Abstract
To obtain a complete picture of subcellular nanostructures, cells must be imaged with high resolution in all three dimensions (3D). Here, we present tilted light sheet microscopy with 3D point spread functions (TILT3D), an imaging platform that combines a novel, tilted light sheet illumination strategy with engineered long axial range point spread functions (PSFs) for low-background, 3D super localization of single molecules as well as 3D super-resolution imaging in thick cells. TILT3D is built upon a standard inverted microscope and has minimal custom parts. The axial positions of the single molecules are encoded in the shape of the PSF rather than in the position or thickness of the light sheet, and the light sheet can therefore be formed using simple optics. The result is flexible and user-friendly 3D super-resolution imaging with tens of nm localization precision throughout thick mammalian cells. We validated TILT3D for 3D super-resolution imaging in mammalian cells by imaging mitochondria and the full nuclear lamina using the double-helix PSF for single-molecule detection and the recently developed Tetrapod PSF for fiducial bead tracking and live axial drift correction. We envision TILT3D to become an important tool not only for 3D super-resolution imaging, but also for live whole-cell single-particle and single-molecule tracking.
Keywords: Super-resolution microscopy, light sheet illumination, single-molecule imaging, point spread function engineering, 3D imaging
1. INTRODUCTION
The development of imaging techniques beyond the diffraction limit has over the last decades paved the way for detailed studies of nanostructures and molecular mechanisms in biological systems. Several methods have been developed to extend the imaging capability of single-molecule super-resolution microscopy1–3 to three dimensions (3D)4, including multiplane imaging5–8, interferometric approaches9–11, and point spread function (PSF) engineering (for a review, see 4). In this work, we have used PSF engineering, where the shape of the PSF of the microscope is modified to encode information about the axial (z) position of each single emitter directly in its image. This is accomplished by modifying the phase pattern of the emitted light in the Fourier plane of the microscope. PSF engineering typically only requires the addition of a small number of optical elements to a standard microscope, making the method relatively simple to implement while exhibiting high precision for 3D single-molecule localization. This method has been used to create astigmatic PSFs12, 13 and the bisected pupil PSF14 with axial ranges of 1–2 μm, the corkscrew15, self-bending16, and double-helix (DH) PSF17–21 with axial ranges of ~2–3 μm, and the recently developed Tetrapod PSFs22–24 which have a tunable range of up to 20 μm.
Imaging thicker samples, such as mammalian cells, in 3D is challenging due to the increased background and volume to image. Light sheet illumination25 is a method that allows for selective irradiation of the focal plane, and its inherent optical sectioning capability allows for imaging of biological samples with reduced background, photobleaching, and photodamage. Several different light sheet designs have been implemented for single-molecule tracking and super-resolution imaging 26–33, but these designs have drawbacks in certain situations. Some designs are incompatible with imaging of fluorophores close to the coverslip using a high numerical aperture (NA) detection objective. High-NA detection objectives are required in order to increase the photon collection efficiency, since this will improve the localization precision of single molecules. Some designs require dipping of the illumination or the detection objective into the sample medium, which increases the risk of biological and fluorescent contamination. Some previous designs are constructed using complicated optics and electronics, or many custom-made parts. Such designs are often expensive and difficult to build and operate, and thus may not be easily accessible to the general research community.
We alleviate many of these issues by tilting the light sheet illumination plane and detecting the 3D position of single molecules using engineered long axial range PSFs, with a design termed tilted light sheet microscopy with 3D point spread functions (TILT3D) (details can be found in Ref.34). TILT3D yields high localization precision of single molecules in 3D over the entire axial range of a mammalian cell via a stack of light sheet slices combined with imaging with engineered PSFs in each slice. The imaging platform offers the reduced background, photodamage, and photobleaching of light sheet microscopy, while being simple and cost-efficient to construct and operate. The tilt of the light sheet allows for optical sectioning throughout entire adherent mammalian cells cultured on conventional coverslips. Since the axial positions of the single molecules are encoded in the shape of the PSF rather than in the position or thickness of the light sheet, the light sheet need not be extremely thin and can be formed using simple optics. The design allows for use of a high-NA detection objective for imaging of single molecules with high photon efficiency, and there is no need for dipping of objectives into the sample chamber, which reduces the risk of sample contamination. We validated the performance of TILT3D for 3D single-molecule super-resolution imaging of whole mammalian cells by imaging of mitochondria and the entire nuclear lamina using the DH-PSF with 2 μm axial range for single-molecule imaging within each light sheet slice and a Tetrapod PSF with 6 μm axial range for detection of fiducial beads.
2. METHODOLOGY
2.1 Light sheet formation and characterization
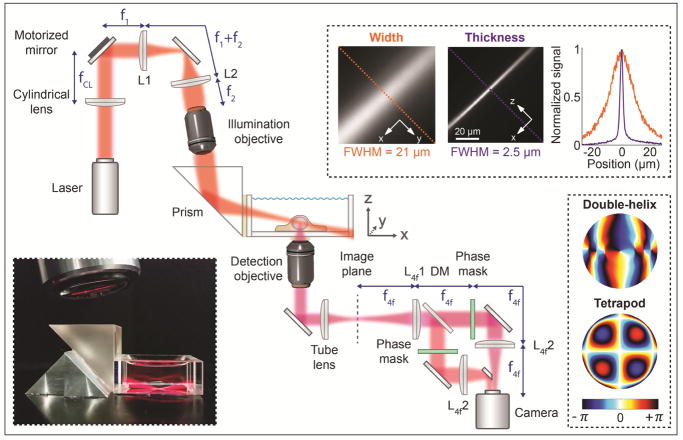
The optical setup was built around a conventional inverted microscope (IX71, Olympus). Illumination lasers (200 mW 561 nm and 120 mW 647 nm, both CW, from Coherent) were spectrally filtered (561 nm: ff01-554/23-25 excitation filter, 647 nm: ff01-631/36-25 excitation filter, both Semrock), circularly polarized (LPVISB050-MP2 polarizers, Thorlabs, and 561 nm: WPQ05M-561 quarter-wave plate, Thorlabs, 647 nm: WPQ05M-633 quarter-wave plate, Thorlabs), and expanded and collimated using lens telescopes. The toggling of the lasers was controlled with shutters (VS14S2T1 with VMM-D3 three-channel driver, Vincent Associates Uniblitz) and synchronized with the detection optics via MATLAB. The 561 nm laser was introduced into the back port of the microscope through a Köhler lens to allow for wide field epi-illumination. The 647 nm laser was either introduced into the epi-illumination pathway or sent to the light sheet illumination pathway; the pathway was easily switched with a flip mirror. The light sheet module was built on an optical bread board and mounted as an add-on to a conventional epi-fluorescence microscope, and the entire light sheet bread board could be translated in xyz using a translation stage (460P, Newport) for easy alignment. The light sheet was formed by a cylindrical lens (LJ1558L2-A, f = 300 mm, Thorlabs, or ACY254-200-A, f = 200 mm, Thorlabs) and focused in one direction onto a motorized mirror (8821 mirror mount with 8742 Picomotor controller, Newport) (Fig. 1). The motorized mirror plane was then relayed by two lenses in 4f-configuration onto the back aperture of a long working distance illumination objective lens (378-803-3, 10x, NA 0.28, Mitutoyo). After the illumination objective, the light sheet was reflected at an angle of about 10° by a glass prism (PS908L-A, Thorlabs) into a glass-walled sample chamber (704-00-20-10, Hellma), and focused at the bottom of the chamber where the sample was positioned on a conventional coverslip (Fisher Premium Cover Glass, no. 1.5). The tilt of the light sheet allowed for illumination of entire adherent cells all the way down to the coverslip, without the need for complicated optics. The long working distance illumination objective in combination with the prism enabled introduction of the light sheet into the sample chamber without dipping of the objective into the sample medium, which reduced the risk of biological and fluorescent contamination. Tilting of the motorized mirror was performed using the New Focus Picomotor Application software (Newport), and allowed for translation of the light sheet both in y and z (using the coordinate system defined in Fig. 1). The resulting light sheet had a width (in y) of 21 μm FWHM, a thickness (in z) of 2.5 μm FWHM, and a confocal parameter of 73 μm (the distance along the direction of beam propagation for which the beam thickness remains within a factor of ). These parameters are well suited for imaging of mammalian cells using the DH-PSF. They can also easily be adjusted for imaging of different samples by adjusting the choice of lenses accordingly.
Figure 1.
Simplified schematic of the optical setup (not to scale). The top right inset shows measured images of the light sheet width and thickness, and a graph with line scans at the corresponding dashed lines in the images. The thickness was measured by rotating the cylindrical lens by 90°. The bottom left inset shows a photo of the light sheet being focused by the illumination objective, reflected by the prism into the glass-walled sample chamber, and focused at the bottom of the chamber where the sample was positioned. The bottom right inset shows the phase masks encoding for the double-helix PSF and the 6-μm axial range Tetrapod PSF. Figure adapted from Ref. 34 with permission.
2.2 3D detection of single molecules and fiducial beads
Light emitted from the sample was collected by a high-NA detection objective (UPLSAPO100XO, x100, NA 1.4, Olympus) mounted on a piezoelectric objective scanner (Nano-F100, C-Focus, Mad City Labs), spectrally filtered (Di01-R405/488/561/635 dichroic, for far red detection: Semrock, ET700/75m bandpass filter, Chroma, ZET647NF notch filter, Chroma, 3RD650LP longpass filter, Omega Optical, and for red detection: ZET647NF notch filter, Chroma, et610/60 bandpass filter, Chroma, and FF01-593/40 bandpass filter, Semrock), and focused by the tube lens. The intermediate image plane was relayed by lenses in 4f-configuration (f = 90 mm, G322389000, Qioptiq) to allow for phase modulation of the light in the Fourier plane of the microscope (Fig. 1). Two spectrally separated channels were created by placing a dichroic mirror (T660lpxrxt, Chroma) after the first 4f-lens, and the two channels were aligned using a D-shaped mirror (BBD1-E02, Thorlabs) before detection with a single EM-CCD camera (iXon3 897, Andor). Transmissive dielectric phase masks of choice were placed in the Fourier plane, one focal length from the first 4f-lens, in one or both channels of the emission pathway.
In this work, we used phase masks encoding for the DH-PSF with a ~2-μm axial range (Double-Helix Optics, LLC) and for a Tetrapod PSF with 6-μm axial range (fabricated at the Stanford Nanofabrication Facility as described in Ref.34). The DH-PSF consists of two lobes that revolves around each other, where the lateral and axial position of the emitter is encoded in the mid-point and in the angle between the two lobes, respectively. The Tetrapod PSF consists of a more complex pattern, roughly tracing out the shape of a tetrapod when envisioned in 3D. Conventional 2D epi- or light sheet imaging was achieved by not inserting any phase masks in the emission channels. Single-slice 3D imaging was achieved by adding the DH phase mask in one of the emission channels, which yielded a useful axial range of about 2 μm. Whole-cell imaging was performed by inserting the DH phase mask in one channel for single-molecule detection and the Tetrapod phase mask in the second channel for fiducial bead detection (see also section 2.3). This versatile design with two channels where the phase masks can be easily changed thus enabled imaging where the axial detection range could be optimized separately for imaging of single molecules and of fiducial beads.
2.3 Interleaved illumination for live axial drift correction
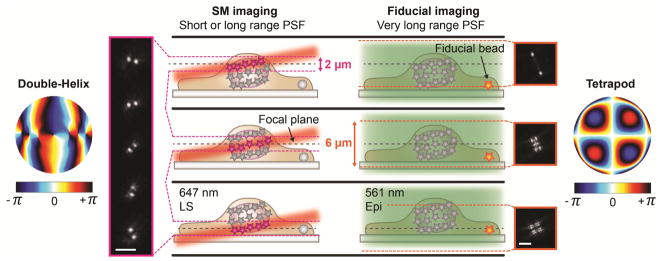
To accurately reconstruct nanoscale features in cells, drift occurring during the course of image acquisition must be measured and corrected for. A common way to achieve this is to image a fiducial bead in the sample. To facilitate fiducial bead detection during whole-cell imaging, we used the 6-μm axial range Tetrapod PSF in the second imaging channel for fiducial bead detection. The illumination and detection was interleaved between 647 nm light sheet illumination for sectioned illumination of single molecules and detection using the DH-PSF, and 561 nm epi-illumination and the Tetrapod PSF for excitation and detection of fiducial beads (Fig. 2). This scheme offered two major advantages: (1) using a PSF that covered the entire axial range of the cell together with epi-illumination allowed for detection of the fiducial bead independent of its axial position in the sample and of the position of the light sheet and the focal plane. (2) The excitation and emission wavelengths of the fiducial bead could be separated from the single-molecule channel. This allowed for optimization of the signal from the bead, which resulted in good localization precision, without the risk of saturating the single-molecule channel with light emitted from the fiducial bead. The position of the fiducial bead was analyzed in real time and potential axial drift was corrected for in real time using a piezoelectric objective scanner. Lateral drift was corrected during post-processing, but could in principle have been corrected in real time by using a translation stage. This interleaved illumination scheme was implemented for imaging of the entire nuclear lamina (see section 3.2).
Figure 2.
Schematic showing the interleaved illumination scheme used for whole-cell imaging with live axial drift correction. For single-molecule imaging, the focal plane was moved together with the light sheet in 1 μm steps to sample the entire cell in thick, overlapping slices. The 3D position of the individual molecules were detected using the double-helix PSF, which has an axial range of about 2 μm. Imaging of the single molecules was interleaved with imaging of fiducial beads every twentieth frame. Here, the fiducial bead was excited at a different wavelength than the single molecules using epi-illumination, and detected using a 6-μm range Tetrapod PSF. This allowed for excitation and detection of the fiducial bead independent of the position of the bead in the sample and of the position the light sheet and the focal plane. The position of the fiducial bead was analyzed and potential axial drift was corrected in real time. Figure adapted from Ref. 34 with permission.
2.4 Sample preparation
HeLa cells (CCL-2, ATCC) were cultured at 37°C and 5% CO2 in high-glucose Dulbecco’s modified Eagle’s medium (DMEM, HyClone) supplemented with 10% (v/v) FBS (HyClone). Two days before imaging, cultured cells were plated onto plasma-etched coverslips (Fisher Premium Cover Glass, no. 1.5) spun coat with a 1% (w/v) polyvinyl alcohol (PVA, Polysciences Inc.) layer containing far red (mitochondria) (625/645 nm, F8806, Invitrogen) or red (lamin B1) (580/605 nm, F8810, Invitrogen) fluorescent microspheres, cultured for 24 hours in high-glucose DMEM supplemented with 10% FBS, and subsequently cultured for 24 hours in high-glucose, phenol-red free DMEM (HyClone) supplemented with 10% FBS. During this period, some of the microspheres were endocytosed.
For immunolabeling of mitochondria, the cells were washed two times in PBS (HyClone), fixed in chilled 4% paraformaldehyde (PFA) (Electron Microscopy Sciences) in PBS for 20 min, washed once in PBS, and incubated with 10 mM NH4Cl (Sigma-Aldrich) in PBS for 10 min. Next, the cells were permeabilized with three washing steps with 0.2% (v/v) Triton X-100 (Sigma-Aldrich) in PBS with 5 min incubation between each wash, and blocked with 3% (w/v) BSA (Sigma-Aldrich) in 0.1% (v/v) Triton X-100 in PBS for 1 hour. The cells were labeled with Alexa Fluor 647-conjugated primary rabbit anti-TOMM20 (ab209606, Abcam) using a 1:100 dilution in 1% (w/v) BSA in 0.1% (v/v) Triton X-100 in PBS for 2 hours. The cells were then washed five times with 0.1% (v/v) Triton X-100 in PBS with 3 min incubation between each wash, post-fixed in 4% PFA in PBS for 5 min, and washed three times in PBS.
For immunolabeling of lamin B1, the cells were washed three times in PBS, fixed in chilled 4% PFA in PBS for 20 min, washed once in PBS, and incubated with 10 mM NH4Cl in PBS for 10 minutes. Next, the cells were permeabilized with three washing steps with 0.2% (v/v) Triton X-100 in PBS with 5 min incubation between each wash, and blocked with 3% (w/v) BSA in PBS for 1 hour. The cells were then labeled with primary rabbit anti-lamin B1 (ab16048, Abcam) using a 1:1000 dilution in 1% (w/v) BSA in PBS for 2 hours, washed three times with 0.1% (v/v) Triton X-100 in PBS with 3 min incubation between each wash, and labeled with secondary donkey anti-rabbit conjugated with Alexa Fluor 647 (ab150067, Abcam) using a 1:1000 dilution in 1% (w/v) BSA in PBS for 1 hour. The cells were finally washed five times with 0.1% (v/v) Triton X-100 in PBS with 1 min incubation between each wash.
Immunolabeling steps were performed with coverslips placed on parafilm, while all other steps were performed with coverslips placed in 6-well plates. After the immunolabeling, all samples were protected from light and stored in PBS at 4°C. The samples were imaged within 48 hours of labeling.
Before imaging, the sample coverslips were mounted on the bottom of the four transparent walls of a sliced commercial glass cuvette (704-000-20-10, Hellma) using two-part silicone rubber (Ecoflex® 5, Reynolds Advanced Materials). During imaging, the sample chamber could be left open on the top to allow for easy sample access, or it could be easily sealed with a second coverslip placed on top of the four walls and attached with silicone rubber. This reduced the risk of sample contamination, decreased the rate of evaporation of the medium, and limited the access of oxygen to the sample. The glass surfaces between the coverslip and the imaging objective, and between the sample chamber and the glass prism, were brought into optical contact with immersion oil (Zeiss Immersol 518 F, n = 1.518). The glass prism used for reflecting the light sheet into the sample chamber was attached to the microscope stage with silicone rubber using a custom-made right triangular aluminum prism for support (Fig. 1). The aluminum prism was hollow at the back of the glass prism to allow for total internal reflection of the light sheet.
2.5 Imaging methodology
For diffraction-limited imaging, cells were imaged in PBS using ~1 W/cm2 647 nm excitation. Comparisons between light sheet and epi-illumination were performed by manually switching illumination light paths using a flip mirror during image acquisition.
For single-molecule imaging, the PBS was replaced by a reducing and oxygen-scavenging buffer35 comprising 100 mM Tris-HCl (Invitrogen), 10% (w/v) glucose (BD Difco), 560 μg/ml glucose oxidase (Sigma-Aldrich), 2 μl/ml catalase (Sigma-Aldrich), and cysteamine (Sigma-Aldrich) with a concentration of 20 mM (mitochondria) or 40 mM (lamin B1). At the beginning of the measurement, a large fraction of the Alexa Fluor 647 molecules were converted into a dark state using 647 nm epi-illumination at 5 kW/cm2. For all single-molecule measurements, an exposure time of 50 ms and a calibrated EM gain of 186 was used. For each imaging experiment, at least 300 dark frames were acquired when the shutter of the camera was closed. The mean of these dark frames was subtracted from the images before analysis. A calibration of each PSF was carried out by axial scanning of the fiducial beads in the sample over the full axial range of the PSF using the piezoelectric objective scanner.
For 3D single-molecule super-resolution imaging of mitochondria, ~25 kW/cm2 647 nm light sheet illumination was used for imaging of single Alexa Fluor 647 molecules and of far red fiducial beads using the DH-PSF. This scheme was very easy to implement and requires no scripts to control the setup and only a single laser for illumination.
Acquisition of 3D super-resolution images of lamin B1 in HeLa cells was performed using interleaved illumination and live axial drift correction, as described in section 2.3. The DH-PSF was used together with ~25 kW/cm2 647 nm light sheet illumination for imaging of single Alexa Fluor 647 molecules in the far red channel, while for every twentieth frame fiducial beads were imaged using the Tetrapod PSF and 10 W/cm2 561 nm epi-illumination in the red channel. Several slices were aqcuired and the focal plane was moved together with the light sheet in 1 μm steps to sample the entire nuclear lamina. The positions of the fiducial beads, which were detectable in all slices owing to the very long axial range of the Tetrapod PSF, were used during post-processing to correct lateral and axial drift within each slice and to stitch together the different slices. The final z-position of each slice was corrected using cross-correlation between two adjacent slices.
2.6 Data analysis
Calibration and fitting analysis of DH and Tetrapod images were performed using modified versions of the open-source software packages Easy-DHPSF36 (https://sourceforge.net/projects/easy-dhpsf/) and Easy-Pupil-Finder37 (https://sourceforge.net/projects/easy-pupil-finder/), respectively. Detailed description of the calibration and analysis methodology, and of the post-processing filtering, can be found in Ref.34. In brief, when analyzing images of the DHPSF, the two lobes of each PSF were fitted using non-linear least-squares functions in MATLAB with a pair of identical, radially symmetric 2D Gaussians as the objective function. When localizing the Tetrapod PSF data, an imaging model based on the Gibson-Lanni scalar diffraction approximation for mismatched refractive indices38 was used. Here, the PSF was calculated from the pupil plane electric field, and aberrations in the PSF caused by the optical setup were corrected for using phase retrieval37. All localization using the phase-retrieved PSF model was done in MATLAB using a maximum-likelihood estimation routine assuming a Poisson noise model.
3. RESULTS
3.1 Contrast and localization precision improvement by light sheet illumination
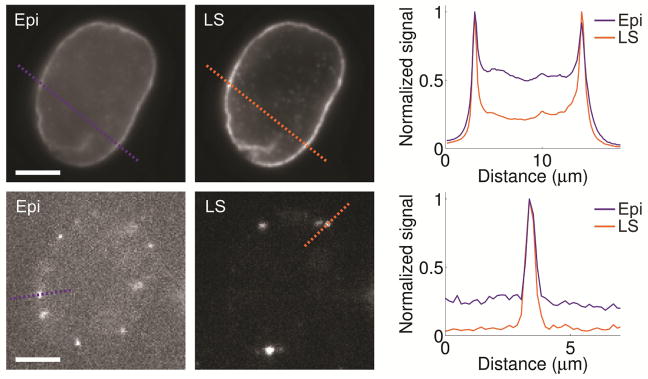
The contrast improvement by light sheet illumination was measured by imaging of lamin B1 in HeLa cells immunolabeled with Alexa Fluor 647. Diffraction limited and single-molecule imaging using light sheet illumination yielded an up to five-fold improvement in the signal-to-background ratio relative to conventional epi-illumination (Fig. 3). This resulted in an improved localization precision for single-molecules from 23 nm to 16 nm in xy and from 35 nm to 24 nm in z when switching from epi- to light sheet illumination using the DH-PSF, as demonstrated for imaging of mitochondria.
Figure 3.
Comparison between epi-illumination (Epi) and light sheet illumination (LS) for diffraction limited bulk (top row) and single-molecule (bottom row) imaging of lamin B1 in a HeLa cell immunolabeled with Alexa Fluor 647. The graphs show line scans at the dashed lines in the images, demonstrating the improved contrast when using light sheet illumination. Compared images are shown with the same linear grayscale, respectively. Scale bars are 5 μm. Figure adapted from Ref. 34 with permission.
3.2 3D super-resolution imaging of mitochondria and the nuclear lamina
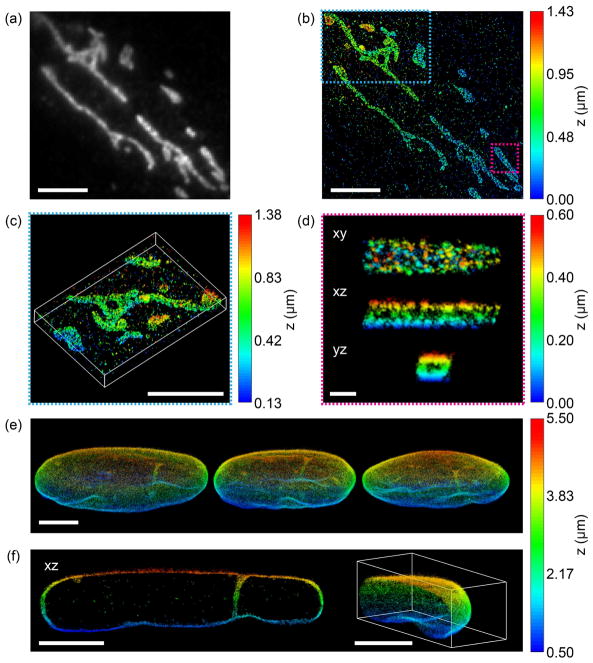
The performance of TILT3D for 3D single-molecule super-resolution imaging was first demonstrated by imaging of mitochondria in HeLa cells. The DH-PSF was used to image both single molecules and a fiducial bead over a 2 μm range using a single position of the light sheet and of the focal plane (Fig. 4a–c). This resulted in a localization precision for single molecules of 13 nm in xy and 20 nm in z. Cross sections of individual mitochondria revealed the hollow structure of the mitochondrial outer membrane (Fig. 4d).
Figure 4.
Performance of TILT3D for 3D single-molecule super-resolution imaging in mammalian cells. (a) 2D diffraction limited image of mitochondria in a HeLa cell immunolabeled with Alexa Fluor 647. (b) 3D super-resolution reconstruction of the sample shown in (a) acquired using the double-helix PSF. (c) 3D view of the mitochondria shown in the blue dashed rectangle in (b). (d) xy, xz, and yz views of a single mitochondrion shown in the magenta rectangle in (b). (e) Three different views of a 3D super-resolution reconstruction of the entire nuclear lamina in a HeLa cell. Here, lamin B1 was immunolabeled with Alexa Fluor 647. Single molecules were detected in several thick, overlapping slices using the double-helix PSF, while fiducial beads were detected using the 6-μm Tetrapod PSF. (f) The xz view shows a 1.3-μm thick slice through the cell in the reconstruction in (e), where lamin meshwork enveloping an intranuclear channel is visible. The right panel shows a cap of the reconstruction in (e). Scale bars are 500 nm in (d) and 5 μm in all other panels. Figure adapted from Ref. 34 with permission.
Next, TILT3D was benchmarked for 3D single-molecule super-resolution imaging of whole mammalian cells by imaging of the entire nuclear lamina in HeLa cells (Fig. 4e, f). Single molecules were detected using the DH-PSH, while fiducial beads were detected using the 6-μm Tetrapod PSF. The light sheet was translated axially together with the focal plane in 1 μm steps to sample the entire lamina in thick, overlapping slices. The very long axial range of the Tetrapod PSF allowed the fiducial beads to be detected and localized in all slices. At 3.3 μm above the coverslip, the localization precision was estimated to 16 nm in xy and 23 nm in z for single-molecule detection using the DH-PSF, and 3 nm in xy and 7 nm in z for fiducial bead detection using the 6-μm Tetrapod PSF. The thickness of the lamina at the bottom and the top of the cell was measured to be 113 nm and 101 nm (FWHM), respectively. An xz view reveals the lamina meshwork enveloping an intranuclear lamina channel (Fig. 4f).
4. CONCLUSIONS
We have developed TILT3D, a simple and versatile platform that combines a tilted light sheet with PSF engineering for 3D single-molecule super-resolution imaging of thick mammalian cells. TILT3D can be added to any conventional epi-fluorescence microscope, and it allows for imaging using a high-NA detection objective. The nanoscale 3D positions of single molecules are determined from the shape of the PSFs, rather than from the thickness or position of the light sheet. This allows for creation of the light sheet using very simple optics. Using a tilted light sheet for illumination enables excitation of single molecules throughout entire adherent cells cultured on conventional coverslips. The long working distance illumination objective allows for introduction of the light sheet without a need for dipping of the objective into the sample chamber, which reduces the risk of biological and fluorescent contamination. The inherent optical sectioning capability of the light sheet reduces background from out-of-focus fluorophores, photobleaching in regions of the sample not currently imaged, and the risk of photodamage from excessive illumination of the sample. The reduced background, in combination with the use of a high-NA detection objective, improves the precision of the single-molecule localizations. The dimensions of the light sheet and the axial range of the detection PSFs can conveniently be adjusted for the sample of interest by the choice of lenses and phase masks used. TILT3D is thus both versatile, simple, and user-friendly in its design and implementation, and we think that TILT3D in the future will become an important and wide-spread tool for both 3D super-resolution imaging and single-molecule tracking in whole mammalian cells.
Acknowledgments
This work was supported in part by the National Institute of Biomedical Imaging and Bioengineering (Grant No. U01EB021237) to W.E.M. and by the National Institute of General Medical Sciences (Grant No. R35GM118067) to W.E.M. A.-K.G. acknowledges partial financial support from the Swedish Research Council (Grant No. 2016-00130), and from the Foundation BLANCEFLOR Boncompagni-Ludovisi, née Bildt. M.Y.L. is supported by a National Science Scholarship (PhD) from A*STAR, Singapore. Y.S. is supported in part by a Career Advancement Chairship from the Technion. The authors also thank Dr. Carl G. Ebeling, Worldwide Application Scientist for Bruker Fluorescence Microscopy, for his support and for the use of the Bruker SRX visualization and analysis software for rendering localization data, and Dr. Steffen Sahl for early discussions. Fabrication of dielectric phase masks was performed at the Stanford Nanofabrication Facility, which is supported by the National Science Foundation as part of the National Nanotechnology Coordinated Infrastructure under award ECCS-1542152.
References
- 1.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313(5793):1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 2.Hess ST, Girirajan TPK, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91(11):4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3(10):793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Diezmann A, Shechtman Y, Moerner WE. Three-Dimensional Localization of Single Molecules for Super-Resolution Imaging and Single-Particle Tracking. Chem Rev. 2017;117(11):7244. doi: 10.1021/acs.chemrev.6b00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ram S, Prabhat P, Ward ES, Ober RJ. Improved single particle localization accuracy with dual objective multifocal plane microscopy. Opt Express. 2009;17(8):6881–6898. doi: 10.1364/oe.17.006881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juette MF, Gould TJ, Lessard MD, Mlodzianoski MJ, Nagpure BS, Bennett BT, Hess ST, Bewersdorf J. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat Methods. 2008;5(6):527–529. doi: 10.1038/nmeth.1211. [DOI] [PubMed] [Google Scholar]
- 7.Abrahamsson S, Chen J, Hajj B, Stallinga S, Katsov AY, Wisniewski J, Mizuguchi G, Soule P, Mueller F, Darzacq CD, Darzacq X, Wu C, Bargmann CI, Agard DA, Dahan M, Gustafsson MGL. Fast multicolor 3D imaging using aberration-corrected multifocus microscopy. Nat Methods. 2013;10(1):60–63. doi: 10.1038/nmeth.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hajj B, Wisniewski J, Beheiry ME, Chen J, Ravyakin A, Wu C, Dahan M. Whole-cell, multicolor super resolution imaging using volumetric multi focus microscopy. Proc Natl Acad Sci U S A. 2014;111:17480–17485. doi: 10.1073/pnas.1412396111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang F, Sirinakis G, Allgeyer E, Schroeder L, Duim W, Kromann E, Phan T, Rivera-Molina F, Myers J, Irnov I, Lessard M, Zhang Y, Handel M, Jacobs-Wagner C, Lusk C, Rothman J, Toomre D, Booth M, Bewersdorf J. Ultra-High Resolution 3D Imaging of Whole Cells. Cell. 2016;166(4):1028–1040. doi: 10.1016/j.cell.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM, Manley S, Sougrat R, Waterman CM, Kanchanawong P, Davidson MW, Fetter RD, Hess HF. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci U S A. 2009;106(9):3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao X, Antonello J, Allgeyer ES, Bewersdorf J, Booth M. Aberrations in 4Pi Microscopy. Opt Express. 2017;25:14049–14058. doi: 10.1364/OE.25.014049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang B, Jones SA, Brandenburg B, Zhuang X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat Methods. 2008;5(12):1047–1052. doi: 10.1038/nmeth.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jungmann R, Avendano MS, Woehrstein JB, Dai M, Shih WM, Yin P. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat Methods. 2014;11:313–318. doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Backer AS, Backlund MP, von Diezmann AR, Sahl SJ, Moerner WE. A bisected pupil for studying single-molecule orientational dynamics and its application to 3D super-resolution microscopy. Appl Phys Lett. 2014;104(19):193701–193705. doi: 10.1063/1.4876440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lew MD, Lee SF, Thompson MA, Lee HD, Moerner WE. Single-Molecule Photocontrol and Nanoscopy. Springer; Berlin Heidelberg: 2012. pp. 1–24. [Google Scholar]
- 16.Jia S, Vaughan JC, Zhuang X. Isotropic three-dimensional super-resolution imaging with a self-bending point spread function. Nat Photonics. 2014;8:302–306. doi: 10.1038/nphoton.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pavani SRP, Thompson MA, Biteen JS, Lord SJ, Liu N, Twieg RJ, Piestun R, Moerner WE. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc Natl Acad Sci U S A. 2009;106(9):2995–2999. doi: 10.1073/pnas.0900245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson MA, Lew MD, Badieirostami M, Moerner WE. Localizing and tracking single nanoscale emitters in three dimensions with high spatiotemporal resolution using a double-helix point spread function. Nano Lett. 2010;10(1):211–218. doi: 10.1021/nl903295p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lew MD, Lee SF, Ptacin JL, Lee MK, Twieg RJ, Shapiro L, Moerner WE. Three-dimensional superresolution colocalization of intracellular protein superstructures and the cell surface in live Caulobacter crescentus. Proc Natl Acad Sci U S A. 2011;108(46):E1102–E1110. doi: 10.1073/pnas.1114444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu B, Yu J, Li W, Cao B, Li H, Chen D, Niu H. Nanoscale three-dimensional single particle tracking by light-sheet-based double-helix point spread function microscopy. Appl Opt. 2016;55(3):449–453. doi: 10.1364/AO.55.000449. [DOI] [PubMed] [Google Scholar]
- 21.Carr AR, Ponjavic A, Basu S, McColl J, Santos AM, Davis S, Laue ED, Klenerman D, Lee SF. Three-Dimensional Super-Resolution in Eukaryotic Cells Using the Double-Helix Point Spread Function. Biophys J. 2017;112:1444–1454. doi: 10.1016/j.bpj.2017.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shechtman Y, Sahl SJ, Backer AS, Moerner WE. Optimal Point Spread Function Design for 3D Imaging. Phys Rev Lett. 2014;113(13):133902. doi: 10.1103/PhysRevLett.113.133902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shechtman Y, Weiss LE, Backer AS, Sahl SJ, Moerner WE. Precise 3D scan-free multiple-particle tracking over large axial ranges with Tetrapod point spread functions. Nano Lett. 2015;15(6):4194–4199. doi: 10.1021/acs.nanolett.5b01396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shechtman Y, Gustavsson A-K, Petrov PN, Dultz E, Lee MY, Weis K, Moerner WE. Observation of live chromatin dynamics in cells via 3D localization microscopy using Tetrapod point spread functions. Biomed Opt Express. 2017;8(12):5735–5748. doi: 10.1364/BOE.8.005735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305(5686):1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 26.Ritter JG, Veith R, Veenendaal A, Siebrasse JP, Kubitscheck U. Light sheet microscopy for single molecule tracking in living tissue. PLoS ONE. 2010;5(7):e11639. doi: 10.1371/journal.pone.0011639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods. 2008;5(2):159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 28.Zanacchi CF, Lavagnino Z, Donnorso MP, Del Bue A, Furia L, Faretta M, Diaspro A. Live-cell 3D super-resolution imaging in thick biological samples. Nat Methods. 2011;8:1047–1049. doi: 10.1038/nmeth.1744. [DOI] [PubMed] [Google Scholar]
- 29.Gebhardt JC, Suter DM, Roy R, Zhao ZW, Chapman AR, Basu S, Maniatis T, Xie XS. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat Methods. 2013;10(5):421–426. doi: 10.1038/nmeth.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Y, Zhu Q, Elkins K, Tse K, Li Y, Fitzpatrick J, Verma I, Cang H. Light-sheet Bayesian microscopy enables deep-cell super-resolution imaging of heterochromatin in live human embryonic stem cells. Optical Nanoscopy. 2013;2(1):7. doi: 10.1186/2192-2853-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA, 3rd, Liu Z, English BP, Mimori-Kiyosue Y, Romero DP, Ritter AT, Lippincott-Schwartz J, Fritz-Laylin L, Mullins RD, Mitchell DM, Bembenek JN, Reymann AC, Bohme R, Grill SW, Wang JT, Seydoux G, Tulu US, Kiehart DP, Betzig E. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346(6208):1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galland R, Grenci G, Aravind A, Viasnoff V, Studer V, Sibarita J. 3D high- and super-resolution imaging using single-objective SPIM. Nat Methods. 2015;12(7):641–644. doi: 10.1038/nmeth.3402. [DOI] [PubMed] [Google Scholar]
- 33.Meddens MM, Liu S, Finnegan PS, Edwards TL, James CD, Lidke KA. Single objective light-sheet microscopy for high-speed whole-cell 3D super-resolution. Biomed Opt Express. 2016;7:2219–2236. doi: 10.1364/BOE.7.002219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gustavsson A-K, Petrov PN, Lee MY, Shechtman Y, Moerner WE. 3D single-molecule super-resolution microscopy with a tilted light sheet. Nat Commun. 2018;9(123):1–8. doi: 10.1038/s41467-017-02563-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halpern AR, Howard MD, Vaughan JC. Point by Point: An Introductory Guide to Sample Preparation for Single-Molecule, Super-Resolution Fluorescence Microscopy. Curr Protoc Chem Biol. 2015;7:103–120. doi: 10.1002/9780470559277.ch140241. [DOI] [PubMed] [Google Scholar]
- 36.Lew MD, von Diezmann ARS, Moerner WE. Easy-DHPSF open-source software for three-dimensional localization of single molecules with precision beyond the optical diffraction limit. Protocol Exchange. 2013;026 [Google Scholar]
- 37.Petrov PN, Shechtman Y, Moerner WE. Measurement-based estimation of global pupil functions in 3D localization microscopy. Opt Express. 2017;25(7):7945–7959. doi: 10.1364/OE.25.007945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibson SF, Lanni F. Experimental test of an analytical model of aberration in an oil-immersion objective lens used in three-dimensional light microscopy. J Opt Soc Am A. 1991;8:1601–1613. doi: 10.1364/josaa.9.000154. [DOI] [PubMed] [Google Scholar]