Abstract
Despite well-documented evidence for lifespan extension by methionine restriction (MR), underlying mechanisms remain unknown. As methionine can alter S-adenosylmethionine (SAM) and S-adenosylhomocysteine (SAH), the substrate and product of DNA methyltransferase-1 (DNMT1), we hypothesized that MR diet alters DNA methylation. Young (8-week-old) and adult (1-year-old) male C57BL/6J mice were fed diets with different levels of methionine (0.12%-MR, 0.84%-CD) for 12 weeks. Functional indicators of DNA methylation, including global methylation (GM), gene-specific methylation (GSM) and LINE-1 methylation; and biochemical factors affecting DNA methylation, SAH, SAM, and DNMT1 were assessed in different tissues. MR altered DNA methylation depending on the age of intervention. While MR had no effect on hepatic GM in young animals, it increased GM by 27% over CD in adults (P<0.01). In comparison with young animals, hepatic GM levels were 17% lower in CD adults (P<0.05), but not different in MR adults. The MR-induced increase in hepatic GM was associated with a 38% decrease in SAH levels in adults (P<0.001), with SAH and GM levels being negatively correlated (r2=0.33, p<0.001). No changes were observed in DNMT protein levels in liver. In adipose tissue, MR caused a 6% decline in GM in adults (p<0.05), a corresponding 2-fold increase in SAH (p<0.05), and a 2-fold decrease in DNMT1 (p<0.01). MR caused both increases and decreases in GSM of liver and adipose. No changes were observed in LINE-1. Together, these findings provide evidence for protective effects of MR diet on hepatic DNA hypomethylation in adults, apparently mediated by SAH. These findings also indicate that altered DNA methylation might be playing a role in benefits conferred by MR diet.
Keywords: Aging, DNA methylation, methionine, caloric restriction, one-carbon cycle, sulfur amino acids
1. Introduction
Methionine restriction (MR) is one of the few dietary regimens which extend lifespan in rodents [1, 2]. Beneficial effects induced by MR in glucose metabolism, lipid metabolism, and oxidative stress have been identified as some contributory phenomena [3, 4]. To date the mechanism(s) by which MR exerts its beneficial effect on the aging process remain unknown. Many of its effects are associated with altered levels of intermediates of methionine metabolism such as glycine, serine, choline, cysteine, and glutathione. But, little is known about MR-induced changes in other intermediates such as S-adenosylmethionine (SAM) and S-adenosylhomocysteine (SAH) and their effect on MR phenotype. SAM and SAH regulate methyl group metabolism by transferring methyl moieties from methionine to several cellular substrates.
Deficiency of methyl groups leads to several pathologies. Liver is the primary organ for methyl group metabolism and a major source of SAM. Up to 80% of methionine in liver is converted to SAM [5]. Decreased levels of methyl group donors such as methionine, folate, choline, and betaine are associated with non-alcoholic fatty liver disease, cancer, and cardiovascular diseases [6–8]. Surprisingly, MR diet, despite having 80% lower methionine does not cause symptoms that are typical of methyl donor deficiency [9–12]. A possible reason for the safe nature of MR diet could be the presence of other fungible methyl donors such as choline and folate. MR diet can still alter methyl group metabolism as the carbon skeleton of methionine is essential for the synthesis of SAM and SAH, the substrate and product of transmethylation reactions [13]. While methylation of several substrates can affect cell metabolism, changes in methylation of DNA can result in a cascade effect impacting multiple events. In this study, we hypothesized that MR diet alters DNA methylation through SAH and SAM.
A gradual decline in DNA methylation is observed during aging [14]. Unlike genetically programmed changes in early development, changes in DNA methylation during later life are driven primarily by biochemical factors (SAH, DNMT, and SAM) which are susceptible to dietary interventions. Regulation of methionine metabolism and the levels of its intermediates is also age-dependent [15]. In vivo studies in rats show that hepatic concentrations of SAH increase with aging, suggesting that DNA hypomethylation during aging could be due to the inhibitory effect of SAH on DNMT [16]. Although a general decline in global DNA methylation is observed during aging, specific regions of genome show both hypomethylation and hypermethylation, which could affect gene expression [17]. Previous studies demonstrate that MR induces a transcriptional shift resulting in altered expression of genes involved in glucose and lipid metabolism [18]. However, it is not known whether altered GSM contributes to these transcriptional changes. In this study, we sought to determine the impact of MR on SAH, SAM and DNMT levels as well as both GM and GSM in liver and other relevant tissues in young (8-week-old) and adult (1-year-old) mice. To this end, C57BL/6J mice were fed an isocaloric MR diet (0.12% methionine) and control diet (CD, 0.84% methionine) for 12 weeks and tissue levels of DNA methylation (global methylation, GM; gene-specific methylation, GSM; and methylation of long interspersed nuclear elements, LINE-1), and SAH, SAM and DNMT1 were determined.
2. Methods
2.1. Animals, diets, and tissue harvesting
Animal use was approved by the Institutional Animal Care and Use Committee of Orentreich Foundation for the Advancement of Science. Two cohorts of male C57BL/6J mice (Jackson Laboratories, Bar Harbor, ME, USA), young (8-week-old) and adult (1-year-old) were housed in a conventional facility at 20±2°C and 50±10% relative humidity with a 12h dark/12h light cycle. One week after quarantine, each cohort was divided into two weight-matched diet groups (n=8/group) and fed either CD (0.84% methionine) or MR (0.12% methionine, Research Diets, New Brunswick, NJ, USA) diet ad libitum for 12 weeks. Both diets were isocaloric and isonitrogenous, and contained no cysteine (Table 1). Growth rates were monitored weekly. At the end of the study period, the mice were sacrificed by cervical dislocation, and tissues of interest [liver, epididymal adipose (white adipose tissue), quadriceps muscle, lung, brain, heart, and kidneys] were harvested, immediately snap frozen, and stored at −80°C until processed.
Table 1.
Dietary composition
| Ingredient | Amount (g %) |
|---|---|
| L-Arginine | 1.09 |
| L-Histidine-HCl-H2O | 0.32 |
| L-Isoleucine | 0.80 |
| L-Leucine | 1.08 |
| L-Lysine | 1.40 |
| DL-Methionine | 0.84 (0.12)a |
| L-Phenylalanine | 1.13 |
| L-Threonine | 0.80 |
| L-Tryptophan | 0.17 |
| L-Valine | 0.80 |
| L-Glutamic acid | 2.62 (3.34)a |
| Glycine | 2.26 |
| Corn starch | 41.24 |
| Maltodextrin | 12.14 |
| Dextrose | 4.86 |
| Sucrose | 14.57 |
| Cellulose | 4.86 |
| Lard | 0.00 |
| Corn oil | 4.47 |
| Mineral mix S10001 | 3.40 |
| Choline bitartrate | 0.19 |
| Vitamin mix 10001b | 0.97 |
| B6 | 7.00 |
| Folic acid | 2.00 |
| B12 | 10.00 |
Numbers in parenthesis represent amounts in MR diet
B6 and folic acid are expressed as mg/kg diet. B12 is expressed as μg/kg diet.
2.2 Determination of DNA methylation
2.2.1. DNA extraction
Genomic DNA was extracted by the phenol-chloroform method. Frozen tissues (250 mg) were thawed and homogenized in a TissueLyser (Qiagen, Valencia, CA, USA) after adding 1mL of ice-cold lysis buffer (20mM Tris-HCl, 1mM EDTA, 10mM NaCl, and 10% SDS, Sigma-Aldrich, St. Louis, MO, USA). Homogenates were incubated with proteinase K (0.1mg/mL, Sigma-Aldrich) at 55°C for 3h. DNA was obtained by two repeated extractions with phenol followed by chloroform-isoamyl alcohol. DNA in the aqueous phase was purified by precipitation with 0.1 volume of 3M sodium acetate and 2 volumes of ice-cold absolute ethanol. Pellets were dissolved in DNAse-free water, checked for quality (absorbance at 260nm, 280nm, and 230nm), and used in methylation analyses.
2.2.2. Global DNA Methylation
GM was determined by UPLC/MS quantification of methylated (5-meC) and unmethylated (5-C) cytosines. DNA from different tissues was degraded by nuclease (DNA Degradase Plus, Zymo Research, Irvine, CA, USA) according to the manufacturer’s protocol and diluted with 5% methanol to a final concentration of 25 ng/μL. A mixed standard curve of 5-dC and 5-medC (Berry & Associates, Dexter, MI, USA) was constructed with concentrations ranging from 0.25ng/mL to 500ng/mL. Separation of 5-dC and 5-medC was achieved on a Waters Acquity UPLC separation system coupled to an ABSciex 4000 Q Trap mass spectrometry system. HPLC conditions were as follows: column - Acquity UPLC BEH C18 column, 1.7μm (Waters, Ireland), gradient elution at a flow rate of 0.3mL/min, 2.5minutes in 20% mobile phase B (0.1% formic acid in methanol) and 80% solvent A (0.1% formic acid in water) followed by a linear gradient to 95% mobile phase B in 30s, and column flushing with 95% mobile phase B. The ABSciex 4000 Q Trap mass spectrometer was equipped with an electrospray ionization probe operated in positive mode. The decluster potential was 41V for 5-dC and 51V for 5-medC; the entrance potential was 10V, the collision energy was 17V and the collision cell exit potential was 10V for both compounds, while the curtain gas was 25h/L, the collision gas was 5h/L for both 5-dC and 5-medC. The ionspray voltage was 5500V, the temperature was 400°C, the ion source gas 1 was 70h/L and the ion source gas 2 was 50h/L. The multiple reaction monitoring mode was used to analyze and quantify 5-dC and 5-medC, with the transitions of m/z 228.2 > 112.2 for 5-dC and 242.2 > 126.1 for 5-medC. All peaks were integrated and quantified by ABSciex MultiQuant 3.0 software. 5-medC was expressed as percent total cytosines.
2.2.3. LINE-1 methylation
LINE-1 methylation was determined by sodium bisulfite conversion of unmethylated cytosines to uracils followed by PCR amplification as thymines and pyrosequencing (assays were performed at EpigenDx, Hopkinton, MA, USA). Genomic DNA (500ng) from liver, adipose, and skeletal muscle was denatured by adding 5μL of 3N NaOH in a total reaction volume of 50μL and incubated at 42°C for 30min. Denatured DNA was bisulfite treated by adding 100μL of proprietary Bisulfite salt solution and incubated at 50°C for 14h followed by column purification as recommended by the manufacturer (Zymo DNA columns, Zymo Research, Irvine, CA) and was eluted with 20μL of T1E0.2 pH 8.0. Eluted DNA was PCR-amplified using a proprietary primer set (EpigenDx assay no. ADS685) with one biotinylated primer (0.2μM). The amplicons were purified by immobilization on sepharose beads (Streptavidin Sepharose HP, Amersham Biosciences, Uppsala, Sweden), according to the manufacturer’s recommendations. 10μL of the purified single-stranded PCR product was sequenced by using 0.2μM pyrosequencing primer on a Pyrosequencing PSQ96 HS System (Biotage AB) according to the manufacturer’s instructions (Pyrosequencing, Qiagen). The methylation status of 5 CpG loci in the product was analyzed individually as a T/C SNP using QCpG software (Pyrosequencing, Qiagen). LINE-1 methylation is expressed as the average percent methylation of the 5 CpG loci.
2.2.4. Screening for gene-specific methylation
GSM was determined by quantifying the degree of methylation in known differentially-methylated regions of selected genes by targeted next generation bisulfite sequencing (EpigenDx). A local database of genomic sequences was established for a panel of genes involved in glucose metabolism, lipid metabolism, oxidative stress, and the methionine cycle. Genomic sequences and differentially-methylated CpG regions were derived using three publicly available databases, Ensembl (http://www.ensembl.org/), NCBI Epigenomics database (http://www.ncbi.nlm.nih.gov/epigenomics), and UCSC Encode database (http://genome.ucsc.edu/ENCODE/). Bisulfite PCR primers to amplify 100 to 350 bases covering the target region were designed using PyroMark Assay Design Software (Qiagen). Optimization and validation of multiplexing resulted in 6 multiplexing reactions with a total of 40 amplicons. Genomic DNA (500ng) from liver, adipose, and skeletal muscle was bisulfite treated using the EZ DNA Methylation Kit (Zymo Research) and eluted in 40μL buffer. Multiplex PCRs were performed on 3μL of eluted bisulfite-treated DNA using 0.75 unit of TaKaRa EpiTaq HS polymerase (Takara Bio, Mountain View, CA, USA) and master mix (EpigenDx). The primer sequences are listed in Supplementary Table 1. Libraries were prepared using the KAPA Library Preparation Kit for Ion Torrent. The Ion PGM Template OT2 200 Kit and Ion PGM Sequencing 200 Kit v2 were used with an Ion 318 Chip (Life Technologies, Carlsbad, CA, USA) on the Ion Torrent PGM. The methylation percentage of each sequenced CpG site was calculated by dividing the number of methylated reads by the number of total reads (sum of methylated and unmethylated), considering all CpG sites covered by a minimum of 20 reads. A local alignment database for the listed genes was generated upon assay design. FASTQ files from the Ion Torrent PGM were aligned to the local database using open source Bismark Bisulfite Read Mapper with the Bowtie2 alignment algorithm. Bismark was used for methylation calling, and Genomics Suite (Partek, St. Louis, MO, USA) was used for annotation. Four samples were analyzed from each treatment group, and statistical analysis was done by ANOVA.
2.3. Determination of SAM and SAH
Methionine cycle intermediates were measured by LC/QE-MS. Detailed procedures for metabolite extraction, LC-MS, and data analysis have been described elsewhere [19]. Briefly, 5–10 mg of tissue was ground in liquid nitrogen and dissolved in 250 μL ice-cold methanol. The homogenate was incubated on ice for 10 min and centrifuged at 20,000g, 4°C for 10min. Solvent was evaporated in a speed vacuum and the pellet dissolved in 15 μL water and 15 μL methanol/acetonitrile (1:1 v/v) (LC-MS optima grade, Thermo Scientific, Waltham, MA, USA). Samples were centrifuged at 20,000g for 10min at 4°C and 5μL of the supernatants were transferred to liquid chromatography vials. Samples were separated on an Xbridge amide column (100 × 2.1 mm, i.d.3.5 μm; Waters) utilizing liquid chromatography (Ultimate 3000 UHPLC), and then injected into the qExactive Mass Spectrometer (QE-MS) equipped with a heated electrospray ionization probe. The LC method and QE-MS parameters are described elsewhere [19]. Raw data collected from LC-Q Exactive MS was processed for peak alignment and detection using thermo software Sieve 2.0 (Thermo Scientific) using the method peak alignment and frame extraction.
2.4. DNMT1 protein levels
DNMT1 protein was quantified by an immunoaffinity-based colorimetric assay using a DNMT1 assay kit (Epigentek, Farmingdale, NY, USA). The method involves incubating nuclear protein in wells coated with a substrate and detecting the amount of DNMT1 bound to the substrate with a specific antibody. Nuclear extracts from liver, adipose, and muscle were prepared using EpiQuik Nuclear Extraction Kit according to the manufacturer’s protocol (Epigentek), and total protein was determined by BCA assay (Thermo Fisher). Total DNMT1 was quantified based on a standard curve for known amounts of DNMT1 according to the manufacturer’s instructions.
2.5. Statistical analysis
Data were analyzed by two-way ANOVA. P-values for interaction between age and diet are represented as pint in text and by § in figures. Post-hoc analyses were done by Sidak’s multiple comparison tests. Trends for independent effects of age and diet were tested by the unpaired two-tailed Student’s t-test (p-values represented as p2t). In all cases, differences were considered significant if p<0.05.
3. Results
3.1. Growth rates
Effects of MR on growth rates and food intake in both young and adult mice were similar to those previously observed in rodents [20]. Young mice on the CD continued to grow until the end of the study; those on MR also continued to grow but at a slower rate (Figure 1). Adult mice on the CD grew until 8 weeks on the diet, while mice on MR, lost 10% of body weight by the 3rd week and maintained it thereafter. After 12 weeks on the MR diet, young and adult mice weighed 27% and 25% less than controls, respectively (p<0.0001). Mice on CD and MR ate similar amounts of food (g/week) regardless of age (Supplementary Figure 1a). But, on bodyweight basis young and adult mice on MR ate 9% (p=0.007) and 16% (p=0.0002), respectively, more than their controls (Supplementary Figure 1b). No differences were observed in the organ weights in relation to body weight as a result of MR except for lungs where a 15% and 28% increase (p=0.01, p<0.0001) was observed in young and adult mice on MR, respectively (data not shown). Despite a change in percent weights of lung, no signs of respiratory distress were apparent during general animal husbandry activities.
Figure 1. Growth rates of C57BL/6J mice on an MR diet.
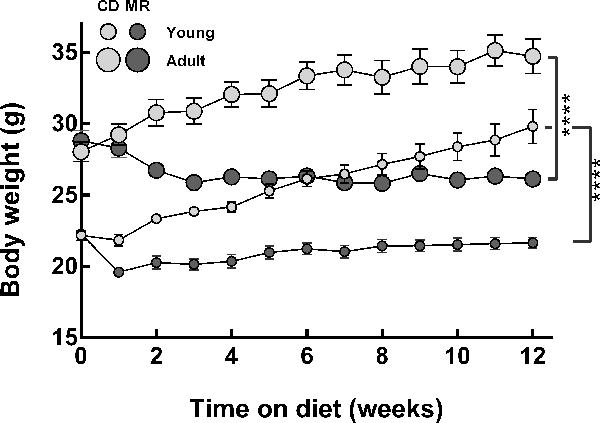
Young (8-week-old) and adult (1-year-old) male C57BL/6J mice (n=8/group) were fed ad libitum with diets containing either 0.12% methionine (MR) or 0.84% methionine (CD) for 12 weeks. Body weights were monitored weekly. After 12 weeks on MR diet, both young and adult mice weighed 25% and 27% lower than their respective controls (p<0.0001). Error bars represent standard errors.
3.2 Effect of MR on GM, SAH, and DNMT in liver
A large proportion of methionine metabolism occurs in liver. MR diet affected GM in liver depending on the age of intervention (pint=0.015). MR had no effect on GM in young mice but increased it in adult mice by 26% (p=0. 001, Figure 2a). On CD, as observed in previous studies, an age-associated decline in GM was found [21]. Adult mice on CD had 17% lower GM levels than in young mice (p=0. 018) [22]. However, levels in adult MR mice were similar to those in young animals in both diet groups. MR also induced changes in hepatic SAH, which were opposite and concomitant to changes in GM (Figure 2b). Changes induced in SAH were also dependent on age of intervention (pint=0.012). MR had no effect on SAH in young mice but decreased it in adult mice by 50% (p<0.001). Similar to previous observations, an age-related increase in SAH of 38% was observed in adults compared to young CD animals (p<0.0001) [15]. SAH levels in adult MR mice were similar to those in young mice on either diet. A significant negative correlation was found between SAH and GM (r2=0.33; p<0.001, Figure 2c). No age-related or diet-related changes were observed in DNMT protein levels (data not shown). MR decreased liver SAM in young mice by 64% (p<0.001, Figure 2d), but no changes were observed in adults (p<0.001). No correlation was found between GM and SAM levels (Figure 2e)
Figure 2. MR diet protected C57BL6/J mice from age-related hepatic global DNA hypomethylation and increase in S-adenosylhomocysteine.
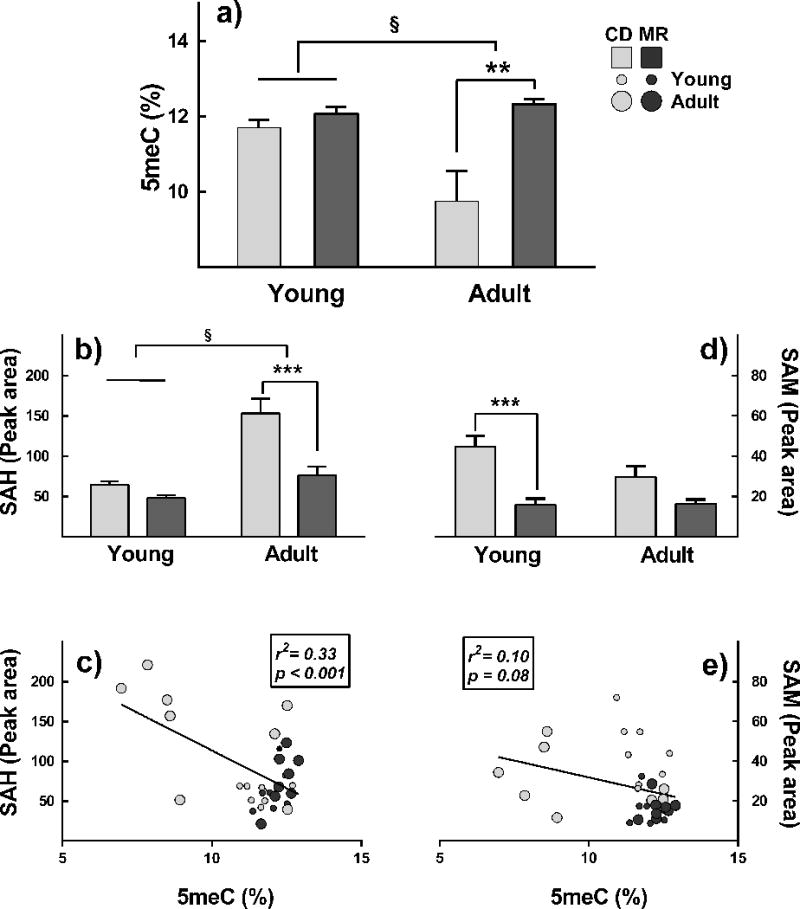
Young (8-week-old) and adult (1-year-old) male C57BL/6J mice (n=8/group) were either 0.12% methionine (MR) or 0.84% methionine (CD) diets for 12 weeks. Unlike CD, MR did not cause DNA hypomethylation (panel a) and did not increase SAH (panel b) in livers of adult mice. No differences were found in young mice. MR decreased SAM levels in young mice but no changes were found in adults (Figure 2d). A significant correlation was found between GM and SAH (panel c) but not SAM (panel e) Note: §, p<0.05 for interaction between diet and age of intervention; *p≤0.05, **p≤0.01, and ***p≤0.001.
3.3. Effect of MR on GM, SAH, and DNMT in adipose
Changes induced by MR in epididymal adipose were significant but less robust than those in liver. Even though no interaction between age and diet was found, MR-induced changes in adipose were limited to adult mice (Figure 3). Adult mice on MR had 6% lower GM levels in adipose compared to adult mice on CD (p=0.025, Figure 3a). Changes observed in SAH and DNMT levels in adult mice complemented the changes in GM. MR increased SAH levels in adult mice by 16% (p=0.025, Figure 3b) and lowered DNMT1 protein levels by 49% (p=0. 03, Figure 3c). MR had no effect on GM, SAH and DNMT in young mice. Except for an increasing trend in GM of skeletal muscle of young mice, no differences were observed in other organs investigated (p2t< 0.001, Supplementary Figure 2)
Figure 3. Effect of MR on global DNA methylation, S-adenosylhomocysteine, and DNMT in adipose.
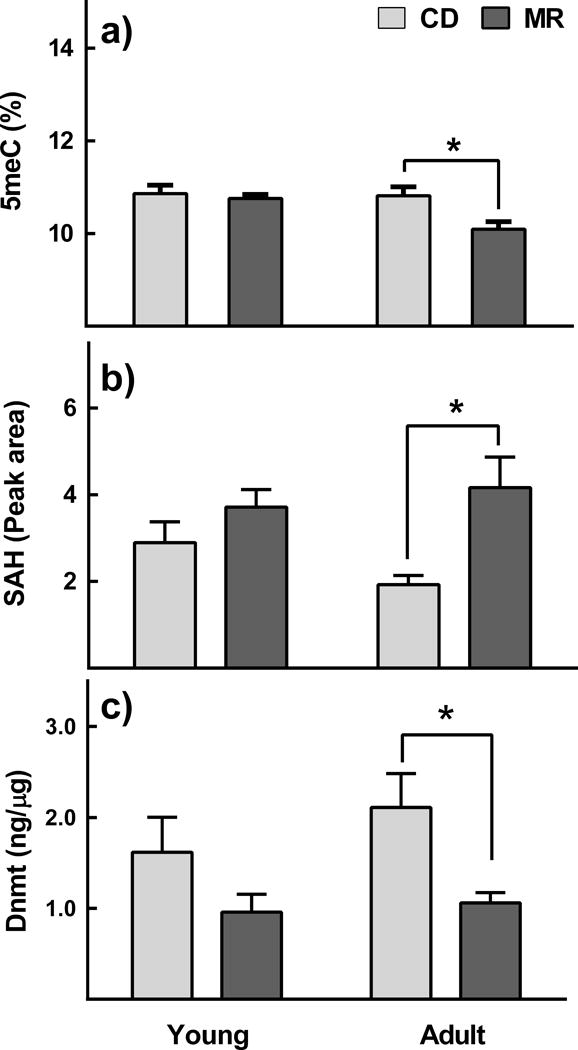
Young (8-week-old) and adult (1-year-old) male C57BL/6J mice (n=8/group) were ad libitum fed with diets containing either 0.12% methionine (MR) or 0.84% methionine (CD) for 12 weeks. MR decreased global DNA methylation in adult mice (panel a). A complementary increase in SAH (panel b) and decrease in DNMT (panel c) was also found. MR had no effect in young mice. Note: *p≤0.05.
3.4. Gene-specific methylation
Unlike GM, GSM can be either increased or decreased with age, dependent on the specific gene locus being investigated [23]. We investigated the status of GSM in a selected panel of genes, whose mRNA expression was previously reported to be effected by the MR diet [24]. Diet and age caused both increases and decreases in GSM in both liver and adipose, with liver exhibiting the more number of changes, followed by adipose. CpG sites with more than a 2-fold change in methylation are presented in Figure 4 and Supplementary Figure 3 (p<0.05). To determine if the amplitude of change induced by MR depends on natural levels of methylation at each locus, we preformed correlation analyses between the methylation levels at each CpG site on CD and the ratio of methylation. No significant correlations were observed in any of the tissues examined. However, it is noteworthy that in general changes induced by MR in liver of both adult (Figure 4b) and young mice (Figure 4c) are opposite to changes induced by aging (Figure 4a).
Figure 4. Effect of MR on gene-specific methylation in liver.
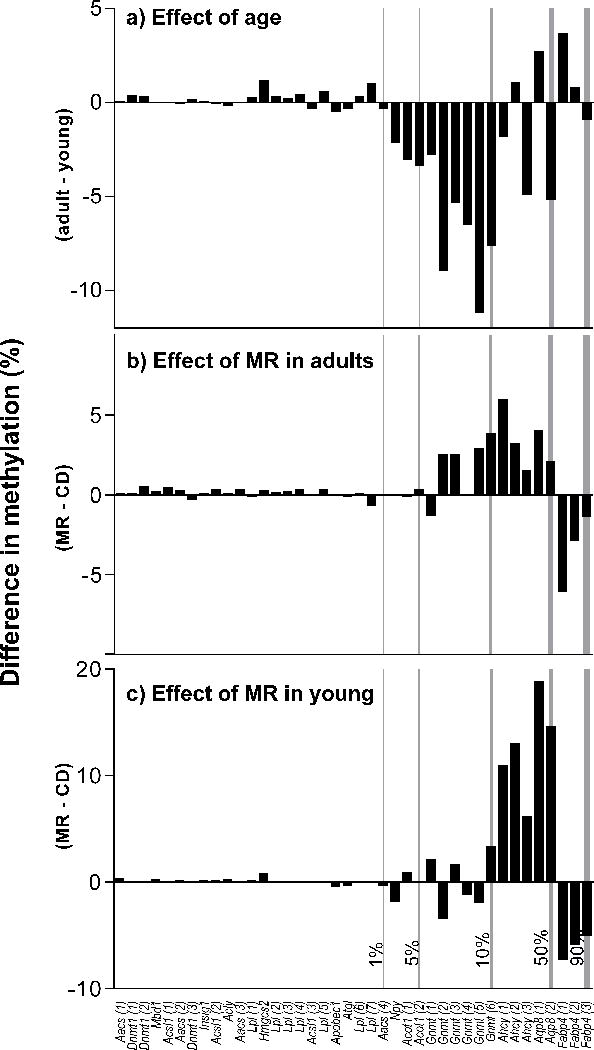
Young (8-week-old) and adult (1-year-old) male C57BL/6J mice were fed ad libitum with diets containing either 0.12% methionine (MR) or 0.84% methionine (CD) for 12 weeks. Changes induced by MR in GSM of liver in adult (panel b) and young mice (panel c) are opposite to those changes occurred during aging on CD (panel a). Note: CpG sites with at least a 2-fold difference by 2-way ANOVA are presented (p<0.05). CpG sites on x-axis are ranked based on the natural levels of methylation in young mice on CD (methylation levels are indicated at the bottom of vertical lines). A positive value indicates higher methylation in adult mice (panel a) and mice on MR (panels b and c). Bars represent average (n=4/group) difference in percent methylation at each CpG site.
3.5. LINE-1 methylation
Altered LINE-1 methylation is associated with carcinogenesis [25]. Since MR is known to prevent cancer, we investigated if MR alters LINE-1 methylation status. MR did not alter LINE-1 methylation levels either in liver or adipose (Figure 5).
Figure 5. MR diet does not alter LINE-1 methylation.
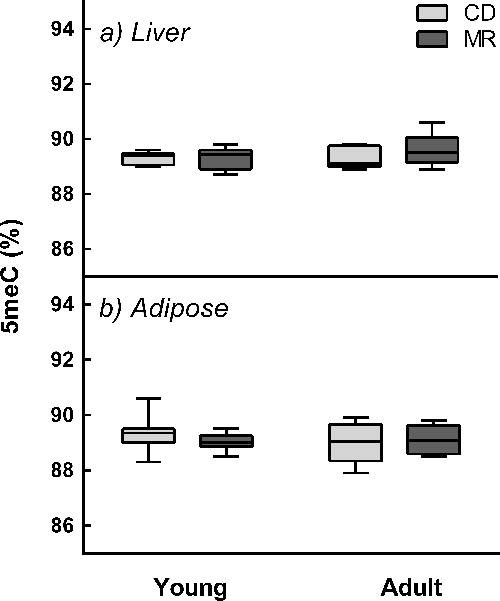
Young (8-week-old) and adult (1-year-old) male C57BL/6J mice (n=8/group) were fed ad libitum with diets containing either 0.12% methionine (MR) or 0.84% methionine (CD) for 12 weeks. LINE-1 methylation (5meC) in liver and adipose was determined by pyrosequencing of bisulfite treated DNA. Lines in boxes represent 25th percentile, median, and 75th percentile. Whiskers represent range of minimum and maximum values.
4. Discussion
MR diet extends health span and life span in rodents, but mechanisms remain unknown [1]. Because methionine is a direct precursor for SAM and SAH, we hypothesized that MR diet alters DNA methylation by modulating SAM and SAH levels. We found that MR diet protects adult mice from age-related DNA hypomethylation in liver. While MR diet altered both SAM and SAH, DNA methylation levels correlated with SAH only. In addition, the effects of MR were age-dependent, reflecting age-dependent changes in SAH levels. These findings suggest that maintenance of DNA methylation and integrity, by modulating SAH levels, might be playing a role in MR-induced benefits. DNA hypomethylation is associated with lifespan and several age-related diseases such as cancer, Alzheimer disease, and diabetes [26]. Additional studies are required to investigate if changes in DNA methylation contribute to lifespan extension by MR diet.
In vivo studies indicate that global DNA methylation levels are associated with changes in the levels of SAM, SAH, or the ratio of two with GM [27, 28]. Whether SAM and SAH can independently predict GM levels has been controversial. Previous studies show that changes in SAM, SAH and DNA methylation occur as soon as 6–8 weeks after dietary intervention [29–31]. In our 12-week intervention, we found a significant negative correlation between GM in liver and SAH alone, but not with either SAM alone or the SAM:SAH ratio. This is in direct agreement with the enzyme kinetics of DNMT at physiological conditions. For many methyltransferases, including DNMT, the ki for SAH is much lower than the km for SAM, suggesting that DNA methylation is much more sensitive to changes in SAH than to changes in SAM [32]. This also explains the lack of effect on GM in liver of young mice on MR despite a decrease in SAM levels (Figures 1a and 1d) and lack of correlation between SAM levels and GM (Figure 1e). Furthermore, previous studies have demonstrated that a decrease in SAM alone is not sufficient to alter DNA methylation, where as an increase in SAH alone or in combination with a decrease in SAM was able to decrease GM [27].
The effect and mechanisms of MR diet on DNA methylation appear to be unique. Methyl deficient diets are known to induce DNA hypomethylation in LINE-1 elements and cause cancer [8]. But, the MR diet in our study, despite having 80% less methionine, increased GM in adult livers and maintained LINE-1 methylation [33]. This could be due to the availability of methyl groups from choline, which is present in the MR diet at sufficient levels (0.2%). The number of methyl groups provided by choline ((CH3)3N(CH2)2OH) is also higher than those provided by methionine (CH3-S-(CH2)2-CH(NH2)-COOH), three and one, respectively. This suggests that the changes in DNA methylation are not due to the role of methionine as a methyl donor, but due to its ability to alter the levels of SAH. This is in contrast to the mechanisms known for other dietary regimens used in lifespan studies. Caloric restriction, resveratrol, and other dietary polyphenols are reported to alter methylation of specific genes and histones, mostly mediated by sirtuins and DNMTs, rather than by inducing changes in biochemical mediators such as SAH and SAM [34, 35]. As it is difficult to measure enzyme activity under in vivo conditions, we quantified DNMT protein extracted from liver (data not presented). While no differences were found in protein levels, we cannot rule out MR-induced changes in DNMT activity.
The age cohorts chosen in this study were intended to differentiate the effects of MR on active growth phase of life (young) with that in which growth and development slows and senescent changes start to appear (adult). MR had greater effect on the GM and SAH, but lesser effect on SAM in adult mice compared to young mice (Figure2b and 2d, and Supplementary Figure2). These differences are probably rooted in the age-dependent metabolism of methionine. Under higher growth rates and low methionine availability, young mice might be diverting more methionine to protein synthesis, at the expense of SAM synthesis than adults, thus, accentuating the decrease in SAM levels. Conversely, adults would need more glutathione, a metabolite of transsulfuration pathway, to combat environmental and biological challenges, compared to young. Previous studies demonstrated the importance of preserving the transsulfuration pathway in the elderly at the expense of transmethylation, particularly at times of stress such as inflammation [36]. Mild nutritional stress under MR and adolescence could be driving the greater flux of SAH into the transsulfuration pathway in the adult mice than in young mice.
DNA methylation is a tissue-specific phenomenon [37]. Factors such as cell proliferation and rate of metabolic activity determine tissue-specificity. We observed that MR-induced changes in GM were greatest in the liver followed by adipose and skeletal muscle, consistent with the extent of overall methyl group metabolism in these tissues [38]. The greater impact of MR on liver is not surprising considering that 85% of transmethylation reaction ns and 45% of methionine metabolism occurs in this tissue [39]. MR-induced changes in GM of adipose tissue were significant but smaller than in liver. This could be due to limited proliferative capacity and decreased need for DNA replication compared to liver. Adipose, compared to liver, is also known to undergo fewer epigenetic changes during the aging process [40]. Previous studies demonstrate that among 40,000 unique genomic sites evaluated for DNA methylation levels, liver had 50% more significant changes than adipose tissue [40, 41].
Considering the growing evidence for altered mRNA expression by individual CpG sites, we analyzed methylation status in intragenic regions with one or more CpG sites [42]. MR changed methylation status in several CpG sites in liver and adipose. But for many genes reported changes in mRNA expression and GSM are not congruent [24]. While it is difficult to explain the lack of association from the current study, we speculate that changes in gene expression can be better explained by considering the changes in histone methylation together. Previous studies show that methylation of H3K4me3 is inversely associated with DNA methylation [42]. We recently demonstrated that MR-induced decrease in H3K4me3 methylation in gene bodies is associated with increased mRNA expression [43]. Thus it is possible that changes in mRNA expression of genes we selected could rather be driven by changes in histone methylation. Also, unlike GM, GSM can be either increased or decreased with age, dependent on the specific gene locus being investigated [23]. For some genes, MR diet complemented, while for others it counteracted age-induced changes in GSM. The differential effect could be due to the fact that DNA methylation is partly governed by access to DNA by DNMT1 [23]. Types of histones present in the vicinity of gene, post-translation modification of histone tails, and the effect of aging on histone marks might be playing a contributory role [44].
Based on our findings, we conclude that an MR diet improves the efficiency of DNA methylation maintenance systems in adult mice but had no effect in young mice. The effect of MR on DNA methylation is tissue-specific and is driven by changes in SAH. More comprehensive and genome-wide studies on the GSM of genes involved in aging and aging-associated diseases such as cancer would provide additional insights into the mechanistic basis of lifespan extension by MR diet.
Supplementary Material
Young (8-week-old) and adult (1-year-old) male C57BL/6J mice (n=8/group) were fed ad libitum with diets containing either 0.12% methionine (MR) or 0.84% methionine (CD) for 12 weeks. No difference between CD and MR mice was observed in total food intake/animal in either age group (a). On body weight basis, MR mice in both young and adult groups had higher food intake than CD mice (b). Note: Error bars represent standard errors. For young group, bars represent data from last 5 weeks. BW, body weight.
Young (8-week-old) and adult (1-year-old) male C57BL/6J mice were fed ad libitum with diets containing either 0.12% methionine (MR) or 0.84% methionine (CD) for 12 weeks. A trend for an increase in GM was fond in young mice, but no changes were observed in adult mice (p2t<0.05). Error bars represent standard errors.
Young (8-week-old) and adult (1-year-old) male C57BL/6J mice were fed ad libitum with diets containing either 0.12% methionine (MR) or 0.84% methionine (CD) for 12 weeks. Both in adult (panel b) and young mice (panel c), MR accentuated changes in GSM that occurred during aging (Panel a).
Primer sequences used for gene-specific methylation.
Genomic coordinates of CpG sites with at least 2-fold change in methylation.
Acknowledgments
This study was funded by Orentreich Foundation for the Advancement of Science.
Abbreviations
- MR
Methionine restriction
- SAM
S-adenosylmethionine
- SAH
S-adenosylhomocysteine
- DNMT1
DNA methyltransferase-1
- CD
control diet
- GM
global DNA methylation
- GSM
gene-specific methylation
- LINE-1
long interspersed elements
- 5-meC
methylated cytosine in DNA
- 5-C
unmethylated cytosines in DNA
Footnotes
Author contributions:
DM, JS, GA, SNN: Animal husbandry, laboratory analysis
SM, JL: Metabolomic analysis
DS, JR: Global DNA methylation
SNN: Concept, study design, data analysis, and manuscript writing.
All authors read and commented on the manuscript.
References
- 1.Orentreich N, et al. Low methionine ingestion by rats extends life span. J Nutr. 1993;123(2):269–74. doi: 10.1093/jn/123.2.269. [DOI] [PubMed] [Google Scholar]
- 2.Sun L, et al. Life-span extension in mice by preweaning food restriction and by methionine restriction in middle age. J Gerontol A Biol Sci Med Sci. 2009;64(7):711–22. doi: 10.1093/gerona/glp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richie JP, Jr, et al. Methionine restriction increases blood glutathione and longevity in F344 rats. FASEB J. 1994;8(15):1302–7. doi: 10.1096/fasebj.8.15.8001743. [DOI] [PubMed] [Google Scholar]
- 4.Miller RA, et al. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell. 2005;4(3):119–25. doi: 10.1111/j.1474-9726.2005.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mudd SH, Poole JR. Labile methyl balances for normal humans on various dietary regimens. Metabolism. 1975;24(6):721–35. doi: 10.1016/0026-0495(75)90040-2. [DOI] [PubMed] [Google Scholar]
- 6.Olthof MR, et al. Low dose betaine supplementation leads to immediate and long term lowering of plasma homocysteine in healthy men and women. J Nutr. 2003;133(12):4135–8. doi: 10.1093/jn/133.12.4135. [DOI] [PubMed] [Google Scholar]
- 7.Barak AJ, Beckenhauer HC, Tuma DJ. Betaine, ethanol, and the liver: a review. Alcohol. 1996;13(4):395–8. doi: 10.1016/0741-8329(96)00030-4. [DOI] [PubMed] [Google Scholar]
- 8.Pogribny IP, James SJ, Beland FA. Molecular alterations in hepatocarcinogenesis induced by dietary methyl deficiency. Mol Nutr Food Res. 2012;56(1):116–25. doi: 10.1002/mnfr.201100524. [DOI] [PubMed] [Google Scholar]
- 9.Ables GP, et al. Dietary methionine restriction in mice elicits an adaptive cardiovascular response to hyperhomocysteinemia. Sci Rep. 2015;5:8886. doi: 10.1038/srep08886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malloy VL, et al. Methionine restriction prevents the progression of hepatic steatosis in leptin-deficient obese mice. Metabolism. 2013;62(11):1651–61. doi: 10.1016/j.metabol.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 11.Sinha R, et al. Dietary methionine restriction inhibits prostatic intraepithelial neoplasia in TRAMP mice. Prostate. 2014;74(16):1663–73. doi: 10.1002/pros.22884. [DOI] [PubMed] [Google Scholar]
- 12.Orentreich JRHISFPTCRSJPCEPD. Methionine-restricted diet inhibits growth of MCF10AT1-derived mammary tumors by increasing cell cycle inhibitors in athymic nude mice. BMC Cancer. 2016 doi: 10.1186/s12885-016-2367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkelstein JD, Martin JJ. Methionine metabolism in mammals. Adaptation to methionine excess. J Biol Chem. 1986;261(4):1582–7. [PubMed] [Google Scholar]
- 14.Bollati V, et al. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mechanisms of ageing and development. 2009;130(4):234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stramentinoli G, et al. Tissue levels of S-adenosylmethionine in aging rats. J Gerontol. 1977;32(4):392–4. doi: 10.1093/geronj/32.4.392. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman DR, Cornatzer WE, Duerre JA. Relationship between tissue levels of S-adenosylmethionine, S-adenylhomocysteine, and transmethylation reactions. Can J Biochem. 1979;57(1):56–65. doi: 10.1139/o79-007. [DOI] [PubMed] [Google Scholar]
- 17.Jones MJ, Goodman SJ, Kobor MS. DNA methylation and healthy human aging. Aging Cell. 2015;14(6):924–32. doi: 10.1111/acel.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasek BE, et al. Remodeling the integration of lipid metabolism between liver and adipose tissue by dietary methionine restriction in rats. Diabetes. 2013;62(10):3362–72. doi: 10.2337/db13-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, et al. A strategy for sensitive, large scale quantitative metabolomics. J Vis Exp. 201487 doi: 10.3791/51358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouattara A, et al. Methionine restriction alters bone morphology and affects osteoblast differentiation. Bone Reports. doi: 10.1016/j.bonr.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pogribny IP, Vanyushin BF. Epigenetics of Aging. Springer; 2010. Age-related genomic hypomethylation; pp. 11–27. [Google Scholar]
- 22.Singhal RP, Mays-Hoopes LL, Eichhorn GL. DNA methylation in aging of mice. Mech Ageing Dev. 1987;41(3):199–210. doi: 10.1016/0047-6374(87)90040-6. [DOI] [PubMed] [Google Scholar]
- 23.Jjingo D, et al. On the presence and role of human gene-body DNA methylation. Oncotarget. 2012;3(4):462–74. doi: 10.18632/oncotarget.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perrone CE, et al. Genomic and metabolic responses to methionine-restricted and methionine-restricted, cysteine-supplemented diets in Fischer 344 rat inguinal adipose tissue, liver and quadriceps muscle. J Nutrigenet Nutrigenomics. 2012;5(3):132–57. doi: 10.1159/000339347. [DOI] [PubMed] [Google Scholar]
- 25.Chalitchagorn K, et al. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23(54):8841–6. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 26.Johnson AA, et al. The role of DNA methylation in aging, rejuvenation, and age-related disease. Rejuvenation Res. 2012;15(5):483–94. doi: 10.1089/rej.2012.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caudill MA, et al. Intracellular S-adenosylhomocysteine concentrations predict global DNA hypomethylation in tissues of methyl-deficient cystathionine beta-synthase heterozygous mice. J Nutr. 2001;131(11):2811–8. doi: 10.1093/jn/131.11.2811. [DOI] [PubMed] [Google Scholar]
- 28.Sibani S, et al. Studies of methionine cycle intermediates (SAM, SAH), DNA methylation and the impact of folate deficiency on tumor numbers in Min mice. Carcinogenesis. 2002;23(1):61–5. doi: 10.1093/carcin/23.1.61. [DOI] [PubMed] [Google Scholar]
- 29.Brown-Borg HM, et al. Altered dietary methionine differentially impacts glutathione and methionine metabolism in long-living growth hormone-deficient Ames dwarf and wild-type mice. Longev Healthspan. 2014;3(1):10. doi: 10.1186/2046-2395-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amaral CL, et al. The effects of dietary supplementation of methionine on genomic stability and p53 gene promoter methylation in rats. Mutat Res. 2011;722(1):78–83. doi: 10.1016/j.mrgentox.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 31.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23(15):5293–300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng X, Blumenthal RM. S-adenosylmethionine-dependent methyltransferases: structures and functions. World Scientific; 1999. [Google Scholar]
- 33.Pogribny IP, et al. Breaks in genomic DNA and within the p53 gene are associated with hypomethylation in livers of folate/methyl-deficient rats. Cancer Res. 1995;55(9):1894–901. [PubMed] [Google Scholar]
- 34.Munoz-Najar U, Sedivy JM. Epigenetic control of aging. Antioxid Redox Signal. 2011;14(2):241–59. doi: 10.1089/ars.2010.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Daniel M, Tollefsbol TO. Epigenetic regulation of caloric restriction in aging. BMC Med. 2011;9:98. doi: 10.1186/1741-7015-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mercier S, et al. Methionine kinetics are altered in the elderly both in the basal state and after vaccination. The American Journal of Clinical Nutrition. 2006;83(2):291–298. doi: 10.1093/ajcn/83.2.291. [DOI] [PubMed] [Google Scholar]
- 37.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Obeid R. The metabolic burden of methyl donor deficiency with focus on the betaine homocysteine methyltransferase pathway. Nutrients. 2013;5(9):3481–95. doi: 10.3390/nu5093481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mato JM, et al. S-Adenosylmethionine: a control switch that regulates liver function. FASEB J. 2002;16(1):15–26. doi: 10.1096/fj.01-0401rev. [DOI] [PubMed] [Google Scholar]
- 40.Thompson RF, et al. Tissue-specific dysregulation of DNA methylation in aging. Aging Cell. 2010;9(4):506–18. doi: 10.1111/j.1474-9726.2010.00577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altmann S, et al. Maternal dietary protein restriction and excess affects offspring gene expression and methylation of non-SMC subunits of condensin I in liver and skeletal muscle. Epigenetics. 2012;7(3):239–52. doi: 10.4161/epi.7.3.19183. [DOI] [PubMed] [Google Scholar]
- 42.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466(7303):253–7. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mentch SJ, et al. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab. 2015 doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCauley BS, Dang W. Histone methylation and aging: lessons learned from model systems. Biochim Biophys Acta. 2014;1839(12):1454–62. doi: 10.1016/j.bbagrm.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Young (8-week-old) and adult (1-year-old) male C57BL/6J mice (n=8/group) were fed ad libitum with diets containing either 0.12% methionine (MR) or 0.84% methionine (CD) for 12 weeks. No difference between CD and MR mice was observed in total food intake/animal in either age group (a). On body weight basis, MR mice in both young and adult groups had higher food intake than CD mice (b). Note: Error bars represent standard errors. For young group, bars represent data from last 5 weeks. BW, body weight.
Young (8-week-old) and adult (1-year-old) male C57BL/6J mice were fed ad libitum with diets containing either 0.12% methionine (MR) or 0.84% methionine (CD) for 12 weeks. A trend for an increase in GM was fond in young mice, but no changes were observed in adult mice (p2t<0.05). Error bars represent standard errors.
Young (8-week-old) and adult (1-year-old) male C57BL/6J mice were fed ad libitum with diets containing either 0.12% methionine (MR) or 0.84% methionine (CD) for 12 weeks. Both in adult (panel b) and young mice (panel c), MR accentuated changes in GSM that occurred during aging (Panel a).
Primer sequences used for gene-specific methylation.
Genomic coordinates of CpG sites with at least 2-fold change in methylation.


