | ||
| Spodoptera frugiperda sf9 | Invitrogen | B82501 |
| Experimental Models: Organisms/Strains | ||
| MAX Efficiency® DH10Bac™ Competent Cells | Gibco | Cat # 11496015 |
| Recombinant DNA | ||
| Plasmid: pFastBac 1 | Invitrogen | 10359016 |
| Plasmid: pFastBac 1-Gag1-M1-GFLGL-MVP | Genbank | MH020171 |
| Software and Algorithms | ||
| Relion | (Scheres, 2012) | http://www2.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page |
| Coot | (Emsley et al., 2010) | http://www2.mrc-lmb.cam.ac.uk/Personal/pemsley/coot/ |
| PHENIX | (Adams et al., 2010) | http://www.phenix-online.org/ |
| UCSF Chimera | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera/ |
| Resmap | (Kucukelbir et al., 2014) | http://resmap.sourceforge.net/ |
Highlights.
We found different conformations of the vault, an 13MDa organelle
Visualization of engineered vault with fused peptide encapsulated
Engineering vault near shoulder region to improve vaccine carrying ability
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (GM071940, AI094386, AI043203, AI106528, and DE025567). We acknowledge the use of instruments at the Electron Imaging Center for Nanomachines supported by UCLA and by grants from NIH (1S10RR23057, 1S10OD018111, U24GM116792) and NSF (DBI-338135). K.D. was supported by UCLA Molecular Biology Whitcome Pre-Doctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions Z.H.Z. and L.H.R. designed and supervised research; O.O.Y. and J.M. designed the vaults; J.M., H.L.N. and V.A.K. prepared samples; K.D., X.Z. and Z.H.Z. recorded and analyzed the cryoEM data; K.D. and M.L. built atomic models; Z.H.Z. and K.D. wrote the paper; all authors reviewed and finalized the paper.
Declaration of Interests L.H.R., O.O.Y., and V.A.K. declare that they have a financial interest in Vault Nano Inc. and that the Regents of the University of California have licensed intellectual property invented by L.H.R. and V.A.K. to Vault Nano Inc. V.A.K. is currently working for Vault Nano Inc. J.M. declares he has financial interest in Aukera, Inc.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DH, Kickhoefer VA, Sievers SA, Rome LH, Eisenberg D. Draft crystal structure of the vault shell at 9-angstrom resolution. Plos Biol. 2007;5:2661–2670. doi: 10.1371/journal.pbio.0050318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger W, Steiner E, Grusch M, Elbling L, Micksche M. Vaults and the major vault protein: Novel roles in signal pathway regulation and immunity. Cell Mol Life Sci. 2009;66:43–61. doi: 10.1007/s00018-008-8364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehler DC, Toso DB, Kickhoefer VA, Zhou ZH, Rome LH. Vaults Engineered for Hydrophobic Drug Delivery. Small. 2011;7:1432–1439. doi: 10.1002/smll.201002274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanas A, Querol-Audi J, Guerra P, Pous J, Tanaka H, Tsukihara T, Verdaguer N, Fita I. New features of vault architecture and dynamics revealed by novel refinement using the deformable elastic network approach. Acta Crystallogr D. 2013;69:1054–1061. doi: 10.1107/S0907444913004472. [DOI] [PubMed] [Google Scholar]
- Champion CI, Kickhoefer VA, Liu GC, Moniz RJ, Freed AS, Bergmann LL, Vaccari D, Raval-Fernandes S, Chan AM, Rome LH, et al. A Vault Nanoparticle Vaccine Induces Protective Mucosal Immunity. Plos One. 2009;4 doi: 10.1371/journal.pone.0005409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Miquel MC, Bittner D, Rome LH. Vaults .2. Ribonucleoprotein Structures Are Highly Conserved among Higher and Lower Eukaryotes. J Cell Biol. 1990;110:895–901. doi: 10.1083/jcb.110.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Rome LH. Isolation and characterization of a novel ribonucleoprotein particle: large structures contain a single species of small RNA. J. Cell Biol. 1986;103:699–709. doi: 10.1083/jcb.103.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kickhoefer VA, Han M, Raval-Fernandes S, Poderycki MJ, Moniz RJ, Vaccari D, Silvestry M, Stewart PL, Kelly KA, Rome LH. Targeting Vault Nanoparticles to Specific Cell Surface Receptors. Acs Nano. 2009;3:27–36. doi: 10.1021/nn800638x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong LB, Siva AC, Rome LH, Stewart PL. Structure of the vault, a ubiquitous cellular component. Struct Fold Des. 1999;7:371–379. doi: 10.1016/s0969-2126(99)80050-1. [DOI] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EMEM density maps. Nat Methods. 2014;11 doi: 10.1038/nmeth.2727. 63-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XM, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng YF. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10 doi: 10.1038/nmeth.2472. 584-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Brilot AF, Theobald DL, Grigorieff N. Likelihood-based classification of cryo-EM images using FREALIGN. J Struct Biol. 2013;183:377–388. doi: 10.1016/j.jsb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikyas Y, Makabi M, Raval-Fernandes S, Harrington L, Kickhoefer VA, Rome LH, Stewart PL. Cryoelectron microscopy imaging of recombinant and tissue derived vaults: Localization of the MVP N termini and VPARP. J Mol Biol. 2004;344:91–105. doi: 10.1016/j.jmb.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Momany C, Kovari LC, Prongay AJ, Keller W, Gitti RK, Lee BM, Gorbalenya AE, Tong L, McClure J, Ehrlich LS, et al. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- Mrazek J, Toso D, Ryazantsev S, Zhang X, Zhou ZH, Fernandez BC, Kickhoefer VA, Rome LH. Polyribosomes Are Molecular 3D Nanoprinters That Orchestrate the Assembly of Vault Particles. Acs Nano. 2014;8:11552–11559. doi: 10.1021/nn504778h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Poderycki MJ, Kickhoefer VA, Kaddis CS, Raval-Fernandes S, Johansson E, Zink JI, Loo JA, Rome LH. The vault exterior shell is a dynamic structure that allows incorporation of vault-associated proteins into its interior. Biochemistry-Us. 2006;45:12184–12193. doi: 10.1021/bi0610552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi XY, Huang X, Li H, Wang YS, Xia Y, Natarajan M, Wei J, Venkatraman SS, Zhang H. Vault Protein-Templated Assemblies of Nanoparticles. Nano. 2012;7 [Google Scholar]
- Querol-Audi J, Casanas A, Uson I, Luque D, Caston JR, Fita I, Verdaguer N. The mechanism of vault opening from the high resolution structure of the N-terminal repeats of MVP. Embo J. 2009;28:3450–3457. doi: 10.1038/emboj.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slesina M, Inman EM, Moore AE, Goldhaber JI, Rome LH, Volknandt W. Movement of vault particles visualized by GFP-tagged major vault protein. Cell Tissue Res. 2006;324:403–410. doi: 10.1007/s00441-006-0158-8. [DOI] [PubMed] [Google Scholar]
- Stephen AG, Raval-Fernandes S, Huynh T, Torres M, Kickhoefer VA, Rome LH. Assembly of vault-like particles in insect cells expressing only the major vault protein. J Biol Chem. 2001;276:23217–23220. doi: 10.1074/jbc.C100226200. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Kato K, Yamashita E, Sumizawa T, Zhou Y, Yao M, Iwasaki K, Yoshimura M, Tsukihara T. The Structure of Rat Liver Vault at 3.5 Angstrom Resolution. Science. 2009;323:384–388. doi: 10.1126/science.1164975. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Tsukihara T. Structural studies of large nucleoprotein particles, vaults. P Jpn Acad B-Phys. 2012;88:416–433. doi: 10.2183/pjab.88.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang OO, Ali A, Kasahara N, Faure-Kumar E, Bae JY, Picker LJ, Park H. Short Conserved Sequences of HIV-1 Are Highly Immunogenic and Shift Immunodominance. J Virol. 2015;89:1195–1204. doi: 10.1128/JVI.02370-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Conformation change of vaults in solution, Related to Figure 3
Data Availability Statement
3D cryoEM density maps have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-7126 (conformation 1), EMD-7125 (conformation 2) and their corresponding coordinates of atomic models have been deposited in the Protein Data Bank under the accession number 6BP8 and 6BP7, respectively. The data that support the findings of this study are available from the corresponding author upon request.