Abstract
Liver tissue engineering has emerged as a promising approach in organ transplantation, but has been hampered by the lack of a reliable and readily available cell source. Induced pluripotent cells (hiPSC) have been highlighted a desirable source, due to their differentiation potential, ability to self-renew and the possibility of making patient specific cells. We developed a decellularization protocol that efficiently removes cellular material, while retaining extracellular matrix components. Subsequently, hiPSC were differentiated on the decellularized human liver matrix (hDLM) scaffolds using an established hepatic differentiation protocol. We demonstrate that using hDLM leads to upregulation of functional hepatic markers when compared to standard differentiation conditions. In addition, expression of a number of hepatic transcription and nuclear factors were found to be within levels comparable to those of primary human adult hepatocytes. Analysis of progression of differentiation on hDLM demonstrated that hepatic developmental marker expression was consistent with hepatic development. The hDLM-derived cells exhibited key hepatic characteristics that were comparable to those observed in primary neonatal human hepatocytes. We investigated the optimal timing of introduction of hDLM into the differentiation protocol, and found that best results are obtained when cells are plated on hDLM since the earliest stages, and accompanied by a progressive loss of sensitivity to substrate composition at later stages. The significance of this work is that it allows for the development of differentiation protocols that take into account signals from ECM, closely recapitulating of the in-vivo microenvironment and resulting in cells that are phenotypically closer to mature hepatocytes.
1. Introduction
The only definitive treatment for end-stage liver failure is orthotopic liver transplantation. While recent advances in surgical techniques and immunosuppressive therapy improved the survival of recipients, the shortage of good quality donor livers still remains as a limiting factor. Currently, the number of patients waiting for a suitable liver is over 14,000 and only about 7,000 transplants are performed each year. To address this gap, alternative strategies are being investigated including machine resuscitation of marginal livers or extending the preservation time of donor organs through supercooling.
In recent years, whole liver engineering has emerged as a promising approach to create alternative sources of organs for transplantation. Donor organs not found suitable for transplantation are used to prepare whole-liver scaffolds which are subsequently repopulated with healthy cells to create transplantable liver grafts. The scaffolds maintain native liver architecture and extracellular matrix (ECM) composition which allow for proper cell homing and function. Our group and others have shown the feasibility of this technique in small and larger animal models using primary adult or fetal liver cells (Barakat, et al., 2012, Uygun, et al., 2010). Unfortunately, a reliable and readily available source for primary human hepatocytes (hPH) remains to be determined. Several characteristics highlight human induced pluripotent cells (hiPSC) as a desirable source, including their differentiation potential, ability to be propagated indefinitely and the possibility of making patient specific cells. Differentiation protocols that produce pluripotent stem cell derived hepatocytes have been developed, with limitations such as low functionality characterized by cytochrome P450 activity of only up to 0.1% of that found in hPH (Schwartz, et al., 2014). Most existing protocols exploit modulation of known developmental signals through addition of soluble cues in the media. Although it has been established that ECM is critical for maintaining adult primary hepatocyte phenotype in vitro, hepatic differentiation protocols seldom take into account cell-ECM interactions. Most differentiation protocols use laminin rich-Matrigel as differentiation substrate (Mallanna and Duncan, 2013, Noto, et al., 2014) with a few studies investigating other ECM derived substrates. Other studies that utilize collagen or animal derived ECM have been performed, but they concentrate on their effect at later stages of maturation, rather than the entire differentiation process (Toivonen, et al., 2016, Wang, et al., 2016). However, signals from the ECM in the hepatic microenvironment are combinatorial and complex. A recent proteomics analysis by our group analyzed the protein content of biological scaffolds where up to 58 matrisome proteins were identified in the liver ECM, with most abundant being Collagen VI, but also high content of transglutaminase 2, biglycan, fibrinogen, and fibronectin (Li, et al., 2016). This result highlights the difference in composition of decellularized liver matrix from the other commonly used cell culture substrates. Furthermore, the involvement of ECM in processes such as in-vivo liver development (Duncan, 2003), and maintenance of hepatocytes in-vitro (Mooney, et al., 1992), emphasize the importance of recapitulating ECM signals for in-vitro differentiation.
In this study, we investigated the effect of decellularized adult human liver matrix (hDLM) on hepatic differentiation of hiPSC with the ultimate goal of generating cells that would be used to repopulate engineered liver grafts. hDLM obtained from human livers was used as a substrate during differentiation of iPS cells into hepatocytes. When compared to Matrigel or collagen I, hDLM resulted in cells that have higher expression of mature hepatocyte markers, and displayed hepatic functions at similar levels as primary neonatal human hepatocytes. hDLM was the most effective when introduced at early stages of differentiation.
2. Materials and Methods
2.1 hDLM preparation and characterization
Discarded donor human livers were provided by New England Donor Services. Livers from 3 different donors were employed, a 40-year-old male, 28-year-old female and 24-year-old male, all without history of liver disease and no signs of fatty-liver. Livers biopsied using a 2.5-cm punch, and the biopsies were frozen and sectioned into 50-µm slices using a cryotome. Approximately 10 mg of tissue were collected in a 50-ml conical tube and washed overnight in phosphate buffered saline (PBS) under constant agitation. PBS was replaced with 0.01% sodium dodecyl sulfate (SDS) kept for 4 h under constant agitation, followed by 0.1%, 0.2% and 0.5% SDS for 1 h each, and 1% Triton X-100 for 15 min (Figure 1A). Ten 5-min PBS washes were performed under constant agitation to ensure complete detergent removal.
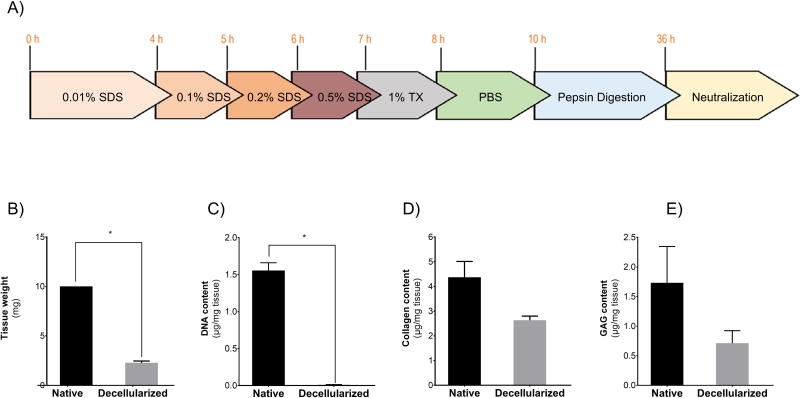
Figure 1. Preparation and characterization of hDLM substrates.
A) Schematic representation of hDLM preparation protocol that consists of decellularization through treatment of liver slices with increasing concentrations of SDS and pepsin digestion followed by neutralization required for gel formation. B) Wet weight of tissue before and after decellularization process shows a 77.1% reduction in tissue mass (p<0.05). C) DNA quantification shows that over 99% of the DNA content is effectively removed from tissue during the decellularization process p<0.05. D) Total collagen content revealed a 40% reduction during decellularization. E) Glycosaminoglycan content analysis showed that 40% of GAGs was preserved in the decellularized tissue. (n = 3, error bars represent Standard Deviation).
For DNA quantification, Purelink Genomic DNA Mini Kit (Invitrogen) was used to extract DNA from the tissue according to manufacturer’s instructions, concentration was estimated by absorbance at 260 nm wavelength using NanoDrop 1000 spectrophotometer. Collagen was quantified using QuickZyme Total Collagen quantification kit (Cedarlane). Glycosaminoglycan (GAG) quantification was performed by hydrolyzing the samples in 6 N HCl for 20 h at 95 °C. Following hydrolysis, 50 µM dimethylene blue solution was added to samples and chondroitin sulfate standards for quantification and the absorbance was measured at 525 nm. For all the assays, normalization was performed with respect to native tissue weight (before recellularization and digestion). For GAG assay, samples were lyophilized in either native state, after decellularization and after pepsin digestion for the analysis.
For differentiation experiments, the slices were lyophilized to obtain the dry weight and 1 mg/ml pepsin in 10 mM HCl was added such that the concentration was 20 mg of hDLM per ml of pepsin solution (pH 2). The hDLM solution was briefly homogenized and pepsin digestion was allowed to proceed for 24 h at room temperature under constant agitation. Undigested hDLM was pelleted by centrifugation and protein concentration of the supernatant was measured using BCA assay (Pierce) using collagen I as standard. hDLM solution was further diluted to desired concentration using 10 mM HCl. hDLM gels were formed neutralization using 10× DMEM at a 1:10 DMEM:DLM ratio, placed in the tissue culture plates at 40 µl/cm2 and allowed to gel at 37°C for 30 min. Collagen I gels were prepared similarly.
2.2 Hepatic differentiation
hIPS-K3 cells (Si-Tayeb, et al., 2010) were received from Dr. Stephen Duncan (Medical College of Wisconsin). Cells were maintained in hESC-qualified Matrigel (Corning-354277) coated plates with mTeSR1 medium (Stemcell Technologies). Once cultures reached 80–90% confluency, differentiation was initiated using a previously established differentiation protocol, with reduced Activin A concentration as most recently reported by the same group (Mallanna and Duncan, 2013, Noto, et al., 2014) (Figure 2A). For experiments involving determination of appropriate timing of hDLM introduction, cells were harvested on the last day of each differentiation stage using ReleSR (Stemcell Technologies) according to manufacturer instructions, and replated at a 1:1 ratio on hDLM, collagen or Matrigel gels without top overlay.
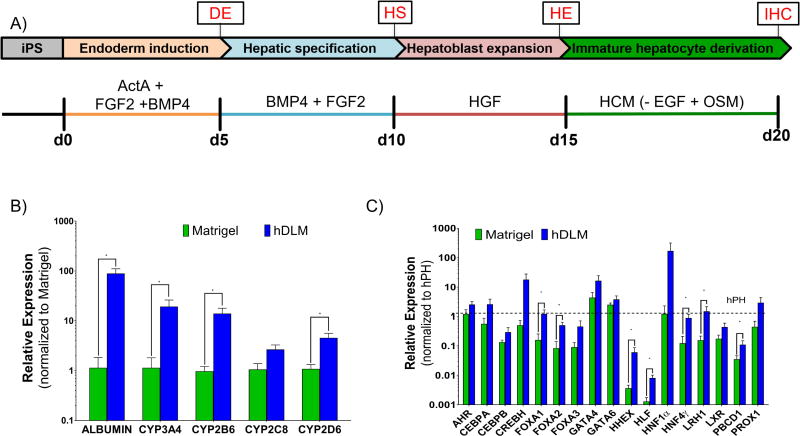
Figure 2. Hepatic differentiation of hiPS cells on hDLM substrates.
A) Schematic representation of differentiation protocol used. Undifferentiated cells were plated on hDLM or hESC qualified Matrigel substrates and were induced to differentiate through addition of soluble factors. B) Expression of mature hepatocyte functional markers on day 20. At the end of the differentiation protocol all functional markers showed higher expression when cells were differentiated on hDLM (*p<0.05, data normalized to Matrigel). C) Expression levels of hepatic nuclear factors and transcription factors on day 20 were consistently higher in cells differentiated on hDLM and at levels comparable to those found in human primary hepatocytes (dashed line). Data normalized to adult hPH expression levels. (*p<0.05), n = 7, error bars represent standard deviation).
2.3 Quantitative polymerase chain reaction
Total RNA of the differentiated cells was extracted using the NucleoSpin kit according to the manufacturer's protocol. Sample absorbance at 280 nm and 260 nm was measured using a NanoDrop 1000 spectrophotometer to obtain concentration and quality. Reverse transcription was performed using ImProm II Promega reverse transcription kit following the manufacturer's recommendations. qRT-PCR analysis was performed in technical triplicates using a ViiA7 instrument and the primers listed in Supplementary Table 1. Relative expression normalized with respect to control groups was obtained using the ΔΔCt method. For microarray experiments, signal transduction pathway finder from SAB Biosciences was acquired, performed and analyzed according to manufacturer’s instructions
2.4 Characterization of mature hepatic functions
Bile canaliculi formation was visualized after the cells were washed with PBS and incubated in fresh media containing 400 nM 5-(and-6)-Carboxy-2',7'-Dichlorofluorescein Diacetate (CDFDA) for 10 min. For glycogen staining, cells were fixed in formalin solution for 1 min and rinsed with deionized water. Periodic acid solution was added to completely cover the surface of the wells and incubated for 5 min at room temperature. The wells were rinsed via several changes of distilled water and Schiff’s Reagent added to cover the wells and incubated for 15 min at room temperature. Wells were rinsed in tap water until water remained clear and finally cells were counterstained in Hematoxylin solution, and once again rinsed until water remained clear.
For immunostaining, cells were fixed in 4% paraformaldehyde, permeabilized in a PBS solution containing 2% BSA and 0.1% Triton-X, blocked with 2% BSA, and subsequently incubated at room temperature for 1 hour in primary antibodies in blocking buffer (Albumin R&D, MAB1455, 25 µg/ml; Hnf4α Santa Cruz 1:200), followed by washing and secondary incubation in secondary antibodies diluted in blocking buffer (AlexaFluor 488 and 633 ThermoFisher, 1:1000). The cells were washed and counterstained with DAPI, and fluorescence images acquired using EVOS FL Cell imaging system.
The differentiated cells were maintained for additional 7 days at the end of the differentiation to evaluate mature hepatocyte functions. The controls were cryopreserved adult and neonatal hPH which were obtained from Cell Resource Core at MGH and plated at a density of 200,000 cells/ cm2. The cells were maintained in hepatocyte maintenance media (Lonza CC-3199 & CC-4182) in sandwich configuration (primary hepatocytes overlaid with collagen, while differentiated cells overlaid with hDLM). Media was sampled daily for measurement of albumin concentration using the in-vitro human albumin ELISA kit (Bethyl Laboratories). CYP3A4 activity was measured using Promega P450-Glo according to manufacturing instructions and results for each sample normalized with respect to total DNA content to account for differences in cell number.
2.5 Statistical analysis
All experiments were performed in at least triplicate. Results are presented as average of different experiments, with error bars illustrating standard deviation. In order to determine statistical significance when comparing groups, non-parametric Mann-Whitney test was used. All graphs and statistics were acquired using Graphpad Prism.
3. Results
3.1 Characterization of hDLM
Livers were decellularized as specified in Figure 1A. The wet weight after decellularization corresponded to 23% of the dry weight in native liver, suggesting that roughly 77% of the liver mass is attributed to cells. In addition, we calculated the weight of the ECM after digestion to quantify the amount of ECM digested by pepsin within 24 hours. For this, the undigested portion was separated by centrifugation and both portions were lyophilized and weighed. We found that about a third of the decellularized ECM is digested by 24 hour pepsin treatment (Figure 1B). Before pepsin digestion removal of cellular material was confirmed by quantification of DNA (Figure 1C). Native liver DNA was estimated to be 1.6 ± 0.18 µg/mg of tissue. After decellularization, this decreased to 13 ± 3.8 ng/mg of tissue representing over 99% reduction of DNA content. GAG and total collagen content were quantified to evaluate retention of ECM components (Figure 1D,E). We found the amount of total collagen after decellularization to be roughly 60% of the collagen in native tissue. We further analyzed the effect of pepsin digestion in GAG content, as this has not been well documented, although several studies have shown abundance of GAGs in pepsin digested ECM from different tissues (DeQuach, et al., 2010, Wolf, et al., 2012), and we found no significant reduction elicited by pepsin treatment The GAG content was 40.7±2.3% vs 37.8±5.0% of the native tissue before digestion and after digestion, respectively (p = 0.1926). Taken together, the results suggest that our decellularization approach efficiently removes cells while retaining a considerable amount of chemical ECM components.
3.2 hDLM leads to improved hepatic differentiation of hiPSC over standard Matrigel culture
hiPS-K3 cells plated on hDLM or hESC qualified Matrigel were differentiated according to a widely used hepatic differentiation protocol (Figure 2A) (Mallanna and Duncan, 2013, Noto, et al., 2014). hDLM was tested for maintenance of primary rat hepatocytes at different concentrations varying from 1.25 to 10 mg/ml (Supplementary Figure 1 and Supplementary Information) with no statistical difference in terms of albumin or urea secretions. For all subsequent experiments, a concentration of 1.25 mg/ml in order to match collagen concentration typically used in collagen sandwich culture. This and other concentrations were tested using primary rat hepatocytes in sandwich configuration and. At the end of differentiation protocol, cells were harvested and analyzed for expression of mature hepatocyte functional markers including ALB and most major hepatic cytochrome P450 enzymes (CYP3A4, CYP2B6, CYP2C8, and CYP2D6). Differentiation of cells on hDLM substrates consistently resulted in significant upregulation of these genes (except CYP2C8 which was not statistically significant) over traditional Matrigel culture (p= 0.0006, 0.007, 0.0006, 0.0728, 0.0175, respectively) (Figure 2B). To further confirm hepatic differentiation, hDLM and Matrigel differentiated cells were evaluated for hepatic nuclear and transcription factors, which consistently showed higher levels in hDLM differentiated cells. These include FOXA1 (p= 0.01), FOXA2 (p=0.01), HHEX (p=0.0175), HLF (p=0.0012), HNF4α (p=0.0023), LHR1 (p= 0.0262), PBDC1 (p= .0262). The expression of these factors was at levels comparable to that of adult hPH (Figure 2C) suggesting that the substrates prepared using human decellularized liver ECM provide morphogenic signals that promote hepatic differentiation of hiPS cells.
3.3 Early exposure to hDLM during differentiation leads to highest hepatic specification
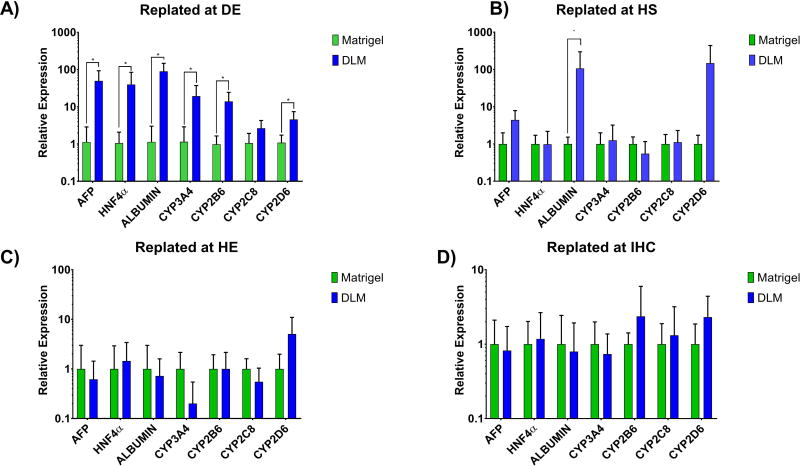
We tested whether introduction of hDLM at different stages of differentiation resulted in higher expression of genes related to essential hepatic functions. Human iPS-cells were differentiated on hESC-qualified Matrigel coated plates under standard conditions (Mallanna and Duncan, 2013, Noto, et al., 2014), and at the end of each stage, cells were harvested and replated. Because the harvesting process may have an effect on the cells, cells were replated on either Matrigel or hDLM (without overlay) to account for the effect of cell harvesting in all groups, making substrate composition the only variable. Cells plated on hDLM since the pluripotent stage are denoted as Definitive endoderm DE. Similarly, cells replated after definite endoderm (DE) was completed (D5) were denoted Hepatic Specification (HS) and so on. Analysis of mature hepatocyte markers at the end of the differentiation protocol showed that the expression levels were the highest when cells were differentiated on hDLM since the definitive endoderm induction stage suggesting that introduction of ECM signals may aid hepatic maturation since very early stages of differentiation (Figure 3A–D). In addition, we found that cells replated at the immature hepatocyte stage (IHC) (Figure 3D) were less sensitive to changes in substrate, with cells resulting in similar levels of expression with highest upregulation being below 2.2-fold for CYP2D6 expression on hDLM. This suggests a loss in sensitivity to substrate composition as differentiation progresses.
Figure 3. Determination of appropriate timing of introduction of hDLM into differentiation protocol.
A) Cells were plated and differentiated on hDLM or matrigel since the beginning of DE or predifferentiated on matrigel and replated onto either matrigel or hDLM at B) beginning of HS stage, C) beginning of HE stage, or E) Beginning of IHC stage. Resulting cells were harvested and analyzed for expression of genes related to essential hepatic functions by qPCR. Expression of most markers was significantly higher (*p<0.05) when cells were plated on hDLM since the beginning of the differentiation protocol. Data normalized with respect to Matrigel results. (n =4, error bars represent standard deviation).
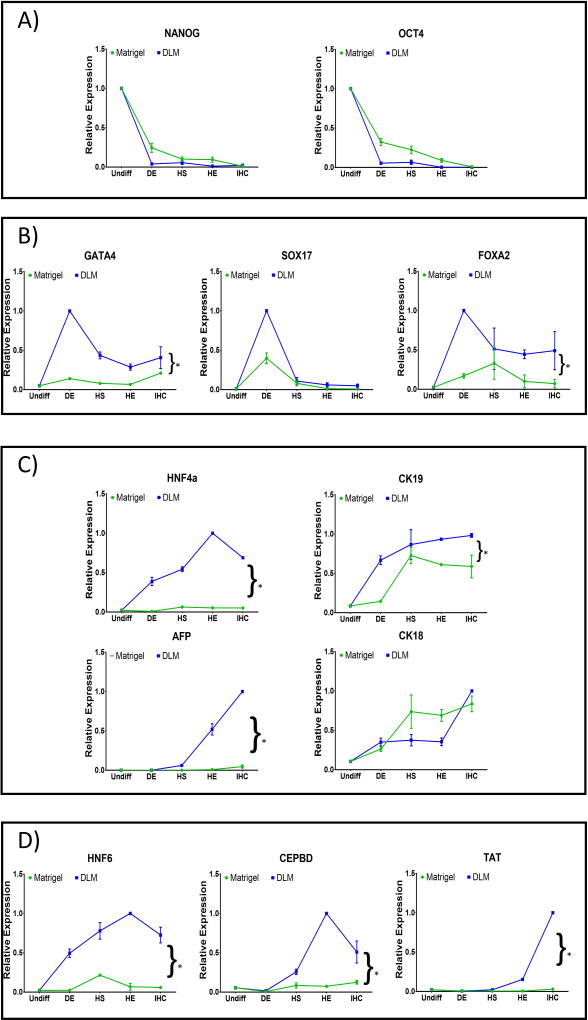
In order to evaluate this sensitivity to substrate without the effect of cell harvesting, differentiation was performed on either hDLM or Matrigel since the beginning of differentiation, but cells were harvested at the end of each stage of differentiation and analyzed for stage specific differentiation markers. Both groups showed rapid loss of pluripotency marker expression, with the most significant drop being at the DE stage (Figure 4A). Definitive endoderm markers (GATA4, SOX17 and FOXA2) show dissimilar expressions in both groups (Figure 4B). GATA4 and FOXA2 are transcriptional factors that play important roles in early stages of development (Gordillo, et al., 2015), however, they are also expressed in the adult liver (Zheng, et al., 2013). In contrast, SOX17 expression is necessary for DE formation, but its expression is absent in the adult liver (Ayatollahi, et al., 2012). In our experiments, SOX17 showed similar patterns of expression in cells differentiated on both Matrigel and hDLM, with its expression peaking at the DE stage and decreasing thereafter, however, expression peak was considerably higher for cells differentiated on hDLM (p = 0.2). Similarly, GATA4 peaked at the DE stage in hDLM differentiated cells, however, expression of this marker was relatively unchanged throughout the protocol for cells differentiated on Matrigel (p = 0.02). Finally, FOXA2 expression peak was earlier and more pronounced for cells on hDLM when compared to cells differentiated on Matrigel (p = 0.001). Hepatoblast markers also showed differences in expression (Figure 4C). HNF4α is considered essential for liver development. Its expression is dispensable at the earlier stages of differentiation, but controls expression of a number of hepatic genes during maturation (Gordillo, et al., 2015), and its expression is abundant in adult hepatocytes. In our experiments, we observed a HNF4α peak at the HE stage for cells plated on hDLM, while its peak was earlier (HS) for cells differentiated on Matrigel, however, the magnitude of this expression was lower even at its peak expression (p=0.0002). It is worth observing that on hDLM, upregulation of HNF4α took place rapidly with high levels being observed even at the DE stage. Expression patterns for CK19, which during development is observed to be expressed in liver progenitor cells, however in the adult liver is restricted to the bile ducts (Gordillo, et al., 2015), were similar in both groups, peaking at the early stages, but once again, magnitude was the highest for the hDLM differentiated cells (p = 0.003), and its expression was observable since the DE stage in these cells. CK18 expression, which is also observed in adult hepatocytes (Gordillo, et al., 2015), appeared to be insensitive to substrate composition in our experiments, with its pattern and expression being similar for both groups at all time points. Most interestingly for this group of markers, AFP expression, which is fundamental for liver development, but lost with maturity (Cantz, et al., 2003), showed very rapid increase after the hepatic specification (HS) stage in the hDLM group, while it started showing a small increase in the control group. Analysis of this marker throughout the differentiation protocol shows the importance of progression analysis, as the AFP level in the resulting cells differentiated on Matrigel is much lower, which could indicate loss of expression, hence, more maturity of cells, however, progression analysis shows that expression to be lower throughout the entire protocol in the Matrigel group (p=0.007), suggesting failure to be highly elicited, which most likely indicates inadequate hepatic differentiation in cells in this group. Finally, mature hepatocyte markers showed interesting trends as well (Figure 4D). HNF6 had overall much higher expression in hDLM differentiated cells at all stages (p=0.0002), and its upregulation was considerable since the DE stage. In contrast, Matrigel differentiated cells showed HNF6 (necessary for hepatoblast differentiation (Gordillo, et al., 2015) peak expression at the HS stage, but its magnitude was much lower than that achieved by hDLM differentiated cells. Expression of tyrosine aminotransferase (TAT), a liver enzyme with metabolic functions in the adult liver, was virtually undetectable on both groups until the Hepatic expansion (HE) stage, but hDLM rapidly increased its expression thereafter, while it remained low on Matrigel cells (p=0.048). CEBPD, an important player in lipid metabolism in mature liver, also remained low in the Matrigel group while its expression was high in hDLM differentiated cells since the HS stage, peaking at the HE stage (p=0.037). Overall, most markers were found to be upregulated on hDLM at all stages of differentiation, however, it is important to note that Hepatoblast and Hepatocyte marker expression appeared earlier when cells were differentiated on hDLM. This is in agreement with the previous observation that there is a higher sensitivity to substrate composition at the earlier stages and that hepatic differentiation on hDLM is best when cells are plated on these substrates since beginning of differentiation. In addition, most of the results show that hDLM differentiated cells show gene expression patterns that are more closely associated with hepatic marker progression during liver organogenesis.
Figure 4. Expression of developmental stage-specific markers.
Cells differentiated on hDLM (blue lines) or Matrigel substrates (green lines) were analyzed at the end of each stage of the differentiation protocol for markers specific to (A) pluripotency (NANOG, OCT4), (B) definitive endoderm (GATA4, SOX17, FOXA2), (C) hepatoblast (HNF4α, CK19, CK18, AFP) and (D) mature hepatocytes (HNF6, CEBDP, TAT). (Results were normalized to the highest expression level of each gene achieved under all conditions)(n = 3, error bars represent standard deviation, *p<0.05).
3.4 Cells differentiated on hDLM are functional but remain immature
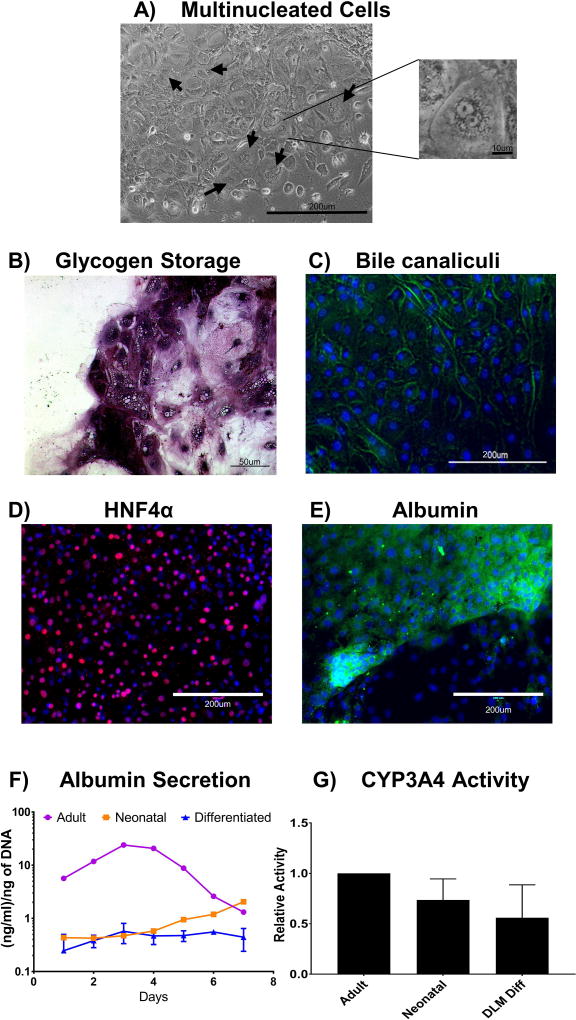
Cells differentiated on hDLM substrates were evaluated for a number of hepatic characteristics. After differentiation was completed, cells resembled primary hepatocytes in terms of shape and size, and we were able to identify multinucleated cells (Figure 5A). PAS assay showed intracellular Schiff staining, suggesting glycogen storage in the cells, a characteristic of mature hepatocytes (Figure 5B). CDFDA assay showed presence of bile canaliculi, a hallmark of hepatocyte polarity (Figure 5C). A large proportion of cells showed expression of HNF4α with nuclear location, as expected since this is a transcription factor (Figure 5D). Similarly, we were able to identify regions of cells positive for cytoplasmic Albumin expression (Figure 5E). To compare their function to primary human hepatocytes, cells were overlaid with a top layer of hDLM gel and maintained in culture in hepatocyte maintenance media in parallel to human adult and neonatal hepatocytes that had been cryopreserved. This additional culture time was done to mimic the conditions used for culture of primary hepatocytes, including sandwich configuration, hepatocyte maintenance media (which differs from IHC media) and the additional time, which is necessary for cryopreserved hepatocytes to recover their function. This was extended to show that we were capturing the peak function, and that hepatocytes lose their functionality in an in-vitro setting. During the 7 d culture, albumin secretion was detected in culture media (Figure 5F) at levels that were lower than those of adult primary hepatocytes, but comparable to those of neonatal hepatocytes on the first 4 days of culture. It is important to note that we observed an increase in albumin secretion in the neonatal hepatocytes in culture after 4 days, suggesting the possibility of in-vitro maturation of these cells, which was not observed in the differentiated cells. Analysis of CYP3A4 activity on day 4 of culture (peak albumin secretion for primary hepatocytes) showed lower levels of activity on both fetal and differentiated cells, representing 73% and 56%, respectively compared to adult hPH (Figure 5G).
Figure 5. Characterization of hDLM-differentiated cells post differentiation.
A) Morphological examination of hDLM differentiated cells revealed cell size to be consistent with that of hPH (not shown), and appearance of multinucleated cells (arrows and insert image), a hallmark characteristic of hepatocytes. B) Glycogen storage in hDLM differentiated cells. Periodic Acid Schiff (PAS) staining was performed to evaluate glycogen storage showing populations of cells with high intracellular glycogen content. C) Bile canaliculi staining, CDFDA was added to the cells and fluorescent images show localization to the cell membranes, indicative of bile canaliculi formation and hepatocyte polarity. Green: CDFDA, blue: DAPI. D) Staining for HNF4α showing nuclear expression in a large percentage of cells. Red: HNF4α, blue: DAPI. E) Staining for Albumin shows cytoplasmic expression in populations of cells, while other populations did not show albumin expression. Green: Albumin, blue: DAPI. F) Albumin secretion during 7 day culture post differentiation. The results were compared to human primary adult and neonatal hepatocytes cultured under same conditions. hDLM differentiated cells showed lower levels of albumin secretion than the primary adult human hepatocytes, however, levels were comparable to those by neonatal hepatocytes in the first 4 days of culture. G) CYP3A4 on day 5 post completion of differentiation. Activity in neonatal and differentiated cells was 74% and 56% of the activity found in adult primary hepatocytes. (n =3, error bars represent standard deviation. No statistical significance between neonatal and differentiated groups).
3.5 Specificity to hDLM composition
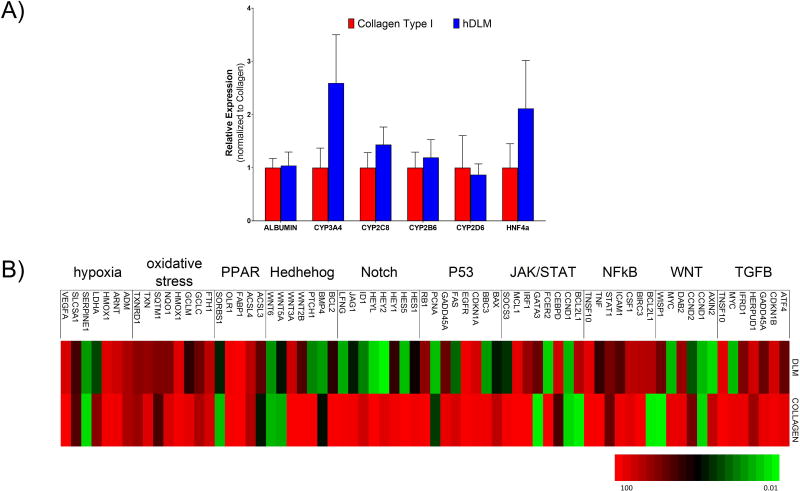
Because collagen I is the main component of the liver ECM, we compared hepatic differentiation of hiPSC on collagen I gels with that on hDLM (Figure 6A). The resulting cells showed a significantly higher expression of hepatic functional markers and main transcription factors, than when cells were differentiated on Matrigel, however, some of these (e.g., CYP3A4 and HNF4α) were lower than when cells were differentiated on hDLM. This indicates that while collagen I has a strong effect on hepatic differentiation, other unique components of the liver ECM have an additive effect on differentiation. In order to further analyze differentiation status, we performed a PCR array to profile the expression of key genes responsive to signal transduction pathway activation or inhibition. We compared cells differentiated on Matrigel, hDLM, collagen and found a number of pathways to be upregulated similarly on hDLM and Collagen I compared to the Matrigel, including TGFB, NFKB, Hypoxia and PPAR (Figure 6B). Interestingly, some pathways showed to be differentially regulated in the collagen and hDLM groups. Amongst these were NOTCH, P53, HEDGEHOG and WNT pathway (CCND2, AXIN2, MYC, WISP1). This is all in agreement with previous studies that have found expression of these genes to be correlated with hepatic maturation (Chen, et al., 2016, Li, et al., 2015b, Matsui, et al., 2002, Tanimizu and Miyajima, 2004). These results further suggest that while collagen plays a role in hepatic differentiation, other components of the ECM, which remain to be identified, play additive roles, highlighting the importance of using complex ECM signals in differentiation protocols.
Figure 6. Specificity of substrate-mediated differentiation to substrate composition.
A) Cells differentiated on Collagen I showed upregulation of Hepatic functionality markers compared to cells differentiated on Matrigel, however, upregulation of most markers was lower compared to cells differentiated on hDLM. B) Signal transduction pathway finder PCR array performed to elucidate some of the possible mechanisms involved with hDLM mediated hepatic differentiation.
4. Discussion
hiPSC are an attractive source of cells for cell-based liver regenerative therapies due to their potential to generate unlimited numbers of patient specific adult hepatocytes. The current hepatic differentiation protocols rely on sequential addition of soluble factors as developmentally relevant signaling cues to drive the differentiation of cells while paying little attention to insoluble factors that are present in the hepatic microenvironment. Most differentiation protocols are performed on Matrigel substrates, a mouse tumor derived, laminin rich ECM which doesn’t represent the hepatic ECM. In this work, we used decellularized human liver ECM as a complex liver specific substrate for hepatic differentiation of hiPS cells and found that unlike Matrigel, the hDLM results in cells that have drastically higher hepatic marker expression which were comparable to adult hPH.
Since the earliest reports of hepatic differentiation of hiPSC, Matrigel is still used as the preferred substrate for differentiation (Carpentier, et al., 2016, Yanagihara, et al., 2016). However, recapitulation of the native environment of developing tissues has long been the working paradigm in the differentiation field, and unlike the native liver, Matrigel is laminin-rich, hence, motivating the question of whether substrates closely recapitulating the chemical composition of the hepatic niche would be more conducive to hepatic differentiation. Upon differentiation of hiPSC on hDLM substrates, we saw a significant upregulation of genes related to hepatic functions as well as hepatic nuclear factors and transcription factors, suggesting that recapitulation of the chemical cues in the liver microenvironment is indeed conducive to hepatic differentiation.
Because it has been suggested that during hepatic organogenesis, the composition of the developing liver ECM undergoes some changes (Martinez-Hernandez and Amenta, 1993, Mooney, et al., 1992), we sought to determine whether there was an optimal timing for introduction of hDLM during differentiation. In order to do this, cells were pre-differentiated under standard conditions and at the end of each differentiation stage, replated onto hDLM or Matrigel substrates. Replating onto Matrigel was performed to cancel out any effects brought out by cell harvesting, making substrate composition the only variable between groups. The results suggest highest expression of markers of hepatic functionality to be achieved when the entirety of the hepatic differentiation program was performed in the hDLM substrates. In addition we observed that as cells were replated onto different substrates further down the differentiation program, they seemed to lose sensitivity to the substrate composition. The superiority of hDLM in terms of hepatic differentiation of hiPSC was further confirmed by stage-specific marker progression analysis, which showed that not only the magnitude of expression of different markers was affected by substrate, but also that when cells were plated on hDLM for the entirety of the protocol, appearance of hepatoblast and hepatocyte markers appeared earlier. In addition, the trends for expression patterns were in agreement with in-vivo progression of these markers during organogenesis,. The significance of these results is that they provide the groundwork for optimization of timing of introduction of cells into whole-organ scaffolds for the development of stem cell derived engineered livers.
The hepatocyte-like cells resulting from differentiating hIPSC on hDLM substrates were characterized, and while they did show morphological and functional features characteristic of hepatocytes, the results, including high levels of AFP, CK19 and albumin secretion and CYP3A4 activity that resembles levels found in fetal hepatocytes suggest that the cells are still immature. From our comparison of hepatic nuclear factors and transcription factors, we saw HHEX and HLF to be expressed at the lowest levels when compared to adult primary hepatocytes. This opens avenues for investigation of strategies to improve hepatic differentiation protocols through targeting activation of expression of these factors. In terms of HHEX, it has been found that its expression in Xenopous models is directly activated by nodal, and indirectly modulated by Wnt/β-Catenin pathway, but also suppressed by Bmp signaling (Rankin, et al., 2011). It is therefore worth investigating the possibility of introducing Wnt signaling into the current differentiation protocol. BMP signaling has been found to be fundamental for hepatic specification (Duncan and Watt, 2001), hence, removal of BMP4 from the current protocol may have negative effects, however, determining the specific timing at which BMP is required and inhibiting at other stages may be a plausible strategy.
Collagen I sandwich culture is currently the gold standard for maintenance of hPH in-vitro. This led us to investigate whether Collagen I has positive effects on hepatic differentiation and whether the results compare to those obtained using hDLM. The results show a significant upregulation of functionality markers compared to Matrigel control, however the magnitude of upregulation in most cases, was lower than that achieved by cells differentiated on hDLM. This suggests that being a major component of the liver microenvironment, collagen has a large positive effect on differentiation, however, hDLM possess additional cues that provide additive differentiation effects.
We analyzed the expression of genes involved in major signal transduction pathways in order to gain more in depth information about the differentiation of cells on substrates of different composition. This analysis shows a number of pathways that are upregulated in both the collagen and hDLM group compared to the control. These include the PPAR pathway, which is involved in metabolism of lipids and carbohydrates, suggesting a role of ECM in increase of these functions in hiPSC-derived hepatocytes (Burri, et al., 2010). In addition, a number of members of the PPAR family are involved in hepatic differentiation of a variety of stem cells, hepatic maturation and mitochondriogenesis (Esmaeli, et al., 2014, Zhu, et al., 2010) which further reinforces the observation that ECM and collagen lead to improved differentiation. While the exact mechanism involved in the activation of this pathway is yet to be investigated, previous studies have shown collagen promotion of FABP1 (PPAR related gene) expression in rat epithelial cells (Hallden, et al., 1994). TGFβ is another pathway that was found to be upregulated in both collagen and hDLM differentiated cells, including upregulation of ATF4, which regulates lipid and glucose metabolism (Seo, et al., 2009). CDKN1B, whose expression is lower in fetal hepatocytes and reduced with loss of hepatocyte maturation (Bhate, et al., 2015), and TNSF10 which has also been found to be upregulated in differentiated liver progenitor cells (Parent and Beretta, 2008).
Interestingly, some signal transduction pathways showed very dissimilar expression in hDLM and collagen differentiated cells, including NOTCH, HEDGEHOG and P53 pathway. Previous studies have shown that NOTCH signaling favors cholangiocyte differentiation of hepatic progenitor cells (Tanimizu and Miyajima, 2004). Our results show hDLM differentiated cells to have considerably lower levels of HEYL, a NOTCH pathway effector involved in biliary regeneration, than Matrigel differentiated cells, while collagen differentiated cells show upregulation of the same gene. This has been implicated with prior studies showing that laminin induces cholangiocyte differentiation (Tanimizu, et al., 2012). Those observations are in agreement with our observation involving Matrigel and hDLM, however, fail to explain why collagen leads to higher expression of notch in collagen than both hDLM and Matrigel. However, a study by Yani et al (Yanai, et al., 2008) showed similar observations, where matrigel induced albumin expression in biliary epithelial cells, which was not stimulated by collagen, and, addition of FGFs repressed expression of albumin when cells were cultured on Matrigel, while in collagen it increased expression of hepatic markers and nuclear factors. This can be translated in our results by the fact that the Matrigel used is growth factor reduced, and the collagen used lacks growth factors, while hDLM contains some additional proteins and growth factors that, along with hepatic collagen, have combinatorial effects on differentiation.
In terms of P53, which was also found to be upregulated in the collagen culture, while downregulated in the hDLM culture, it has been reported that the differentiation of mesenchymal stem cells into hepatocytes was marked by a decrease in P53 pathway gene expression (Khajeniazi, et al., 2013). Similarly, WNT related genes including CCND2 were downregulated by hDLM, but upregulated by Collagen, consistent with reports that downregulation of this is observed when maturation of fetal hepatocytes is induced by exposure to OSM (Matsui, et al., 2002). Within this same signal transduction pathway, the same trend was observed for MYC, which has been previously described as a marker of hepatocyte immaturity (Li, et al., 2015a). Overall, signal transduction pathway analysis results further suggest increased hepatic maturation of hiPSC on hDLM substrates when compared to both Matrigel and collagen I, however, the exact mechanism by which this takes places is yet to be elucidated. Based on previous studies, a possible factor is the presence of growth factors in the hDLM, however, further studies are needed to confirm both presence and involvement of these.
It is important to note that, while indicative of hepatic cell immaturity, dysregulated expression of some of pathways and genes, such as AFP and p53, is also a hallmark of tumorgenicity (Lee, et al., 1999). For this reason, translatability of the work presented here into regenerative technologies is highly dependent in further investigation of the tumorigenic status of these cells by in-depth evaluation of cancer-related gene and pathway expression, and ensure that these return to physiologically normal levels upon complete maturation of cells. Future directions of this research includes investigation of signals that would aide with further cell maturation that results in decrease of AFP expression and increase in P53 pathway activity. One possibility is exploring interactions with non-parenchymal cell types, which have been implicated to modulate full maturation after birth, through secretion of factors, including FGF4, HGF and EGF (Behbahan, et al., 2011).
Finally, we would like to acknowledge that the cell line used for this experiments, hiPSC-K3, has been reported to be vulnerable to gene mutation due to reprogramming. This is an important concern, however, a previous study has shown that aneuploidy is permissive for hepatocyte-like cell differentiation of human induced pluripotent stem cells (Noto, et al., 2014), and hiPSC-K3 cell line is one of the cell lines used in the mentioned report. We decided to use these cells, despite their vulnerability, because they are widely used for hepatic differentiation. The effect can be disregarded from the analysis because ultimately, we are comparing the differentiation in the same cells when using the hDLM and standard conditions, so, the only variable here is the substrate. Similarly, we considered another drawback of our study to be the possibility of population heterogeneity affecting the differentiation results. In previously published reports using the same differentiation protocol and the same cell line cultured on Matrigel, majority of cells were found to express FOXA2 and SOX17 at the DE stage, HNF4 and AFP at the immature hepatocyte stage and HNF4 and Albumin at the end of differentiation (Noto 2014). It is still possible that hDLM may favor the differentiation of a subpopulation of cells, which we are carefully tracking in follow-up studies, however, even if this were the case, it would not undermine the result presented here, as it would suggest a very strong ECM-induced hepatic differentiation in a sub-population of cells, which could be analyzed after further purification with methods, such as recently reported selection through surface markers (Mallanna, et al., 2016), with the possibility of these cells showing functional profiles with higher resemblance to that of fully mature hepatocytes.
While the results presented here show an improvement in hepatic differentiation in the presence of liver derived ECM, some challenges still remain. As previously mentioned, cells remain immature, making them inadequate for clinical settings. In addition, the use of hDLM brings forward some concerns, including patient to patient heterogeneity, hence, batch to batch variability. This suggests the importance of systematic studies to identify specific components of the ECM that promote differentiation, and possibly, development of mimetic scaffolds that closely recapitulate these complex signals in a controlled fashion.
5. Conclusion
The results presented in this study suggest that liver ECM provides specific cues that aid with hepatic differentiation of pluripotent stem cells. The broad significance of this work is that it allows for the development of differentiation protocol that take into account signals from ECM, therefore, closer recapitulating of the in-vivo microenvironment and resulting in cells that are phenotypically closer to mature hepatocytes. We determined the appropriate timing for introduction of these signals, which is relevant for future studies involving creation of whole-organ/pluripotent stem cell derived liver grafts, as it provides the rationale for attempting perfusion-based differentiation of hiPSC seeded in whole liver scaffolds.
Supplementary Material
Acknowledgments
We would like to acknowledge Dr. Stephen A. Duncan for the generous donation of hIPS-K3 cells and New England Donor Services for human livers. We thank the Cell Resource Core at MGH for assistance with primary hepatocyte isolations. We acknowledge funding from the National Institutes of Health (R01DK084053 M.L.Y., R00DK088962 B.E.U.), Shriners Hospitals for Children (M.J.)
Footnotes
Contributions:
Maria Jaramillo: Conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing
Heidi Yeh: Provision of study materials, final approval of manuscript
Martin Yarmush: Financial support, final approval of manuscript
Basak Uygun: Financial support, conception and design, manuscript writing, Final approval of manuscript
Disclosures
B.E.U. has a financial interest in Organ Solutions, LLC, that is reviewed and arranged by MGH and Partners HealthCare in accordance with their conflict of interest policies.
References
- Ayatollahi M, M HS, M KS, et al. Differential Expression Pattern of the Human Endoderm-Specific Transcription Factor Sox17 in Various Tissues and Cells. International Journal of Organ Transplantation Medicine. 2012;3(4):183–187. [PMC free article] [PubMed] [Google Scholar]
- Barakat O, Abbasi S, Rodriguez G, et al. Use of Decellularized Porcine Liver for Engineering Humanized Liver Organ. Journal of Surgical Research. 2012;173(1):e11–e25. doi: 10.1016/j.jss.2011.09.033. [DOI] [PubMed] [Google Scholar]
- Behbahan IS, Duan Y, Lam A, et al. New Approaches in the Differentiation of Human Embryonic Stem Cells and Induced Pluripotent Stem Cells toward Hepatocytes. Stem Cell Reviews. 2011;7(3):748–759. doi: 10.1007/s12015-010-9216-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhate A, Parker DJ, Bebee TW, et al. ESRP2 controls an adult splicing programme in hepatocytes to support postnatal liver maturation. Nature communications. 2015;6:8768. doi: 10.1038/ncomms9768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri L, Thoresen GH, Berge RK. The Role of PPARalpha Activation in Liver and Muscle. PPAR research. 2010;2010 doi: 10.1155/2010/542359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantz T, Zuckerman DM, Burda MR, et al. Quantitative Gene Expression Analysis Reveals Transition of Fetal Liver Progenitor Cells to Mature Hepatocytes after Transplantation in uPA/RAG-2 Mice. The American Journal of Pathology. 2003;162(1):37–45. doi: 10.1016/S0002-9440(10)63796-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier A, Nimgaonkar I, Chu V, et al. Hepatic differentiation of human pluripotent stem cells in miniaturized format suitable for high-throughput screen. Stem Cell Research. 2016;16(3):640–650. doi: 10.1016/j.scr.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Gao W, Zhou P, et al. Enhancement of hepatocyte differentiation from human embryonic stem cells by Chinese medicine Fuzhenghuayu. Scientific Reports. 2016;6:18841. doi: 10.1038/srep18841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeQuach JA, Mezzano V, Miglani A, et al. Simple and High Yielding Method for Preparing Tissue Specific Extracellular Matrix Coatings for Cell Culture. PLOS ONE. 2010;5(9):e13039. doi: 10.1371/journal.pone.0013039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SA. Mechanisms controlling early development of the liver. Mechanisms of Development. 2003;120(1):19–33. doi: 10.1016/s0925-4773(02)00328-3. [DOI] [PubMed] [Google Scholar]
- Duncan SA, Watt AJ. BMPs on the road to hepatogenesis. Genes & development. 2001;15(15):1879–1884. doi: 10.1101/gad.920601. [DOI] [PubMed] [Google Scholar]
- Esmaeli S, Allameh A, Soleimani M, et al. The role of albumin and PPAR-alpha in differentiation-dependent change of fatty acid profile during differentiation of mesenchymal stem cells to hepatocyte-like cells. Cell biochemistry and function. 2014;32(5):410–419. doi: 10.1002/cbf.3031. [DOI] [PubMed] [Google Scholar]
- Gordillo M, Evans T, Gouon-Evans V. Orchestrating liver development. Development. 2015;142(12):2094. doi: 10.1242/dev.114215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallden G, Holehouse EL, Dong X, et al. Expression of intestinal fatty acid binding protein in intestinal epithelial cell lines, hBRIE 380 cells. The American journal of physiology. 1994;267(4 Pt 1):G730–743. doi: 10.1152/ajpgi.1994.267.4.G730. [DOI] [PubMed] [Google Scholar]
- Khajeniazi S, Allameh A, Soleimani M, et al. Changes in COX-2 and oxidative damage factors during differentiation of human mesenchymal stem cells to hepatocyte-like cells is associated with downregulation of p53 gene. Biological chemistry. 2013 doi: 10.1515/hsz-2013-0355. [DOI] [PubMed] [Google Scholar]
- Lee KC, Crowe AJ, Barton MC. p53-Mediated Repression of Alpha-Fetoprotein Gene Expression by Specific DNA Binding. Molecular and Cellular Biology. 1999;19(2):1279–1288. doi: 10.1128/mcb.19.2.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chanrion M, Sawey E, et al. Reciprocal Interaction of Wnt and RXR-α Pathways in Hepatocyte Development and Hepatocellular Carcinoma. PLOS ONE. 2015a;10(3):e0118480. doi: 10.1371/journal.pone.0118480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Uygun BE, Geerts S, et al. Proteomic Analysis of Naturally-Sourced Biological Scaffolds. Biomaterials. 2016;75:37–46. doi: 10.1016/j.biomaterials.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu D, Zong Y, et al. Developmental Stage-Specific Hepatocytes Induce Maturation of HepG2 Cells by Rebuilding the Regulatory Circuit. Molecular Medicine. 2015b;21(1):285–295. doi: 10.2119/molmed.2014.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallanna SK, Cayo MA, Twaroski K, et al. Mapping the Cell-Surface N-Glycoproteome of Human Hepatocytes Reveals Markers for Selecting a Homogeneous Population of iPSC-Derived Hepatocytes. Stem cell reports. 2016;7(3):543–556. doi: 10.1016/j.stemcr.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallanna SK, Duncan SA. Differentiation of hepatocytes from pluripotent stem cells. Current protocols in stem cell biology. 2013;26(Unit 1G.4) doi: 10.1002/9780470151808.sc01g04s26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Hernandez A, Amenta PS. The hepatic extracellular matrix. I. Components and distribution in normal liver. Virchows Archiv A, Pathological anatomy and histopathology. 1993;423(1):1–11. doi: 10.1007/BF01606425. [DOI] [PubMed] [Google Scholar]
- Matsui T, Kinoshita T, Hirano T, et al. STAT3 down-regulates the expression of cyclin D during liver development. The Journal of biological chemistry. 2002;277(39):36167–36173. doi: 10.1074/jbc.M203184200. [DOI] [PubMed] [Google Scholar]
- Mooney D, Hansen L, Vacanti J, et al. Switching from differentiation to growth in hepatocytes: control by extracellular matrix. Journal of cellular physiology. 1992;151(3):497–505. doi: 10.1002/jcp.1041510308. [DOI] [PubMed] [Google Scholar]
- Noto FK, Determan MR, Cai J, et al. Aneuploidy is permissive for hepatocyte-like cell differentiation from human induced pluripotent stem cells. BMC Research Notes. 2014;7:437–437. doi: 10.1186/1756-0500-7-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent R, Beretta L. Translational control plays a prominent role in the hepatocytic differentiation of HepaRG liver progenitor cells. Genome biology. 2008;9(1):R19. doi: 10.1186/gb-2008-9-1-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SA, Kormish J, Kofron M, et al. A gene regulatory network controlling hhex transcription in the anterior endoderm of the organizer. Developmental biology. 2011;351(2):297–310. doi: 10.1016/j.ydbio.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz RE, Fleming HE, Khetani SR, et al. Pluripotent stem cell-derived hepatocyte-like cells. Biotechnology Advances. 2014;32(2):504–513. doi: 10.1016/j.biotechadv.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J, Fortuno ES, 3rd, Suh JM, et al. Atf4 regulates obesity, glucose homeostasis, and energy expenditure. Diabetes. 2009;58(11):2565–2573. doi: 10.2337/db09-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K, Noto FK, Sepac A, et al. Generation of human induced pluripotent stem cells by simple transient transfection of plasmid DNA encoding reprogramming factors. BMC Developmental Biology. 2010;10:81–81. doi: 10.1186/1471-213X-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimizu N, Kikkawa Y, Mitaka T, et al. alpha1- and alpha5-containing laminins regulate the development of bile ducts via beta1 integrin signals. The Journal of biological chemistry. 2012;287(34):28586–28597. doi: 10.1074/jbc.M112.350488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimizu N, Miyajima A. Notch signaling controls hepatoblast differentiation by altering the expression of liver-enriched transcription factors. Journal of cell science. 2004;117(Pt 15):3165–3174. doi: 10.1242/jcs.01169. [DOI] [PubMed] [Google Scholar]
- Toivonen S, Malinen MM, Küblbeck J, et al. Regulation of Human Pluripotent Stem Cell-Derived Hepatic Cell Phenotype by Three-Dimensional Hydrogel Models. Tissue Engineering Part A. 2016;22(13–14):971–984. doi: 10.1089/ten.TEA.2016.0127. [DOI] [PubMed] [Google Scholar]
- Uygun BE, Soto-Gutierrez A, Yagi H, et al. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nature medicine. 2010;16(7):814–820. doi: 10.1038/nm.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Jakus AE, Baptista PM, et al. Functional Maturation of Induced Pluripotent Stem Cell Hepatocytes in Extracellular Matrix—A Comparative Analysis of Bioartificial Liver Microenvironments. Stem Cells Translational Medicine. 2016;5(9):1257–1267. doi: 10.5966/sctm.2015-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf MT, Daly KA, Brennan-Pierce EP, et al. A Hydrogel Derived From Decellularized Dermal Extracellular Matrix. Biomaterials. 2012;33(29):7028–7038. doi: 10.1016/j.biomaterials.2012.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagihara K, Liu Y, Kanie K, et al. Prediction of Differentiation Tendency Toward Hepatocytes from Gene Expression in Undifferentiated Human Pluripotent Stem Cells. Stem cells and development. 2016;25(24):1884–1897. doi: 10.1089/scd.2016.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai M, Tatsumi N, Hasunuma N, et al. FGF signaling segregates biliary cell-lineage from chick hepatoblasts cooperatively with BMP4 and ECM components in vitro. Developmental Dynamics. 2008;237(5):1268–1283. doi: 10.1002/dvdy.21520. [DOI] [PubMed] [Google Scholar]
- Zheng R, Rebolledo-Jaramillo B, Zong Y, et al. Function of GATA Factors in the Adult Mouse Liver. PLOS ONE. 2013;8(12):e83723. doi: 10.1371/journal.pone.0083723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D-y, Wu J-y, Li H, et al. PPAR-β facilitating maturation of hepatic-like tissue derived from mouse embryonic stem cells accompanied by mitochondriogenesis and membrane potential retention. Journal of Cellular Biochemistry. 2010;109(3):498–508. doi: 10.1002/jcb.22426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.