Abstract
RNA regulation and maintenance are critical for proper cell function. Small molecules that specifically alter RNA sequence would be exceptionally useful as probes of RNA structure and function or as potential therapeutics. Here, we demonstrate a photochemical approach for altering the trinucleotide expanded repeat causative of myotonic muscular dystrophy type 1 (DM1), r(CUG)exp. The small molecule, 2H-4-Ru, binds to r(CUG)exp and converts guanosine residues to 8-oxo-7,8-dihydroguanosine upon photochemical irradiation. We demonstrate targeted modification upon irradiation in cell culture and in Drosophila larvae provided a diet containing 2H-4-Ru. Our results highlight a general chemical biology approach for altering RNA sequence in vivo by using small molecules and photochemistry. Furthermore, these studies show that addition of 8-oxo-G lesions into RNA 3′ untranslated regions does not affect its steady state levels.
Keywords: RNA, small molecules, chemical probes, photochemistry, myotonic dystrophy
Graphical abstract
RNA plays important roles in cellular biology that are dictated by sequence and structure.1 One way to interrogate RNA function is by using chemical probes.2, 3 Although antibacterials have been useful for probing the structure and function of the ribosome,4 routine identification of ligands for diverse RNA targets has proven difficult. We previously developed a lead identification strategy in which the secondary structure of a disease-causing RNA is queried against an RNA motif-small molecule database.2,3, 5 We used this approach to identify a dimeric ligand (2H-4) that binds repeating secondary structural elements in r(CUG)exp—an expanded (“exp”) trinucleotide repeat implicated in myotonic muscular dystrophy type 1 (DM1).6-8 2H-4 targets r(CUG)exp in cells and improves various aspects of DM1-associated defects.9, 10
In subsequent work, we conjugated hydroxythiopyridine (HPT) to 2H-4 (2H-4-HPT) to deliver hydroxyl radicals to r(CUG)exp and cleave it upon irradiation.11 Although this approach was promising, only a single reactive hydroxyl radical is produced per HPT moiety. A potentially superior approach for photochemical control of RNA function would be to use Tris(bipyridine)ruthenium(II), which enables the photosensitized and stimulated production of singlet oxygen upon irradiation (Figure 1A).12 In a cell-free environment, singlet oxygen converts RNA guanosine (G) bases to 8-oxo-7,8-dihydroguanosine (8- oxoG) (Figure 1B).13, 14 This approach would facilitate studies on the fate of oxidized RNA transcripts, which could include targeted degradation, as well as provide additional means to affect RNA function in vivo.
Figure 1.
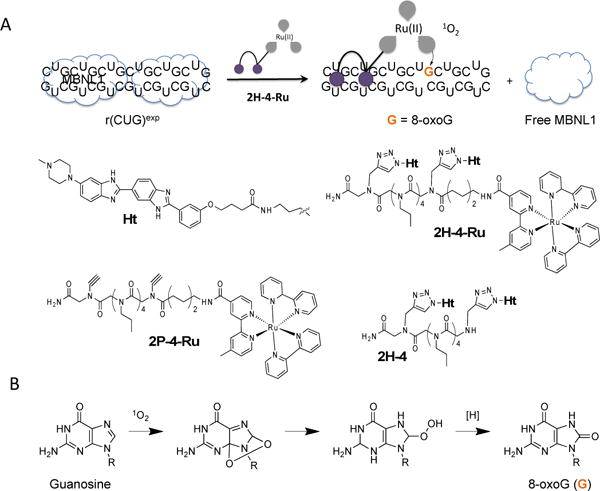
A designer small molecule that targets r(CUG)exp, the causative agent of DM1, was appended with Tris(bipyridine)ruthenium(II) [Ru(bipy)3] to generate reactive oxygen species that can oxidize G residues to 8-oxoG. A, scheme showing binding and oxidation of RNA by 2H-4-Ru and subsequent release of MBNL1. The Ru(bipy)3 warhead produces singlet oxygen upon photoactivation. Other controls tested in this study include 2H-4, the binding module alone, and 2P-4-Ru, which lacks the RNA-binding component (Ht). The RNA binding modules are defined by the purple circles, and the linkers that span them and Ru(bipy)3 indicated with gray lines. B, singlet oxygen oxidizes guanine and produces 8-oxoG lesions.15 The complete synthesis of these compounds is described in Scheme S-1 and S-2 (Supporting Information).
The trinucleotide repeat disorder DM1 is an incurable neuromuscular disease that is caused by an RNA gain-of-function mechanism in which non-coding r(CUG)exp binds and sequesters proteins such as muscleblind-like 1 protein (MBNL1), knocking out its function and triggering disease.6, 8, 16 To determine if we could alter the sequence of r(CUG)exp by using a small molecule, we appended 2H-4 with Tris(bipyridine)ruthenium(II) [Ru(bipy)3] to generate 2H-4-Ru (Figures 1A and S1). Briefly, an aminohexanoate linker was added to 2H-4 to provide a reactive amine handle onto which an activated ester of Ru(bipy)3 could be attached. Exclusion of the RNA-binding module, Ht, produced the control compound, 2P-4-Ru (Figures 1A and S2), which does not bind to r(CUG)exp. The 2H-4 compound has been shown to avidly recognize RNAs with two consecutive copies of 5′CUG/3′GUC 1×1 motifs that is present in r(CUG)exp and shortened models thereof such as r(CUG)10 as previoiusly described.9
To study the ability of 2H-4-Ru to photochemically alter r(CUG)exp sequences in vitro, the compound was incubated with 5′-end–labeled r(CUG)10, irradiated, and treated with aniline, which cleaves RNA at positions containing 8-oxoG.14 Polyacrylamide gel (PAGE) analysis showed aniline-mediated cleavage occurs at Gs when r(CUG)10 was treated with 2H-4-Ru and irradiated, but not with the control (2P-4-Ru) in the presence or absence of light (Figure 2A), suggesting that Gs were specifically oxidized to 8-oxoG by 2H-4-Ru upon photoactivation.14 To further confirm that 8-oxoG was installed into r(CUG)10, an ELISA-based approach with an anti-8-oxoG antibody was used. Biotinylated r(CUG)10 was bound to a streptavidin-coated microtiter plate and treated with compound in the presence or absence of light. Upon probing with anti-8-oxoG antibody conjugated with horseradish peroxidase (HRP), 8-oxoG bases were only present in samples that were incubated with 2H-4-Ru and irradiated (Figure 2B). We also completed ESI-MS-MS analysis of r(CUG)10 after treatment with 2H-4-Ru or 2P-4-Ru and nuclease digestion, using a previously studied RNA as a standard.17 Indeed, G was only oxidized to 8-oxoG with 2H-4-Ru (or riboflavin as positive control18) upon photoactivation (Figure S-3 to S-6). Thus, all three independent studies show that small molecules can be used in vitro to alter the base composition of RNA transcripts.
Figure 2.
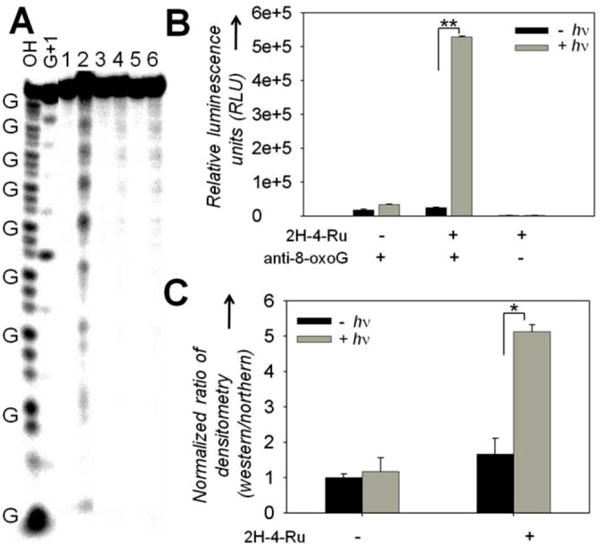
In vitro studies of 8-oxoG formation in r(CUG)exp by 2H-4-Ru upon photoactivation. A, gel electrophoresis of r(CUG)10 shows 8-oxoG formation after incubation with 2H-4-Ru and photoactivation in the presence of 50-fold excess total tRNA. Lanes: OH, alkaline hydrolysis; G+1, RNase T1 digest; 1, 2H-4-Ru and light; 2, 2H-4-Ru and light followed by aniline treatment; 3, 2P-4-Ru and light; 4, 2P-4-Ru and light followed by aniline treatment; 5, RNA only and light; 6, RNA only and light followed by aniline treatment. B, ELISA using an anti-8-oxoG antibody confirms the formation of 8-oxoG in r(CUG)10 by 2H-4-Ru upon photoactivation. C, Northwestern blot analysis shows an increase in the amount of 8-oxoG bases in r(CUG)exp when cells were treated with 2H-4-Ru and light (see also Figure S3). *, p < 0.05; **, p < 0.01.
After confirming that 2P-4-Ru and 2H-4-Ru are non-toxic and cell permeable (Figure S-7), we tested the ability of 2H-4-Ru to install 8-oxoG into r(CUG)exp using a cellular model of DM1 (HeLa cells that express r(CUG)960 repeats). After treatment with compound and irradiation, total RNA was isolated and analyzed via Northwestern blot. That is, a Northern blot was used to detect r(CUG)exp and was then also probed with an 8-oxo-G antibody (Western blot) to detect 8-oxoG modifications in r(CUG)960. Indeed, 8-oxo-G lesions were found in r(CUG)exp after 2H-4-Ru treatment and irradiation (Figures 2C and S-8). To further verify that r(CUG)exp was selectively oxidized, immunoprecipitation (IP) experiments were carried out using anti-8-oxoG antibody-coated magnetic beads, which capture 8- oxoG-containing transcripts harvested from cells (Figure S9). Quantification of captured RNA by RT-qPCR showed enrichment of r(CUG)960 only after drug and light treatment (Figure S6). Apparently, photochemistry can be used to alter RNA sequence in a cellular system. RT-qPCR of the r(CUG)960- containing mRNA shows that its steady state levels were stable in cells (Figure S10). Interestingly, a previous study showed that 8- oxo-G lesions induce No-Go decay of the corresponding transcript.17 In those studies, the lesions were present in a coding region. It is possible that levels of the r(CUG)960-containing mRNA were unaffected because it is present in a UTR. Collectively, RNA repeat expansions in coding regions, such as the expanded r(CAG) repeats that cause Huntington’s disease, could be targeted and destroyed by introduction of 8-oxo-G lesion and subsequent induction of No-Go decay.
Invertebrates such as Drosophila melanogaster, the fruit fly, have emerged as powerful models for studying the genetic mechanisms and pathophysiology of neurodegenerative diseases.19 Their small body size, low cost of maintenance, and short lifespan also make them attractive platforms for in vivo testing of potential therapeutics. Previously, a Drosophila model of DM1 was developed by expression of r(CUG)250 in the 3′ untranslated region (UTR) of a control gene, DsRed.20 We took advantage of this model to test whether 2H-4-Ru would show efficacy in vivo. Drosophila 1st instar larvae developed on food dosed once with 2H-4-Ru. After several days, 3rd instar live larvae were irradiated once and total RNA was harvested. Immunoprecipitation of 8-oxoG-containing transcripts showed that DsRed is enriched upon 2H-4-Ru treatment and irradiation, suggesting that r(CUG)250 was selectively modified (Figure 3). Again, the r(CUG)250 transcript upon photoactivation is stable without significant alteration in RNA levels (Figure S11).
Figure 3.
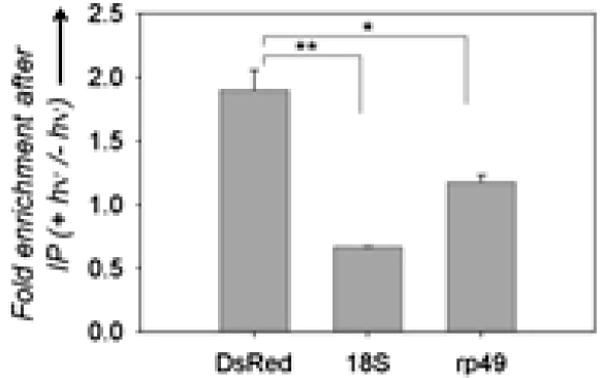
2H-4-Ru modified the sequence of r(CUG)250 in a Drosophila model of DM1. RT-qPCR analysis of DsRed-r(CUG)250 and the control genes, 18S and rp49, after anti–8-oxoG immunoprecipitation of RNA from fly larvae treated with 2H-4-Ru. Greater enrichment of DsRed with irradiation suggests that DsRed-r(CUG)250 is selectively modified. Pull-down of 18S and rp49 may suggest a lack of exquisite selectivity of the antibody.
Importantly, these approaches also further demonstrate another approach to validate the cellular and animal targets of small molecules directed at RNA. For example, Chemical cross linking and isolation by pull down (Chem-CLIP) and small–molecule nucleic acid profiling by cleavage applied to RNA (Ribo-SNAP) have provided cross-linking and cleavage approaches to read out the RNA targets of small molecules.11, 21-23 The ability to iummunopreciptate RNAs with 8-oxo-G lesions as introduced via small molecules offers a further approach to target validation and also validates r(CUG)exp as a target for small molecules in animals for the first time.
In conclusion, we have demonstrated that small molecules that bind to RNA targets can alter RNA sequence when a photosensitizer is added to the compound. In addition, these studies have facilitated an understanding of the fate of oxidized RNAs in cells and animals, showing that abundance of modified RNAs do not significantly change, at least with the overexpressed targets used in these cases. This approach may have application to other targets for studying and manipulating RNA function both in vitro and in vivo. Since many neurological disorders produce reactive oxygen species that modify RNA bases, our approach may inform future studies to determine the specific effect of these modifications on disease-associated RNAs and to establish whether they are associative or causative of disease.
Supplementary Material
Acknowledgments
This work was funded by the US National Institutes of Health (grants DP1NS096898, MDD, and R01AG045036, WWJ) and the Muscular Dystrophy Association (grant 380467).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yeung ML, Bennasser Y, Jeang KT. Curr Med Chem. 2007;14:191. doi: 10.2174/092986707779313417. [DOI] [PubMed] [Google Scholar]
- 2.Guan L, Disney MD. ACS Chem Biol. 2012;7:73. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- 3.Thomas JR, Hergenrother PJ. Chem Rev. 2008;108:1171. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 4.Poehlsgaard J, Douthwaite S. Nat Rev Microbiol. 2005;3:870. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- 5.Parkesh R, Childs-Disney JL, Nakamori M, Kumar A, Wang E, Wang T, Hoskins J, Tran T, Housman DE, Thornton CA, Disney MD. J Am Chem Soc. 2012;134:4731. doi: 10.1021/ja210088v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Science. 2000;289:1769. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 7.Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF, Thornton CA. Nature. 2012;488:111. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Science. 2009;325:336. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, Disney MD. J Am Chem Soc. 2009;131:9767. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Childs-Disney JL, Hoskins J, Rzuczek SG, Thornton CA, Disney MD. ACS Chem Biol. 2012;7:856. doi: 10.1021/cb200408a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan L, Disney MD. Angew Chem Int Ed Engl. 2013;52:1462. doi: 10.1002/anie.201206888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winterle JS, Kliger DS, Hammond GS. J Am Chem Soc. 1976;98:3719. [Google Scholar]
- 13.Tibodeau JD, Fox PM, Ropp PA, Theil EC, Thorp HH. Proc Natl Acad Sci U S A. 2006;103:253. doi: 10.1073/pnas.0509744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Odom DT, Barton JK. Biochemistry. 2001;40:8727. doi: 10.1021/bi0102961. [DOI] [PubMed] [Google Scholar]
- 15.Cadet J, Douki T, Gasparutto D, Ravanat JL. Mutat Res. 2003;531:5. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Philips AV, Timchenko LT, Cooper TA. Science. 1998;280:737. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 17.Simms CL, Hudson BH, Mosior JW, Rangwala AS, Zaher HS. Cell Rep. 2014;9:1256. doi: 10.1016/j.celrep.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto F, Nishimura S, Kasai H. Biochem Biophys Res Commun. 1992;187:809. doi: 10.1016/0006-291x(92)91268-u. [DOI] [PubMed] [Google Scholar]
- 19.Lu B, Vogel H. Ann Rev Pathol. 2009;4:315. doi: 10.1146/annurev.pathol.3.121806.151529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Z, Teng X, Bonini NM. PLoS Genet. 2011;7:e1001340. doi: 10.1371/journal.pgen.1001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rzuczek SG, Colgan LA, Nakai Y, Cameron MD, Furling D, Yasuda R, Disney MD. Nat Chem Biol. 2017;13:188. doi: 10.1038/nchembio.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang WY, Wilson HD, Velagapudi S, Disney MD. J Am Chem Soc. 2015 doi: 10.1021/ja507448y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Childs-Disney JL, Disney MD. Ann Rev Pharmacol Toxicol. 2016;56:123. doi: 10.1146/annurev-pharmtox-010715-103910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


