We appreciate the interest and the editorial comments of Luo J and Fan S on our article “The mutant p53-ID4 complex controls VEGFA isoforms by recruiting lncRNA MALAT1”, recently published in EMBO Reports (1). We agree with the first criticism raised by the authors. Indeed, the use of in vivo tumor experimental systems would strongly strengthen the relevance of the results presented in our study and would clarify the extent of the contribution of the ribonucleoprotein complex comprising mutant p53, ID4, SRSF1 proteins and lncRNA MALAT1 to the modulation of VEGFA isoforms and to tumor angiogenesis.
It is well established that the induction of a tumor vasculature is required for the progression of tumors and for the acquisition of malignant traits. It has been reported that an efficient tumor angiogenesis may be modulated by the activity of a variety of cell types that replete the tumor stroma. In particular, tumor-associated macrophages play a crucial role in regulating angiogenesis and wound healing. On this basis, we believe that the best experimental model to evaluate the contribution of the various components of the RNP complex comprising mutant p53, ID4, SRSF1 proteins and lncRNA MALAT1, would be the use of immunocompetent mouse models of breast carcinogenesis or breast cancer cells orthotopically injected in syngeneic mouse models. Xenograft models unfortunately are established in immunocompromised mice and this prevents the correct evaluation of the contribution of cells of the immune system to the development and spreading of tumors.
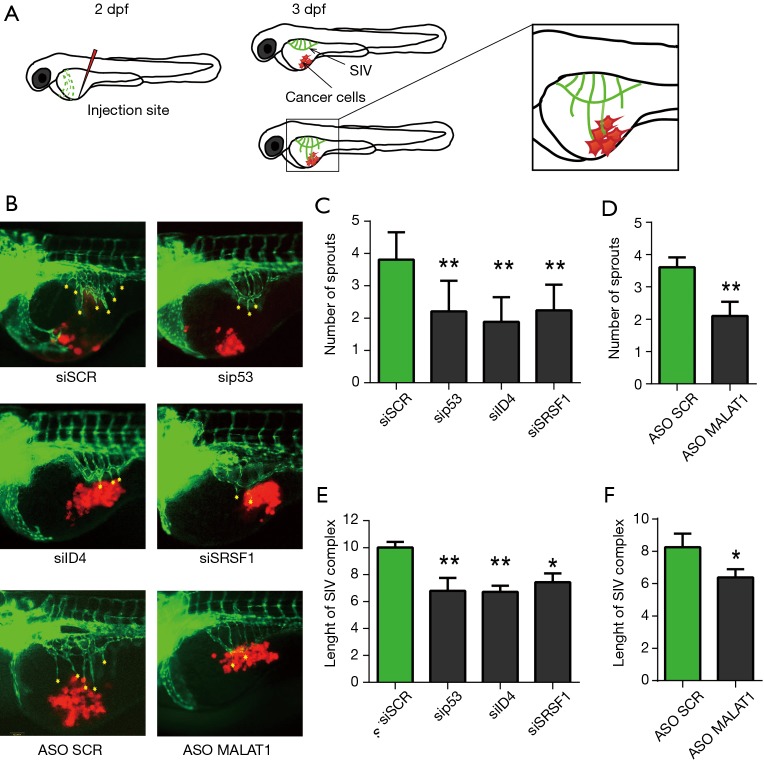
Despite mouse models represent the ideal system for the evaluation of associations with various tumor pathological parameters, we obtained a first indication of the in vivo relevance of the RNP complex described in Pruszko et al. (1) using Zebrafish (Danio rerio) as experimental model. Zebrafish has emerged recently as a potent model to study cancer cells invasion, metastasis and angiogenesis. Using zebrafish embryos for research presents multiple advantages over other models due to its rapid development, high number of offspring and transparency of embryo and larvae (2). Moreover, there are a few transgenic zebrafish lines expressing fluorescent proteins in endothelial cells, which make it possible to trace blood vessels development in living organism (3). Importantly, molecular pathways involved in vasculature development are conserved between fish and mammals; hence the obtained results are translatable into information applicable in human cancer treatment (4). At an early stage of development xenotransplantation of human cancer cells is possible because of an immature immune system. To test the influence of mutant p53, ID4, SRSF1 and MALAT1 on proangiogenic properties of cancer cells, SKBR3 breast cancer cells, carrying endogenous mutant p53R175H, were transfected with specific siRNA or ASO oligonucleotides before injection in zebrafish embryos. The number and length of sprouts growing from SIV (subintestinal vessel) complex were measured 24 h post injection. In line with previous results, in control embryos injected with cells transfected with si-SCR or ASO-SCR, growth of additional branches of the SIV complex was observed (Figure 1). Depletion of p53, ID4, SRSF1 or MALAT1 in SKBR3 cells resulted in reduced number of sprouts and in decreased length of the SIV complex (Figure 1). These observations confirmed that p53, ID4, SRSF1 or MALAT1 expressed by cancer cells are involved in induction of angiogenesis in vivo. However, it is undeniable that the establishment of mouse models with depletion of ID4, as well as of other components of the identified RNP complex, will allow a more complete evaluation of the effects observed on the tumor vasculature and on the various cell components of the tumor stroma.
Figure 1.
Silencing of p53, ID4, SRSF1, MALAT1 genes attenuates the proangiogenic properties of breast cancer cells in zebrafish embryos. (A) The scheme shows the injection site indicated by a red “needle”. Cancer cells (red) are injected into zebrafish embryo 2 days post fertilization (2 dpf). At this stage of development big blood vessel called duct of Couvier (green) is visible. At 3 dpf duct of Couvier disappears and a SIV complex is formed. Magnification shows a fragment of a fish presented in the pictures below; (B) representative pictures presenting SIV complex (green) growth in response to injection of cancer cells SKBR3 (red) depleted of p53, ID4, SRSF1 or MALAT1; (C-F) the graphs present a mean number of sprouts (C,D) and SIV complex length in arbitrary units (E,F) calculated from three independent experiments. In every experiment 20 embryos per condition were analyzed. *, P≤0.05; **, P≤0.01; one way ANOVA.
As mentioned in the Editorial by Luo and Fan, in our study we assessed the consequences of the presence of high ID4 and/or VEGFA signature expression by performing Kaplan-Meier analysis in basal-like breast cancers (BLBC). This analysis evidenced that tumors from patients characterized by worse survival show concomitant high expression of ID4 and VEGFA signature. This analysis has been performed in BLBC because ID4 shows prognostic power specifically in this breast cancer subtype. Certainly, it would be interesting to evaluate the VEGFA isoforms, and downstream targets of VEGFA signaling, in relation to the clinical and pathological parameters in breast cancer. This could be evaluated also by using the TCGA dataset, for which the information about expression level of the splicing isoforms can be also investigated using the SpliceSeq Database (5), and this is one of our current interests.
Much importantly, it would be very relevant to evaluate the nuclear pattern of MALAT1 staining to assess whether also in tissues the delocalization of MALAT1 from nuclear speckles associates with the presence of mutant p53 and high ID4 expression level, and with high microvessel density. This would also enable evaluating whether MALAT1 intranuclear localization pattern correlates with specific VEGFA isoforms expression and to the downstream targets of VEGFA activity. Importantly, as not all p53 missense mutations lead to proteins with gain-of-function (GOF) activity (6), it is possible that the presence of a MALAT1 expression pattern delocalized from speckles together with repression of VEGFA anti-angiogenic isoforms could act as indicators of the presence of a GOF mutant p53. If this is the case, RNA FISH for MALAT1 detection coupled with VEGFA immunohistochemistry could be considered as a promising marker in breast cancer pathology.
Concerning the translational aspect of our study, it’s worth underlining that molecules, which interfere with the activity of SRPK1 kinase, are currently under deep investigation as potential therapeutics for various angiogenesis-related diseases. These highly potent and selective inhibitors inhibit SRPK1 activity and phosphorylation of serine/arginine splicing factor 1 (SRSF1), resulting in alternative splicing of VEGFA from pro-angiogenic to antiangiogenic isoforms. This property results in potent inhibition of blood vessel growth in in vivo models of angiogenesis (7). We believe that these molecules could represent an important therapeutic option for those BLBCs characterized by missense mutation of TP53 and high ID4 expression. The efficacy of such compounds on the modulation of the angiogenic potential and on the abundance of the various VEGFA isoforms thus merits to be evaluated in the context of cell lines and in vivo models characterized by different TP53 status (wild-type vs. mutated) and ID4 expression levels.
Acknowledgements
Funding: This work was supported by Italian Ministry of Health grant (GR-2011-02348567) and AIRC (MFAG10728) to G Fontemaggi; MIUR Epigen (13/05/R/42) and AIRC (IG14455) to G Blandino; a MAESTRO NZ1/00089 grant from the National Science Center of Poland to M Pruszko, A Zylicz, and M Zylicz.
Provenance: This is an invited article commissioned by the Section Editor Dr. Chunlin Ou (Cancer Research Institute of Central South University, Changsha, China).
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Pruszko M, Milano E, Forcato M, et al. The mutant p53-ID4 complex controls VEGFA isoforms by recruiting lncRNA MALAT1. EMBO Rep 2017;18:1331-51. 10.15252/embr.201643370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tulotta C, He S, van der Ent W, et al. Imaging Cancer Angiogenesis and Metastasis in a Zebrafish Embryo Model. Adv Exp Med Biol 2016;916:239-63. 10.1007/978-3-319-30654-4_11 [DOI] [PubMed] [Google Scholar]
- 3.Chávez MN, Aedo G, Fierro FA, et al. Zebrafish as an Emerging Model Organism to Study Angiogenesis in Development and Regeneration. Front Physiol 2016;7:56. 10.3389/fphys.2016.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicoli S, Ribatti D, Cotelli F, et al. Mammalian tumor xenografts induce neovascularization in zebrafish embryos. Cancer Res 2007;67:2927-31. 10.1158/0008-5472.CAN-06-4268 [DOI] [PubMed] [Google Scholar]
- 5.Ryan M, Wong WC, Brown R, et al. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res 2016;44:D1018-22. 10.1093/nar/gkv1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 2014;25:304-17. 10.1016/j.ccr.2014.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batson J, Toop HD, Redondo C, et al. Development of Potent, Selective SRPK1 Inhibitors as Potential Topical Therapeutics for Neovascular Eye Disease. ACS Chem Biol 2017;12:825-32. 10.1021/acschembio.6b01048 [DOI] [PubMed] [Google Scholar]