Abstract
Background
The epidermal growth factor receptor (EGFR) T790M mutation is associated with resistance to EGFR tyrosine kinase inhibitors (EGFR-TKIs) in non-small cell lung cancer (NSCLC). However, tissues for the genotyping of the EGFR T790M mutation can be difficult to obtain in a clinical setting. The aims of this study were to evaluate a blood-based, non-invasive approach to detecting the EGFR T790M mutation in advanced NSCLC patients using the PointMan™ EGFR DNA enrichment kit, which is a novel method for the selective amplification of specific genotype sequences.
Methods
Blood samples were collected from NSCLC patients who had activating EGFR mutations and who were resistant to EGFR-TKI treatment. Using cell-free DNA (cfDNA) from plasma, EGFR T790M mutations were amplified using the PointMan™ enrichment kit, and all the reaction products were confirmed using direct sequencing. The concentrations of plasma DNA were then determined using quantitative real-time PCR.
Results
Nineteen patients were enrolled, and 12 patients (63.2%) were found to contain EGFR T790M mutations in their cfDNA, as detected by the kit. T790M mutations were detected in tumor tissues in 12 cases, and 11 of these cases (91.7%) also exhibited the T790M mutation in cfDNA samples. The concentrations of cfDNA were similar between patients with the T790M mutation and those without the mutation.
Conclusions
The PointMan™ kit provides a useful method for determining the EGFR T790M mutation status in cfDNA.
Keywords: Cell-free DNA (cfDNA), epidermal growth factor receptor (EGFR), T790M, lung cancer
Introduction
The high mortality rate of patients with lung cancer remains a global problem that requires innovative approaches. A considerable number of non-small cell lung cancer (NSCLC) patients have benefited from the development of molecularly targeted therapy. The epidermal growth factor receptor (EGFR) mutation is a strong oncogene in NSCLC, and EGFR-targeted therapy using EGFR tyrosine kinase inhibitors (TKIs), such as gefitinib, erlotinib and afatinib, has enabled a dramatic improvement in the prognosis of patients with EGFR mutations according to randomized, large-scale trials (1-6). The aforementioned EGFR-TKIs are now used as a standard therapy in patients with EGFR mutations. Unfortunately, almost all patients ultimately develop a recurrence of disease either because of the progression of the primary lesion or distant metastasis. The mechanisms of EGFR-TKI resistance have been investigated, and several reports have shown that the T790M mutation in the EGFR gene was present in approximately one-half of the patients who developed resistance to EGFR-TKI treatment (7-9). The T790M mutation causes a structural change in the ATP binding pocket of the EGFR protein. This alteration inhibits EGFR-TKI molecules from passing through the gate to the ATP binding pocket (7,10,11). Recently, large scale, randomized studies compared third generation EGFR-TKI agents, osimertinib and rociletinib, with platinum-based chemotherapy in patients with the T790M mutation. The data showed that the new drugs overcame T790M resistance, shrinking tumors and resulting in good clinical outcomes (12,13). In Japan, osimertinib monotherapy is a standard therapy for patients with the T790M mutation appearing after resistance to EGFR-TKI. Therefore, confirmation of the T790M mutation is required for treatment selection; mutation analyses are generally performed using re-biopsy specimens. In clinical practice, however, we have found that we cannot obtain tumor samples repeatedly in patients with resistance to EGFR-TKIs because of difficulty obtaining tumor samples, patients’ status and rejection of the patients for the invasive testing. A retrospective study analyzed patients eligible for third generation EGFR-TKIs treatment after resistance to EGFR-TKIs. Only 63% of the enrolled patients were able to undergo rebiopsies and testing for the T790M mutation analysis using tissue or cytology samples (14).
Cell-free DNA (cfDNA) extracted from plasma contains tumor-derived DNA. The concentration of cfDNA is reportedly higher in patients with malignant tumors than in healthy volunteers (15,16). Additionally, tumor-specific mutations, such as those in EGFR and KRAS, can be detected in cfDNA samples (17,18). Utilizing cfDNA is appealing because it can be collected easily, non-invasively and repeatedly. Tumor-derived mutations are not easily detected with cfDNA because of DNA fragmentation and the small quantities available. Several reports have demonstrated the detection of EGFR mutations in cfDNA using highly sensitive detection assays, such as scorpion-ARMS, BEAMS, and droplet digital PCR (ddPCR). These high-sensitivity assays have increased the detection of EGFR mutations, with reported detection rates ranging between 65% and 81% (17,19-24).
The PointMan™ assay is a highly sensitive method for amplifying a gene with specific mutations while inhibiting amplification of wild-type DNA. Two sets of primer pairs are used in this assay: an enriching primer pair specific for the target mutation site of wild-type DNA, and an amplifying primer pair identical to the primer used in conventional PCR reactions. The amplifying primer amplifies PCR products containing a targeted mutation site, whereas the enriching primer binds to the targeted mutation site in the wild-type gene and blocks its amplification. The enriching primer has a higher avidity for the wild-type sequence than for the mutant sequence at the targeted mutation site. Thus, only the mutant gene is amplified exponentially by the extension reaction of the amplification primers, since it is not inhibited by the enriching primers, whereas the extension reaction for the wild-type gene is inhibited by the enriching primers that have annealed to the targeted mutation site in the wild-type gene.
The aim of the present study was to evaluate the PointMan™ assay and its detection of T790M in cfDNA from patients who had developed resistance to EGFR-TKIs and to compare the mutation status with that of resistant tumor tissues obtained by re-biopsy. Additionally, we investigated the relationships between the T790M mutation status in cfDNA and clinical characteristics.
Methods
Patient selection and sample collection
Nineteen NSCLC patients harboring an EGFR mutation were enrolled. We had samples of tumor tissues from re-biopsy as well as plasma at the time of resistance to EGFR-TKIs. The samples were collected at Kanazawa University Hospital between January 2006 and June 2015 with full consent and ethical board approval from Kanazawa University (No. 344-1). We collected data from clinical records as follows: age at diagnosis, sex, smoking status, histology, disease stage, EGFR mutation status at initial diagnosis, agents used as the first EGFR-TKI therapy, overall response to the first EGFR-TKI therapy according to the Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST) and treatment regimen after re-biopsy.
EGFR mutation assay of tissue samples
All the patients had pathologically proven NSCLC. The EGFR mutation status of a tissue sample was tested at the time of the initial diagnosis and after disease progression following EGFR-TKI therapy as part of routine practice using commercial methods.
Plasma collection
Patients’ blood samples were collected in test tubes containing EDTA at the time of disease progression after EGFR-TKI therapy. The plasma samples were isolated by centrifugation at 700 ×g for 10 min and were then either used immediately or stored at −80 °C until use.
DNA was extracted from 1 mL of plasma using the QIAamp Circulating Nucleic Acid Kit (QIAGEN, Hilden, Germany) and stored at −20 °C until use. The DNA concentrations were calculated using a previously reported method (15).
Detection of T790M mutation of EGFR in plasma DNA
We used the PointMan™ EGFR DNA enrichment kit (provided by EKF Molecular Diagnostics Ltd, Cardiff, UK) for the enrichment of EGFR mutations in a 5-µL DNA sample. Two µL of PointMan primer mix or control primer mix, 10 µL of master mix, 3 µL of RNAse/DNase-free water and 5 µL of template were thoroughly mixed and real-time PCR was performed using AB StepOnePlus™ (Applied Biosystems, Foster City, CA, USA) under the following conditions: enzyme activation at 95 °C for 2 min, followed by 50 cycles of 95 °C for 10 s, 50 °C for 20 s, 70 °C for 1 s, and 60 °C for 30 s, followed by heating from 60 °C to 95 °C to analyze the melt curve. The PCR reaction solutions were visualized using agarose gel electrophoresis to confirm that the target genes had been amplified. After purification of the PCR products, direct sequencing was performed using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and 4 µL of BigDye terminator, v3.1. A sequence of T790M alteration was visually confirmed by the wave form (Figure 1). If the amount of the amplified product was too small to determine the mutation status, gene amplification-nested PCR using the PointMan™ products was performed using KOD -Plus- (TOYOBO Life Science, Tokyo, Japan). The method was as follows: 0.5 µL of KOD -Plus-, 2.5 µL of 10× PCR buffer, 2.5 µL of 2 mM dNTPs, 1 µL of 25 mM MgSO4, 0.75 µL of each primer, 1.5 µL of the template, and 16.7 µL of pure water were mixed and amplification was performed with pre-denaturing at 94 °C for 2 min, followed by 30 cycles of 94 °C for 15 s, 55 °C for 30 s, and 68 °C for 20 s. The primer sequences were as follows, forward: 5'-TCCAGGAAGCCTACGTGATG-3' and reverse: 5'-CCCTGATTACCTTTGCGATCTG-3' (201 base pairs) for the T790M mutation. The PCR products were purified once again, and direct sequencing was conducted using the method described above. Additionally, we performed analysis for the presence of the T790M mutation using the cobas® EGFR Mutation Test v2 (Roche Molecular Systems, Inc.).
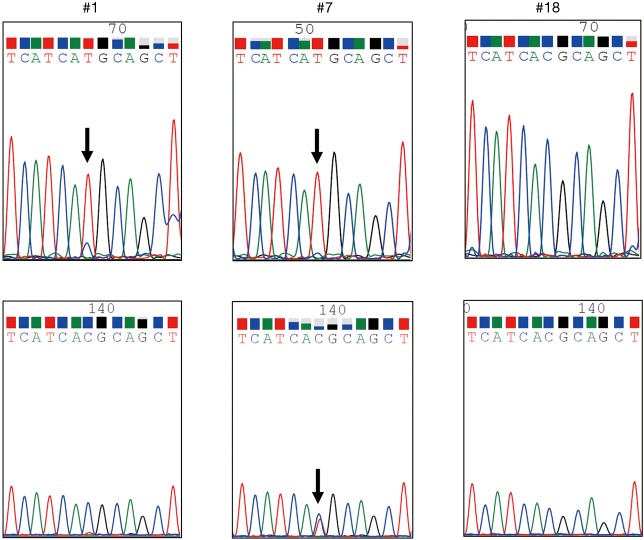
Figure 1.
A wave form illustration of the T790M mutation. Direct sequencing was performed using a PCR product amplified by the PointMan® assay (upper column). The arrow indicates a substitution mutation (2369C>T) of T790M. T790M was detected in two samples (#1, #7) by the PointMan® assay. In #1, T790M was not detected by the usual method, not using the PointMan® assay (lower column). In #7, a waveform indicating T790M was emphasized in the PointMan® assay, although double waveforms indicating both wild-type and T790M were overlapping in the usual method.
Survival analysis
Progression-free survival (PFS) was defined as the interval between the initiation of the EGFR-TKI therapy and the first manifestation of progressive disease (PD) or death from any cause. The post-progression survival (PPS) was defined as the interval between the first EGFR-TKI treatment failure and death from any cause; patients who were still alive at the time of the analysis were censored at the time of their last follow-up examination.
Statistical analysis
The present study was a retrospective observational study. The χ2 test was used to analyze the relationships between the T790M mutation status in plasma extracted after disease progression following EGFR-TKI therapy and several patient characteristics. The age at diagnosis was compared using the Student’s t-test. Kaplan-Meier curves were used to analyze PFS and PPS, and the log-rank test was used for comparisons. The statistical analyses were performed using JMP software, version 9. All the statistical tests were two-sided, and a P value <0.05 was considered significant.
Results
Patient characteristics
The characteristics of all the patients and the T790M status of the post-treatment cfDNA samples are shown in Table 1. The median age was 62 years, 12 patents (63.2%) were female, 9 patients (47.4%) had never smoked and all the patients had adenocarcinoma. Sixteen patients (84.2%) had stage IV disease. Regarding the EGFR mutation status, 14 patients (73.7%) had an exon 19 deletion, 4 patients (21.1%) had an L858R mutation, and one patient had both an L858R and an A859S mutation. None of the patients harbored a T790M mutation at the time of the initial diagnosis when examined with a commercial method, such as the cobas®, PCR clamp method and Scorpion ARMS method. Nine patients (47.4%) were treated with gefitinib, 9 patients (47.4%) were treated with erlotinib, and 1 patient (5.3%) was treated with afatinib. Fourteen patients (73.7%) obtained a partial response to the first EGFR-TKI therapy.
Table 1. Patient characteristics.
| Characteristics | All patients (n=19) |
|---|---|
| Age, median [range] (years) | 62 [48–78] |
| Sex, n (%) | |
| Men | 7 (36.8) |
| Women | 12 (63.2) |
| Smoking history, n (%) | |
| Never | 9 (47.4) |
| ES/CS | 10 (52.6) |
| Histology—adenocarcinoma, n (%) | 19 (100.0) |
| Stage, n (%) | |
| III | 3 (15.8) |
| IV | 16 (84.2) |
| EGFR mutation status, n (%) | |
| Ex19 deletion | 14 (73.7) |
| L858R | 4 (21.1) |
| L858R, A859S | 1 (5.3) |
| Prior EGFR-TKIs, n (%) | |
| Gefitinib | 9 (47.4) |
| Erlotinib | 9 (47.4) |
| Afatinib | 1 (5.3) |
EGFR-TKI, epidermal growth factor receptor tyrosine kinase inhibitor; ES, ex-smoker; CS, current smoker.
Mutation status in cfDNA
T790M mutations in cfDNA were detected in 12 patients (63.2%) after progression following the first EGFR-TKI therapy (Table 2). No statistically significant differences in the characteristics were observed between patients with the T790M mutation and those without the mutation. The incidence of T790M mutations in the cfDNA tended to be higher in women and in patients with an exon 19 deletion although not significantly. There was no significant difference in the response rate to prior EGFR-TKIs between patients with the T790M mutation (9/12, 75.0%) and those without the mutation (5/7, 71.4%).
Table 2. Patients’ list related to T790M mutate status.
| Patient # | Sex | EGFR status at diagnosis | Prior EGFR-TKIs | Response to prior EGFR-TKIs | T790M status | Concentrations in cfDNA (ng/L) | ||
|---|---|---|---|---|---|---|---|---|
| Re-biopsy tissue (cobas®) | cfDNA (PointMan) | cfDNA (cobas®) | ||||||
| 1 | F | Exon19 deletion | Gefitinib | PR | T790M | T790M | Undecidable | 0.407 |
| 2 | M | Exon19 deletion | Erlotinib | SD | T790M | T790M | Undecidable | 0.391 |
| 3 | M | Exon19 deletion | Erlotinib | PR | T790M | T790M | T790M | 1.316 |
| 4 | F | Exon19 deletion | Erlotinib | PR | T790M | T790M | Undecidable | 1.781 |
| 5 | F | L858R | Gefitinib | PR | T790M | T790M | T790M | 1.083 |
| 6 | F | Exon19 deletion | Erlotinib | PR | T790M | T790M | T790M | 0.396 |
| 7 | F | Exon19 deletion | Erlotinib | SD | T790M | T790M | T790M | 2.623 |
| 8 | M | L858R | Gefitinib | PR | T790M | T790M | T790M | 1.150 |
| 9 | F | Exon19 deletion | Erlotinib | PR | T790M | T790M | Wild | 1.162 |
| 10 | F | Exon19 deletion | Gefitinib | PR | T790M | T790M | T790M | 0.840 |
| 11 | F | Exon19 deletion | Gefitinib | PR | T790M | T790M | Wild | 0.156 |
| 12 | F | Exon19 deletion | Gefitinib | SD | T790M | Wild | Wild | 0.068 |
| 13 | F | Exon19 deletion | Erlotinib | PD | Wild | T790M | Wild | 0.351 |
| 14 | F | Exon19 deletion | Afatinib | PR | Wild | Wild | Wild | 0.273 |
| 15 | M | Exon19 deletion | Erlotinib | SD | Wild | Wild | Wild | 0.416 |
| 16 | M | L858R, A859S | Gefitinib | PR | Wild | Wild | Wild | 0.690 |
| 17 | F | L858R | Gefitinib | PR | Wild | Wild | Wild | 0.391 |
| 18 | M | Exon19 deletion | Erlotinib | PR | Wild | Wild | Wild | 7.520 |
| 19 | M | L858R | Gefitinib | PR | Wild | Wild | Wild | 6.688 |
EGFR-TKI, epidermal growth factor receptor tyrosine kinase inhibitor; cfDNA, cell-free DNA; PR, partial response; PD, progressive disease; SD, stable disease.
T790M mutations in tumor DNA from re-biopsy specimens were detected in 12 (63.2%) of the 19 patients (Table 3). The relationships between the T790M mutation status in the tumor DNA and that in the cfDNA are shown in Table 3. T790M mutations in cfDNA were detected in 11 of the 12 patients with the mutation in tumor DNA and in 1 of the 7 patients without the mutation in tumor DNA. The sensitivity of T790M detection from cfDNA was 91.7% (11/12) and the overall concordance rate was 89.5% (17/19).
Table 3. Relationships of T790M mutation between re-biopsy tissue and plasma sample using PointMan kit.
| Mutation status | cfDNA | Total | |
|---|---|---|---|
| Positive | Negative | ||
| Tissue | |||
| Positive | 11 | 1 | 12 |
| Negative | 1 | 6 | 7 |
| Total | 12 | 7 | 19 |
χ2 test, P<0.001; sensitivity, 91.7%; specificity, 91.7%; concordance rate, 89.5%. cfDNA, cell-free DNA.
Using a cobas® analysis of cfDNA, the T790M mutation status was confirmed in 16 of the 19 samples (84.2%). The other 3 samples were undecidable. The analyses of the other three samples were indeterminate. In 6 of the 12 patients (50.0%) with the T790M mutation in tumor tissues, T790M mutations were detected in their cfDNA using the cobas® analysis. These results indicate that the detectability using cobas® was lower than that using the PointMan™ method.
Survival analysis
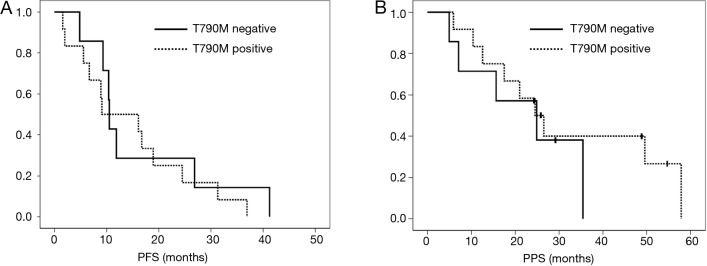
In the survival analyses, the median PFS of patients with the T790M mutation in their cfDNA was 9.1 months, while that of patients without the T790M mutation was 10.5 months. No significant difference was observed between the two groups (P=0.58) (Figure 2A). The median PPS were 24.5 months for the patients with the T790M mutation and 24.9 months for the patients without the T790M mutation. No significant difference was observed between the two groups (P=0.46) (Figure 2B). Nine of the 12 patients with T790M mutation in tumor tissues received osimertinib treatment. Six of the 9 patients achieved PR with osimertinib and the response rate to osimertinib was 66.7%. None of the patients with T790M in cfDNA alone received osimertinib treatment.
Figure 2.
Survival analysis using Kaplan-Meier curves. (A) The progression-free survival after first EGFR-TKI therapy in patients with T790M (solid line) and those without T790M (dashed line) in plasma samples; (B) the post-progression survival of patients with T790M (solid line) and those without T790M (dashed line) in plasma samples. No statistically significant difference in the plasma T790M mutation status was observed in either analysis. PFS, progression-free survival; PPF, post-progression survival.
DNA concentrations in cfDNA
The mean concentration of DNA extracted from the plasma in all enrolled patients was 1.458 (0.068–7.520) ng/µL. The mean concentration in patients with the T790M mutation in cfDNA was 0.971 (0.156–2.623) ng/µL and that in patients without T790M mutation was 2.292 (0.068–7.520) ng/µL. No correlation was observed between the concentration of DNA and the T790M mutation status (P=0.52). Concentration ratios of ALU247 and ALU157 (ALU247/ALU157), which indicated the degree of cfDNA fragmentation, were 0.65 in patients with T790M mutation in cfDNA and 1.01 in patients without the T790M mutation. The ratio in patients without T790M tended to be higher compared to patients with the T790M mutation, but the difference was not statistically significant (P=0.06).
Discussion
We showed that the T790M mutation, which causes resistance to EGFR-TKIs, could be detected in cfDNA from patients with EGFR-TKI resistance using the PointMan™ EGFR DNA enrichment kit, a method for selective mutant gene amplification. The PointMan™ method was previously established as a useful kit for research purposes and readily enables the determination of a patient’s mutation status. Recently, a third generation EGFR-TKI, osimertinib, has produced good clinical outcomes among patients with the T790M mutation in some clinical studies (12,13). We suggest that all NSCLC patients with EGFR mutations should be screened to determine whether the T790M mutation has occurred in tumor cells following resistance to first- or second-generation EGFR-TKIs as a part of routine testing for treatment decisions. Therefore, accurate methods for detecting the T790M mutation need to be established so that the survival of patients with EGFR mutations can be prolonged. Molecular testing of peripheral blood samples (using cfDNA) can be highly sensitive. Moreover, this approach has the advantage of being less invasive than a biopsy. Our data suggest that the PointMan™ assay is likely to become an important tool for the detection of tumor-derived mutations in cfDNA. Additionally, the results showed a higher detection rate than the cobas® EGFR Mutation Test v2, an approved method from in vitro diagnostics (IVD). The results obtained with the cobas® EGFR Mutation Test v2 in this study appeared less successful than usual. One possible explanation was that the plasma volumes used in this study were smaller than the amounts in the IVD. Although this is a limitation of this study, the data indicated that the PointMan™ method was superior to the cobas® EGFR Mutation Test v2.
The most meaningful finding in this study was that the T790M detection rate in cfDNA was extremely high (91.7%, 11 of 12 patients with T790M detected in tumor tissues) compared with previous reports, in which it ranged from 46.0–81.8% (21,24-27). This concordance is likely the result of the mutant-specific enrichment enabled by the PointMan™ technology and the additional amplification that enabled the mutation status to be identified even if the amount of cfDNA was below the detectable limit. We ultimately confirmed the mutation status visually by evaluating the direct sequences produced using BigDye® terminator after the selective amplification of the target mutant gene. Confirmation by direct sequencing permitted us to avoid amplification errors caused by the non-specific amplification of the wild-type gene.
The T790M mutation was not detected in cfDNA in only 1 of the 12 patients carrying a T790M mutation. Two possible explanations for this discordance can be considered. The first explanation is a false-negative result. The T790M mutant allele was present at very low levels in the circulation of the patient, and additionally might also have been fragmented. As a result, the PointMan™ Assay may have failed to detect the T790M mutation. The second explanation is that although tumor-derived DNA existed in the circulation, the mutant gene sequence was not efficiently amplified because of the low T790M allelic burden.
Of the seven patients without a T790M mutation in the re-biopsied samples, one was found to have the T790M mutation in their cfDNA. The T790M mutation might not have been detected in some tissue samples if the re-biopsy collected only inappropriate samples. In other words, the amounts of DNA from the re-biopsied samples might have been insufficient to detect the T790M mutation, or DNA with the T790M mutation might not have been present in the re-biopsy samples because of intratumoral and/or intertumoral heterogeneity (28,29). De Bruin et al. (28) reported the presence of intratumoral heterogeneity in terms of copy number alterations, translocations and mutations in multiple NSCLCs that had been surgically resected. If the T790M mutation is detected only in cfDNA and not in tumor tissue, it is not clear whether third-generation EGFR-TKIs targeting T790M should be used. According to a previous clinical study, in a limited subgroup of T790M-negative tumors, the response rate to osimertinib did not differ between the patients with or without T790M cfDNA (30). The T790M status of a tumor should be respected to prospect the outcome of osimertinib. On the other hand, the T790M status of cfDNA could be considered as the alternative prospective factor when tumor re-biopsy cannot be obtained.
In our survival analysis, neither the PFS nor the OS differed according to the plasma T790M mutation status. Uramoto et al. showed that the OS of a T790M-positive population in a tissue sample was longer than that of a T790M-negative population, and our study showed only a similar tendency to that of the previous report (31). Wang et al. evaluated the T790M mutation in matched pre- and post-TKI plasma samples, and the PFS and OS values were inferior for patients with de novo plasma T790M mutations identified using digital PCR, compared with patients without the T790M mutation (32).
The limitations of the present study include the enrollment of a small number of patients at a single facility and the retrospective design. Additionally, we do not have any information regarding alternative mechanisms of resistance to EGFR-TKI therapy, such as MET and EGFR amplification, small cell carcinoma transformation, and PIK3CA mutation. Furthermore, the present reported method cannot be used to quantify the allele frequency of T790M or other variants.
In conclusion, the selective mutation gene amplification assay is a useful means of detecting T790M mutations in plasma as an indicator of third generation EGFR-TKIs. As a future study, we plan to evaluate the clinical significance of the T790M mutation status in cfDNA in a larger prospective study.
Acknowledgements
The authors would like to thank Dr. Andrew Webb, Dr. Gary Dowthwaite and Dr. Chris Booth at EKF Molecular Diagnostics Ltd. for their technical supporting of the PointMan™ EGFR DNA enrichment kit, Dr. Miki Abo at Kanazawa University Hospital for suggestion on the manuscript.
Funding: This work was financially supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI, Grant-in-Aid for Scientific Research (C) (grant number: 26430142).
Ethical Statement: This study was approved by the Ethics Committee of Kanazawa University (No. 344-1).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. 10.1056/NEJMoa0810699 [DOI] [PubMed] [Google Scholar]
- 2.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. 10.1056/NEJMoa0909530 [DOI] [PubMed] [Google Scholar]
- 3.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. 10.1016/S1470-2045(11)70184-X [DOI] [PubMed] [Google Scholar]
- 4.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 5.Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327-34. 10.1200/JCO.2012.44.2806 [DOI] [PubMed] [Google Scholar]
- 6.Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 2014;15:213-22. 10.1016/S1470-2045(13)70604-1 [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786-92. 10.1056/NEJMoa044238 [DOI] [PubMed] [Google Scholar]
- 8.Oxnard GR, Arcila ME, Sima CS, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res 2011;17:1616-22. 10.1158/1078-0432.CCR-10-2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arcila ME, Oxnard GR, Nafa K, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res 2011;17:1169-80. 10.1158/1078-0432.CCR-10-2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem 2002;277:46265-72. 10.1074/jbc.M207135200 [DOI] [PubMed] [Google Scholar]
- 11.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005;2:e73. 10.1371/journal.pmed.0020073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. 10.1056/NEJMoa1411817 [DOI] [PubMed] [Google Scholar]
- 13.Sequist LV, Soria JC, Goldman JW, et al. Rociletinib in EGFR-mutated non-small-cell lung cancer. N Engl J Med 2015;372:1700-9. 10.1056/NEJMoa1413654 [DOI] [PubMed] [Google Scholar]
- 14.Kawamura T, Kenmotsu H, Taira T, et al. Rebiopsy for patients with non-small-cell lung cancer after epidermal growth factor receptor-tyrosine kinase inhibitor failure. Cancer Sci 2016;107:1001-5. 10.1111/cas.12963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Umetani N, Giuliano AE, Hiramatsu SH, et al. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol 2006;24:4270-6. 10.1200/JCO.2006.05.9493 [DOI] [PubMed] [Google Scholar]
- 16.Hao TB, Shi W, Shen XJ, et al. Circulating cell-free DNA in serum as a biomarker for diagnosis and prognostic prediction of colorectal cancer. Br J Cancer 2014;111:1482-9. 10.1038/bjc.2014.470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura H, Kasahara K, Kawaishi M, et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res 2006;12:3915-21. 10.1158/1078-0432.CCR-05-2324 [DOI] [PubMed] [Google Scholar]
- 18.Huang MY, Liu HC, Yen LC, et al. Detection of activated KRAS from cancer patient peripheral blood using a weighted enzymatic chip array. J Transl Med 2014;12:147. 10.1186/1479-5876-12-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura H, Suminoe M, Kasahara K, et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA). Br J Cancer 2007;97:778-84. 10.1038/sj.bjc.6603949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Douillard JY, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014;9:1345-53. 10.1097/JTO.0000000000000263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuang Y, Rogers A, Yeap BY, et al. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin Cancer Res 2009;15:2630-6. 10.1158/1078-0432.CCR-08-2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taniguchi K, Uchida J, Nishino K, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res 2011;17:7808-15. 10.1158/1078-0432.CCR-11-1712 [DOI] [PubMed] [Google Scholar]
- 23.Zhu G, Ye X, Dong Z, et al. Highly Sensitive Droplet Digital PCR Method for Detection of EGFR-Activating Mutations in Plasma Cell-Free DNA from Patients with Advanced Non-Small Cell Lung Cancer. J Mol Diagn 2015;17:265-72. 10.1016/j.jmoldx.2015.01.004 [DOI] [PubMed] [Google Scholar]
- 24.Ishii H, Azuma K, Sakai K, et al. Digital PCR analysis of plasma cell-free DNA for non-invasive detection of drug resistance mechanisms in EGFR mutant NSCLC: Correlation with paired tumor samples. Oncotarget 2015;6:30850-8. 10.18632/oncotarget.5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 2008;359:366-77. 10.1056/NEJMoa0800668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sequist, LV, Goldman JW, Wakelee, HA, et al. Efficacy of rociletinib (CO-1686) in plasma-genotyped T790M-positive non-small cell lung cancer (NSCLC) patients (pts). J Clin Oncol 2015;33:abstr 8001.
- 27.Sun W, Yuan X, Tian Y, et al. Non-invasive approaches to monitor EGFR-TKI treatment in non-small-cell lung cancer. J Hematol Oncol 2015;8:95. 10.1186/s13045-015-0193-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014;346:251-6. 10.1126/science.1253462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piotrowska Z, Niederst MJ, Karlovich CA, et al. Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discov 2015;5:713-22. 10.1158/2159-8290.CD-15-0399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3375-82. 10.1200/JCO.2016.66.7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uramoto H, Yano S, Tanaka F. T790M is associated with a favorable prognosis in Japanese patients treated with an EGFR-TKI. Lung Cancer 2012;76:129-30. 10.1016/j.lungcan.2012.01.010 [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Chen R, Wang S, et al. Quantification and dynamic monitoring of EGFR T790M in plasma cell-free DNA by digital PCR for prognosis of EGFR-TKI treatment in advanced NSCLC. PLoS One 2014;9:e110780. 10.1371/journal.pone.0110780 [DOI] [PMC free article] [PubMed] [Google Scholar]