Abstract
Background
We previously observed that high serum 25-hydroxyvitamin D [25(OH)D] (>38.0 ng/mL) was inversely associated with breast cancer. Here, we examined effect modification by single nucleotide polymorphisms (SNPs) in vitamin D-related genes.
Methods
The Sister Study enrolled 50,884 US women who had a sister with breast cancer, but who had never had breast cancer themselves. Using a case-cohort design, we compared 1,524 women who developed breast cancer within 5 years to 1,810 randomly selected participants. We estimated ratios of hazards ratios (RHRs) for the 25(OH)D-breast cancer association per copy of the minor allele using Cox proportional hazards models. We considered 82 SNPs in 7 vitamin D-related genes (CYP24A1, CYP27B1, CYP2R1, GC, DHCR7/NADSYN1, RXRA, and VDR). We also tested gene-based interactions with 25(OH)D.
Results
The SNP with the smallest interaction p-value was rs4328262 in VDR (p=0.0008); the 25(OH)D hazard ratio (HR) was 0.92 (95% confidence interval [CI]: 0.68–1.24) among those homozygous for the common allele, and the minor allele was estimated to decrease the HR by 33% per copy (RHR=0.67, 95% confidence interval [CI]=0.53–0.85). Five other VDR SNPs showed evidence of interaction at p<0.05, as did one SNP in CYP2R1 and one in RXRA. As a group, the 82 SNPs showed evidence of multiplicative interaction with 25(OH)D (p=0.04). In gene-based tests, only VDR showed strong evidence of interaction (p=0.04).
Conclusions
SNPs in vitamin D-related genes may modify the association between serum 25(OH)D and breast cancer.
Impact
This work strengthens the evidence for protective effects of vitamin D.
Keywords: Breast cancer, vitamin D, 25-hydroxyvitamin D, single nucleotide polymorphisms, gene-by-environment interaction
Introduction
Vitamin D is a prohormone with known anti-carcinogenic properties, including the ability to regulate cell growth and proliferation, stimulate apoptosis, and bolster immune response. Both vitamin D2 and D3 can be acquired through dietary sources and supplements, but most vitamin D3 is produced by a reaction between ultraviolet B radiation and cutaneous 7-dehydrocholesterol (1). D2 and D3 are subsequently metabolized into 25-hydroxyvitamin D [25(OH)D] by the liver. 25(OH)D is then converted to the biologically active form of vitamin D, 1,25-dihydroxyvitamin D [1,25(OH)2D], by the kidney (1). This conversion also occurs in other tissues, including the breast. Excess 25(OH)D and 1,25(OH)2D are catabolized into a biologically inactive form, calcitroic acid [24,25(OH)2D], and excreted (2).
Genes directly involved in this metabolic process include DHCR7, CYP24A1, CYP27B1, CYP2R1, GC, VDR, and RXRA. DHCR7 encodes 7-dehydrocholesterol reductase, an enzyme that converts 7-dehydrocholesterol to cholesterol. Cytochrome P450 enzymes facilitate each of the three metabolic conversions: D2 or D3 to 25(OH)D (CYP2R1), 25(OH)D to 1,25(OH)2D (CYP27B1) and 25(OH)D or 1,25(OH)2D into 24,25(OH)2D (CYP24A1).
While circulating in serum, 25(OH)D and 1,25(OH)2D are usually bound to vitamin D binding proteins or, to a lesser degree, albumin (3). Vitamin D binding proteins are transcribed from the GC gene. Unbound 1,25(OH)2D is considered “bioavailable” and is free to attach to a vitamin D receptor (VDR), encoded by the VDR gene. VDR plays a vital role in the regulation of many genes with diverse functions, as it can activate transcription at binding sites throughout the genome (4, 5). VDR can bind to these vitamin D response elements as a homodimer, but in the presence of 1,25(OH)2D it preferentially forms heterodimers with retinoid X receptors (the gene product of RXRA) (6), which may affect its affinity for certain sites.
Most previous studies of gene-by-environment interaction for vitamin D and breast cancer have focused on SNPs in VDR with putative functional roles. Primary candidates have included rs2228570 (also known as FokI), rs731236 (TaqI), and rs1544410 (BsmI). Some of these studies observed evidence of multiplicative interactions between these SNPs and vitamin D (measured as 25(OH)D, dietary intake, or sunlight exposure) on breast cancer risk (7–11), but others observed no noteworthy interactions for these or other vitamin D-related gene variants (12–18).
In a recent prospective observational study of vitamin D and incident breast cancer, we reported that women with 25(OH)D levels in the highest quartile (>38.0 ng/mL) had a reduced risk of breast cancer over the subsequent 5 years, relative to women with 25(OH)D levels in the lowest quartile (≤24.6 ng/mL; adjusted HR=0.79; CI: 0.63–0.98) (19). The association may be limited to post-menopausal women. Our study was unique in that we examined prospectively-measured, recent vitamin D levels and used liquid chromatography-mass spectrometry to measure total 25(OH)D [25(OH)D3 + 25(OH)D2 + 3-epi-25(OH)D3]. Although many longer-term prospective cohort studies have reported null findings (8, 20–29), some have reported modestly protective (though often not statistically significant) associations (30–35), and a recent meta-analysis of prospective studies reported a protective association between plasma vitamin D levels and breast cancer in analyses limited to post-menopausal women (36). Further, many previous case-control studies have reported strong and statistically-significant inverse associations of similar magnitude to our reported results (37–41). This epidemiologic evidence suggests that recent 25(OH)D levels are associated with reduced risk of post-menopausal breast cancer.
Here we explore how inherited genetic variants may affect individual responses to vitamin D and modify the association between 25(OH)D and breast cancer risk. Identification of such factors would help us understand the biological mechanisms behind vitamin D’s protective effects and identify individuals with altered susceptibility to those effects. We are particularly interested in the influence of single nucleotide polymorphisms (SNPs) in genes involved in vitamin D metabolism. For our main analysis we considered the vitamin D metabolism genes selected a priori and discussed here – DHCR7 (and the adjacent gene, NADSYN1), CYP24A1, CYP27B1, CYP2R1, GC, VDR, and RXRA – but in secondary exploratory analyses we also examined 1,439 SNPs in 89 other potentially related genes, including those selected from previously published breast cancer gene-by-environment interaction or vitamin D genome-wide association studies (13, 42, 43), those involved in other vitamin D-related pathways (44–46), and genes regulated through vitamin D response elements (47).
Materials and Methods
To explore how SNPs in vitamin D-related genes might modify the association between total 25(OH)D and incident breast cancer, we used data from the Sister Study (data release 4.1, updated 7/2014), a prospective cohort of 50,884 women who had a sister with breast cancer, but had never had breast cancer themselves at enrollment (2003–2009). To be eligible, participants had to be between 35 and 74 years old and reside in the United States or Puerto Rico. The Sister Study was approved by the institutional review boards of the National Institute of Environmental Health Sciences and the Copernicus Group.
All Sister Study participants completed a computer-assisted telephone interview at baseline and were visited by trained examiners who obtained written informed consent and took measurements and blood samples. Participants are regularly asked to complete annual follow-up questionnaires, and those who go on to develop breast cancer are then asked to provide detailed information about their diagnosis. We obtained information about breast cancers from medical records, whenever possible, but otherwise relied on patient self-report. To date, we have retrieved medical records for 82% of self-reported breast cancer cases, 99% of whom were confirmed as true cases. Because the false self-report rate is so low, we consider all self-reported cases to be true cases.
We used a case-cohort design to investigate the association between serum 25(OH)D and incident breast cancer. Cases were Sister Study participants diagnosed with invasive breast cancer or ductal carcinoma in situ within 5 years of their baseline interview (n=1611). The comparison group was 1,843 women randomly selected from the Sister Study cohort (68 of whom were included among the 1,611 because they had developed breast cancer within 5 years of baseline).
Serum 25(OH)D assessment
As described previously (19), we used liquid chromatography-mass spectrometry to measure each of three vitamin D metabolites – 25(OH)D3, 25(OH)D2, and 3-epi-25(OH)D3. The concentration sum of these metabolites was used to estimate overall available serum vitamin D. 25(OH)D3 was the most prevalent metabolite, making up approximately 83% of the total.
We adjusted the total 25(OH)D values for time of year of blood collection using loess regression (48), allowing seasonal effects to vary by race/ethnicity, supplement use, and latitude. We also adjusted for assay batch effects using a random effects model. Based on our previous findings that 38.0 ng/mL was a risk-related cut-point for breast cancer, we categorized women based on whether their season and batch-adjusted total 25(OH)D levels were above or below this threshold.
Genotype analysis
The vitamin D sub-study was nested within a slightly larger case-cohort sample that was originally selected for genotype analysis. Genotyping of a subset of Sister Study participants was conducted using Illumina’s Infinium OncoArray-500K beadchip platform via participation in the GAME-ON consortium (49). The OncoArray panel includes a genome-wide backbone of 230,000 tag SNPs. The remaining SNPs were selected because they were: shown to be associated with breast, colorectal, lung, prostate or ovarian cancer; located in fine-mapped regions around those variants; used to assess ancestry or quantitative traits; or because they had a putative functional role or association with important functions, such as DNA repair.
Data cleaning and quality control filtering were conducted by the GAME-ON consortium, with further details described elsewhere (49). After these exclusions, 3,363 Sister Study participants (1829 in sub-cohort [including 67 cases] and 1534 additional cases) provided both genotype and 25(OH)D data. The final analyses sample consisted of 1810 random sub-cohort members (including 66 cases) and 1524 additional cases who had complete data for the key covariates, listed below.
For this specific candidate gene study, we identified SNPs located within 2000 base pairs of the gene transcription start and termination sites, as defined by University of California Santa Cruz Genome Browser (50) (GRCh37/hg19; RefSeq notation). We then excluded SNPs with minor allele frequencies less than 2% and one of each pair of SNPs that were in very high linkage disequilibrium (r2>0.95) with one another. No exclusions were made based on Hardy-Weinberg equilibrium. In total, we evaluated the gene-by-environment interaction effect estimates of 82 SNPs in 7 genes (35 in VDR, 16 in RXRA, 4 in CYP2R1, 14 in CYP24A1, 8 in GC, 4 in DHCR7/NADSYN1, and 1 in CYP27B1).
Ancestry proportions for all Sister Study participants were defined using 2,294 ancestry informative markers (AIMs) selected by the GAME-ON consortium. Genotypes for these markers were compared to those from three HapMap populations (CEU, YRI and CHB) using Bayesian clustering methods, allowing for admixture. Using STRUCTURE (v2.3.4), we ran the clustering analysis for 35,000 iterations, discarding the first 10,000 as burn-in. We included proportion CEU ancestry and proportion YRI ancestry as covariates in all models, as well as self-reported race/ethnicity. As a sensitivity analysis, we adjusted the multivariate models for the first 10 principal components, defined using the selected AIMs, rather than ancestry. The results were nearly identical and we have not included them here.
Statistical Analysis
We used Cox proportional hazards models, with age as the primary time scale, to evaluate the joint association between total 25(OH)D and each candidate SNP on the risk of breast cancer. The model included terms for the main effect of vitamin D exposure (dichotomous: total 25(OH)D levels >38.0 ng/mL versus ≤38.0 ng/mL), the main effects of each genotype (assuming a co-dominant model, so that the main SNP effects are saturated) and a 25(OH)D-by-SNP interaction term (with genotype coded as number of copies of the minor allele). The primary effect estimate of interest was the exponentiated beta coefficient for the multiplicative interaction term, which can be interpreted as the ratio of hazard ratios (RHR), or the relative increase in the hazard ratio (HR) for the 25(OH)D-breast cancer association per copy of the minor allele. We calculated 95% confidence intervals (CIs) for this parameter using robust variance estimators to account for the case-cohort design (51, 52).
Models were adjusted for the following covariates, as determined at baseline and selected a priori: ancestry (proportion CEU and YRI), self-reported race/ethnicity (categorical), education (categorical), menopausal status (pre or postmenopausal), physical activity during the preceding year (categorical), current hormone therapy use (none, estrogen plus progestin, or unopposed estrogen), current hormonal birth control use (yes/no), history of osteoporosis (yes/no), BMI (continuous), alcohol consumption in the previous year (never/former drinker, current drinker <1 drink/day, current drinker ≥1 drink/day), parity (0, 1, 2, or ≥3 births) and a BMI-by-menopausal-status interaction term. Women were considered post-menopausal if they had experienced natural menopause and not had a menstrual period within the last year (63% of post-menopausal women), had previously had both ovaries removed (22%), or had had a hysterectomy with retained ovaries but were older than 55 (15%). As we previously found that the 25(OH)D-breast cancer association was stronger in post-menopausal women, we conducted analyses within this subgroup. We also examined modification by first-degree family history of breast cancer (1 first-degree relative vs. >1 first-degree relatives). All statistical analyses were carried out in SAS (v9.3, Cary, NC) or R (v3.2.1).
For each selected candidate SNP, we estimated hazard ratios for SNP-breast cancer associations. These associations were assessed using a Cox proportional hazards model, adjusting for self-reported race/ethnicity and ancestry only, with genotype coded log-additively (n=1,829 from the subcohort and 1,600 cases, after exclusions for missing covariates). We additionally conducted multi-SNP, gene-based gene-by-environment tests using the gene-environment set association test (GESAT) (53). GESAT utilizes a variance component score test within a generalized linear model framework to test the association of a vector of SNP-by-environment interaction terms. Because this test was developed for case-control studies rather than a case-cohort design, we modified our analysis to compare genotypes and 25(OH)D levels of women who developed breast cancer within five years of enrollment (n=1590 cases with complete covariate information) to those who did not develop breast cancer during that time period (n=1,744 non-cases/controls with complete covariate information). We included the same covariates as in the single SNP interaction analyses, as well as age at blood draw.
Our primary analysis was a candidate gene study of 82 non-independent SNPs from 7 genes known to be involved in vitamin D metabolism. We did not correct our main analysis for multiple comparisons, but note that based on the correlation structure of the included SNPs, the number of effective tests was 56 (54). As we considered our secondary analysis of the remaining 1,439 SNPs to be exploratory, we present both uncorrected p-values and false-discovery rate q-values (55). Here, a q-value<0.05 was considered noteworthy.
Results
As expected, cases were slightly older than those in the randomly sampled subcohort (mean of 57.4 years versus 55.3 years) and had lower average 25(OH)D levels (31.0 ng/mL versus 31.8 ng/mL) (Table 1). Compared to the subcohort, cases were more highly educated, more likely to be post-menopausal, more likely to be obese, more likely to be current hormone therapy users, and more likely to have two or more first-degree relatives with breast cancer. Cases were less likely to have a history of osteoporosis.
Table 1.
Characteristics of participants included in vitamin D and genetic sub-studies (Sister Study, 2003–2009)
| Characteristic | Random sub-cohort (n=1829); N (%) | Breast cancer casesa (n=1601); N (%) |
|---|---|---|
| Age at blood draw; mean (std) | 55.3 (8.9) | 57.4 (8.9) |
| Total 25(OH)D level; mean (std) | 31.8 (10.5) | 31.0 (10.1) |
| Total 25(OH)D >38 ng/mL | 460 (25) | 330 (21) |
| Race/Ethnicity | ||
| Non-Hispanic White | 1,576 (86) | 1,365 (85) |
| Non-Hispanic Black | 134 (7) | 122 (8) |
| Hispanic | 81 (4) | 63 (4) |
| Other | 38 (2) | 50 (3) |
| Education level | ||
| High school or less | 292 (16) | 245 (15) |
| Some college | 639 (35) | 514 (32) |
| Bachelor’s degree | 468 (26) | 421 (26) |
| Graduate degree | 430 (24) | 420 (26) |
| Menopausal Status | ||
| Premenopausal | 613 (34) | 461 (29) |
| Post-menopausal | 1,216 (66) | 1,140 (71) |
| Current body mass index (BMI) | ||
| <25.0 kg/m2 | 702 (38) | 582 (36) |
| 25–29.9 kg/m2 | 581 (32) | 506 (32) |
| ≥30 kg/m2 | 543 (30) | 513 (32) |
| Current hormonal birth control use | ||
| Yes | 75 (4) | 66 (4) |
| No | 1,750 (96) | 1,531 (96) |
| Current hormone therapy use | ||
| Current, estrogen plus progestin | 65 (4) | 84 (5) |
| Current, unopposed estrogen | 125 (7) | 133 (8) |
| No current use | 1,634 (90) | 1,379 (86) |
| Physical Activity (in last year) | ||
| 0 – 1 hours/week | 635 (35) | 539 (34) |
| 1.1 – 3 hours/week | 557 (30) | 516 (32) |
| >3 hours/week | 637 (35) | 546 (34) |
| Alcohol consumption in last year | ||
| Never/former drinker | 341 (19) | 300 (19) |
| Current drinker, <1 drink/day | 1230 (67) | 1069 (67) |
| Current drinker, ≥1 drink/day | 253 (14) | 231 (14) |
| Parity | ||
| 0 births | 339 (19) | 295 (18) |
| 1 birth | 276 (15) | 235 (15) |
| 2 births | 664 (36) | 576 (36) |
| ≥3 births | 549 (30) | 495 (31) |
| History of osteoporosis | ||
| Yes | 431 (24) | 343 (21) |
| No | 1,397 (76) | 1,258 (79) |
| Regular vitamin D supplement use (≥4 times/week) | ||
| None | 840 (47) | 731 (46) |
| Multivitamin, no extra vitamin D | 732 (41) | 657 (42) |
| Multivitamin and vitamin D | 131 (7) | 122 (8) |
| Vitamin D and calcium | 76 (4) | 54 (3) |
| Vitamin D only | 10 (1) | 9 (1) |
| Family history of breast cancer | ||
| Affected sister or half-sister only | 1,359 (74) | 1,047 (65) |
| >1 first degree relative | 470 (26) | 554 (35) |
Includes 67 women also selected as part of sub-cohort
Missing values: Race (1 case), Education (1 case), Current BMI (3 in sub-cohort), Current hormonal birth control use (4 in sub-cohort, 4 cases), Current hormone therapy use (5 in sub-cohort, 5 cases), Alcohol (5 in sub-cohort, 1 case), Parity (1 in sub-cohort), Osteoporosis (1 in sub-cohort), Supplement use (40 in sub-cohort, 28 cases)
Eight of the 82 SNPs in the 7 candidate vitamin D metabolism genes had a statistically significant interaction with 25(OH)D in relation to breast cancer (based on an uncorrected p<0.05; Table 2). The SNP with the lowest p-value for interaction was rs4328262 in VDR (p=0.0008). Here, the HR for the 25(OH)D effect in non-carriers of the variant allele was 0.92 (95% CI: 0.68–1.24) and each copy of the minor allele was associated with an 33% decrease in the HR between 25(OH)D and breast cancer (RHR=0.67, 95% CI: 0.53–0.85).
Table 2.
Interacting effects of 25(OH)D and single nucleotide polymorphisms (SNPs) in vitamin D metabolism genes on the 5-year risk of breast cancer: Ratio of hazard ratios and 95% confidence intervals (N=3,334a; 1810 in sub-cohort [66 cases] and 1,524 additional cases)
| Rank | SNP | Location | Gene | Minor Allele Frequencyb | 25(OH)D-breast cancer HR in non-carriers | Ratio of Hazard Ratios (95% CI)c | p-value |
|---|---|---|---|---|---|---|---|
| 1 | rs4328262 | 12: 48285648 | VDR | 0.41 | 0.92 (0.68, 1.24) | 0.67 (0.53, 0.85) | 0.0008 |
| 2 | rs11168287 | 12: 48285414 | VDR | 0.47 | 0.92 (0.64, 1.31) | 0.71 (0.56, 0.90) | 0.004 |
| 3 | rs117913124 | 11: 14900931 | CYP2R1 | 0.02 | 0.71 (0.59, 0.84) | 5.96 (1.71, 20.8) | 0.005 |
| 4 | rs4237855 | 12: 48287203 | VDR | 0.37 | 0.83 (0.63, 1.10) | 0.73 (0.58, 0.92) | 0.007 |
| 5 | rs17883984 | 12: 48293716 | VDR | 0.32 | 0.60 (0.46, 0.78) | 1.34 (1.05, 1.71) | 0.02 |
| 6 | rs4516035 | 12: 48299826 | VDR | 0.4 | 0.56 (0.42, 0.76) | 1.30 (1.04, 1.64) | 0.02 |
| 7 | rs2239182 | 12: 48255411 | VDR | 0.5 | 0.53 (0.37, 0.76) | 1.27 (1.01, 1.58) | 0.04 |
| 8 | rs3132301 | 9: 137294030 | RXRA | 0.19 | 0.65 (0.52, 0.81) | 1.36 (1.01, 1.84) | 0.04 |
| 9 | rs3132296 | 9: 137302631 | RXRA | 0.34 | 0.68 (0.52, 0.89) | 1.28 (1.00, 1.65) | 0.05 |
| 10 | rs3118536 | 9: 137308462 | RXRA | 0.17 | 0.66 (0.53, 0.81) | 1.35 (0.98, 1.84) | 0.06 |
| 11 | rs1989969 | 12: 48278010 | VDR | 0.4 | 0.69 (0.52, 0.91) | 1.25 (0.98, 1.60) | 0.07 |
| 12 | rs35079168 | 9: 137280939 | RXRA | 0.39 | 0.62 (0.47, 0.83) | 1.25 (0.98, 1.59) | 0.07 |
| 13 | rs11102986 | 9: 137285503 | RXRA | 0.18 | 0.66 (0.53, 0.81) | 1.32 (0.97, 1.78) | 0.07 |
| 14 | rs7129781 | 11: 14912417 | CYP2R1 | 0.08 | 0.70 (0.58, 0.85) | 1.44 (0.93, 2.23) | 0.10 |
| 15 | rs12794714 | 11: 14913575 | CYP2R1 | 0.42 | 0.98 (0.74, 1.30) | 0.81 (0.62, 1.06) | 0.12 |
| 16 | rs886441 | 12: 48262964 | VDR | 0.21 | 0.62 (0.50, 0.77) | 1.25 (0.94, 1.65) | 0.12 |
| 17 | rs2853564 | 12: 48278487 | VDR | 0.37 | 0.69 (0.53, 0.91) | 1.20 (0.95, 1.53) | 0.13 |
| 18 | rs2228572 | 12: 48272840 | VDR | 0.02 | 0.76 (0.64, 0.90) | 0.52 (0.22, 1.22) | 0.13 |
| 19 | rs7967152 | 12: 48244184 | VDR | 0.46 | 0.59 (0.42, 0.82) | 1.19 (0.95, 1.51) | 0.14 |
| 20 | rs2239179 | 12: 48257766 | VDR | 0.42 | 0.76 (0.56, 1.04) | 0.84 (0.67, 1.06) | 0.14 |
| 21 | rs4917354 | 9: 137237661 | RXRA | 0.31 | 0.64 (0.50, 0.82) | 1.20 (0.93, 1.55) | 0.16 |
| 22 | rs1155563 | 4: 72643488 | GC | 0.27 | 0.65 (0.52, 0.82) | 1.22 (0.92, 1.64) | 0.17 |
| 23 | rs1805343 | 9: 137328286 | RXRA | 0.38 | 0.66 (0.50, 0.87) | 1.18 (0.93, 1.49) | 0.18 |
| 24 | rs4588 | 4: 72618323 | GC | 0.27 | 0.64 (0.51, 0.80) | 1.22 (0.91, 1.65) | 0.19 |
| 25 | rs2181874 | 20: 52784478 | CYP24A1 | 0.25 | 0.66 (0.52, 0.85) | 1.18 (0.91, 1.54) | 0.21 |
| 26 | rs11574143 | 12: 48234917 | VDR | 0.11 | 0.78 (0.64, 0.94) | 0.80 (0.55, 1.16) | 0.24 |
| 27 | rs2239186 | 12: 48269410 | VDR | 0.18 | 0.68 (0.55, 0.85) | 1.19 (0.89, 1.60) | 0.24 |
| 28 | rs2248098 | 12: 48253356 | VDR | 0.48 | 0.62 (0.44, 0.88) | 1.15 (0.91, 1.45) | 0.24 |
| 29 | rs2296241 | 20: 52786219 | CYP24A1 | 0.48 | 0.84 (0.62, 1.15) | 0.87 (0.69, 1.10) | 0.24 |
| 30 | rs2762934 | 20: 52771261 | CYP24A1 | 0.18 | 0.68 (0.56, 0.85) | 1.19 (0.89, 1.61) | 0.25 |
| 31 | rs9729 | 12: 48236623 | VDR | 0.46 | 0.61 (0.44, 0.85) | 1.14 (0.90, 1.43) | 0.28 |
| 32 | rs757343 | 12: 48239675 | VDR | 0.12 | 0.77 (0.63, 0.94) | 0.82 (0.58, 1.17) | 0.28 |
| 33 | rs2228570 | 12: 48272895 | VDR | 0.36 | 0.70 (0.53, 0.92) | 0.89 (0.71, 1.11) | 0.30 |
| 34 | rs2238136 | 12: 48277713 | VDR | 0.24 | 0.79 (0.62, 1.00) | 0.87 (0.66, 1.14) | 0.31 |
| 35 | rs16999116 | 20: 52777467 | CYP24A1 | 0.14 | 0.77 (0.63, 0.94) | 0.85 (0.61, 1.17) | 0.31 |
| 36 | rs35873579 | 20: 52788190 | CYP24A1 | 0.03 | 0.75 (0.63, 0.90) | 0.68 (0.31, 1.47) | 0.33 |
| 37 | rs10898193 | 11: 71197083 | NADSYN1 | 0.17 | 0.69 (0.56, 0.85) | 1.17 (0.85, 1.61) | 0.35 |
| 38 | rs1044535 | 11: 71145778 | DHCR7 | 0.08 | 0.70 (0.58, 0.84) | 1.25 (0.78, 1.99) | 0.35 |
| 39 | rs4809958 | 20: 52782438 | CYP24A1 | 0.16 | 0.78 (0.64, 0.96) | 0.86 (0.62, 1.19) | 0.37 |
| 40 | rs35603635 | 9: 137323202 | RXRA | 0.03 | 0.75 (0.63, 0.90) | 0.73 (0.37, 1.45) | 0.37 |
| 41 | rs7975128 | 12: 48245828 | VDR | 0.39 | 0.76 (0.57, 1.02) | 0.90 (0.71, 1.14) | 0.38 |
| 42 | rs3819545 | 12: 48265006 | VDR | 0.37 | 0.77 (0.58, 1.02) | 0.90 (0.71, 1.14) | 0.39 |
| 43 | rs2238135 | 12: 48278190 | VDR | 0.25 | 0.78 (0.62, 0.98) | 0.88 (0.67, 1.17) | 0.39 |
| 44 | rs61749689 | 20: 52790005 | CYP24A1 | 0.02 | 0.75 (0.63, 0.90) | 0.73 (0.33, 1.58) | 0.42 |
| 45 | rs1570669 | 20: 52774427 | CYP24A1 | 0.37 | 0.85 (0.65, 1.11) | 0.91 (0.72, 1.16) | 0.45 |
| 46 | rs11168275 | 12: 48272275 | VDR | 0.23 | 0.75 (0.60, 0.95) | 0.90 (0.68, 1.18) | 0.45 |
| 47 | rs1045570 | 9: 137332311 | RXRA | 0.16 | 0.71 (0.58, 0.88) | 1.13 (0.83, 1.53) | 0.45 |
| 48 | rs2254210 | 12: 48273714 | VDR | 0.36 | 0.73 (0.56, 0.96) | 1.10 (0.86, 1.40) | 0.46 |
| 49 | rs1352844 | 4: 72647749 | GC | 0.12 | 0.77 (0.63, 0.94) | 0.89 (0.64, 1.24) | 0.48 |
| 50 | rs12717991 | 12: 48259126 | VDR | 0.37 | 0.69 (0.52, 0.91) | 1.08 (0.85, 1.37) | 0.54 |
| 51 | rs731236 | 12: 48238757 | VDR | 0.39 | 0.76 (0.57, 1.01) | 0.93 (0.74, 1.17) | 0.54 |
| 52 | rs11185660 | 9: 137259992 | RXRA | 0.25 | 0.70 (0.56, 0.88) | 1.09 (0.83, 1.43) | 0.56 |
| 53 | rs705120 | 4: 72614140 | GC | 0.42 | 0.66 (0.50, 0.88) | 1.08 (0.84, 1.38) | 0.57 |
| 54 | rs12785878 | 11: 71167449 | NADSYN1 | 0.3 | 0.71 (0.56, 0.89) | 1.08 (0.83, 1.41) | 0.57 |
| 55 | rs6537998 | 9: 137292505 | RXRA | 0.03 | 0.73 (0.61, 0.87) | 1.22 (0.60, 2.50) | 0.58 |
| 56 | rs222020 | 4: 72636272 | GC | 0.19 | 0.80 (0.65, 0.99) | 0.92 (0.67, 1.26) | 0.59 |
| 57 | rs1540339 | 12: 48257326 | VDR | 0.36 | 0.67 (0.51, 0.89) | 1.06 (0.84, 1.34) | 0.61 |
| 58 | rs67816242 | 9: 137268441 | RXRA | 0.24 | 0.71 (0.57, 0.88) | 1.07 (0.81, 1.42) | 0.62 |
| 59 | rs2248359 | 20: 52791518 | CYP24A1 | 0.41 | 0.62 (0.46, 0.84) | 1.05 (0.84, 1.32) | 0.68 |
| 60 | rs7038018 | 9: 137309959 | RXRA | 0.14 | 0.74 (0.60, 0.90) | 1.07 (0.77, 1.48) | 0.68 |
| 61 | rs3733359 | 4: 72649774 | GC | 0.09 | 0.75 (0.62, 0.90) | 0.92 (0.58, 1.44) | 0.71 |
| 62 | rs2239180 | 12: 48256046 | VDR | 0.12 | 0.71 (0.59, 0.87) | 1.07 (0.75, 1.51) | 0.72 |
| 63 | rs143304420 | 11: 71144815 | DHCR7 | 0.03 | 0.73 (0.61, 0.88) | 1.13 (0.56, 2.28) | 0.73 |
| 64 | rs2248461 | 20: 52792202 | CYP24A1 | 0.39 | 0.63 (0.47, 0.84) | 1.04 (0.83, 1.30) | 0.76 |
| 65 | rs222009 | 4: 72631735 | GC | 0.02 | 0.74 (0.62, 0.88) | 0.82 (0.21, 3.22) | 0.78 |
| 66 | rs4364228 | 4: 72623347 | GC | 0.1 | 0.75 (0.62, 0.90) | 0.95 (0.63, 1.41) | 0.78 |
| 67 | rs10877012 | 12: 58162085 | CYP27B1 | 0.3 | 0.68 (0.52, 0.88) | 0.97 (0.76, 1.23) | 0.79 |
| 68 | rs2283342 | 12: 48255859 | VDR | 0.14 | 0.75 (0.61, 0.92) | 0.96 (0.69, 1.33) | 0.81 |
| 69 | rs7136534 | 12: 48294626 | VDR | 0.25 | 0.73 (0.58, 0.93) | 1.03 (0.79, 1.35) | 0.82 |
| 70 | rs11907350 | 20: 52770439 | CYP24A1 | 0.04 | 0.73 (0.61, 0.87) | 1.07 (0.57, 2.02) | 0.82 |
| 71 | rs11574026 | 12: 48288246 | VDR | 0.11 | 0.74 (0.61, 0.90) | 1.04 (0.70, 1.55) | 0.85 |
| 72 | rs34312136 | 9: 137269456 | RXRA | 0.4 | 0.70 (0.52, 0.95) | 1.02 (0.81, 1.29) | 0.86 |
| 73 | rs10741657 | 11: 14914878 | CYP2R1 | 0.37 | 0.71 (0.52, 0.97) | 1.02 (0.81, 1.29) | 0.86 |
| 74 | rs9615 | 9: 137331525 | RXRA | 0.04 | 0.73 (0.61, 0.88) | 0.96 (0.50, 1.82) | 0.90 |
| 75 | rs2296239 | 20: 52775528 | CYP24A1 | 0.24 | 0.79 (0.63, 0.98) | 0.99 (0.75, 1.30) | 0.92 |
| 76 | rs11168267 | 12: 48251542 | VDR | 0.09 | 0.72 (0.60, 0.87) | 1.02 (0.68, 1.53) | 0.92 |
| 77 | rs3782905 | 12: 48266167 | VDR | 0.32 | 0.72 (0.55, 0.94) | 0.99 (0.78, 1.26) | 0.93 |
| 78 | rs6022987 | 20: 52770596 | CYP24A1 | 0.29 | 0.75 (0.58, 0.97) | 1.01 (0.78, 1.31) | 0.94 |
| 79 | rs10875693 | 12: 48269650 | VDR | 0.33 | 0.75 (0.57, 0.98) | 0.99 (0.78, 1.26) | 0.94 |
| 80 | rs927650 | 20: 52772741 | CYP24A1 | 0.46 | 0.73 (0.53, 1.00) | 1.00 (0.80, 1.26) | 0.97 |
| 81 | rs78077736 | 9: 137257165 | RXRA | 0.05 | 0.74 (0.62, 0.88) | 1.00 (0.57, 1.73) | 0.99 |
| 82 | rs2107301 | 12: 48255570 | VDR | 0.27 | 0.72 (0.56, 0.91) | 1.00 (0.78, 1.29) | 0.99 |
After excluding those with missing covariate information
Among sub-cohort members
The minor allele is the index allele, so a value >1.00 indicates that 25(OH)D has a stronger protective association with breast cancer among those with no copies of the minor allele than those with 1 or 2, assuming a log-additive genetic model. Similarly, an RHR <1.00 indicates that 25(OH)D has a stronger inverse association with breast cancer among those 1 or 2 copies of the minor allele than those none.
Five other VDR SNPs also showed evidence of gene-by-vitamin D interaction (rs11168287, rs4237855, rs17883984, rs4516035, and rs2239182). For half of the six significant VDR SNPs, the RHR was below 1.00, indicating that 25(OH)D had a stronger inverse association with breast cancer among carriers of the variant allele. The other three SNPs had RHRs that exceeded 1.00, meaning that 25(OH)D had a stronger inverse association with breast cancer among non-carriers. These 6 SNPs were not in high linkage disequilibrium with one another within our sub-cohort sample (r2 <0.80; as seen in Supplementary Table 1), though some showed moderate correlations with one another (0.50≤ r2 <0.80), including rs4328262 and rs4237855 (r2=0.61) and rs17883984 and rs4516035 (r2=0.64).
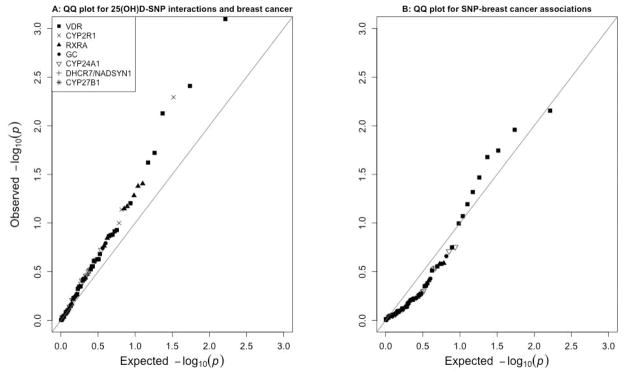
Other SNPs with statistically significant interactions were rs117913124 (CYP2R1; p=0.005) and rs3132301 (RXRA; p=0.04). The RHRs exceeded 1.00 for both of these SNPs. Overall, the observed p-values were smaller than would have been expected by chance under the null hypothesis of no association, as shown by the quantile-quantile plot in Figure 1a. We formally tested the combined results for all 82 candidate SNPs relative to the global no-interaction null hypothesis. We did so by using Fisher’s method to combine p-values and then comparing the observed test statistic (a sum of the −2 log p-values) to those from each of 1000 data sets with permuted case statuses. The estimated overall p-value was 0.04.
Figure 1. Quantile-quantile plots.
Figure 1 shows the observed versus expected p-values for the associations between each 25(OH)D-SNP interaction and incident breast cancer (A) and between each SNP and incident breast cancer (B).
Results from analyses restricted to post-menopausal person-time were generally similar to those from the main analyses, with the same top SNP (rs4328262; RHR=0.65, 95% CI: 0.50, 0.84; Supplementary Table 2). Four of the 8 statistically-significant SNPs from the overall analysis still showed significant associations among post-menopausal women as well as two other SNPs (rs7967152 in VDR and rs3118536 in RXRA).
In an exploratory analysis, we examined whether the number of first-degree relatives with breast cancer modified the gene-by-vitamin-D interaction. Despite having similar minor allele frequencies in both groups, some of the top 8 SNPs showed evidence of RHR modification (Supplementary Table 3). For example, rs17883984 in VDR (the fifth ranked SNP in the overall analysis), had an RHR of 1.69 (95% CI: 1.24, 2.29) in those with only one affected sister, but an RHR of 0.80 (95% CI: 0.52, 1.24) in those with >1 first-degree relatives with breast cancer (p-value for 3-way interaction=0.008). In general, however, the observed associations for these 8 SNPs tended to be in the same direction for the two groups.
Only six of the 82 candidate SNPs were independently associated with breast cancer risk at p<0.05 (Table 3). All of these were located in VDR (rs2238135, rs11168287, rs1989969, rs2238136, rs4237855, rs4516035). Of these six, only rs2238135 and rs2238136 were moderately correlated with one another (r2=0.73; Supplementary Table 1). Three of the six also showed evidence of statistically-significant gene-by-environment interaction with 25(OH)D (rs11168287, rs4237855, and rs4516035). Overall, the p-values observed for the SNP-breast cancer association analysis were consistent with what would be expected by chance under the null hypothesis (Figure 1b).
Table 3.
SNP-breast cancer associations for SNPs in selected candidate genes (N=3,334; 1810 in sub-cohort [66 cases] and 1,524 additional cases)
| SNP-Breast Cancer association rank | Overall rank for GxE | SNP | Location | Gene | Hazard Ratio (95% CI) | p-value |
|---|---|---|---|---|---|---|
| 1 | 43 | rs2238135 | 12: 48278190 | VDR | 1.17 (1.04, 1.30) | 0.007 |
| 2 | 2 | rs11168287 | 12: 48285414 | VDR | 1.13 (1.03, 1.25) | 0.01 |
| 3 | 11 | rs1989969 | 12: 48278010 | VDR | 0.89 (0.81, 0.98) | 0.02 |
| 4 | 34 | rs2238136 | 12: 48277713 | VDR | 1.14 (1.02, 1.28) | 0.02 |
| 5 | 4 | rs4237855 | 12: 48287203 | VDR | 1.11 (1.01, 1.23) | 0.03 |
| 6 | 6 | rs4516035 | 12: 48299826 | VDR | 0.90 (0.82, 1.00) | 0.05 |
| 7 | 17 | rs2853564 | 12: 48278487 | VDR | 0.91 (0.83, 1.01) | 0.06 |
| 8 | 69 | rs7136534 | 12: 48294626 | VDR | 0.91 (0.81, 1.01) | 0.09 |
| 9 | 1 | rs4328262 | 12: 48285648 | VDR | 1.09 (0.98, 1.20) | 0.10 |
| 10 | 64 | rs2248461 | 20: 52792202 | CYP24A1 | 1.07 (0.97, 1.17) | 0.18 |
| 11 | 48 | rs2254210 | 12: 48273714 | VDR | 0.93 (0.85, 1.03) | 0.18 |
| 12 | 59 | rs2248359 | 20: 52791518 | CYP24A1 | 1.06 (0.97, 1.17) | 0.20 |
| 13 | 49 | rs1352844 | 4: 72647749 | GC | 1.09 (0.95, 1.25) | 0.22 |
| 14 | 47 | rs1045570 | 9: 137332311 | RXRA | 1.08 (0.95, 1.23) | 0.26 |
| 15 | 26 | rs11574143 | 12: 48234917 | VDR | 0.91 (0.78, 1.07) | 0.26 |
| 16 | 81 | rs78077736 | 9: 137257165 | RXRA | 1.13 (0.91, 1.40) | 0.27 |
| 17 | 5 | rs17883984 | 12: 48293716 | VDR | 0.94 (0.85, 1.05) | 0.28 |
| 18 | 25 | rs2181874 | 20: 52784478 | CYP24A1 | 1.06 (0.95, 1.18) | 0.29 |
| 19 | 29 | rs2296241 | 20: 52786219 | CYP24A1 | 0.95 (0.86, 1.05) | 0.29 |
| 20 | 62 | rs2239180 | 12: 48256046 | VDR | 0.92 (0.79, 1.08) | 0.31 |
| 21 | 61 | rs3733359 | 4: 72649774 | GC | 0.92 (0.78, 1.10) | 0.37 |
| 22 | 74 | rs9615 | 9: 137331525 | RXRA | 0.90 (0.70, 1.15) | 0.39 |
| 23 | 32 | rs757343 | 12: 48239675 | VDR | 0.94 (0.81, 1.09) | 0.42 |
| 24 | 28 | rs2248098 | 12: 48253356 | VDR | 1.04 (0.94, 1.14) | 0.44 |
| 25 | 46 | rs11168275 | 12: 48272275 | VDR | 1.04 (0.93, 1.17) | 0.45 |
| 26 | 30 | rs2762934 | 20: 52771261 | CYP24A1 | 1.04 (0.92, 1.18) | 0.50 |
| 27 | 67 | rs10877012 | 12: 58162085 | CYP27B1 | 1.04 (0.93, 1.15) | 0.51 |
| 28 | 19 | rs7967152 | 12: 48244184 | VDR | 1.03 (0.94, 1.13) | 0.53 |
| 29 | 18 | rs2228572 | 12: 48272840 | VDR | 0.91 (0.67, 1.24) | 0.55 |
| 30 | 76 | rs11168267 | 12: 48251542 | VDR | 0.95 (0.80, 1.13) | 0.56 |
| 31 | 21 | rs4917354 | 9: 137237661 | RXRA | 0.97 (0.88, 1.08) | 0.58 |
| 32 | 37 | rs10898193 | 11: 71197083 | NADSYN1 | 0.97 (0.85, 1.09) | 0.59 |
| 33 | 40 | rs35603635 | 9: 137323202 | RXRA | 0.93 (0.71, 1.21) | 0.59 |
| 34 | 44 | rs61749689 | 20: 52790005 | CYP24A1 | 0.92 (0.66, 1.26) | 0.60 |
| 35 | 65 | rs222009 | 4: 72631735 | GC | 1.11 (0.76, 1.62) | 0.60 |
| 36 | 71 | rs11574026 | 12: 48288246 | VDR | 0.96 (0.82, 1.12) | 0.61 |
| 37 | 7 | rs2239182 | 12: 48255411 | VDR | 0.98 (0.89, 1.07) | 0.61 |
| 38 | 3 | rs117913124 | 11: 14900931 | CYP2R1 | 0.92 (0.67, 1.27) | 0.62 |
| 39 | 66 | rs4364228 | 4: 72623347 | GC | 1.04 (0.89, 1.22) | 0.63 |
| 40 | 10 | rs3118536 | 9: 137308462 | RXRA | 1.03 (0.91, 1.17) | 0.64 |
| 41 | 50 | rs12717991 | 12: 48259126 | VDR | 0.98 (0.89, 1.08) | 0.66 |
| 42 | 31 | rs9729 | 12: 48236623 | VDR | 1.02 (0.93, 1.12) | 0.68 |
| 43 | 57 | rs1540339 | 12: 48257326 | VDR | 0.98 (0.89, 1.09) | 0.72 |
| 44 | 60 | rs7038018 | 9: 137309959 | RXRA | 0.98 (0.85, 1.12) | 0.72 |
| 45 | 8 | rs3132301 | 9: 137294030 | RXRA | 0.98 (0.87, 1.11) | 0.74 |
| 46 | 9 | rs3132296 | 9: 137302631 | RXRA | 0.98 (0.89, 1.09) | 0.74 |
| 47 | 45 | rs1570669 | 20: 52774427 | CYP24A1 | 0.98 (0.89, 1.09) | 0.75 |
| 48 | 78 | rs6022987 | 20: 52770596 | CYP24A1 | 1.02 (0.92, 1.12) | 0.75 |
| 49 | 77 | rs3782905 | 12: 48266167 | VDR | 1.02 (0.92, 1.13) | 0.76 |
| 50 | 20 | rs2239179 | 12: 48257766 | VDR | 1.01 (0.92, 1.12) | 0.77 |
| 51 | 13 | rs11102986 | 9: 137285503 | RXRA | 0.98 (0.87, 1.11) | 0.78 |
| 52 | 14 | rs7129781 | 11: 14912417 | CYP2R1 | 1.02 (0.86, 1.21) | 0.80 |
| 53 | 80 | rs927650 | 20: 52772741 | CYP24A1 | 0.99 (0.90, 1.08) | 0.80 |
| 54 | 53 | rs705120 | 4: 72614140 | GC | 1.01 (0.92, 1.11) | 0.80 |
| 55 | 22 | rs1155563 | 4: 72643488 | GC | 0.99 (0.89, 1.10) | 0.80 |
| 56 | 68 | rs2283342 | 12: 48255859 | VDR | 0.98 (0.86, 1.13) | 0.81 |
| 57 | 27 | rs2239186 | 12: 48269410 | VDR | 1.02 (0.90, 1.15) | 0.81 |
| 58 | 16 | rs886441 | 12: 48262964 | VDR | 1.01 (0.90, 1.14) | 0.83 |
| 59 | 38 | rs1044535 | 11: 71145778 | DHCR7 | 0.98 (0.83, 1.17) | 0.84 |
| 60 | 33 | rs2228570 | 12: 48272895 | VDR | 0.99 (0.90, 1.09) | 0.85 |
| 61 | 35 | rs16999116 | 20: 52777467 | CYP24A1 | 0.99 (0.86, 1.13) | 0.87 |
| 62 | 63 | rs143304420 | 11: 71144815 | DHCR7 | 0.98 (0.74, 1.29) | 0.87 |
| 63 | 72 | rs34312136 | 9: 137269456 | RXRA | 1.01 (0.91, 1.11) | 0.87 |
| 64 | 73 | rs10741657 | 11: 14914878 | CYP2R1 | 0.99 (0.90, 1.09) | 0.88 |
| 65 | 36 | rs35873579 | 20: 52788190 | CYP24A1 | 0.98 (0.72, 1.32) | 0.88 |
| 66 | 54 | rs12785878 | 11: 71167449 | NADSYN1 | 1.01 (0.91, 1.12) | 0.89 |
| 67 | 70 | rs11907350 | 20: 52770439 | CYP24A1 | 0.98 (0.77, 1.26) | 0.89 |
| 68 | 79 | rs10875693 | 12: 48269650 | VDR | 1.01 (0.91, 1.12) | 0.89 |
| 69 | 42 | rs3819545 | 12: 48265006 | VDR | 0.99 (0.90, 1.10) | 0.90 |
| 70 | 58 | rs67816242 | 9: 137268441 | RXRA | 1.01 (0.90, 1.13) | 0.90 |
| 71 | 55 | rs6537998 | 9: 137292505 | RXRA | 1.02 (0.74, 1.40) | 0.90 |
| 72 | 41 | rs7975128 | 12: 48245828 | VDR | 0.99 (0.90, 1.10) | 0.90 |
| 73 | 56 | rs222020 | 4: 72636272 | GC | 0.99 (0.88, 1.12) | 0.91 |
| 74 | 15 | rs12794714 | 11: 14913575 | CYP2R1 | 0.99 (0.90, 1.09) | 0.91 |
| 75 | 75 | rs2296239 | 20: 52775528 | CYP24A1 | 1.01 (0.90, 1.12) | 0.92 |
| 76 | 23 | rs1805343 | 9: 137328286 | RXRA | 1.00 (0.90, 1.10) | 0.94 |
| 77 | 12 | rs35079168 | 9: 137280939 | RXRA | 1.00 (0.91, 1.11) | 0.94 |
| 78 | 52 | rs11185660 | 9: 137259992 | RXRA | 1.00 (0.90, 1.12) | 0.95 |
| 79 | 24 | rs4588 | 4: 72618323 | GC | 1.00 (0.90, 1.11) | 0.97 |
| 80 | 39 | rs4809958 | 20: 52782438 | CYP24A1 | 1.00 (0.88, 1.14) | 0.97 |
| 81 | 51 | rs731236 | 12: 48238757 | VDR | 1.00 (0.91, 1.10) | 0.97 |
| 82 | 82 | rs2107301 | 12: 48255570 | VDR | 1.00 (0.90, 1.11) | 0.99 |
Of the 1,439 additional candidate SNPs with potential links to vitamin D, 77 showed evidence of a statistically significant interaction at an uncorrected p-value of 0.05 (Supplementary Table 4), and none met the false discovery rate threshold (q≤0.05).
In gene-based analyses, the only primary candidate gene to show strong evidence of multiplicative interaction was VDR (35 SNPs, p=0.04; Table 4). Six genes from the secondary list of 89 genes had gene-by-environment interaction p-values less than 0.05 (Supplementary Table 5), including AMZ1 (8 SNPs, p=0.001), GPR114 (5 SNPs, p=0.01), EGFR (85 SNPs, p=0.01), TRIM24 (9 SNPs, p=0.02), CYP3A4 (10 SNPs, p=0.03), and ITGB3 (15 SNPs, p=0.04).
Table 4.
Gene-environment set association tests (GESAT) for 25(OH)D and vitamin D-related genes on the 5-year risk of breast cancer (N=3,334; 1,590 cases, 1,744 controls/non-cases)
| Gene | Number of included SNPs | GESAT p-value |
|---|---|---|
| VDR | 35 | 0.04 |
| RXRA | 16 | 0.19 |
| CYP2R1 | 4 | 0.22 |
| GC | 8 | 0.38 |
| DHCR7/NADSYN1 | 4 | 0.53 |
| CYP24A1 | 14 | 0.65 |
Gene-level effects were not calculated for CYP27B1 (1 SNP).
Discussion
Using a prospective, case-cohort design, we examined gene-by-25(OH)D interaction in relation to breast cancer risk over 5 years of follow-up. We examined 82 SNPs in 7 candidate vitamin D metabolism genes (CYP24A1, CYP27B1, CYP2R1, GC, DHCR7/NADSYN1, RXRA, and VDR), and 8 SNPs showed evidence of multiplicative interactions. Most of these were in VDR, with other hits appearing in CYP2R1 and RXRA. As a group, the candidate SNPs were stronger modifiers of the 25(OH)D-breast cancer association than would have been expected by chance. In gene-level tests, 25(OH)D and VDR showed evidence of multiplicative interaction with breast cancer incidence over five years of follow-up. These findings were generally similar for analyses restricted to post-menopausal women.
Six SNPs in VDR were nominally associated with breast cancer risk, including several of the SNPs that modified the 25(OH)D-breast cancer association. For a few of the top SNPs in the gene-by-environment interaction analysis, the observed RHR was in the opposite direction for those who had one first-degree relative with breast cancer versus more than one first-degree relative with breast cancer. There was negligible evidence that any of the secondary candidate SNPs modified the 25(OH)D-breast cancer association.
Most of the eight SNPs that significantly modified the 25(OH)D-breast cancer association are intronic with no known function (56, 57). Exceptions included the SNP in CYP2R1 (rs117913124), which is a synonymous substitution that does not result in an amino acid change, and one SNP upstream of VDR (rs4516035), which could affect transcription binding sites in the gene’s promotor region. Similarly, aside from rs4516035, all of the VDR SNPs associated with breast cancer risk were in intronic regions (56, 57). SNPs in introns can have important regulatory roles, such as affecting protein splicing (58) or encoding micro-RNA binding sites (50, 59).
The two SNPs with the smallest gene-by-environment interaction p-values (rs4328262 and rs11168287 in VDR) were previously shown to be associated with breast cancer risk in premenopausal European women and post-menopausal Asian women, respectively (60), though Engel et al. found no evidence of an interaction between rs11168287 and sunlight exposure in relation to breast cancer risk (15). Another of the statistically-significant VDR SNPs, rs4516035, was not associated with breast cancer in a previous case-control study (61). We did not corroborate any of the previously observed gene-by-environment interaction effects for the putative functional SNPs in VDR (p=0.30, p=0.54, and p=0.38, for rs2228570/FokI, rs731236/TaqI, and rs7975128 respectively, where rs7975128 is highly correlated with rs1544410/BsmI [r2=0.97]).
Although difficult to study because of its rarity (2% minor allele frequency), the CYP2R1 SNP identified here (rs117913124) was shown to be associated with 25(OH)D levels in a recent genome-wide meta-analysis (62). To the best of our knowledge, rs3132301 (RXRA) has not been previously assessed as a possible effect modifier. Clendenen et al. (14) previously examined the association between various RXRA SNPs (including several also included our analysis) and breast cancer risk, but identified no significant associations. Replication of our findings in an independent study is needed.
The VDR SNP with the lowest p-value for the breast cancer risk analysis, rs2238135, has not been previously reported to be associated with breast cancer risk, but has been linked to esophageal (63), oral cavity (64), and prostate cancer (65). The second lowest p-value was seen for rs11168287, which also showed evidence of modifying the 25(OH)D-breast cancer association and is discussed above. None of the remaining four VDR SNPs associated with breast cancer in our sample has been previously linked to the disease.
With the exception of rs117913124, few of the SNPs in the vitamin D metabolism genes (CYP24A1, CYP2R1, or CYP27B1) showed evidence of interacting with 25(OH)D to affect breast cancer risk. However, these genes may still play a key role in breast cancer prevention by mediating how individuals process and make use of UV-B radiation and dietary vitamin D intake. These relationships should be better explored in future analyses.
All of the women in our study have a first-degree family history of breast cancer. While this does not affect the validity of our hypothesis tests, it may limit the generalizability of our estimates (66). More specifically, the genetic background could matter: the RHRs measured in our sample could be larger than those assessed in a population-based sample if the SNP of interest interacted with or was in linkage disequilibrium with one or more breast cancer risk variants. This could explain the RHR modification we observed for some SNPs when we stratified by the number of first-degree family members with a history of breast cancer. Of note, minor allele frequencies were very similar across those two strata and 25(OH)D levels were slightly lower in women with >1 affected first-degree relative (23% of sub-cohort in top quartile versus 26% of subcohort restricted to women with only 1 first-degree relative). We are interested in assessing possible interactions between BRCA1/2 and vitamin D-related genes, but we currently only have self-reported data on BRCA1/2 mutation status and we estimate that despite over-selecting those with a strong family history, only a small fraction of our participants carry deleterious mutations (66).
Our generalizability could also be limited by the fact that the majority of our participants are white and non-Hispanic. On the other hand, our unique study design of including sisters of women diagnosed with breast cancer presumably ensured that we had some enrichment for both risk alleles and factors associated with a healthier lifestyle, such as increased supplement use. This enrichment for both genetic and environmental risk factors enhances power for this prospective study to detect gene-by-environment interactions (66).
Although this is one of the largest gene-by-environment interaction studies of vitamin D and breast cancer to date, the sample size is limited and some of our effect estimates are imprecise. Results for SNPs with low minor allele frequencies and for subgroup-specific analyses should be interpreted with caution. Because we selected specific candidate genes instead of conducting an agnostic search, we did not adjust for multiple comparisons. Consequently, some of the reported statistically-significant interactions may be false positives rather than true associations. That being said, we observed more significant associations than would be expected under the null hypothesis. We calculated false discovery rate q-values when conducting more exploratory analyses among the secondary candidate SNPs, finding none that met the criteria for statistical significance.
The gene-based tests are both a strength and limitation of this study. While we believe it can be useful to consider the group-level effects of multiple SNPs within a gene, we were not able to identify any gene-based tests that could accommodate our case-cohort design. We instead chose to adapt our data to approximate a case-control study. Although the results should be roughly the same, some discrepancies may exist.
As discussed previously, the main strengths of our study include prospective collection of serum specimens, detailed covariate information, use of LC/MS to measure 25(OH)D levels (including 3-epi-25(OH)D3), our use of a cohort with higher than average breast cancer risk, and data from a highly motivated and committed cohort of women (19). Most other studies of this topic have relied on either 25(OH)D levels in samples collected after cases’ diagnoses, which may have been altered by the disease or disease-related behavioral changes, or on prospectively collected 25(OH)D levels that might have reflected levels from a time period not relevant to breast cancer risk.
We have conducted a large and comprehensive analysis of gene-by-environment interactions between recent serum 25(OH)D levels, genetic variants in vitamin D-related genes, and breast cancer risk. We found evidence of interactions for SNPs in VDR, CYP2R1, and RXRA. This research aims to advance our understanding of the biologic mechanisms involved in breast cancer etiology and the anti-carcinogenic properties of vitamin D. If our findings are replicated in other prospective studies, they may ultimately help to personalize prevention by identifying individuals who would most benefit from interventions to modify vitamin D levels.
Supplementary Material
Acknowledgments
Funding: This work was supported by an Office of Dietary Supplement Research Scholars Program Grant (to KMO) and the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (project Z01-ES044005 to DPS and Z01-ES102245 to CRW).
The authors would like to thank Min Shi and Alexandra White for their helpful comments on an early version of this manuscript.
Abbreviations
- 1,25(OH)2D
1,25-dihydroxyvitamin D
- 25(OH)D
25-hydroxyvitamin D
- AIMs
Ancestry Informative Markers
- BMI
Body Mass Index
- CI
95% Confidence Interval
- HR
Hazard Ratio
- LC/MS
Liquid Chromatography-Mass Spectrometry
- RHR
Ratio of Hazard Ratios
- SNP
Single Nucleotide Polymorphism
Footnotes
Financial Disclosure: The authors declare they have no actual or potential competing financial interests.
References
- 1.Holick MF. Vitamin D deficiency. N Engl J Med. 2007;357(3):266–81. doi: 10.1056/NEJMra070553. [DOI] [PubMed] [Google Scholar]
- 2.DeLuca HF. Overview of general physiologic features and functions of vitamin D. Am J Clin Nutr. 2004;80(supp):1689S–96S. doi: 10.1093/ajcn/80.6.1689S. [DOI] [PubMed] [Google Scholar]
- 3.Welsh J. Vitamin D and prevention of breast cancer. Acta Pharmacol Sin. 2007;28(9):1373–82. doi: 10.1111/j.1745-7254.2007.00700.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J, Chalmers MJ, Stayrook KR, Burris LL, Wang Y, Busby SA, et al. DNA binding alters coactivator interaction surfaces of the intact VDR-RXR complex. Nat Struct Mol Biol. 2011;18(5):556–63. doi: 10.1038/nsmb.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pike JW, Meyer MB. Fundamentals of vitamin D hormone-regulated gene expression. J Steroid Biochem Mol Biol. 2014;144(Pt A):5–11. doi: 10.1016/j.jsbmb.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeshita A, Ozawa Y, Chin WW. Nuclear receptor coactivators facilitate vitamin D receptor homodimer action on direct repeat hormone response elements. Endocrinology. 2000;141(3):1281–63. doi: 10.1210/endo.141.3.7441. [DOI] [PubMed] [Google Scholar]
- 7.Chen WY, Bertone-Johnson ER, Hunter DJ, Willett WC, Hankinson SE. Associations between polymorphisms in the vitamin D receptor and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14(10):2335–9. doi: 10.1158/1055-9965.EPI-05-0283. [DOI] [PubMed] [Google Scholar]
- 8.Deschasaux M, Souberbielle JC, Latino-Martel P, Sutton A, Charnaux N, Druesne-Pecollo N, et al. Weight Status and Alcohol Intake Modify the Association between Vitamin D and Breast Cancer Risk. J Nutr. 2016 doi: 10.3945/jn.115.221481. [DOI] [PubMed] [Google Scholar]
- 9.Fuhrman BJ, Freedman DM, Bhatti P, Doody MM, Fu YP, Chang SC, et al. Sunlight, polymorphisms of vitamin D-related genes and risk of breast cancer. Anticancer Research. 2013;33:543–52. [PMC free article] [PubMed] [Google Scholar]
- 10.Lowe LC, Guy M, Mansi JL, Peckitt C, Bliss J, Wilson RG, et al. Plasma 25-hydroxy vitamin D concentrations, vitamin D receptor genotype and breast cancer risk in a UK Caucasian population. Eur J Cancer. 2005;41(8):1164–9. doi: 10.1016/j.ejca.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Rollison DE, Cole AL, Tung KH, Slattery ML, Baumgartner KB, Byers T, et al. Vitamin D intake, vitamin D receptor polymorphisms, and breast cancer risk among women living in the southwestern. US Breast Cancer Res Treat. 2012;132(2):683–91. doi: 10.1007/s10549-011-1885-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abbas S, Nieters A, Linseisen J, Slanger T, Kropp S, Mutschelknauss EJ, et al. Vitamin D receptor gene polymorphisms and haplotypes and postmenopausal breast cancer risk. Breast Cancer Res. 2008;10(2):R31. doi: 10.1186/bcr1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson LN, Cotterchio M, Cole DE, Knight JA. Vitamin D-related genetic variants, interactions with vitamin D exposure, and breast cancer risk among Caucasian women in Ontario. Cancer Epidemiol Biomarkers Prev. 2011;20(8):1708–17. doi: 10.1158/1055-9965.EPI-11-0300. [DOI] [PubMed] [Google Scholar]
- 14.Clendenen TV, Ge W, Koenig KL, Axelsson T, Liu M, Afanasyeva Y, et al. Genetic Polymorphisms in Vitamin D Metabolism and Signaling Genes and Risk of Breast Cancer: A Nested Case-Control Study. PLoS One. 2015;10(10):e0140478. doi: 10.1371/journal.pone.0140478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engel LS, Satagopan J, Sima CS, Orlow I, Mujumdar U, Coble J, et al. Sun exposure, vitamin D receptor genetic variants, and risk of breast cancer in the Agricultural Health Study. Environ Health Perspect. 2014;122(2):165–71. doi: 10.1289/ehp.1206274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCullough ML, Stevens VL, Diver WR, Feigelson HS, Rodriguez C, Bostick RM, et al. Vitamin D pathway gene polymorphisms, diet, and risk of postmenopausal breast cancer: a nested case-control study. Breast Cancer Res. 2007;9(1):R9. doi: 10.1186/bcr1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nemenqani DM, Karam RA, Amer MG, Abd El Rahman TM. Vitamin D receptor gene polymorphisms and steroid receptor status among Saudi women with breast cancer. Gene. 2015;558(2):215–9. doi: 10.1016/j.gene.2014.12.065. [DOI] [PubMed] [Google Scholar]
- 18.Reimers LL, Crew KD, Bradshaw PT, Santella RM, Steck SE, Sirosh I, et al. Vitamin D-related gene polymorphisms, plasma 25-hydroxyvitamin D, and breast cancer risk. Cancer Causes Control. 2015;26(2):187–203. doi: 10.1007/s10552-014-0497-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Brien KM, Sandler DP, Taylor JA, Weinberg CR. Serum Vitamin D and Risk of Breast Cancer within Five Years. Environ Health Perspect. 2017;125(7):077004. doi: 10.1289/EHP943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almquist M, Bondeson AG, Bondeson L, Malm J, Manjer J. Serum levels of vitamin D, PTH and calcium and breast cancer risk-a prospective nested case-control study. Int J Cancer. 2010;127(9):2159–68. doi: 10.1002/ijc.25215. [DOI] [PubMed] [Google Scholar]
- 21.Amir E, Cecchini RS, Ganz PA, Costantino JP, Beddows S, Hood N, et al. 25-Hydroxy vitamin-D, obesity, and associated variables as predictors of breast cancer risk and tamoxifen benefit in NSABP-P1. Breast Cancer Res Treat. 2012;133(3):1077–88. doi: 10.1007/s10549-012-2012-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eliassen AH, Spiegelman D, Hollis BW, Horst RL, Willett WC, Hankinson SE. Plasma 25-hydroxyvitamin D and risk of breast cancer in the Nurses’ Health Study II. Breast Cancer Res. 2011;13(3):R50. doi: 10.1186/bcr2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freedman DM, Chang SC, Falk RT, Purdue MP, Huang WY, McCarty CA, et al. Serum levels of vitamin D metabolites and breast cancer risk in the prostate, lung, colorectal, and ovarian cancer screening trial. Cancer Epidemiol Biomarkers Prev. 2008;17(4):889–94. doi: 10.1158/1055-9965.EPI-07-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn T, Kaaks R, Becker S, Eomois PP, Clavel-Chapelon F, Kvaskoff M, et al. Plasma 25-hydroxyvitamin D and the risk of breast cancer in the European prospective investigation into cancer and nutrition: a nested case-control study. Int J Cancer. 2013;133(7):1689–700. doi: 10.1002/ijc.28172. [DOI] [PubMed] [Google Scholar]
- 25.McCullough ML, Stevens VL, Patel R, Jacobs EJ, Bain EB, Horst RL, et al. Serum 25-hydroxyvitamin D concentrations and postmenopausal breast cancer risk: a nested case control study in the Cancer Prevention Study-II Nutrition Cohort. Breast Cancer Res. 2009;11(4):R64. doi: 10.1186/bcr2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neuhouser ML, Manson JE, Millen A, Pettinger M, Margolis K, Jacobs ET, et al. The influence of health and lifestyle characteristics on the relation of serum 25-hydroxyvitamin D with risk of colorectal and breast cancer in postmenopausal women. Am J Epidemiol. 2012;175(7):673–84. doi: 10.1093/aje/kwr350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ordonez-Mena JM, Schottker B, Haug U, Muller H, Kohrle J, Schomburg L, et al. Serum 25-hydroxyvitamin d and cancer risk in older adults: results from a large German prospective cohort study. Cancer Epidemiol Biomarkers Prev. 2013;22(5):905–16. doi: 10.1158/1055-9965.EPI-12-1332. [DOI] [PubMed] [Google Scholar]
- 28.Scarmo S, Afanasyeva Y, Lenner P, Koenig KL, Horst RL, Clendenen TV, et al. Circulating levels of 25-hydroxyvitamin D and risk of breast cancer: a nested case-control study. Breast Cancer Res. 2013;15(1):R15. doi: 10.1186/bcr3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skaaby T, Husemoen LL, Thuesen BH, Pisinger C, Jorgensen T, Roswall N, et al. Prospective population-based study of the association between serum 25-hydroxyvitamin-D levels and the incidence of specific types of cancer. Cancer Epidemiol Biomarkers Prev. 2014;23(7):1220–9. doi: 10.1158/1055-9965.EPI-14-0007. [DOI] [PubMed] [Google Scholar]
- 30.Bertone-Johnson ER, Chen WY, Holick MF, Hollis BW, Colditz GA, Willett WC, et al. Plasma 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(8):1991–7. doi: 10.1158/1055-9965.EPI-04-0722. [DOI] [PubMed] [Google Scholar]
- 31.Chlebowski RT, Johnson KC, Kooperberg C, Pettinger M, Wactawski-Wende J, Rohan T, et al. Calcium plus vitamin D supplementation and the risk of breast cancer. J Natl Cancer Inst. 2008;100(22):1581–91. doi: 10.1093/jnci/djn360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engel P, Fagherazzi G, Boutten A, Dupre T, Mesrine S, Boutron-Ruault MC, et al. Serum 25(OH) vitamin D and risk of breast cancer: a nested case-control study from the French E3N cohort. Cancer Epidemiol Biomarkers Prev. 2010;19(9):2341–50. doi: 10.1158/1055-9965.EPI-10-0264. [DOI] [PubMed] [Google Scholar]
- 33.Kim Y, Franke AA, Shvetsov YB, Wilkens LR, Cooney RV, Lurie G, et al. Plasma 25-hydroxyvitamin D3 is associated with decreased risk of postmenopausal breast cancer in whites: a nested case-control study in the multiethnic cohort study. BMC Cancer. 2014;14:29. doi: 10.1186/1471-2407-14-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohr SB, Gorham ED, Alcaraz JE, Kane CI, Macera CA, Parsons JK, et al. Serum 25-hydroxyvitamin D and breast cancer in the military: a case-control study utilizing pre-diagnostic serum. Cancer Causes Control. 2013;24(3):495–504. doi: 10.1007/s10552-012-0140-6. [DOI] [PubMed] [Google Scholar]
- 35.Rejnmark L, Tietze A, Vestergaard P, Buhl L, Lehbrink M, Heickendorff L, et al. Reduced prediagnostic 25-hydroxyvitamin D levels in women with breast cancer: a nested case-control study. Cancer Epidemiol Biomarkers Prev. 2009;18(10):2655–60. doi: 10.1158/1055-9965.EPI-09-0531. [DOI] [PubMed] [Google Scholar]
- 36.Bauer SR, Hankinson SE, Bertone-Johnson ER, Ding EL. Plasma vitamin D levels, menopause, and risk of breast cancer: dose-response meta-analysis of prospective studies. Medicine (Baltimore) 2013;92(3):123–31. doi: 10.1097/MD.0b013e3182943bc2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abbas S, Chang-Claude J, Linseisen J. Plasma 25-hydroxyvitamin D and premenopausal breast cancer risk in a German case-control study. Int J Cancer. 2009;124(1):250–5. doi: 10.1002/ijc.23904. [DOI] [PubMed] [Google Scholar]
- 38.Abbas S, Linseisen J, Slanger T, Kropp S, Mutschelknauss EJ, Flesch-Janys D, et al. Serum 25-hydroxyvitamin D and risk of post-menopausal breast cancer--results of a large case-control study. Carcinogenesis. 2008;29(1):93–9. doi: 10.1093/carcin/bgm240. [DOI] [PubMed] [Google Scholar]
- 39.Chen P, Li M, Gu X, Liu Y, Li X, Li C, et al. Higher blood 25(OH)D level may reduce the breast cancer risk: evidence from a Chinese population based case-control study and meta-analysis of the observational studies. PLoS One. 2013;8(1):e49312. doi: 10.1371/journal.pone.0049312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colston KY, Lowe LC, Mansi JL, Campbell MJ. Vitamin D status and breast cancer risk. Anticancer Res. 2006;26(4A):2573–2580. [PubMed] [Google Scholar]
- 41.Crew KD, Gammon MD, Steck SE, Hershman DL, Cremers S, Dworakowski E, et al. Association between plasma 25-hydroxyvitamin D and breast cancer risk. Cancer Prev Res (Phila) 2009;2(6):598–604. doi: 10.1158/1940-6207.CAPR-08-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahn J, Yu K, Stolzenberg-Solomon R, Simon KC, McCullough ML, Gallicchio L, et al. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet. 2010;19(13):2739–45. doi: 10.1093/hmg/ddq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engelman CD, Meyers KJ, Ziegler JT, Taylor KD, Palmer ND, Haffner SM, et al. Genome-wide association study of vitamin D concentrations in Hispanic Americans: the IRAS family study. J Steroid Biochem Mol Biol. 2010;122(4):186–92. doi: 10.1016/j.jsbmb.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43(Database issue):D1049–56. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welsh J, Wietzke JA, Zinser GM, Byrne B, Smith K, Narvaez CJ. Vitamin D-3 receptor as a target for breast cancer prevention. J Nutr. 2003;133:2425S–33S. doi: 10.1093/jn/133.7.2425S. [DOI] [PubMed] [Google Scholar]
- 47.Goeman F, De Nicola F, D’Onorio De Meo P, Pallocca M, Elmi B, Castrignano T, et al. VDR primary targets by genome-wide transcriptional profiling. J Steroid Biochem Mol Biol. 2014;143:348–56. doi: 10.1016/j.jsbmb.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 48.Borkowf CB, Albert PS, Abnet CC. Using lowess to remove systematic trends over time in predictor variables prior to logistic regression with quantile categories. Stat Med. 2003;22(9):1477–93. doi: 10.1002/sim.1507. [DOI] [PubMed] [Google Scholar]
- 49.Amos CI, Dennis J, Wang Z, Byun J, Schumacher FR, Gayther SA, et al. The OncoArray Consortium: a Network for Understanding the Genetic Architecture of Common Cancers. Cancer Epidemiol Biomarkers Prev. 2016 doi: 10.1158/1055-9965.EPI-16-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.UCSC Genome Browser Home. University of California Santa Cruz; [accessed June 29, 2016]. https://genome.ucsc.edu/ [Google Scholar]
- 51.Prentice RL. A case-cohort design for epidemiologic cohort studies and disease prevention trials. Biometrika. 1986;73:1–11. [Google Scholar]
- 52.Barlow WE, Ichikawa L, Rosner D, Izumi S. Analysis of case-cohort designs. J Clin Epidemiol. 1999;52:1165–72. doi: 10.1016/s0895-4356(99)00102-x. [DOI] [PubMed] [Google Scholar]
- 53.Lin X, Lee S, Christiani DC, Lin X. Test for interactions between a genetic marker set and environment in generalized linear models. Biostatistics. 2013;14(4):667–81. doi: 10.1093/biostatistics/kxt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 2005;95(3):221–7. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 55.Benjamini Y, Hochberg Y. Controlling the false discover rate: A practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57(1):289–300. [Google Scholar]
- 56.The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chelala C, Khan A, Lemoine NR. SNPnexus: a web database for functional annotation of newly discovered and public domain single nucleotide polymorphisms. Bioinformatics. 2009;25(5):655–61. doi: 10.1093/bioinformatics/btn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pagini F, Baralle FE. Genomic variants in exons and introns: Identifying the splicing spoilers. Nature Reviews Genetics. 2004;5:389–96. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 59.Tombolan L, Zampini M, Casara S, Boldrin E, Zin A, Bisogno G, et al. MicroRNA-27a Contributes to Rhabdomyosarcoma Cell Proliferation by Suppressing RARA and RXRA. PLoS One. 2015;10(4):e0125171. doi: 10.1371/journal.pone.0125171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi J, Grundy A, Richardson H, Burstyn I, Schuetz JM, Lohrisch CA, et al. Genetic variation in vitamin D-related genes and risk of breast cancer among women of European and East Asian descent. Tumour Biol. 2016;37(5):6379–87. doi: 10.1007/s13277-015-4417-8. [DOI] [PubMed] [Google Scholar]
- 61.Gapska P, Scott RJ, Serrano-Fernandez P, Huzarski T, Byrski T, Kladny J, et al. Vitamin D receptor variants and breast cancer risk in the Polish population. Breast Cancer Res Treat. 2009;115(3):629–33. doi: 10.1007/s10549-008-0107-1. [DOI] [PubMed] [Google Scholar]
- 62.Manousaki D, Dudding T, Haworth S, Hsu YH, Liu CT, Medina-Gomez C, et al. Low-Frequency Synonymous Coding Variation in CYP2R1 Has Large Effects on Vitamin D Levels and Risk of Multiple Sclerosis. Am J Hum Genet. 2017 doi: 10.1016/j.ajhg.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zgaga L, O’Sullivan F, Cantwell MM, Murray LJ, Thota PN, Coleman HG. Markers of Vitamin D Exposure and Esophageal Cancer Risk: A Systematic Review and Meta-analysis. Cancer Epidemiol Biomarkers Prev. 2016;25(6):877–86. doi: 10.1158/1055-9965.EPI-15-1162. [DOI] [PubMed] [Google Scholar]
- 64.Malodobra-Mazur M, Paduch A, Lebioda A, Konopacka M, Rogolinski J, Szymczyk C, et al. VDR gene single nucleotide polymorphisms and their association with risk of oral cavity carcinoma. Acta Biochim Pol. 2012;59(4):627–30. [PubMed] [Google Scholar]
- 65.Holick CN, Stanford JL, Kwon EM, Ostrander EA, Nejentsev S, Peters U. Comprehensive association analysis of the vitamin D pathway genes, VDR, CYP27B1, and CYP24A1, in prostate cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(10):1990–9. doi: 10.1158/1055-9965.EPI-07-0487. [DOI] [PubMed] [Google Scholar]
- 66.Weinberg CR, Shore DL, Umbach DM, Sandler DP. Using risk-based sampling to enrich cohorts for endpoints, genes, and exposures. Am J Epidemiol. 2007;166(4):447–55. doi: 10.1093/aje/kwm097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.