Abstract
Emerging studies have suggested that the Hippo pathway is involved in the tumorigenesis of hepatocellular carcinoma (HCC). However, the key regulator of the Hippo pathway in liver tumor metabolic reprogramming remains elusive. Here, we provide evidence to support that high mobility group box 1 (HMGB1), a chromosomal protein, plays a role in the regulation of the Hippo pathway during liver tumorigenesis. Cre/loxP recombination-mediated HMGB1 depletion in hepatocytes blocks diethylnitrosamine-induced liver cancer initiation in mice, whereas shRNA-mediated gene silencing of HMGB1 inhibits HCC cell proliferation. Mechanistically, the binding of HMGB1 to GA-binding protein alpha (GABPα) promotes the expression of yes-associated protein (YAP), a major downstream effector of the Hippo pathway, which contributes to liver tumorigenesis by inducing hypoxia-inducible factor 1α (HIF1α)-dependent aerobic glycolysis. Like wild type YAP-cDNA, YAP-5SA-S94A can restore HIF1α DNA binding activity, glycolysis-associated gene expression, and HIF1α-YAP complex formation in YAP-knockdown HCC cell lines. In contrast, verteporfin, a reagent targeting the interface between YAP and TEA domain transcription factor (TEAD), has the ability to block YAP-HIF1α complex formation. Notably, genetic or pharmacological inhibition of the HMGB1-YAP-HIF1α pathway confers protection against excessive glycolysis and tumor growth in mice. Conclusion: These findings uncover that HMGB1 plays a novel role in modulating the YAP-dependent HIF1α pathway and sheds light on the development of metabolism-targeting therapeutics for HCC chemoprevention.
Keywords: HMGB1, YAP, hepatocellular carcinoma, Warburg effect
Introduction
The Hippo pathway was first discovered in Drosophila as an evolutionarily conserved regulator of organ size and regeneration. The loss of Hippo leads to tissue overgrowth due to increased proliferation and decreased apoptosis. Similarly, dysregulation of the Hippo pathway can also trigger tumorigenesis in Drosophila and mice through multiple mechanisms (1–3). The core components of the Hippo pathway are composed of a regulatory serine-threonine kinase module (e.g., STE20-like protein kinase 1/2 [MST1/2] and large tumor suppressor 1/2 [LATS1/2]), a transcriptional module (e.g., yes-associated protein [YAP], and transcriptional co-activator with PDZ-binding motif [TAZ]). In particular, the YAP-mediated gene transcriptional output is essential for hepatocellular carcinoma (HCC) initiation. Liver-specific YAP overexpression in transgenic mice leads to hepatomegaly and subsequent tumor formation (4–6). Additionally, genetic knockdown of YAP reduces subcutaneous tumor growth of HCC lines (7). The widespread expression and activation of YAP in human liver cancer further implicates YAP as an important HCC therapeutic target (8).
High mobility group protein B1 (HMGB1) belongs to a family of highly conserved chromosome proteins that contain HMG box domains. In addition to nuclear function as a DNA chaperone, HMGB1 can be released into the extracellular environment and act as an immune mediator in infectious and sterile inflammation. Abnormal expression and release of HMGB1 are implicated in multiple human diseases (9). HMGB1 has been reported to play dual roles in tumorigenesis depending on tumor type and its subcellular distribution (10). Multiple clinical studies have shown that HMGB1 expression and release is increased in human HCC (11). Extracellular HMGB1 promotes the growth, migration, and metastasis of HCC cells (12), whereas intracellular HMGB1 mediates mitochondrial biogenesis in liver tumor growth (13). However, the specific role of intracellular HMGB1 in metabolic reprogramming during liver tumorigenesis remains largely unknown.
In the current study, we provide transgenic animal evidence that intracellular HMGB1 plays an oncogenic role in primary liver cancer. We observed that conditional knockout of HMGB1 in hepatocytes delayed diethylnitrosamine (DEN)-induced liver cancer initiation in mice. Mechanistically, we demonstrated that HMGB1 is a novel regulator of the Hippo pathway and that HMGB1-mediated YAP expression contributes to aerobic glycolysis in tumor growth. Our data therefore identify a key role for HMGB1 action in the modulation of the Hippo pathway and metabolic reprogramming in liver tumorigenesis. Thus, targeting this HMGB1-YAP-dependent metabolic pathway holds promise as a novel anticancer strategy.
Materials and Methods
Antibodies and reagents
The antibodies to HMGB1 (#3935), YAP (#14074), TAZ (#4883), Axin1 (#2087), β-catenin (#9587), Notch1 (#3608), Notch2 (#5732), HDAC2 (#5113), PCNA (#2586), cyclin D1 (#2922), cleaved-caspase 3 (#9661), γH2AX (#9718), GAPDH (#2118), and actin (#3700) were obtained from Cell Signaling Technology. The antibody to GABPα (#21542-1-AP) was obtained from Proteintech Group. The antibody to p73 (#ab26123) was obtained from Abcam. The antibody to HIF1α (#NB100-105) was obtained from NOVUS. Diethylnitrosamine (#N0258), glycyrrhizin (#CDS020796), verteporfin (#SML0534), glucose (#G8270), oligomycin (#O4876), and 2-deoxy-D-glucose (“2-DG”, #D8375) were obtained from Sigma.
Cell lines and culture
Hepa1-6, HepaG2, and Hep3B cells were obtained from American Type Culture Collection. HuH7 cells were a gift from Dr. Allan Tsung. HMGB1−/− mouse embryonic fibroblasts (MEFs) were a gift from Dr. Marco E. Bianchi. These cells were grown in Eagle's Minimum Essential Medium (HepaG2 and Hep3B) or Dulbecco's Modified Eagle's Medium (Hepa1-6, HuH7, and MEFs) with 10% fetal bovine serum, 2 mM L-glutamine, and 100 U/ml of penicillin and streptomycin.
Hepatocyte isolation
Hepatocytes were isolated from tissue samples taken from HCC patients who had undergone hepatic resections or from livers of mice. Sample collection from patients was approved by the Institutional Review Board. Hepatocyte isolation was carried out using a modified “two-stage” collagenase procedure developed by Berry and Friend (14).
Cell viability and clonogenic cell survival assay
Cell viability was assayed using Cell Counting Kit-8 kits (Dojindo Laboratories). Long-term cell survival was monitored in a colony formation assay. In brief, 1,000 cells were reseeded into 24-well plates after treatment with indicated drugs for 24 hours. The cells were allowed to grow for the next 10 to 12 days to allow colony formation. The colonies were visualized using crystal violet staining.
Western blot analysis
Western blot was used to analyze protein expression as described previously (15). In brief, after extraction, proteins in the cell lysate were resolved on 4%–12% Criterion XT Bis-Tris gels (Bio-Rad) and transferred to a nitrocellulose membrane. After blocking with 5% milk, membranes were incubated for two hours at 25°C with various primary antibodies. After incubation with peroxidase-conjugated secondary antibodies for one hour at room temperature, the signals were visualized using enhanced or super chemiluminescence and exposure to X-ray films.
Immunoprecipitation analysis
Cells were lysed at 4°C in ice-cold radioimmunoprecipitation assay buffer and cell lysates were cleared using brief centrifugation. Concentrations of proteins in the supernatant were determined using BCA assay. Prior to immunoprecipitation, samples containing equal amounts of proteins were pre-cleared with protein G agarose (4°C, 3 h), and subsequently incubated with various irrelevant IgG or specific antibodies (2–4 µg/mL) in the presence of protein G agarose beads for overnight at 4°C with gentle shaking. Following incubation, agarose beads were washed extensively with phosphate buffered saline (PBS) and proteins were eluted by boiling in 2 × sodium dodecyl sulfate (SDS) sample buffer before SDS polyacrylamide gel electrophoresis.
RNAi and gene transfection
The human HMGB1-shRNA-1 (5’- CCGGCCCAGATGCTTCAGTCAACTTCTCGAGAAGTTGACTGAAGCATCTGGGTTTTT-3’), human HMGB1-shRNA-2 (5’- CCGGCCGTTATGAAAGAGAAATGAACTCGAGTTCATTTCTCTTTCATAACGGTTTTT-3’), mouse HMGB1-shRNA-1 (5’- CCGGGAAGATGATGATGATGAATAACTCGAGTTATTCATCATCATCATCTTCTTTTTG-3’), mouse HMGB1-shRNA-2 (5’- CCGGTGACAAGGCTCGTTATGAAAGCTCGAGCTTTCATAACGAGCCTTGTCATTTTTG-3’), human YAP-shRNA (5’- CCGGGCCACCAAGCTAGATAAAGAACTCGAGTTCTTTATCTAGCTTGGTGGCTTTTTG-3’), mouse YAP-shRNA (5’- CCGGGAAGCGCTGAGTTCCGAAATCCTCGAGGATTTCGGAACTCAGCGCTTCTTTTTG-3’), human GLUT1-shRNA (5’- CCGGGCCACACTATTACCATGAGAACTCGAGTTCTCATGGTAATAGTGTGGCTTTTTG-3’), mouse GLUT1-shRNA (5’- CCGGGCTGAGAACTTAACTGCTGAACTCGAGTTCAGCAGTTAAGTTCTCAGCTTTTTG-3’), human HK2-shRNA (5’- CCGGCCAAAGACATCTCAGACATTGCTCGAGCAATGTCTGAGATGTCTTTGGTTTTTTG-3’), mouse HK2-shRNA (5’- CCGGCGGTACAGAGAAAGGAGACTTCTCGAGAAGTCTCCTTTCTCTGTACCGTTTTTG-3’) and control empty shRNA (pLKO.1) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Stable knockdown cells were selected by adding puromycin. Expression plasmids for mouse HMGB1-cDNA and YAP-cDNA (pCMV6) were purchased from OriGene Technologies, Inc. YAP-5SA-S94A mutant (pCMV) and PGC1α-cDNA (pcDNA3.1) were obtained from Addgene. For the rescue experiments, wild type HMGB1-cDNA and YAP-cDNA were transfected into a stable shRNA cell line by Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions.
Quantitative real time polymerase chain reaction assay (Q-PCR)
cDNA from various cell samples was amplified using Q-PCR with specific primers (mouse HMGB1: 5’- CCAAGAAGTGCTCAGAGAGGTG-3’ and 5’- GTCCTTGAACTTCTTTTTGGTCTC-3’; human HMGB1: 5’- GCGAAGAAACTGGGAGAGATGTG-3’ and 5’-GCATCAGGCTTTCCTTTAGCTCG-3’; mouse YAP: 5’- CCAGACGACTTCCTCAACAGTG-3’ and 5’- GCATCTCCTTCCAGTGTGCCAA-3’; human YAP: 5’- TGTCCCAGATGAACGTCACAGC-3’ and 5’-TGGTGGCTGTTTCACTGGAGCA-3’; mouse TAZ: 5’- CCTTATCACCGTCTCCAACCAC-3’ and 5’- CCTTGGTGAAGCAGATGTCTGC-3’; mouse β-catenin: 5’- GTTCGCCTTCATTATGGACTGCC-3’ and 5’-ATAGCACCCTGTTCCCGCAAAG-3’; mouse Axin1: 5’- GTCCAGTGATGCTGACACGCTA-3’ and 5’- GCCCATTGACTTGGATACTCTCC-3’; mouse Notch1: 5’- GCTGCCTCTTTGATGGCTTCGA-3’ and 5’-CACATTCGGCACTGTTACAGCC-3’; mouse Notch2: 5’- CCACCTGCAATGACTTCATCGG-3’ and 5’- TCGATGCAGGTGCCTCCATTCT-3’; mouse BIRC5: 5’- CCTACCGAGAACGAGCCTGATT-3’ and 5’-CCATCTGCTTCTTGACAGTGAGG-3’; human BIRC5: 5’- CCACTGAGAACGAGCCAGACTT-3’ and 5’- GTATTACAGGCGTAAGCCACCG-3’; mouse CCND1: 5’- GCAGAAGGAGATTGTGCCATCC-3’ and 5’-AGGAAGCGGTCCAGGTAGTTCA-3’; human CCND1: 5’- TCTACACCGACAACTCCATCCG-3’ and 5’- TCTGGCATTTTGGAGAGGAAGTG-3’; mouse MYC: 5’- TCGCTGCTGTCCTCCGAGTCC-3’ and 5’-GGTTTGCCTCTTCTCCACAGAC-3’; human MYC: 5’- CCTGGTGCTCCATGAGGAGAC-3’ and 5’- CAGACTCTGACCTTTTGCCAGG-3’; mouse SPP1: 5’- GCTTGGCTTATGGACTGAGGTC-3’ and 5’-CCTTAGACTCACCGCTCTTCATG-3’; human SPP1: 5’- CGAGGTGATAGTGTGGTTTATGG-3’ and 5’- GCACCATTCAACTCCTCGCTTTC-3’; mouse GPC3: 5’- CTGTGCTGGAACGGACAAGAAC-3’ and 5’-GTCAATGATCTGGCTAACCACCG-3’; mouse BAX: 5’- AGGATGCGTCCACCAAGAAGCT-3’ and 5’- TCCGTGTCCACGTCAGCAATCA-3’; mouse PUMA: 5’- ACCGCTCCACCTGCCGTCAC-3’ and 5’-ACGGGCGACTCTAAGTGCTGC-3’; mouse GLUT1: 5’- GCTTCTCCAACTGGACCTCAAAC-3’ and 5’- ACGAGGAGCACCGTGAAGATGA-3’; human GLUT1: 5’- TTGCAGGCTTCTCCAACTGGAC-3’ and 5’-CAGAACCAGGAGCACAGTGAAG-3’; mouse HK2: 5’- CCCTGTGAAGATGTTGCCCACT-3’ and 5’- CCTTCGCTTGCCATTACGCACG-3’; human HK2: 5’- GAGTTTGACCTGGATGTGGTTGC-3’ and 5’-CCTCCATGTAGCAGGCATTGCT-3’; mouse ALDOA: 5’- CACGAGACACTGTACCAGAAGG-3’ and 5’- TTGTCTCGCCATTGGTTCCTGC-3’; human ALDOA: 5’- GACACTCTACCAGAAGGCGGAT-3’ and 5’-GGTGGTAGTCTCGCCATTTGTC-3’; mouse LDHA: 5’-CACGAGACACTGTACCAGAAGG-3’ and 5’- TTGTCTCGCCATTGGTTCCTGC-3’; human LDHA: 5’- ACGCAGACAAGGAGCAGTGGAA-3’ and 5’-ATGCTCTCAGCCAAGTCTGCCA-3’; mouse HIF1α: 5’-CCTGCACTGAATCAAGAGGTTGC-3’ and 5’- CCATCAGAAGGACTTGCTGGCT-3’; mouse 18S RNA (5’- GCAATTATTCCCCATGAACG-3’ and 5’-GGCCTCACTAAACCATCCAA-3’); human 18S RNA (5’-CTACCACATCCAAGGAAGCA-3’ and 5’- TTTTTCGTCACTACCTCCCCG-3’) using a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad) with SsoFast™ EvaGreen® Supermix (#1725201, Bio-Rad). 18S rRNA was used to normalize the relative expression levels of target genes.
Glycolysis assay
Cellular glycolysis was monitored using the Seahorse Bioscience Extracellular Flux Analyzer (XF24, Seahorse Bioscience Inc., North Billerica, MA, USA) by measuring the extracellular acidification rate (ECAR) in real time as previously described (16, 17).
Serum biochemistry
Serum levels of lactate (#MAK064, Sigma), glucagon (#RAB0202, Sigma), and insulin (#EMINS, Thermo Fisher Scientific Inc.) were measured using ELISA per the manufacturer's protocol. For measurement of serum nucleosomes, Cell Death Detection ELISAplus (#11920685001, Roche Diagnostics) was used. Glucose concentration measurements were obtained from whole-blood samples using hand-held whole-blood glucose monitors (Bayer) according to the manufacturer's protocol.
HIF1α activation assay
HIF-1α Transcription Factor Assay (#ab133104, Abcam) is a non-radioactive, sensitive method for detecting HIF1α DNA binding activity to core DNA sequence 5'-[AG]CGTG-3' within the hypoxia response element of target gene promoters in nuclear extracts or whole cell lysate per the manufacturer’s instructions. HIF1α luciferase reporter activity was assayed using Cignal HIF Reporter Kits (#CCS-007L, QIAGEN) according to the manufacturer's protocol.
Secrete-pair luminescence assay
Cells were transfected with pEZX-PG04-YAP-promoter-Gaussia luciferase (GLuc)/secreted alkaline phosphatase (SEAP) (#MPRM35478-PG04, GeneCopoeia). The YAP promoter luciferase activity was measured using a secrete-pair dual luminescence assay kit (#SPDA-D010, GeneCopoeia) in accordance with the manufacturer’s guidelines.
Chromatin immunoprecipitation (ChIP) assay
ChIP was performed according to the protocol of the chromatin immunoprecipitation assay kit (#17-295, EMD Millipore) as previously described (18). Briefly, cells were fixed with 1% formaldehyde for 10 min at room temperature and then quenched with glycine for 5 min. The fixed cells were washed with PBS containing protease inhibitors and lysed in lysis buffer for 10 min on ice prior to the sonication, centrifugation, and addition of dilution buffer. One percent of input was removed and the lysates were immunoprecipitated with 2 mg of anti-GABPα antibody, anti-HMGB1 antibody, or control IgG for 2 hr. Salmon sperm DNA/protein A/G-Sepharose beads were added to these immunoprecipitations for incubation overnight. Immune complex pellets were washed and then eluted. The elutes were heated at 65°C for four hours to reverse the cross-linking and treated with RNase A for 30 min at 37°C, followed by treatment with proteinase K for one hour at 45°C to remove RNA and protein. DNA was recovered, eluted, and then assayed using PCR.
Animals and treatments
All animal experiments were approved by the Institutional Animal Care and Use Committees. Hmgb1flox/flox mice on C57BL/6J (B6) background were obtained from Dr. Eugene B. Chang. Hif1αflox/flox and Alb-Cre mice on B6 background were purchased from Jackson Laboratories.
To generate murine subcutaneous tumors, 2×106 Hepa1-6 or HuH7 cells in 100 µl PBS were injected subcutaneously to the right of the dorsal midline in six- to eight-week-old athymic nude or B6 mice. Once the tumors reached 50–70 mm3 at day seven, mice were randomly allocated into groups and treated with glycyrrhizin (50 or 100 mg/kg, intraperitoneal (i.p.)) or verteporfin (25 or 50 mg/kg, i.p.) every day beginning on the seventh day post xenograft injection for two weeks. Tumors were measured twice weekly and volumes were calculated using the formula length×width2×π/6.
To generate DEN-induced liver tumors, 15-day-old male mice were treated with a single i.p. injection of DEN (5 mg/kg body weight). Mice were then randomly allocated into groups and treated with glycyrrhizin (100 mg/kg, i.p.) or verteporfin (50 mg/kg, i.p.) twice every week for two months. Liver tissues were harvested to determine mRNA and protein levels at three, five, or 10 months after DEN-induced tumorigenesis. Externally and internally, tumors were counted and measured using a combination of visual inspection and hematoxylin and eosin staining as previously described (19).
Statistical analysis
Unless otherwise indicated, data are expressed as means ± SD. Unpaired Student’s t tests were used to compare the means of two groups. One-way ANOVA was used for comparison among the different groups. When ANOVA was significant, post hoc testing of differences between groups was performed using the LSD test. Differences with P values of < 0.05 were considered statistically significant.
Results
HMGB1 deletion inhibits liver cancer initiation
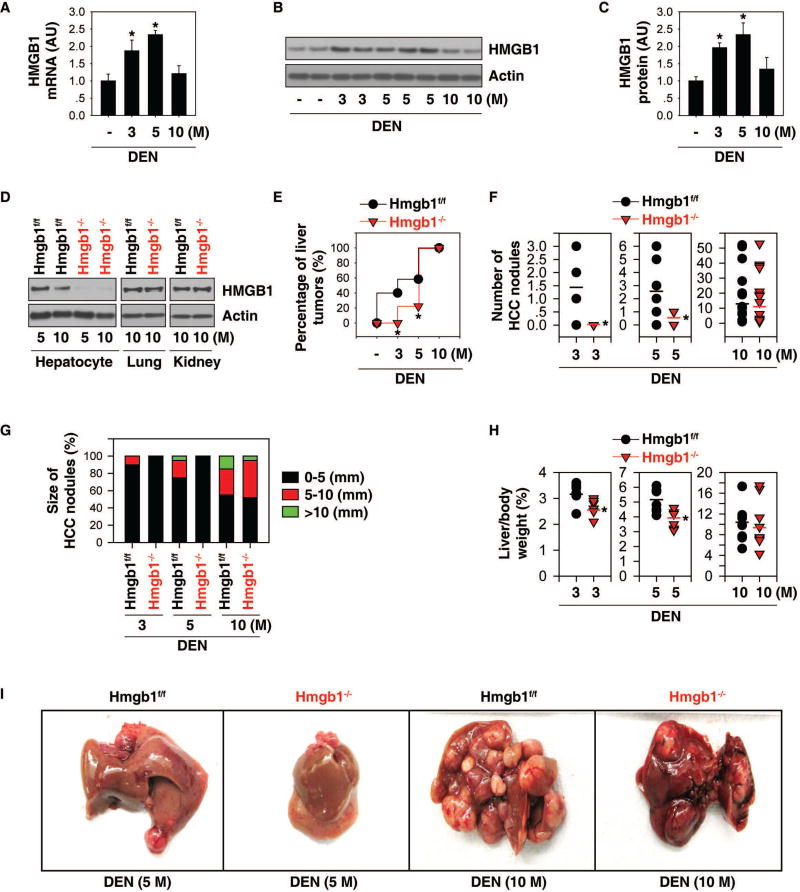
To determine the role of HMGB1 in tumorigenesis, we first analyzed the expression of HMGB1 in the liver after male B6 mice were challenged with DEN. Q-PCR and western blot analysis showed that the hepatic expression levels of HMGB1 mRNA (Fig. 1A) and protein (Fig. 1B and 1C) were initially upregulated at three and five months, and then restored to basal expression levels at 10 months after DEN treatment, suggesting that HMGB1 may play a different role in liver cancer initiation, progression, and advancement.
Fig. 1. HMGB1 deletion inhibits liver cancer initiation.
(A) Q-PCR analysis of HMGB1 mRNA expression in wild-type livers after DEN treatment for three to 10 months (n=3 mice for each time point. *p<0.05). (B, C) Representative western blots and quantitation of HMGB1 levels in wild-type livers after DEN treatment for three to 10 months (n=3 mice for each time point. *p<0.05). (D) Western blot analysis of HMGB1 protein expression in isolated hepatocytes, lungs, and kidneys from hepatocyte-specific HMGB1 knockout mice (“Hmgb1−/−”) and control Hmgb1flox/flox mice (“Hmgb1f/f mice”) at five to 10 months of age. (E) Percentage of liver tumors in Hmgb1−/− and Hmgb1f/f mice following DEN treatment for three to 10 months (three months, n=10 for each group; five months, n=12 for Hmgb1f/f mice and n=10 for Hmgb1−/− mice; 10 months, n=15 for Hmgb1f/f mice and n=14 for Hmgb1−/− mice. (F–H) Number (F) and size (G) of HCC nodules, as well as liver weight (H) in Hmgb1−/− and Hmgb1f/f mice following DEN treatment for three to 10 months (n=8–10 for each group, *p<0.05 versus Hmgb1f/f group). (I) Representative pictures of the livers.
To define the time frame at which HMGB1 is required for liver tumor development, hepatocyte-specific HMGB1 knockout mice (Alb-Cre;Hmgb1flox/flox, termed “Hmgb1−/−” mice) were employed. HMGB1 protein expression was constitutively deleted in hepatocytes of Hmgb1−/− mice compared to control Hmgb1flox/flox mice (termed “Hmgb1f/f mice”) (Fig. 1D). In contrast, HMGB1 protein expression in the kidney and lung did not differ between the Hmgb1−/− and Hmgb1f/f mice (Fig. 1D). Notably, the rate of liver tumorigenesis was significantly reduced by HMGB1 deletion at three and five months, but not at 10 months after DEN treatment (Fig. 1E). Consistently, the number (Fig. 1F)/size (Fig. 1G) of HCC nodules, as well as liver weight (Fig. 1H)/size (Fig. 1I) were significantly reduced in Hmgb1−/− mice at three or five months, but not at 10 months after DEN treatment, supporting an important role for intracellular HMGB1 in the initiation of liver cancer.
HMGB1 regulates the Hippo pathway
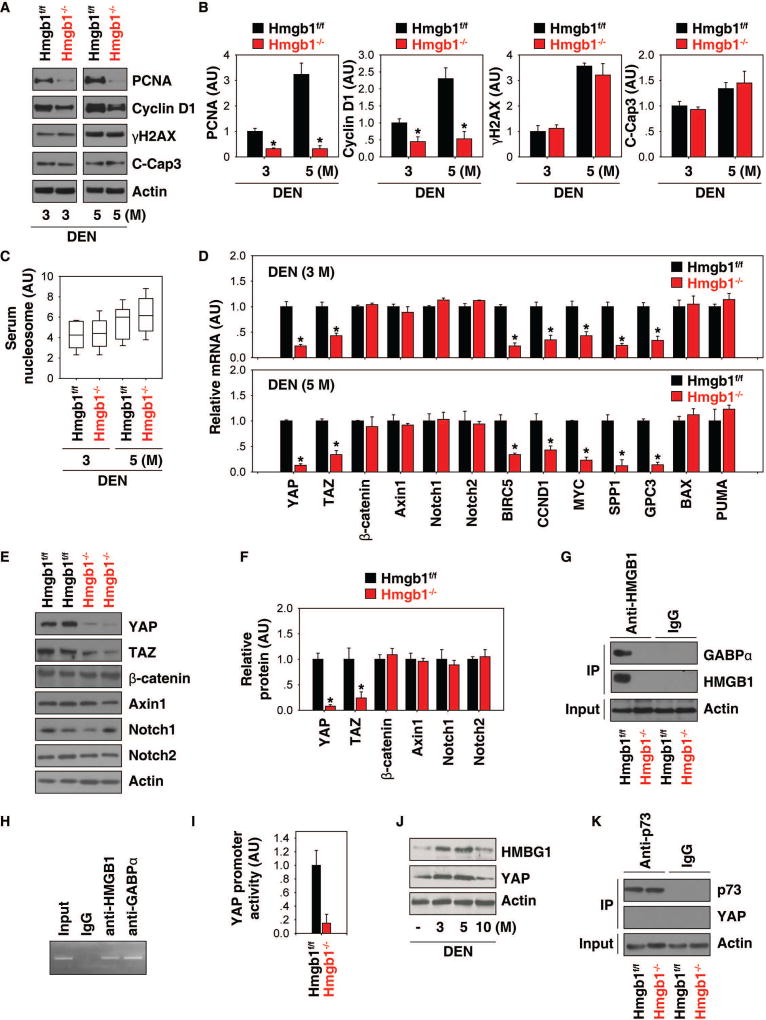
We recently demonstrated that intracellular HMGB1 inhibits pancreatic tumorigenesis partly though blocking DNA damage-mediated proinflammatory nucleosome release (20). Interestingly, the levels of γH2AX (a marker of DNA damage) and cleaved caspase-3 (“C-Cap3,” a marker of cell apoptosis) in livers (Fig. 2A and 2B) as well as circulating nucleosome (Fig. 2C) were not affected by hepatic HMGB1 depletion at three or five months after DEN treatment. In contrast, the expression levels of cell proliferation markers such as the proliferating cell nuclear antigen (PCNA) and cyclin D1 were significantly reduced in livers from Hmgb1−/− mice following DEN treatment (Fig. 2A and 2B). These results demonstrate that HMGB1 is required for cell proliferation, but not cell death and DNA damage during liver cancer initiation.
Fig. 2. HMGB1 regulates the Hippo pathway.
(A, B) Representative western blots and quantitation of indicated protein levels in livers from Hmgb1−/− and Hmgb1f/f mice after DEN treatment for three and five months (n=3 mice for each time point. *p<0.05 versus Hmgb1f/f group). (C) Measurement of serum nucleosome levels in Hmgb1−/− and Hmgb1f/f mice after DEN treatment for three and five months. Data are presented as median value (black line), interquartile range (box), and minimum and maximum of all data (black line) (n=10 mice for each time point). (D) Q-PCR analysis of mRNA expressions of indicated genes in livers from Hmgb1−/− and Hmgb1f/f mice after DEN treatment for three and five months (n=3 mice for each time point. *p<0.05 versus Hmgb1f/f group). (E, F) Representative western blot image and analysis to quantify indicated protein levels in livers from Hmgb1−/− and Hmgb1f/f mice after DEN treatment for five months (n=3 mice for each time point. *p<0.05 versus Hmgb1f/f group). (G) Immunoprecipitation analysis of the interaction between HMGB1 and GABPα in livers from Hmgb1−/− and Hmgb1f/f mice after DEN treatment for five months. (H) ChIP analysis of GABPα and HMGB1 binding to the YAP promoter in primary mouse hepatocytes. (I) Analysis of YAP promoter activity in hepatocytes from Hmgb1−/− and Hmgb1f/f mice after DEN treatment for five months. (J) Western blot analysis of YAP and HMGB1 expression in wild-type livers after DEN treatment for three to 10 months. (K) Immunoprecipitation analysis of the interaction between p73 and YAP in livers from Hmgb1−/− and Hmgb1f/f mice after DEN treatment for five months.
To search for downstream cell proliferation effectors of HMGB1-mediated liver cancer initiation, we examined the expression levels of several signaling molecules (e.g., Wnt/β-catenin, Notch, and Hippo) implicated in the regulation of cell proliferation during liver carcinogenesis. Q-PCR (Fig. 2D) and western blot (Fig. 2E and 2F) analysis confirmed that the expression of core components of the Hippo pathway (e.g., YAP and TAZ) was reduced in livers of DEN-induced Hmgb1−/− mice. In contrast, the expression of core components of the Wnt/β-catenin (e.g., β-catenin and Axin1) and Notch pathways (e.g., Notch1 and Notch2) was not affected by HMGB1 depletion during DEN-induced liver tumorigenesis (Fig. 2D–2F).
Given that GA-binding protein alpha (GABPα) is an important transcription regulator of YAP gene expression (18), we hypothesized that HMGB1 can bind GABPα to regulate YAP promoter activity. Indeed, a direct interaction between HMGB1 and GABPα occurred in liver extracts after DEN treatment (Fig. 2G). The ChIP assay showed that GABPα and HMGB1 bond to the YAP promoter in primary mouse hepatocytes (Fig. 2H). As expected, loss of HMGB1 in hepatocytes significantly inhibited YAP promoter activity (Fig. 2I). Like HMGB1, the hepatic expression levels of YAP protein were initially upregulated at three and five months, and were then restored to basal expression levels at 10 months after DEN treatment (Fig. 2J).
To better understand the role of HMGB1 in the regulation of the Hippo pathway, we further measured the mRNA expression levels of YAP target genes such as baculoviral IAP repeat-containing protein 5 (BIRC5), cyclin D1 (CCND1), V-Myc avian myelocytomatosis viral oncogene homolog (MYC), secreted phosphoprotein 1 (SPP1), and glypican 3 (GPC3). The expressions of these target genes were all downregulated in the livers of Hmgb1−/− mice after DEN treatment (Fig. 2D), supporting HMGB1 as a novel regulator of the Hippo pathway.
In addition to acting as an oncogene in many cases, YAP may occasionally also serve as a tumor suppressor through complex interaction with other transcription factors (2, 3). For example, YAP binds to the tumor suppressor p73, contributing to the transcription of pro-apoptotic genes (e.g., BAX and p53 up-regulated modulator of apoptosis [PUMA]) in several hematological cancers (21). However, the YAP-p73 complex was not observed in DEN-induced liver tumorigenesis (Fig. 2K). Additionally, the mRNA of BAX and PUMA was not affected by HMGB1 in DEN-induced liver tumorigenesis (Fig. 2D).
YAP is responsible for HMGB1-mediated cell growth
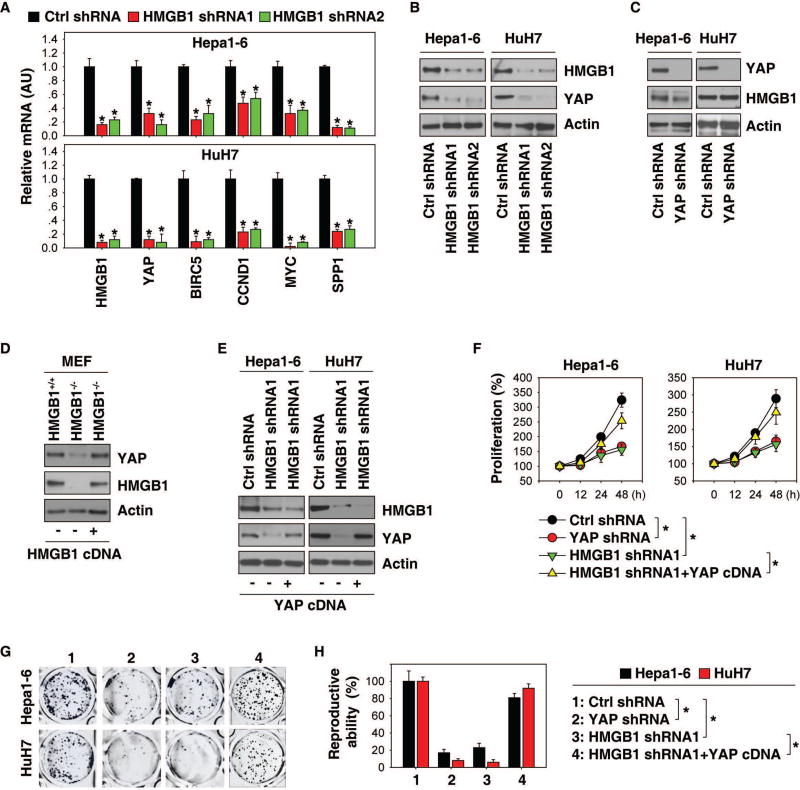
We next examined whether genetic inhibition of HMGB1 affected YAP expression in HCC cells. Knockdown of HMGB1 by two specific shRNAs reduced mRNA expressions of YAP and its target genes (BIRC5, CCND1, MYC, and SPP1) in Hepa1-6 and HuH7 cells (Fig. 3A). Consistently, knockdown of HMGB1 also decreased YAP protein expression (Fig. 3B). In contrast, genetic silencing of YAP did not affect HMGB1 expression, suggesting that HMGB1 expression is not reversely regulated by YAP (Fig. 3C). In addition to HCC cell lines, we also observed that the protein expression of YAP was diminished in HMGB1−/− MEF cell lines (Fig. 3D). Enforced genetic expression of HMGB1-cDNA restored expression of HMGB1 as well as YAP in HMGB1−/− MEFs (Fig. 3D). However, the enforced expression of YAP-cDNA only restored YAP expression, but not HMGB1 expression, in HMGB1-knockdown HCC cells (Fig. 3E). These results clearly indicate that YAP expression is controlled by HMGB1, but not vice versa.
Fig. 3. YAP is responsible for HMGB1-mediated cell growth.
(A) Q-PCR analysis of mRNA expressions of indicated genes in Hepa1-6 and HuH7 cells after knockdown of HMGB1 (n=3, *p<0.05 versus control shRNA group). (B, C) Western blot analysis of indicated protein expression in Hepa1-6 and HuH7 cells after knockdown of HMGB1 or YAP. (D) Forced expression of HMGB1 cDNA restored HMGB1 and YAP expression in HMGB1−/− MEFs. (E) Forced expression of YAP cDNA restored YAP expression, but not HMGB1 expression in indicated HMGB1-knockdown HCC cells. (F) Forced expression of YAP reversed cell proliferation inhibition in HMGB1 knockdown HCC cells (n=3, *p < 0.05). (G–H) Clonogenic cell survival assay determines the reproductive ability of indicated HCC cells. A representative image is shown in panel G. Relative reproductive ability is semi-quantified in panel H (n=3, *p < 0.05).
We next analyzed whether YAP expression is responsible for HMGB1 function in cell survival and proliferation. The direct knockdown of YAP by shRNA recapitulates the HMGB1-deficient phenotype in cell proliferation (Fig. 3F) and colony formation (Fig. 3G and 3H). Conversely, the enforced expression of YAP reversed these phenotypes in HMGB1-knockdown cells (Fig. 3F–3H). These findings suggest that YAP expression is responsible for HMGB1 function in cell proliferation and growth.
HMGB1-YAP pathway mediates the Warburg effect
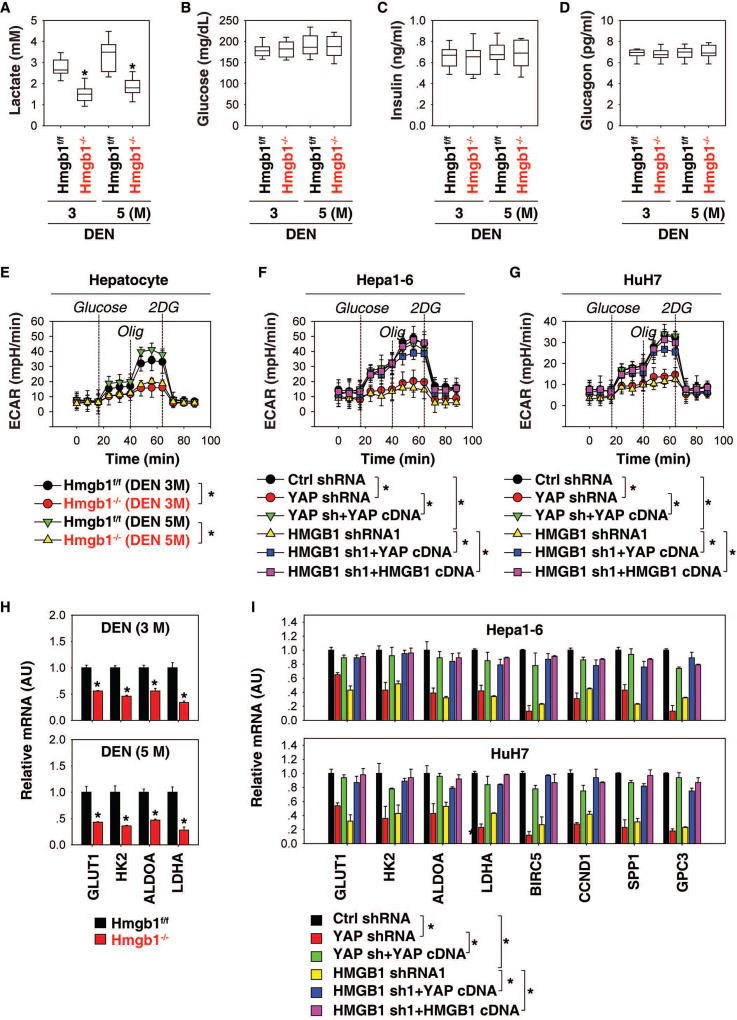
The Warburg effect, also known as aerobic glycolysis, is a metabolic hallmark of most cancer cells, including HCC, characterized by an excessive conversion of glucose to lactate, even with ample oxygen. To assess whether the HMGB1-YAP pathway is responsible for the Warburg effect in liver tumorigenesis, we first measured serum levels of lactate, glucose, insulin, and glucagon in Hmgb1flox/flox and Hmgb1−/− mice after treatment with DEN. The level of lactate was significantly downregulated in Hmgb1−/− mice compared with Hmgb1flox/flox following DEN treatment at three and five months (Fig. 4A). In contrast, serum levels of glucose (Fig. 4B), insulin (Fig. 4C), and glucagon (Fig. 4D) did not differ between DEN-induced Hmgb1−/− and Hmgb1flox/flox mice. Thus, HMGB1 is required for lactate production, but not glucose production, during DEN-induced liver tumorigenesis.
Fig. 4. The HMGB1-YAP pathway mediates the Warburg effect.
(A–D) Serum levels of lactate (A), glucose (B), insulin (C), and glucagon (D) in Hmgb1−/− and Hmgb1f/f mice after DEN treatment for three and five months (n=10 mice for each time point. *p<0.05 versus Hmgb1f/f group). (E–G) Analysis of ECAR in indicated hepatocytes or HCC cells (n=3, *p < 0.05). (H) Q-PCR analysis of mRNA expressions of indicated genes in livers from Hmgb1−/− and Hmgb1f/f mice after DEN treatment for three and five months (n=3 mice for each time point. *p<0.05 versus Hmgb1f/f group). (I) Q-PCR analysis of mRNA expressions of indicated genes in indicated Hepa1-6 and HuH7 cells (n=3, *p<0.05).
To further define the role of HMGB1 in the regulation of lactate production, we measured ECAR in hepatocytes using the Seahorse Bioscience Extracellular Flux Analyzer. This assay was started in the absence of glucose; then glucose, oligomycin, and 2-DG were sequentially added as previously described (17). The level of aerobic glycolysis was significantly reduced in hepatocytes derived from Hmgb1−/− mice compared to hepatocytes from Hmgb1flox/flox mice (Fig. 4E). Genetic inhibition of HMGB1 or YAP by shRNA in Hepa1-6 (Fig. 4F) and HuH7 (Fig. 4G) cells also resulted in a decrease in ECAR. In contrast, the enforced expression of YAP or HMGB1 restored ECAR in HMGB1-knockdown cells (Fig. 4F and 4G). The enforced expression of YAP also restored ECAR in YAP-knockdown cells (Fig. 4F and 4G). These findings support our hypothesis that HMGB1-mediated YAP expression contributes to aerobic glycolysis in HCC cells.
Compared with normal cells, glycolysis is accelerated in HCC cells by preferential expression of glucose transporters (e.g., glucose transporter 1 [GLUT1]) and enzyme isoforms (e.g., hexokinase 2 [HK2], fructose-bisphosphate aldolase A [ALDOA], and lactate dehydrogenase A [LDHA]) that drive glucose flux forward and to adapt to the anabolic demands of cancer cells (22). We therefore explored whether the HMGB1-YAP pathway regulates glycolysis through targeting these glycolysis-associated genes. Indeed, the mRNA levels of GLUT1, HK2, ALDOA, and LDHA were all downregulated in the livers of Hmgb1−/− mice during DEN-induced tumorigenesis (Fig. 4H). The knockdown of HMGB1 or YAP in HCC cells also led to downregulation of glycolysis-associated genes (GLUT1, HK2, ALDOA, and LDHA) and YAP-targeted genes (BIRC5, CCND1, SPP1, and GPC3) (Fig. 4I). Like the enforced expression of YAP or HMGB1 in HMGB1-knockdown cells, the enforced expression of YAP also restored expression of these genes in YAP-knockdown HCC cells (Fig. 4I). Collectively, the changes in expression of genes involved in glycolysis could inform the underlying biological mechanisms by which HMGB1 and YAP contribute to the DEN-induced liver tumorigenesis.
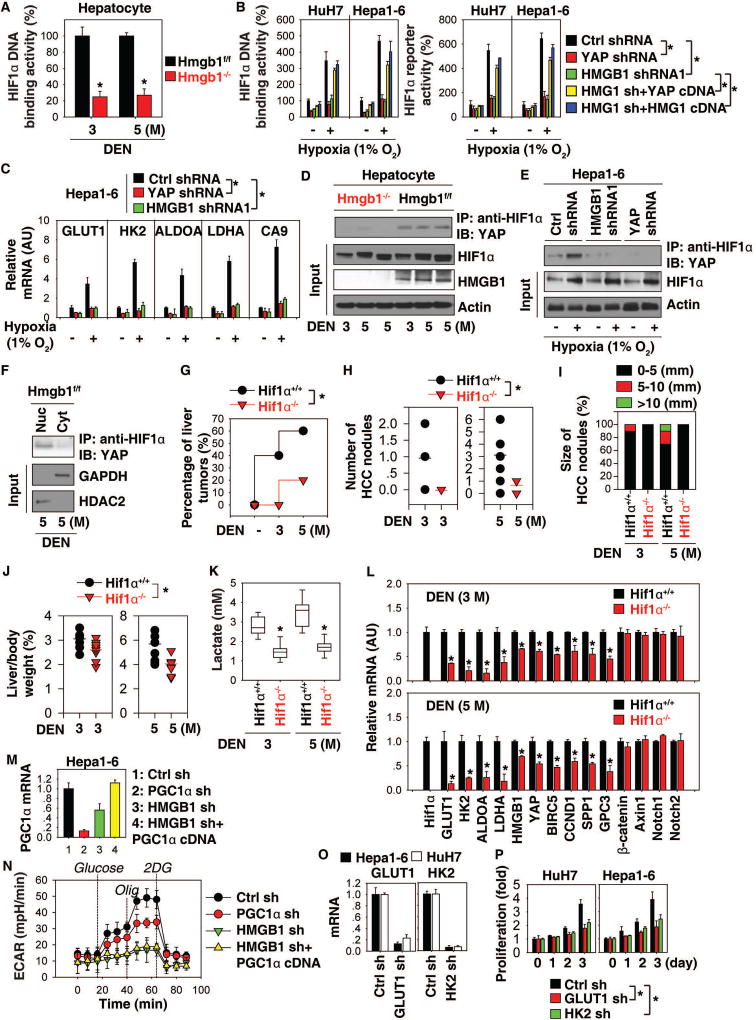
Interplay between HMGB1, YAP, and HIF1α contributes to liver tumorigenesis
Hypoxia-inducible factor 1α (HIF1α) is implicated in modulating key enzymes involved in aerobic glycolysis, as well as key processes required for the Warburg effect. HIF1α DNA binding activity in hepatocytes was decreased in Hmgb1−/− mice following DEN treatment (Fig. 5A). Knockdown of HMGB1 or YAP in HCC cells also decreased HIF1α DNA binding activity and luciferase reporter activity with or without hypoxia (Fig. 5B). In contrast, the enforced expression of YAP or HMGB1 restored HIF1α DNA binding activity and luciferase reporter activity in HMGB1-knockdown cells (Fig. 5B). Consistently, hypoxia-induced expression of GLUT1, HK2, ALDOA, and LDHA, as well as carbonic anhydrase 9 (CA9, a well-known HIF1α-targeted gene), were inhibited in HMGB1- or YAP-knockdown cells (Fig. 5C).
Fig. 5. Interplay between HMGB1, YAP, and HIF1α contributes to liver tumorigenesis.
(A) HIF1α DNA binding activity in hepatocytes from Hmgb1−/− and Hmgb1f/f mice after DEN treatment for three and five months (n=3 mice for each time point. *p<0.05 versus Hmgb1f/f group). (B, C) HIF1α activity and glycolysis-associated gene expression in indicated HCC cells with or without hypoxia (1% O2) treatment for 24 hours (n=3, *p<0.05). (D) Immunoprecipitation (IP) analysis of HIF1α-YAP complex in hepatocytes from Hmgb1−/− and Hmgb1f/f mice after DEN treatment for three and five months. (E) IP analysis of HIF1α-YAP complex in Hepa1-6 cells with or without hypoxia (1% O2) treatment for 24 hours. (F) IP analysis of HIF1α-YAP complex in nuclear (“Nuc”) or cytoplasmic (“Cyt”) extractions of hepatocytes from Hmgb1f/f mice after DEN treatment for five months. (G) Percentage of liver tumors in Hif1α−/− and Hif1α+/+ mice after DEN treatment for three to five months (n=10 for each group, *p<0.05). (H–J) Number (H) and size (I) of HCC nodules as well as liver weight (J) in Hif1α−/− and Hif1α+/+ mice at three to five months of age (n=10 for each group, *p<0.05). (K) Serum level of lactate in Hif1α−/− and Hif1α+/+ mice after DEN treatment for three and five months (n=10 mice for each time point. *p<0.05 versus Hif1α+/+ group). (L) Q-PCR analysis of mRNA expressions of indicated genes in livers from Hif1α−/− and Hif1α+/+ mice after DEN treatment for three and five months (n=3 mice for each time point. *p<0.05 versus Hif1α+/+ group). (M, N) Levels of PGC1α mRNA expression and ECAR in indicated Hepa1-6 cells. (O–P) Knockdown of GLUT1 or HK2 inhibited cell proliferation in indicated HCC cells (n=3, *p<0.05).
We next determined whether the HMGB1-YAP pathway regulates HIF1α activity through modulating its expression and degradation. The protein level of HIF1α was not significantly affected by the depletion of HMGB1 or YAP in hepatocytes (Fig. 5D) or Hepa1-6 cells (Fig. 5E). In contrast, the DEN- or hypoxia-induced interaction between YAP and HIF1α was decreased in hepatocytes from Hmgb1−/− mice (Fig. 5D) or HMGB1/YAP-knockdown cells (Fig. 5E). YAP-HIF1α complex formation was observed in the nucleus, but not in the cytosolic extract of hepatocytes from DEN-induced Hmgb1f/f mice (Fig. 5F). These findings therefore indicate that HMGB1-mediated YAP upregulation contributes to HIF1α activation through formation of nuclear YAP-HIF1α complex.
To elucidate the possible role of HIF1α in the regulation of the Warburg effect in liver tumorigenesis, we tested whether HIF1α depletion impairs lactate production and mRNA expression of aerobic glycolysis genes in vivo. Conditional knockout of HIF1α (termed Hif1α−/−) in liver inhibited DEN-induced liver tumor imitation (Fig. 5G). Consistently, the number (Fig. 5H)/size (Fig. 5I) of HCC nodules as well as liver weight (Fig. 5J) were reduced in DEN-induced Hif1α−/− mice, supporting the notion that a hypoxic microenvironment promotes tumor growth. This process was also associated with decreased serum lactate (Fig. 5K), as well as mRNA expression of GLUT1, HK2, ALDOA, and LDHA in livers from DEN-induced Hif1α−/− mice (Fig. 5L). Interestingly, the mRNA expression of HMGB1, YAP, and YAP targeted genes (BIRC5, CCND1, SPP1, and GPC3) also decreased in livers from DEN-induced Hif1α−/− mice (Fig. 5L), suggesting a possible feedback loop between the expression of these genes in metabolic reprogramming.
A recent study showed that HMGB1-mediated peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC1α) expression contributes to hypoxia-mediated mitochondrial biogenesis in HCC cells (23). Knockdown of PGC1α caused mild downregulation of ECAR compared to knockdown of HMGB1 in Hepa1-6 cells (Fig. 5M and 5N). However, forced expression of PGC1α cDNA did not rescue ECAR in HMGB1-knockdown cells (Fig. 5M and 5N). Thus, in the absence of HMGB1, PGC1α was not sufficient to increase glycolysis in HCC cells.
We next knocked down GLUT1 and HK2 by shRNA in Hepa1-6 and HuH7 cells (Fig. 5O). Suppression of GLUT1 and HK2 expression significantly inhibited cell proliferation (Fig. 5P), indicating that aerobic glycolysis contributes to HCC cell growth.
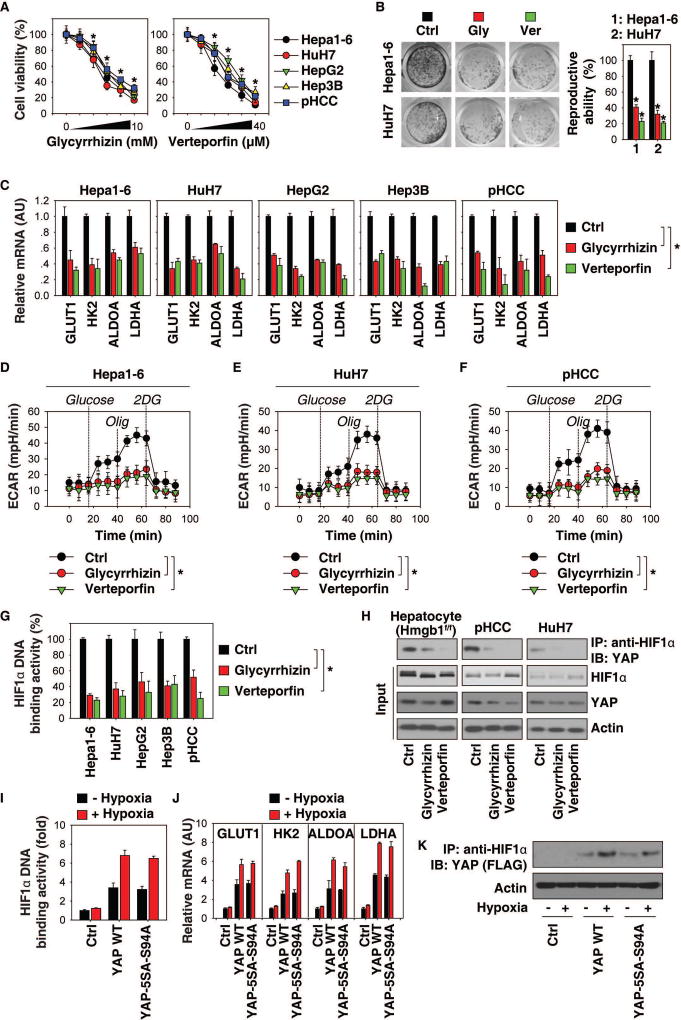
Pharmacological inhibition of the HMGB1-YAP pathway limits liver tumor cell growth in vitro
Glycyrrhizin, a direct HMGB1 inhibitor, has been demonstrated as a promising anti-cancer agent for the treatment of several solid cancers (24). Verteporfin, an FDA-approved drug currently used to treat neovascular macular degeneration, has the ability to block YAP activity to inhibit the growth of breast, pancreas, and colon cancers (25). However, anticancer activity and the mechanism of action of glycyrrhizin and verteporfin in HCC remain obscure.
Thus, we employed four different murine and human HCC cell lines (Hepa1-6, HuH7, HepG2, and Hep3B), as well as primary human HCC (pHCC) cells, to elucidate the tumor suppression mechanisms of glycyrrhizin and verteporfin. First, treatment with glycyrrhizin or verteporfin limited cell proliferation (Fig. 6A) and colony formation (Fig. 6B) in these cells. Second, glycyrrhizin or verteporfin suppressed the mRNA expression levels of glycolysis-associated genes (GLUT1, HK2, ALDOA, and LDHA) (Fig. 6C). Third, ECAR levels were decreased in Hepa1-6 (Fig. 6D), HuH7 (Fig. 6E), and pHCC (Fig. 6F) cells after treatment with glycyrrhizin or verteporfin. Finally, glycyrrhizin or verteporfin remarkably inhibited HIF1α DNA binding activity (Fig. 6G) and HIF1α-YAP complex formation (Fig. 6H). These findings support our hypothesis that the HMGB1-YAP pathway promotes tumor growth in HCC cells through activation of HIF1α-dependent aerobic glycolysis.
Fig. 6. Pharmacological inhibition of the HMGB1-YAP pathway limits tumor cell growth in vitro.
(A) Indicated mouse and human HCC cells were treated with glycyrrhizin (0.625, 1.25, 2.5, 5, 10 mM) or verteporfin (2.5, 5, 10, 20, and 40 µM) for 24 hours. Cell viability was assayed (n=3, *p<0.05 versus control untreated group). (B) Clonogenic cell survival assay determined the reproductive ability of Hepa1-6 and HuH7 cells following treatment with glycyrrhizin (2.5 mM) or verteporfin (10 µM). A representative image is shown in the left panel. Relative reproductive ability is semi-quantified in the right panel (n=3, *p < 0.05 versus control untreated group). (C) Q-PCR analysis of mRNA expressions of indicated genes in HCC cells after glycyrrhizin (2.5 mM) or verteporfin (10 µM) treatment for 24 hours (n=3, *p<0.05). (D–F) ECAR levels in indicated HCC cells (n=3, *p < 0.05). (G) HIF1α DNA binding activity in indicated HCC cells with or without glycyrrhizin (2.5 mM) or verteporfin (10 µM) treatment for 24 hours (n=3, *p<0.05 versus control in the treated group). (H) Immunoprecipitation analysis of HIF1α-YAP complex in hepatocytes from DEN-induced Hmgb1f/f mice or primary human HCC (pHCC) cells or HuH7 cells. (I–K) YAP-knockdown HuH7 cells were transfected with YAP-cDNA or YAP-5SA-S94A mutant for 48 hours, and then treated with hypoxia (1% O2) for 24 hours. HIF1α DNA binding activity (I), gene expression (J), and YAP-HIF1α complex (K) were assayed.
Given that verteporfin can inhibit YAP activity through blocking YAP-TEAD interaction (25), we next determined the effects of TEAD on YAP-HIF1α complex. YAP-5SA-S94A mutant selectively abolishes its ability to bind TEAD, but does not impair its general transcriptional activity (26). Like wild type YAP-cDNA, YAP-5SA-S94A was still able to restore HIF1α DNA binding activity (Fig. 6I), glycolysis-associated gene (GLUT1, HK2, ALDOA, and LDHA) expression (Fig. 6J), and HIF1α-YAP complex formation (Fig. 6K) in YAP-knockdown HuH7 cells with or without hypoxia.
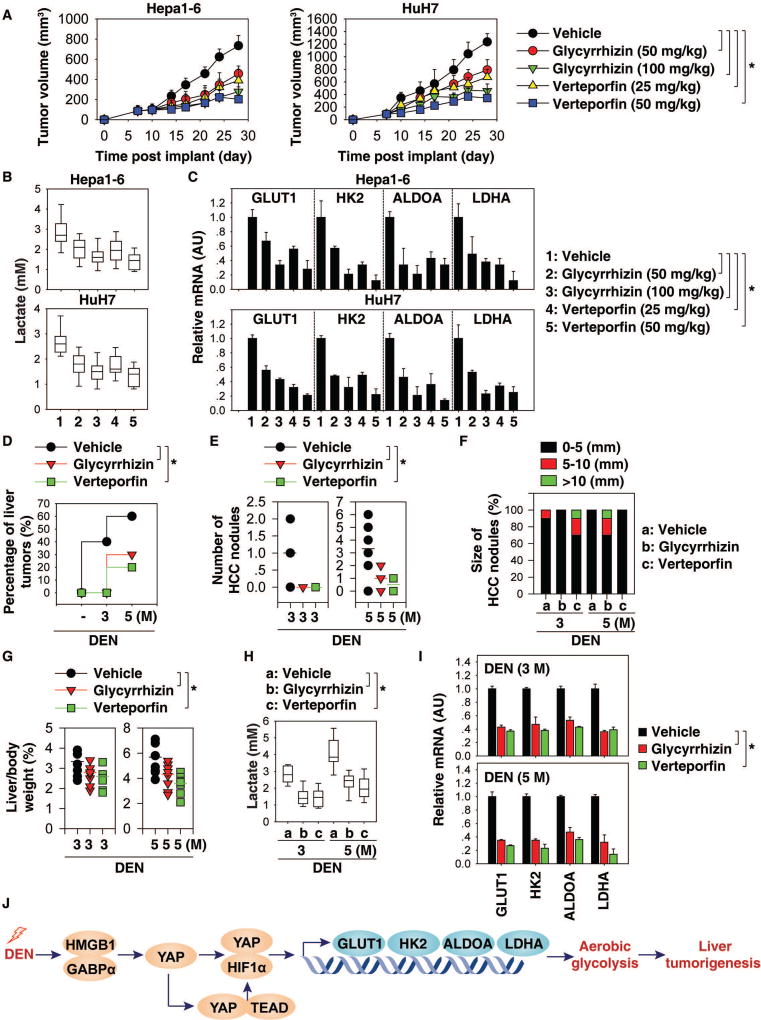
Pharmacological inhibition of the HMGB1-YAP pathway prevents tumor growth in vivo
To evaluate the anticancer activity of glycyrrhizin and verteporfin in vivo, human HuH7 cells or mouse Hepa1-6 cells were implanted into the subcutaneous space of the right flank of immunodeficient nu/nu mice or immunocompetent B6 mice, respectively. Beginning on day seven post-tumor implantation, mice were administered glycyrrhizin (50 and 100 mg/kg, i.p.) or verteporfin (25 and 50 mg/kg, i.p.) for two weeks. Compared to the vehicle control group, administration of glycyrrhizin or verteporfin effectively reduced tumor growth (Fig. 7A) as it decreased serum lactate levels (Fig. 7B) and mRNA expression of GLUT1, HK2, ALDOA, and LDHA (Fig. 7C) in the tumor.
Fig. 7. Pharmacological inhibition of the HMGB1-YAP pathway prevents tumor growth in vivo.
(A) B6 or athymic nude mice were injected subcutaneously with Hepa1-6 cells (2 × 106 cells/mouse) or HuH7 cells (2 × 106 cells/mouse) and treated with glycyrrhizin (50 or 100 mg/kg, i.p., every other day) or verteporfin (25 or 50 mg/kg, i.p., every other day) at day seven for two weeks. Tumor volume was calculated twice weekly. Data is expressed as means ± SD (n=10 mice/group, * p < 0.05). (B) Analysis of serum lactate levels in mice at day 28 after treatment. Data are presented as median value (black line), interquartile range (box), and minimum and maximum of all data (black line) (n=10 mice/group, *p<0.05). (C) Q-PCR analysis of mRNA expressions of indicated genes in livers at day 28 (n=3 mice/group, *p<0.05). (D) Percentage of DEN-induced liver tumors in mice after treatment with glycyrrhizin (100 mg/kg, i.p., twice every week for two months) or verteporfin (50 mg/kg, i.p., twice every week for two months) at three to five months of age (n=10 for each group, *p<0.05). (E–G) Number (E) and size (F) of HCC nodules as well as liver weight (G) in indicated mice (n=10 for each group, *p<0.05). (H) Serum lactate levels in indicated mice (n=10 mice for each time point. *p<0.05). (I) Q-PCR analysis of mRNA expressions of indicated genes in livers from indicated mice (n=3 mice for each time point. *p<0.05). (J) Schematic depicting the role of HMGB1 and YAP in the regulation of aerobic glycolysis during DEN-induced liver tumorigenesis.
We next evaluated the anticancer activity of glycyrrhizin and verteporfin in DEN-induced liver tumorigenesis. Administration of glycyrrhizin (100 mg/kg, i.p.) and verteporfin (50 mg/kg, i.p.) inhibited DEN-induced liver tumor imitation (Fig. 7D). Consistently, the number (Fig. 7E)/size (Fig. 7F) of HCC nodules, as well as liver weight (Fig. 7G), were reduced after glycyrrhizin or verteporfin treatment. This process was also associated with decreased serum lactate (Fig. 7H) as well as mRNA expression of GLUT1, HK2, ALDOA, and LDHA in livers (Fig. 7I). These results further support the notion that pharmacological inhibition of the HMGB1-YAP pathway prevents liver tumor growth through inhibition of aerobic glycolysis.
Discussion
Tumorigenesis has been recognized as not only a process intrinsic to genetic instability, but also a consequence of metabolic reprogramming. Here we identified an HMGB1-dependent regulatory network involving YAP and HIF1α, which controls metabolic reprogramming during liver cancer initiation (Fig. 7J). Targeting either HMGB1 or YAP by glycyrrhizin or verteporfin effectively prevented the proliferation of initiated tumor cells and tumorigenesis, providing a potential strategy for HCC prevention.
While most studies investigating the action of HMGB1 in tumorigenesis have focused on its DNA repair and proinflammatory activity capabilities (27), HMGB1 also plays metabolic roles involved in the regulation of autophagy that may contribute to drug resistance (15). Macrophages can release HMGB1 to trigger an inflammatory response in an aerobic glycolysis-dependent manner (28). In addition, HMGB1 released from cancer cells induces autophagy in the muscle, which mediates the Warburg effect during tumor growth in vitro and in vivo (29). However, extracellular HMGB1 also has the ability to induce cell death through limiting the Warburg effect in clone cancer cells (30). This dual activity of extracellular HMGB1 in the regulation of the Warburg effect may depend on its redox status, cleavage, and receptors.
HMGB1 has also been shown to interact with and modulate the activities of a number of transcription factors (e.g., p53 and estrogen receptor) implicated in tumor development (10). These functions of HMGB1 are mediated by its ability to bind and bend to DNA in a non-sequence-specific manner. We show here that HMGB1 can bind GABPα to activate YAP transcription in hepatocytes. The GABP transcription factor has been linked to the regulation of diverse genes, including YAP, in the Hippo pathway (18). Our findings are consistent with other studies emphasizing the importance of HMGB1 in the regulation of GABP transcription factor activity in cancer cells (31).
In this study, we demonstrated that HMGB1-mediated YAP expression contributes to metabolic reprogramming in liver tumorigenesis. In the initiated liver cancer cells, the expression of YAP is controlled by HMGB1. Disruption of the YAP gene in HCC cells with wild-type HMGB1 recapitulates growth inhibition, deficits in aerobic glycolysis, and lactate production observed with targeted deletion of HMGB1. Forced expression of YAP reverses this phenotype in HMGB1-silencing cells. The Warburg effect mediated by HMGB1 regulation of YAP expression may serve as a metabolic checkpoint, enabling cell proliferation in the hypoxic tumor microenvironment. While increased nucleosome release was observed in HMGB1-deficient pancreas following K-Ras stress (20), circulating nucleosome levels were unchanged in HMGB1-deficient livers following DEN treatment. This may explain the unchanged overall DNA damage and cell death observed in DEN-treated HMGB1-deficient livers compared to HMGB1-wild type livers.
Functionally, our study indicated that the HMGB1-YAP pathway is required for HIF1α transcriptional activity, but not HIF1α expression. We also showed that the formation of YAP-HIF1α complex was not affected by YAP-5SA-S94A mutant. In contrast, verteporfin, a well-known reagent targeting the interface between YAP and TEAD (25), still had the ability to block YAP-HIF1α complex formation. These findings indicate that HMGB1 regulates YAP, which then regulates transcription with TEAD factors, and that all the data on HIF1α are either something happening in parallel or downstream of YAP-TEAD complex. Of note, verteporfin can inhibit YAP function through upregulating 14-3-3σ, which is TEAD-independent (32). The precise role of TEAD in the regulation of HIF1α activity remains to be further explored.
HIF1α is found to be expressed at higher levels in dysplastic nodules and implicated in the progression of hepatocarcinogenesis (33). Consistently, we found that knockout of HIF1α inhibited DEN-induced tumor formation associated with decreased lactate production in mice. The HIF1α-mediated Warburg effect not only supports tumor cell growth, but also limits antitumor T-cell responses (34). The present study suggests a novel link between YAP and HIF1α in liver cancer initiation. YAP binds HIF1α in the nucleus to activate the transcription of glycolysis genes, which in turn mediates the Warburg effect to acquire necessary energy to support proliferation. Our animal study also shows that activation of HIF1α is required for HMGB1 and YAP expression in hepatocytes. Further evidence is needed to determine whether functional crosstalk occurs between both cascades as a metabolism-induced feedback loop in HCC.
Acknowledgments
Financial Support: This work was supported by grants from the US National Institutes of Health (R01GM115366, R01CA160417, R01AT005076, R01GM063075, R01GM44100, and R01CA211070), the Natural Science Foundation of Guangdong Province (2016A030308011), the American Cancer Society (Research Scholar Grant RSG-16-014-01-CDD), the National Natural Science Foundation of China (31671435, 81272253, 81502098, 81772508, and 81400132), and Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2017). This project partly utilized University of Pittsburgh Cancer Institute shared resources supported by award P30CA047904.
We thank Christine Heiner (Department of Surgery, University of Pittsburgh) for her critical reading of this manuscript.
List of Abbreviations
- HCC
hepatocellular carcinoma
- MST1/2
STE20-like protein kinase 1/2
- LATS1/2
large tumor suppressor 1/2
- YAP
yes-associated protein
- TAZ
transcriptional co-activator with PDZ-binding motif
- HMGB1
high mobility group protein B1
- DEN
diethylnitrosamine
- MEFs
mouse embryonic fibroblasts
- PBS
phosphate buffered saline
- SDS
sodium dodecyl sulfate
- ECAR
extracellular acidification rate
- PCNA
proliferating cell nuclear antigen
- BIRC5
baculoviral IAP repeat-containing protein 5
- CCND1
cyclin D1
- MYC
V-Myc avian myelocytomatosis viral oncogene homolog
- SPP1
secreted phosphoprotein 1
- GPC3
glypican 3
- PUMA
p53 up-regulated modulator of apoptosis
- GLUT1
glucose transporter 1
- HK2
hexokinase 2
- ALDOA
fructose-bisphosphate aldolase A
- LDHA
lactate dehydrogenase A
- pHCC
primary human HCC cells
- HIF1α
hypoxia-inducible factor 1α
- PGC1α
peroxisome proliferator-activated receptor gamma coactivator 1 alpha
References
- 1.Yu FX, Zhao B, Guan KL. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perra A, Kowalik MA, Ghiso E, Ledda-Columbano GM, Di Tommaso L, Angioni MM, Raschioni C, et al. YAP activation is an early event and a potential therapeutic target in liver cancer development. J Hepatol. 2014;61:1088–1096. doi: 10.1016/j.jhep.2014.06.033. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Ma L, Weng W, Qiao Y, Zhang Y, He J, Wang H, et al. Mutual interaction between YAP and CREB promotes tumorigenesis in liver cancer. Hepatology. 2013;58:1011–1020. doi: 10.1002/hep.26420. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen HB, Babcock JT, Wells CD, Quilliam LA. LKB1 tumor suppressor regulates AMP kinase/mTOR-independent cell growth and proliferation via the phosphorylation of Yap. Oncogene. 2013;32:4100–4109. doi: 10.1038/onc.2012.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yimlamai D, Fowl BH, Camargo FD. Emerging evidence on the role of the Hippo/YAP pathway in liver physiology and cancer. J Hepatol. 2015;63:1491–1501. doi: 10.1016/j.jhep.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang R, Chen R, Zhang Q, Hou W, Wu S, Cao L, Huang J, et al. HMGB1 in health and disease. Mol Aspects Med. 2014;40:1–116. doi: 10.1016/j.mam.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang R, Zhang Q, Zeh HJ, 3rd, Lotze MT, Tang D. HMGB1 in cancer: good, badm or both? Clin Cancer Res. 2013;19:4046–4057. doi: 10.1158/1078-0432.CCR-13-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Han J, Wu H, Liang X, Zhang J, Li J, Xie L, et al. The association of HMGB1 expression with clinicopathological significance and prognosis in hepatocellular carcinoma: a meta-analysis and literature review. PLoS One. 2014;9:e110626. doi: 10.1371/journal.pone.0110626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan HX, Wu HP, Zhang HL, Ashton C, Tong C, Wu H, Qian QJ, et al. p53 promotes inflammation-associated hepatocarcinogenesis by inducing HMGB1 release. J Hepatol. 2013;59:762–768. doi: 10.1016/j.jhep.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tohme S, Yazdani HO, Liu Y, Loughran P, van der Windt DJ, Huang H, Simmons RL, et al. Hypoxia mediates mitochondrial biogenesis in hepatocellular carcinoma to promote tumor growth via HMGB1 and TLR9 interaction. Hepatology. 2017 doi: 10.1002/hep.29184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berry MN, Friend DS. High-yield preparation of isolated rat liver parenchymal cells: a biochemical and fine structural study. J Cell Biol. 1969;43:506–520. doi: 10.1083/jcb.43.3.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang D, Kang R, Livesey KM, Kroemer G, Billiar TR, Van Houten B, Zeh HJ, 3rd, et al. High-mobility group box 1 is essential for mitochondrial quality control. Cell Metab. 2011;13:701–711. doi: 10.1016/j.cmet.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.TeSlaa T, Teitell MA. Techniques to monitor glycolysis. Methods Enzymol. 2014;542:91–114. doi: 10.1016/B978-0-12-416618-9.00005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H, Xiao Y, Zhang S, Ji S, Wei L, Fan F, Geng J, et al. The Ets transcription factor GABP is a component of the hippo pathway essential for growth and antioxidant defense. Cell Rep. 2013;3:1663–1677. doi: 10.1016/j.celrep.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J, Li J, Salcedo R, Mivechi NF, Trinchieri G, Horuzsko A. The proinflammatory myeloid cell receptor TREM-1 controls Kupffer cell activation and development of hepatocellular carcinoma. Cancer Res. 2012;72:3977–3986. doi: 10.1158/0008-5472.CAN-12-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang R, Xie Y, Zhang Q, Hou W, Jiang Q, Zhu S, Liu J, et al. Intracellular HMGB1 as a Novel Tumor Suppressor of Pancreatic Cancer. Cell Res. 2017 doi: 10.1038/cr.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cottini F, Hideshima T, Xu C, Sattler M, Dori M, Agnelli L, ten Hacken E, et al. Rescue of Hippo coactivator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nat Med. 2014;20:599–606. doi: 10.1038/nm.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hay N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat Rev Cancer. 2016;16:635–649. doi: 10.1038/nrc.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tohme S, Yazdani HO, Liu Y, Loughran P, van der Windt DJ, Huang H, Simmons RL, et al. Hypoxia mediates mitochondrial biogenesis in hepatocellular carcinoma to promote tumor growth through HMGB1 and TLR9 interaction. Hepatology. 2017;66:182–197. doi: 10.1002/hep.29184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hibasami H, Iwase H, Yoshioka K, Takahashi H. Glycyrrhetic acid (a metabolic substance and aglycon of glycyrrhizin) induces apoptosis in human hepatoma, promyelotic leukemia and stomach cancer cells. Int J Mol Med. 2006;17:215–219. [PubMed] [Google Scholar]
- 25.Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bald T, Quast T, Landsberg J, Rogava M, Glodde N, Lopez-Ramos D, Kohlmeyer J, et al. Ultraviolet-radiation-induced inflammation promotes angiotropism and metastasis in melanoma. Nature. 2014;507:109–113. doi: 10.1038/nature13111. [DOI] [PubMed] [Google Scholar]
- 28.Yang L, Xie M, Yang M, Yu Y, Zhu S, Hou W, Kang R, et al. PKM2 regulates the Warburg effect and promotes HMGB1 release in sepsis. Nat Commun. 2014;5:4436. doi: 10.1038/ncomms5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livesey KM, Kang R, Vernon P, Buchser W, Loughran P, Watkins SC, Zhang L, et al. p53/HMGB1 complexes regulate autophagy and apoptosis. Cancer Res. 2012;72:1996–2005. doi: 10.1158/0008-5472.CAN-11-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gdynia G, Sauer SW, Kopitz J, Fuchs D, Duglova K, Ruppert T, Miller M, et al. The HMGB1 protein induces a metabolic type of tumour cell death by blocking aerobic respiration. Nat Commun. 2016;7:10764. doi: 10.1038/ncomms10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiota M, Izumi H, Miyamoto N, Onitsuka T, Kashiwagi E, Kidani A, Hirano G, et al. Ets regulates peroxiredoxin1 and 5 expressions through their interaction with the high-mobility group protein B1. Cancer Sci. 2008;99:1950–1959. doi: 10.1111/j.1349-7006.2008.00912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang C, Zhu X, Feng W, Yu Y, Jeong K, Guo W, Lu Y, et al. Verteporfin inhibits YAP function through up-regulating 14-3-3sigma sequestering YAP in the cytoplasm. Am J Cancer Res. 2016;6:27–37. [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka H, Yamamoto M, Hashimoto N, Miyakoshi M, Tamakawa S, Yoshie M, Tokusashi Y, et al. Hypoxia-independent overexpression of hypoxia-inducible factor 1alpha as an early change in mouse hepatocarcinogenesis. Cancer Res. 2006;66:11263–11270. doi: 10.1158/0008-5472.CAN-06-1699. [DOI] [PubMed] [Google Scholar]
- 34.Siska PJ, Rathmell JC. T cell metabolic fitness in antitumor immunity. Trends Immunol. 2015;36:257–264. doi: 10.1016/j.it.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]