Abstract
The genomic clone encoding an α-tubulin, OsTubA1, has been isolated from rice (Oryza sativa L.). The gene consists of four exons and three introns. RNA-blot analysis showed that the gene is strongly expressed in actively dividing tissues, including root tips, young leaves, and young flowers. Analysis of chimeric fusions between OsTubA1 and β-glucuronidase (GUS) revealed that the intron 1 was required for high-level GUS expression in actively dividing tissues, corresponding with normal expression pattern of OsTubA1. Fusion constructs lacking the intron 1 showed more GUS staining in mature tissues rather than young tissues. When the intron 1 was placed at the distal region from 5′-upstream region or at the 3′-untranslated region, no enhancement of GUS expression was observed. Sequential deletions of the OsTubA1 intron 1 brought about a gradual reduction of GUS activity in calli. These results suggest that tissue-preferential expression of the OsTubA1 gene is mediated by the intron 1 and that it may be involved in a mechanism for an efficient RNA splicing that is position dependent.
Microtubules are the basis of morphogenetic processes that occur by the definition of the plane of cell division and the direction of cell elongation in plant cells. Microtubules are mainly composed of α- and β-tubulin, which have genes that have been conserved through evolution (Little and Seehaus, 1988). α-Tubulin genes have been characterized from several plant species including Chlamydomonas reinhardtii (Silflow et al., 1985), Arabidopsis (Ludwig et al., 1988; Kopczak et al., 1992), maize (Montoliu et al., 1989, 1990, 1992; Villemur et al., 1992), Prunus amygdalus (Stöcker et al., 1993), and rice (Oryza sativa L.; Qin et al., 1997). As shown previously in vertebrates, plant α-tubulin genes form a multigene family; for example, six α-tubulin genes exist in Arabidopsis, and at least seven in maize. Previous studies in Arabidopsis have shown that certain α-tubulin genes are tissue specific, whereas others are constitutively expressed in all tissues (Ludwig et al., 1988; Kim and An, 1992; Kopczak et al., 1992). In maize, two α-tubulin genes are preferentially expressed in the radicular system (Montoliu et al., 1989). It has also been reported in maize that certain α-tubulin genes are preferentially expressed in rapidly dividing tissues such as root tips rather than in more mature tissues (Joyce et al., 1992). Generally, the expression of most α-tubulin genes in plants has been detected preferentially in actively dividing tissues.
Recent studies have shown evidence that the sequences downstream of the transcription initiation site often play important roles on gene expression in plants. The pea ferredoxin (Fed-1) gene requires exon sequences for light response (Dickey et al., 1994). In the pea plastocyanin (PetE) gene, both the coding region and the 5′-untranslated leader region have been found to be required for light regulation (Helliwell et al., 1997). A light-responsive element of the tobacco psaDb gene has been located within the coding region (Yamamoto et al., 1997). The Me1 gene from Flaveria bidentis requires an enhancer-like element in the 3′-flanking region for high-level expression in leaves (Marshall et al., 1997). In Sesbania rostrata, nodule parenchyma-specific expression of SrEnod2 gene is mediated by the 3′-untranslated region (Chen et al., 1998).
It has been reported that some introns in plant genes increase gene expression. Expression enhancements by an intron have been found in maize Adh1 (Callis et al., 1987; Luehrsen and Walbot, 1991), Sh1 (Vasil et al., 1989; Maas et al., 1991), and Ubi1 (Christensen et al., 1992), rice Act1 (McElroy et al., 1990), and the PAT1 gene of Arabidopsis (Rose and Last, 1997). In addition to enhancement effects, cases in which an intron was required for regulated- or tissue-specific gene expression in plant genes have also been found. For example, the PsaD gene of the spinach plant requires an intron sequence for plastid- and light-dependent expression (Bolle et al., 1996). An intron of the AGAMOUS (AG) floral homeotic gene in Arabidopsis contains enhancer sequences for specific gene expression in flowers (Busch et al., 1999).
In this study, we have analyzed β-glucuronidase (GUS) expression in transgenic rice plants carrying various OsTubA1::GUS fusion constructs. From this analysis, we have found that actively dividing tissue-preferential expression of OsTubA1 is controlled by the intron 1.
RESULTS
Isolation of the Rice α-Tubulin Gene OsTubA1
Five rice α-tubulin cDNA clones were isolated from an immature anther cDNA library (Chung et al., 1994) using the Arabidopsis α1-tubulin gene as a probe (Ludwig et al., 1988). Because the nucleotide sequences of the 3′-untranslated regions are not conserved among α-tubulin genes, we categorized the clones into three different groups by sequence analyses of the regions. Three of the clones were identical to OsTubA1 and the other two clones, OsTubA2 and OsTubA3, out of previously published clones (Qin et al., 1997).
As the first step in examining the diversity of expression patterns among the various α-tubulin genes, we isolated a genomic clone corresponding to the OsTubA1 cDNA clone using 3′-untranslated region as the gene-specific probe. The 4.3-kb XbaI-PstI fragment, containing the entire coding region along with the flanking regions, was subcloned into the pBluescript SK(−) vector. Sequencing of the genomic clone revealed that nucleotide sequences of the gene are different from the OsTubA1 cDNA at one site (1,102) of the 5′-untranslated region and at two sites (2,256 and 3,604) of the coding region. Consequently, instead of Ser, the 52nd and 437th amino acids of OsTubA1 were Phe and Ala in the present clone. It is likely that the differences between the two clones may have resulted from the different cultivars used. It can be concluded that our clone is a variant genomic clone of OsTubA1 (Qin et al., 1997).
Comparing OsTubA1 with the other cDNA clones revealed that the gene consisted of four exons and three introns in the coding region (accession no. AF 182523). The first intron is 946 bp long and located between codons 31 and 32. The 86-bp second intron and the 108-bp third intron are located at codon 110 and between codons 233 and 234, respectively. The number and position of these introns were identical to those of previously identified α-tubulin genes, such as Arabidopsis TUA2 (Kopczak et al., 1992) and Zea mays Tubα1 and Tubα2 (Montoliu et al., 1989). All three introns of OsTubA1 present the consensus 5′-GT and 3′-AG recognition signals as reported in the literature (Hanley and Schuler, 1988).
Expression of OsTubA1 in Rice
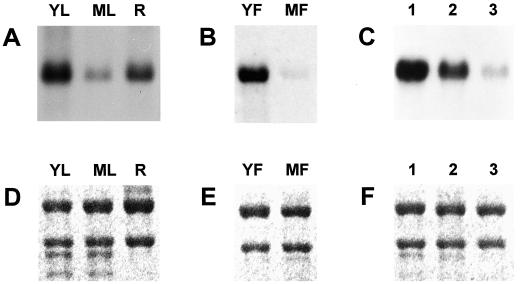
To examine the expression pattern of OsTubA1, RNA gel-blot analysis was carried out using various tissues of rice (Fig. 1). The result showed that OsTubA1 RNA was distributed abundantly in roots and leaves of 7-d-old seedlings and flowers in the early to mature pollen stage and was distributed poorly in mature leaves and flowers at the heading stage (Fig. 1, A and B). This suggests that OsTubA1 may be expressed mainly in rapidly dividing tissues. To confirm the premise, further experiments were carried out using total RNA isolated from manually dissected samples of dividing, elongating, and maturing cells of 7-d-old seedling roots (Fig. 1C). RNA gel-blot analysis demonstrated that OsTubA1 RNA was accumulated mainly at root tips, supporting that the gene is expressed preferentially in actively dividing tissues: root tips, young leaves, and flowers of the meiotic to mitotic pollen stage during rice growth and development.
Figure 1.
RNA gel-blot analyses of the OsTubA1 gene. A, Expression of OsTubA1 mRNA in young leaves (YL) of 7-d-old seedlings, mature leaves (ML) of flowering plants, and roots (R) of 7-d-old seedlings. B, Expression of OsTubA1 mRNA in young flowers (YF) at less than 10-cm panicle size and mature flowers (MF) at the heading stage. C, Expression of OsTubA1 mRNA in roots dissected in dividing (1), elongating (2), and maturing zones (3) of 7-d-old roots. Each 10-μg portion of total RNAs isolated from various tissues was blotted onto a nylon membrane and hybridized to the 32P-labeled OsTubA1 gene-specific probe. D to F, Ethidium bromide staining of 25S and 17S rRNAs demonstrating equal amounts of RNA loading in A to C, respectively.
Effect of Intragenic Region on GUS Expression in Transgenic Rice Plants
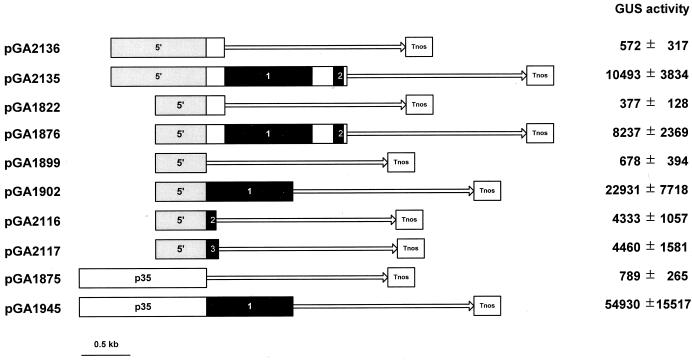
The spatial expression pattern of OsTubA1 was investigated using GUS reporter gene fusions. Two translational fusions, pGA2135 and pGA2136, of OsTubA1 to the GUS gene were made to test the influence of intragenic sequences on gene expression (Fig. 2). The only difference between the two constructs was the length of the included intragenic region. pGA2136 contained only the first 27 codons, whereas pGA2135 included all of exon 1 and 2, intron 1 and 2, and 15 codons of exon 3. The fusions were introduced into rice by Agrobacterium-mediated transformation, and more than 10 lines of each construct were analyzed by histochemical GUS staining.
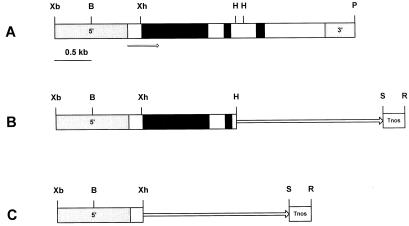
Figure 2.
Restriction map of the OsTubA1 gene and construction of the chimeric plasmid between the OsTubA1 promoter and GUS gene. A, Restriction map of the OsTubA1 gene. The coding regions and introns are represented by white and black bars, respectively. The promoter (5′) and terminator (3′) regions are shown. The arrow indicates direction of transcription. B, Diagram of pGA2135 carrying upstream sequences, exon 1 and 2, intron 1 and 2, a portion of exon 3 of OsTubA1. The HindIII site of the exon 3 of the gene is fused to the GUS gene. C, Diagram of pGA2136 carrying upstream sequences and a portion of exon 1 of OsTubA1. The XhoI site of the exon 1 of the gene is fused to GUS. Tnos signifies the nopaline synthase (nos) terminator. The GUS coding region is indicated by a solid line with an arrow. Letters represent restriction enzyme sites: B, BamHI; H, HindIII; P, PstI; R, EcoRI; S, SacI; Xb, XbaI; Xh, XhoI.
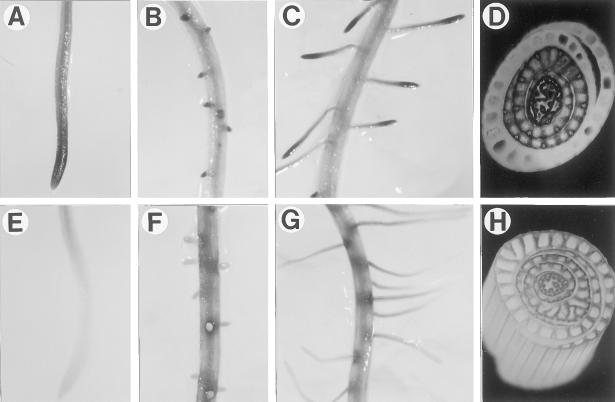
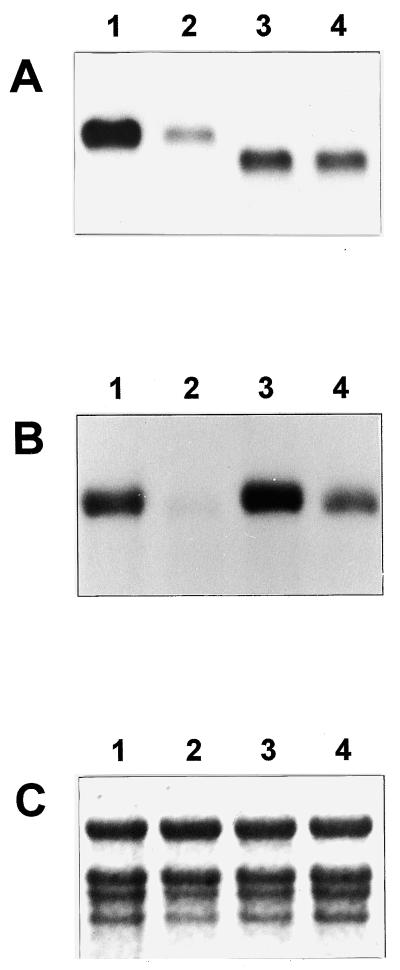
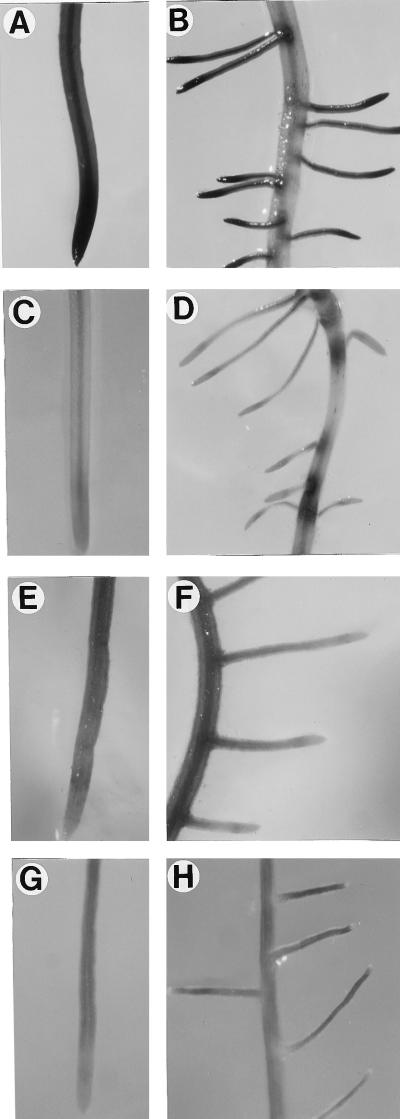
In pGA2135 plants, intense GUS staining was observed from three different parts: actively growing tips of the primary roots, lateral roots at the initiation stage, and actively growing lateral roots (Fig. 3, A–C). This coincided with the distribution pattern of the OsTubA1 RNA in wild-type rice (Fig. 1C). In contrast to the staining pattern of pGA2135, plants transformed with pGA2136 showed more GUS staining in elongation and maturation parts rather than in root tips (Fig. 3, E–G). To determine whether the differences in the GUS staining patterns also appeared in leaves, we stained the shoots of 4- to 6-week-old plants. Histochemical analysis of cross-sectioned tissues of actively growing shoots revealed that the inner young leaves were preferentially GUS-stained in pGA2135 (Fig. 3D). However, in pGA2136 lines, the outer mature leaves were stained as strongly as the inner young ones (Fig. 3H). To determine whether the GUS staining patterns correlated with levels of the GUS RNA, RNA gel-blot analysis was carried out using total RNA isolated from inner young and outer mature leaf tissues (Fig. 4). The results exhibited that in pGA2135 lines, the GUS RNA level was much higher in young leaves than in mature leaves, whereas in pGA2136 lines there was no significant difference between them. These results demonstrated that the 1.3-kb intragenic region of OsTubA1, which was included in the pGA2135 construct, confers young tissue-preferential gene expression.
Figure 3.
Histochemical localization of GUS activity directed by OsTubA1 promoter::GUS fusions in transgenic rice. A to C, Transgenic roots carrying pGA2135. D, Transgenic shoots carrying pGA2135. E to G, Transgenic roots carrying pGA2136. H, Transgenic shoots carrying pGA2136. All reactions were stopped after 12 h.
Figure 4.
RNA gel-blot analyses in shoots of transgenic plants. A, Ten-microgram portions of total RNAs isolated from a total of three independent transgenic shoots were hybridized with the GUS gene. Samples 1 and 2 indicate inner young and outer mature leaves, respectively, of 4- to 6-d-old transgenic plants carrying pGA2135, and samples 3 and 4 indicate inner young and outer mature leaves, respectively, of pGA2136 transgenic plants at the same stage. Due to an additional coding sequence of approximately 0.3-kb, the transcripts in lanes 1 and 2 are larger than those in lanes 3 and 4. B, The filter was washed and reprobed with an OsTubA1 gene-specific probe. C, Ethidium-bromide staining of 25S and 17S rRNAs demonstrating equal amounts of RNA loading.
Effect of the Intron 1 on GUS Expression in Transgenic Rice Plants
The 1.3-kb intragenic region, which was essential for normal expression of the OsTubA1 gene, contained exon 1, exon 2, intron 1, intron 2, and a portion of exon 3. To determine the region that specified the young tissue-preferential expression, we constructed various OsTubA1::GUS fusion molecules that contained different portions of the intragenic region (Fig. 5).
Figure 5.
Structures of the tested GUS fusion constructs and GUS activity in the calli carrying them. Numbers 1, 2, and 3 in black bars indicate introns 1, 2, and 3 of OsTubA1, respectively. Gray bars represent the 5′-upstream sequence of OsTubA1, and white bars indicate the p35 cauliflower mosaic virus 35S promoter. Exons of OsTubA1 are shown in white bars, and the GUS coding region is shown with a solid line with an arrow. Tnos signifies the nos terminator. GUS values are expressed as pmol 4-MU min−1 mg−1 soluble protein.
Because calli are groups of actively dividing cells, we compared the relative GUS expression levels of the different constructs in transformed calli (Fig. 5). The results showed that pGA2135 provided GUS activity 18 times greater compared with that of pGA2136. A similar result was observed with the constructs pGA1822 and pGA1876, which carried a shorter promoter fragment (491 bp; 555–1,045), although overall activity was slightly reduced compared with the longer promoter (1,045 bp; 1–1,045).
Since introns have been known to enhance gene expression, the roles of the three introns of the OsTubA1 gene were examined by placing each intron between the OsTubA1 promoter and GUS reporter. The highest GUS activity was recorded for pGA1902 in which the intron 1 was inserted between the OsTubA1 promoter and the GUS coding sequence. The GUS activity driven by the OsTubA1 promoter and intron 1 was 34-fold higher than that by the OsTubA1 promoter alone (pGA1899), which displayed a low level of GUS expression. Intron 2 (pGA2116) and intron 3 (pGA2117) also increased the expression level to some extent, which were approximately 20% level of that achieved by intron 1. The construct, pGA1876, that contained both intron 1 and intron 2 did not show higher activity compared with the construct carrying the intron 1 alone. The result indicates that the combined presence of intron 1 and intron 2 provided neither synergistic nor additive. Our results agree with the previous reports that the first intron can significantly enhance gene expression in monocots (Callis et al., 1987; Vasil et al., 1989; McElroy et al., 1990).
The enhancing effects of the introns were studied in transgenic roots by histochemical GUS analyses (Fig. 6). In pGA1902 containing the OsTubA1 intron 1, GUS staining was mainly detected at actively dividing cells such as in the tips of primary and secondary roots (Fig. 6, A and B). Unlike the case, in pGA1899, which contained no introns, GUS staining was observed mainly in elongating and maturing regions of roots and was hardly detectable in root tips, which are rich in dividing cells (Fig. 6, C and D). To determine whether the intron 2 and intron 3 of OsTubA1 would exhibit the tissue-preferential enhancer function observed from the intron 1, transgenic roots of the pGA2116 and pGA2117 lines carrying the intron 2 and intron 3, respectively, were analyzed by histochemical GUS staining. The results showed that their GUS-staining patterns were similar to those of pGA1899 (Fig. 6, E–H), indicating that the growing cell-preferential expression is particular to intron 1 of OsTubA1.
Figure 6.
Histochemical localization of GUS activity directed by various OsTubA1::GUS fusions in transgenic roots. A and B, pGA1902. C and D, pGA1899. E and F, pGA2116. G and H, pGA2117. All reactions were stopped after 12 h.
It was observed in pGA1945 that placing the first intron under control of the heterogeneous 35S promoter resulted in a 70-fold increase in GUS expression level over that of pGA1875, the 35S promoter-GUS fusion without the intron 1 (Fig. 5). This result further supports that OsTubA1 intron 1 can significantly enhance gene expression in actively growing cells.
Possible Mechanism of the Growing Cell-Preferential Gene Expression by the OsTubA1 Intron 1
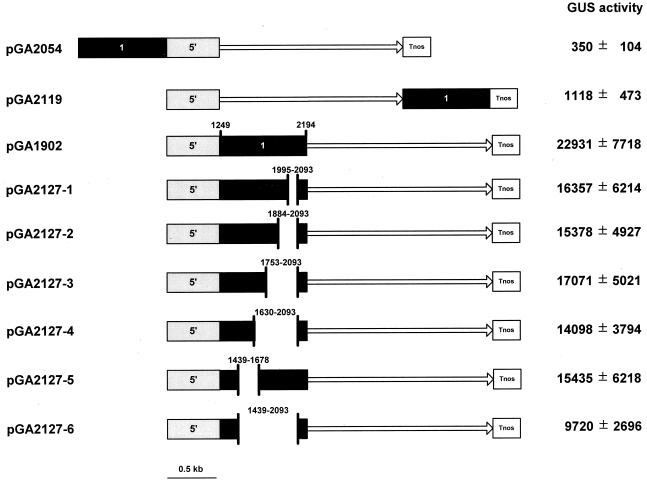
To study the mechanism of enhancing activity of the OsTubA1 intron 1 on gene expression, we constructed additional GUS fusion molecules and examined GUS activities in transformed rice calli (Fig. 7). Placing the intron 1 upstream of the OsTubA1 promoter (pGA2054) or downstream of the GUS coding sequence (pGA2119) did not enhance the gene expression, suggesting that the increased gene expression was position-dependent and was not due to an enhancer-like activity present in the intron 1.
Figure 7.
Diagram of the tested GUS fusion constructs and GUS activity in the calli carrying them. The number 1 contained in a black bar indicates the OsTubA1 intron 1 (nucleotides 1,249–2,194), and gray bars represent the 5′-upstream sequence of OsTubA1. The numbers in truncated black bars represent the positions of deleted intron-1 fragment. The GUS coding region is indicated with a solid line with an arrow. Tnos signifies the nos terminator. GUS values are expressed as pmol 4-MU min−1 mg−1 soluble protein.
To further understand a possible mechanism of the intron-1-mediated enhancement, we constructed six deletion mutants, pGA2127-1 to -6, within the intron-1 sequences and the effects were examined in transgenic calli (Fig. 7). The deletions of the intron-1 sequence resulted in a reduction of GUS activities by approximately 25% to 60%. The largest deletion construct pGA2127-6, which carried 291-bp intron sequence, still retained a significant level of the GUS activity. Further deletion of the 5′ or 3′ region of the intron 1 resulted in severe loss of the GUS activity (data not shown), indicating that the remaining sequences of the intron 1 in pGA2127-6 contained the legitimate splicing sequences, including donor, acceptor, and branch sites. The GUS activity analysis in roots and leaves of pGA2127-1 to -6 lines exhibited nearly identical expression patterns to those of transgenic lines, pGA2135, pGA1876, and pGA1902, with the entire intron 1 (data not shown). These results suggest that the primary effect of the OsTubA1 intron 1 on gene expression is probably mediated by RNA splicing.
DISCUSSION
Regulatory elements for tissue-specific expression have often been found in an intron rather than in the 5′-flanking region of a gene. The leader intron of potato Sus4 was required for high-level tuber expression and Suc inducibility (Fu et al., 1995). The spinach PsaD gene required its intron for correct plastid- and light-dependent regulation (Bolle et al., 1996). Recently it has been reported that the large second intron of AG contains LEAFY-responsive enhancer sites, which are required for proper spatial and temporal expression of AG in Arabidopsis (Busch et al., 1999). In the present study, we report that the first intron of the rice OsTubA1 gene has a strong enhancing effect in actively growing cells such as calli, root tips, and young leaves.
Through histochemical GUS analysis of transgenic rice plants carrying various OsTubA1::GUS fusions, we have shown that the transcribed region of the gene is required for a normal expression of OsTubA1 in rice. Further analysis of each intron revealed that the intron 1 played an important role in tissue-preferential expression of the OsTubA1 gene. Transgenic lines with the intron 1 gave much more intense GUS staining in actively dividing cells such as calli and root tips, whereas transgenic lines with only the intron 2 or intron 3 exhibited major GUS activities in mature regions instead. The enhancer effects were also observed when the intron 1 was placed under the heterogeneous 35S promoter.
It is unlikely that a transcriptional enhancer element is present in the intron 1, since placing the intron-1 sequence in the 5′-upstream region of the OsTubA1 promoter or the 3′-downstream region of the GUS coding region did not result in any enhancing effect. Position-dependent effects of an intron on gene expression have also been observed in maize Adh1 and Sh1 and rice Act1 and triosephosphate isomerase (tpi) genes (Mascarenhas et al., 1990; McElroy et al., 1990; Maas et al., 1991; Snowden et al., 1996). Deletion mutagenesis studies have indicated that a majority of the intron-1 sequence could be deleted without interfering with the tissue-preferential enhancer activity. This result is in agreement with previous reports in which modification of an intron by deletions or substitutions did not significantly affect enhancement effects of maize Adh1 or human factor IX gene expression (Luehrsen and Walbot, 1991, 1994; Kurachi et al., 1995).
Our results suggest that the enhancer activity of the OsTubA1 intron 1 in actively growing tissues may be controlled at a post-transcriptional level, probably by the mediation of tissue-preferential RNA splicing or other RNA-processing events. In higher plants, genes encoding the major subunits of the spliceosome uridylate-rich small nuclear ribonucleoprotein particles consist of a large number of sequence variants. It was shown that the uridylate-rich small nuclear RNA variants exhibit developmental differences in their pattern of expression (Hanley and Schuler, 1991). Thus, it has been suspected that compositionally or functionally distinct uridylate-rich small nuclear ribonucleoprotein particles may affect developmental gene expression patterns (for review, see Simpson and Filipowicz, 1996). In our experiments, unspliced transcripts of the gene were not observed in RNA gel-blot and RT-PCR experiments in all tissues tested (Fig. 1 and 4; data not shown). This indicates that unspliced transcripts of the gene might be rapidly degraded in vivo as suggested by Mascarenhas et al. (1990). Therefore, it can be suspected that tissue-preferential accumulation of the mature OsTubA1 mRNA might depend upon amount of the intron-1-associated splicing factors. We also do not exclude the possibility that other RNA processing events like polyadenylation and nucleocytoplasmic transport might be involved in an increase of the steady-state level of the OsTubA1 mRNA. It was reported in mammals that intron enhancement resulted from an increase in the efficiency of 3′-end processing (Huang and Gorman, 1990). The regions near the 5′- or 3′-splicing site might retain regulatory sites for RNA splicing or other processing-associated factors, which are probably abundant in actively dividing tissues.
Enhancer effects have been found from the introns of several monocot genes, such as Adh1, Sh1, and Bz1 of maize and Act1 and tpi of rice (Callis et al., 1987; Oard et al., 1989; Vasil et al., 1989; McElroy et al., 1990; Maas et al., 1991; Xu et al., 1994). Expression of the rice Act1 gene requires intron 1 to direct constitutive GUS expression in transgenic rice, reflecting an ubiquitous requirement for cytoskeletal components in all plant cell types (Zhang et al., 1991). In the rice tpi gene, the intron 1 was necessary for GUS expression in transgenic rice (Xu et al., 1994). Taken together with the present study, these results suggest that the intron-mediated enhancement of reporter gene expression observed in undifferentiated cells may be due to temporal and/or spatial regulation of gene expression. It would be worthwhile to examine whether these introns also mediate normal expression of their genes in plants.
The OsTubA1 gene is closely related to members of the plant α-tubulin subfamily I, which consists of maize Tubα1 and Tubα2, and Arabidopsis TUA2 and TUA4. The maize Tubα1 promoter is active mainly in root tips and pollen in transgenic tobacco plants (Rigau et al., 1993; Uribe et al., 1998), and the promoter activity of the Arabidopsis TUA2 gene has also been shown to be the highest in tissues with actively dividing cells (Carpenter et al., 1993). The previous studies have demonstrated that promoter regions alone were sufficient to induce the tissue-preferential expression of the genes. These findings differ from the expression pattern of the rice OsTubA1 gene, which appears to be controlled mainly by a post-transcriptional mechanism. The results of this study contribute to the understanding of the cellular mechanisms that regulate the expression of individual members of rice α-tubulin gene families.
MATERIALS AND METHODS
Bacterial Strains
The Escherichia coli strain JM83 (ara, leu, lac, gal, str) was used as the recipient for routine cloning experiments. Agrobacterium tumefaciens LBA4404 containing the Ach5 chromosomal background and a disarmed helper-Ti plasmid pAL4404 was used for transformation of rice (Oryza sativa L.) (Hoekema et al., 1983).
Screening of cDNA and Genomic Libraries
The cDNA library made with mRNA from immature anthers of rice cv Nakdong (Chung et al., 1994) was screened with the [α-32P]dCTP Arabidopsis α1-tubulin gene as a probe (Ludwig et al., 1988). A genomic library constructed in the bacteriophage λ-DASH vector with partial Sau3AI-digested DNA of rice cv IR36 (kindly provided by Steve Kay) was used for isolation of a genomic clone as described by Sambrook et al. (1989). Hybridizations were carried out at 55°C in 6× SSC, 5× Denhardt's reagent, 0.5% (w/v) SDS, and 100 mg L−1 denatured salmon sperm DNA. The final two washes were done in 2 × SSC and 0.1% (w/v) SDS at 45°C for 30 min each. Phage DNA was prepared according to Chisholm (1989). The DNA sequence of a gene was determined by the dideoxynucleotide chain termination method using double-stranded plasmid DNA templates (Sanger et al., 1977).
RNA Gel-Blot Analysis
Total RNA was isolated from leaves, roots, and flowers of rice plants by the RNA isolation kit (TRI reagent, Molecular Research Center, Cincinnati). A 10-μg portion of total RNA was fractionated on a 1.3% (w/v) agarose gel as described previously (Kang et al., 1998). After RNA transfer onto a nylon membrane, the blot was hybridized in a solution containing 0.5 m sodium phosphate (pH 7.2), 1 mm EDTA, 1% (w/v) BSA, and 7% (w/v) SDS for 20 h at 60°C (Church and Gilbert, 1984). After hybridization, the blot was washed twice with a solution containing 0.2× SSPE and 0.1% (w/v) SDS for 5 min at room temperature followed by two washes of the same solution at 60°C for 15 min.
Construction of Binary Vectors
The plasmid pGA2135 was made by fusing the 2.6-kb XbaI-HindIII fragment (nucleotides 1–2,557) of OsTubA1, containing the 5′-upstream region, exon 1 and 2, intron 1 and 2, and a portion of exon 3, to GUS. The plasmid pGA2136 was produced by connecting the 1.2-kb XbaI-XhoI fragment (nucleotides 1–1,231) of OsTubA1, containing the 5′-upstream region and a portion of exon 1, to the GUS coding region. In both constructs, OsTubA1 and GUS existed in the same reading frame, generating translational fusions. The plasmids pGA1822 and pGA1876 were produced from pGA2136 and pGA2135, respectively, by deletion of the 0.55-kb XbaI-BamHI fragment (nucleotides 1–554) at the distal region of the OsTubA1 promoter.
To construct transcriptional fusions between the promoter and GUS, a HindIII site was generated immediately upstream of ATG codon by PCR using the oligomer 5′-GAAGCTTGGCGGAATGGGTT-3′ as a reverse primer, the T3 primer (5′-AATTAACCCTCACTAAAGGG-3′) as a forward primer, and the genomic fragment cloned in pBluescript SK(−) (Stratagene, La Jolla, CA) as a template. The amplified 0.6-kb fragment of OsTubA1 promoter was fused to the GUS gene through the HindIII site to make pGA1899.
Three plasmids, pGA1902, pGA2116, and pGA2117, each with a DNA fragment carrying intron 1, 2, or 3 of OsTubA1, respectively, inserted in the HindIII site of pGA1899 were constructed to examine the effect of three introns of OsTubA1 on gene expression. For construction of pGA1902, HindIII sites were generated at both ends of the fragment containing the intron 1 by PCR, using 5′-GAAGCTTGCATCCAGGTACG-3′ and 5′-GAAGCTTCAGCCTGAAAGGA-3′ (underlined sequences are the exon sequences flanking intron 1) as primers. pGA2116 was constructed byattaching a HindIII site to the 5′ end and a XbaI site to the 3′ end of the fragment containing intron 2 of OsTubA1 by PCR using primers, 5′-TCAAGCTTGTGGTCACTACACCAGTGAG-3′ and 5′-GTTTCTAGACAATCTCCTTGCCAACTG-3′ (underlined sequences are the exon sequences flanking intron 2). Similarly, pGA2117 was constructed by adding HindIII and XbaI sites at the 5′ and 3′ ends of the DNA fragment carrying the intron 3 by PCR using primers, 5′-TCAAGCCTGTGTCTCAGGTACAGACTTG-3′ and 5′-GTTCTAGAGGGATGAGATAACCTGCAC-3′ (underlined sequences are the exon sequences flanking intron 3). To test the effect of OsTubA1 intron 1 on a heterogeneous promoter, the intron-1 fragment was inserted between the 35S promoter and GUS, making pGA1945. Plasmid pGA1875, which contains a fusion between the 35S promoter and GUS, was used as a control.
To determine whether the OsTubA1 intron 1 acts as an enhancer-like element, pGA2054 and pGA2119 were constructed. The plasmid pGA2054 was constructed by inserting the internal region of the intron 1, which lacks both donor and acceptor sites, into the 5′-distal region of the OsTubA1 promoter in a forward orientation. The internal region of intron 1 was generated by PCR using oligomers 5′-GGAAGCTTGCGTCCCATCTCCCTCA-3′ and 5′-GCGTCGACAAAAATACATGTTAGCG-3′. The plasmid pGA2119 was constructed by inserting the DNA fragment containing the intron 1 after the GUS coding region. The PCR fragment was produced using 5′-GGAGCTCGCATCCAGGTACGGATCCGC-3′and 5′-GGGTACCATCAGCCTGAAAGGACAAAA-3′ as primers.
The cis-acting regulatory elements of the OsTubA1 intron 1 were studied by the construction of the truncated intron 1 from pGA1902. These plasmids, pGA2127-1 to -6, have deletion of the nucleotides 1,995 through 2,093, 1,884 through 2,093, 1,753 through 2,093, 1,630 through 2,093, 1,439 through 1,678, and 1,439 through 2,093 from the OsTubA1 intron 1 (nucleotides 1,249–2,194), respectively.
All constructs contain the nos terminator after the GUS gene and the 35S::hygomycin phosphotransferase gene for a plant selectable marker.
Agrobacterium-Mediated Rice Transformation
A japonica rice variety, Dongjin, was used for transformation by the Agrobacterium-mediated cocultivation method as described previously (Jeon et al., 1999; Lee et al., 1999).
GUS Assays
Fluorometric analysis was performed as described by Jefferson (1987) using 4-methylumbelliferyl-β-glucuronide. GUS values were expressed as picomole 4-methyl-umbelliferone (4-MU) min−1 mg−1 soluble protein. Ten independent callus lines cultured for 2 weeks on 2N6 media (N6 medium, 2 mg L−1 2,4-D, 30 g L−1 Suc, 1 g L−1 casamino acids, 2 g L−1 phytagel, pH 5.6–5.7) were used in the quantitative analysis (Lee et al., 1999). Histochemical GUS staining was performed according to Dai et al. (1996) except for addition of 20% (v/v) methanol in the staining solution. The reaction was stopped after 12 h, and samples were stored in 75% (v/v) ethanol.
ACKNOWLEDGMENTS
We thank Gi-Hwan Yi for sharing with us the Dongjin seeds, Chahm An for critical reading of the manuscript, Wen-biao Chen, David Finkel, Min Jung Han, and Woong Suk Yang for technical assistance, and Yong-Hwan Moon, Soon-Kee Sung, Hong-Gyu Kang, and Gyung-Hee Yu for helpful discussions.
Footnotes
This work was supported in part by grants from the Korea Agrobiotech Consortium and National Research Laboratory Program of the Korean Institute of Science and Technology Evaluation and Planning.
LITERATURE CITED
- Bolle C, Herrmann RG, Oelmüller R. Intron sequences are involved in the plastid- and light-dependent expression of the spinach PsaD gene. Plant J. 1996;10:919–924. doi: 10.1046/j.1365-313x.1996.10050919.x. [DOI] [PubMed] [Google Scholar]
- Busch MA, Bomblies K, Weigel D. Activation of a floral homeotic gene in Arabidopsis. Science. 1999;285:585–587. doi: 10.1126/science.285.5427.585. [DOI] [PubMed] [Google Scholar]
- Callis J, Fromm M, Walbot V. Introns increase gene expression in cultured maize cells. Genes Dev. 1987;1:1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- Carpenter JL, Kopczak SD, Snustad DP, Silflow CD. Semi-constitutive expression of an Arabidopsis thaliana α-tubulin gene. Plant Mol Biol. 1993;21:937–942. doi: 10.1007/BF00027126. [DOI] [PubMed] [Google Scholar]
- Chen R, Silver DL, de Bruijn FJ. Nodule parenchyma-specific expression of the Sesbania rostrata early nodulin gene SrEnod2 is mediated by its 3′ untranslated region. Plant Cell. 1998;10:1585–1602. doi: 10.1105/tpc.10.10.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm D. A convenient moderate-scale procedure for obtaining DNA from bacteriophage lambda. Biotechniques. 1989;7:21–23. [PubMed] [Google Scholar]
- Christensen AH, Sharrock RA, Quail PH. Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol. 1992;18:675–689. doi: 10.1007/BF00020010. [DOI] [PubMed] [Google Scholar]
- Chung Y-Y, Kim S-R, Finkel D, Yanofsky MF, An G. Early flowering and reduced apical dominance result from ectopic expression of a rice MADS box gene. Plant Mol Biol. 1994;26:657–665. doi: 10.1007/BF00013751. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1191–1195. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Gao J, An K, Lee JM, Edwards GE, An G. Promoter elements controlling developmental and environmental regulation of a tobacco ribosomal protein gene L34. Plant Mol Biol. 1996;32:1055–1065. doi: 10.1007/BF00041389. [DOI] [PubMed] [Google Scholar]
- Dickey LF, Nguyen T-T, Allen GC, Thompson WF. Light modulation of ferredoxin mRNA abundance requires an open reading frame. Plant Cell. 1994;6:1171–1176. doi: 10.1105/tpc.6.8.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Kim SY, Park WD. High-level tuber expression and sucrose inducibility of a potato Sus4 sucrose synthase gene require 5′ and 3′ flanking sequences and the leader intron. Plant Cell. 1995;7:1387–1394. doi: 10.1105/tpc.7.9.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley BA, Schuler MA. Plant intron sequences: evidence for distinct groups of introns. Nucleic Acids Res. 1988;16:7159–7176. doi: 10.1093/nar/16.14.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley BA, Schuler MA. Developmental expression of plant snRNAs. Nucleic Acids Res. 1991;19:6319–6325. doi: 10.1093/nar/19.22.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Webster CI, Gray JC. Light-regulated expression of the pea plastocyanin gene is mediated by elements within the transcribed region of the gene. Plant J. 1997;12:499–506. doi: 10.1046/j.1365-313x.1997.00499.x. [DOI] [PubMed] [Google Scholar]
- Hoekema A, Hirsch PR, Hooykaas PJJ, Schilperoort RA. A binary vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature. 1983;303:179–180. [Google Scholar]
- Huang MT, Gorman CM. Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990;18:937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Jeon J-S, Chung Y-Y, Lee S, Yi G-H, Oh B-G, An G. Isolation and characterization of an anther-specific gene, RA8, from rice (Oryza sativa L.) Plant Mol Biol. 1999;39:35–44. doi: 10.1023/a:1006157603096. [DOI] [PubMed] [Google Scholar]
- Joyce CM, Villemur R, Snustad DP, Silflow CD. Tubulin gene expression in maize (Zea mays L.): change in isotype expression along the developmental axis of seedling root. J Mol Biol. 1992;227:97–107. doi: 10.1016/0022-2836(92)90684-c. [DOI] [PubMed] [Google Scholar]
- Kang H-G, Jeon J-S, Lee S, An G. Identification of class B and class C floral organ identity genes from rice plants. Plant Mol Biol. 1998;38:1021–1029. doi: 10.1023/a:1006051911291. [DOI] [PubMed] [Google Scholar]
- Kim Y, An G. Pollen-specific expression of the Arabidopsis thaliana α1-tubulin promoter assayed by β-glucuronidase, chloramphenicol acetyltransferase and diphtheria toxin reporter genes. Transgenic Res. 1992;1:188–194. doi: 10.1007/BF02522538. [DOI] [PubMed] [Google Scholar]
- Kopczak SD, Haas NA, Hussey PJ, Silflow CD, Snustad DP. The small genome of Arabidopsis contains at least six expressed α-tubulin genes. Plant Cell. 1992;4:539–547. doi: 10.1105/tpc.4.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurachi S, Hitomi Y, Furukawa M, Kurachi K. Role of intron I in expression of the human factor IX gene. J Biol Chem. 1995;270:5276–5281. doi: 10.1074/jbc.270.10.5276. [DOI] [PubMed] [Google Scholar]
- Lee S, Jeon J-S, Jung K-H, An G. Binary vectors for efficient transformation of rice. J Plant Biol. 1999;42:310–316. [Google Scholar]
- Little M, Seehaus T. Comparative analysis of tubulin sequences. Comp Biochem Physiol. 1988;90:655–670. doi: 10.1016/0305-0491(88)90320-3. [DOI] [PubMed] [Google Scholar]
- Ludwig SR, Oppenheimer DG, Silflow CD, Snustad DP. The α1-tubulin gene of Arabidopsis thaliana: primary structure and preferential expression in flowers. Plant Mol Biol. 1988;10:311–321. doi: 10.1007/BF00029881. [DOI] [PubMed] [Google Scholar]
- Luehrsen KR, Walbot V. Intron enhancement of gene expression and splicing efficiency of introns in maize cells. Mol Gen Genet. 1991;225:81–93. doi: 10.1007/BF00282645. [DOI] [PubMed] [Google Scholar]
- Luehrsen KR, Walbot V. Addition of A- and U-rich sequence increases the splicing efficiency of a deleted form of a maize intron. Plant Mol Biol. 1994;24:449–463. doi: 10.1007/BF00024113. [DOI] [PubMed] [Google Scholar]
- Maas C, Laufs J, Grant S, Korfhage C, Werr W. The combination of a novel stimulatory element in the first exon of the maize Shrunken-1 gene with the following intron 1 enhances reporter gene expression up to 1000-fold. Plant Mol Biol. 1991;16:199–207. doi: 10.1007/BF00020552. [DOI] [PubMed] [Google Scholar]
- Marshall JS, Stubbs JD, Chitty JA, Surin B, Taylor WC. Expression of the C4 Me1 gene from Flaveria bidentis requires an interaction between 5′ and 3′ sequences. Plant Cell. 1997;9:1515–1525. doi: 10.1105/tpc.9.9.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas D, Mettler IJ, Pierce DA, Lowe HW. Intron-mediated enhancement of heterologous gene expression in maize. Plant Mol Biol. 1990;15:913–920. doi: 10.1007/BF00039430. [DOI] [PubMed] [Google Scholar]
- McElroy D, Zhang W, Cao J, Wu R. Isolation of an efficient actin promoter for use in rice transformation. Plant Cell. 1990;2:163–171. doi: 10.1105/tpc.2.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoliu L, Puigdomènech P, Rigau J. The Tubα3 gene from Zea mays: structure and expression in dividing plant tissues. Gene. 1990;94:201–207. doi: 10.1016/0378-1119(90)90388-8. [DOI] [PubMed] [Google Scholar]
- Montoliu L, Rigau J, Puigdomènch P. A tandem of α-tubulin genes preferentially expressed in radicular tissues from Zea mays. Plant Mol Biol. 1989;14:1–15. doi: 10.1007/BF00015650. [DOI] [PubMed] [Google Scholar]
- Montoliu L, Rigau J, Puigdomènch P. Analysis by PCR of the number of homologous genomic sequences to α-tubulin in maize. Plant Sci. 1992;84:179–185. [Google Scholar]
- Oard JH, Paige D, Dvorak J. Chimeric gene expression using maize intron in cultured cells of bread wheat. Plant Cell Rep. 1989;8:156–160. doi: 10.1007/BF00716830. [DOI] [PubMed] [Google Scholar]
- Qin X, Gianì S, Breviario D. Molecular cloning of three rice α-tubulin isotypes: differential expression in tissues and during flower development. Biochim Biophys Acta. 1997;1354:19–23. doi: 10.1016/s0167-4781(97)00110-3. [DOI] [PubMed] [Google Scholar]
- Rigau J, Capellades M, Montoliu L, Torres MA, Romera C, Martínez-Izquierdo JA, Tagu D, Puigdomènech P. Analysis of a maize α-tubulin gene promoter by transient expression and in transgenic tobacco plants. Plant J. 1993;4:1043–1050. [Google Scholar]
- Rose AB, Last RL. Introns act post-transcriptionally to increase expression of the Arabidopsis thaliana tryptophan pathway gene PAT1. Plant J. 1997;11:455–464. doi: 10.1046/j.1365-313x.1997.11030455.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequence with chain-termination inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow CD, Chrisholm RL, Conner TW, Ranum LPW. The two α-tubulin genes of Chlamydomonas reinhardtii code for slightly different proteins. Mol Cell Biol. 1985;5:2389–2398. doi: 10.1128/mcb.5.9.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Filipowicz W. Splicing of precursors to mRNA in higher plants: mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol Biol. 1996;32:1–41. doi: 10.1007/BF00039375. [DOI] [PubMed] [Google Scholar]
- Snowden KC, Buchhholz WG, Hall TC. Intron position affects expression from the tpi promoter in rice. Plant Mol Biol. 1996;31:689–692. doi: 10.1007/BF00042241. [DOI] [PubMed] [Google Scholar]
- Stöcker M, Garcia-Mas J, Arús P, Messeguer R, Puigdomènech P. A highly conserved α-tubulin sequence from Prunus amygdalus. Plant Mol Biol. 1993;22:913–916. doi: 10.1007/BF00027377. [DOI] [PubMed] [Google Scholar]
- Uribe X, Torres MA, Capellades M, Puigdomènech P, Rigau J. Maize α-tubulin genes are expressed according to specific patterns of cell differentiation. Plant Mol Biol. 1998;37:1069–1078. doi: 10.1023/a:1006067710312. [DOI] [PubMed] [Google Scholar]
- Vasil V, Clancy M, Ferl RJ, Vasil I, Hannah LC. Increased gene expression by the first intron of maize Shrunken-1 locus in grass species. Plant Physiol. 1989;91:1575–1579. doi: 10.1104/pp.91.4.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemur R, Joyce CM, Haas NA, Goddard RH, Kopczak SD, Hussey PJ, Snustad DP, Silflow CD. α-Tubulin gene family of maize (Zea mays L.): evidence for two ancient α-tubulin genes in plants. J Mol Biol. 1992;227:81–96. doi: 10.1016/0022-2836(92)90683-b. [DOI] [PubMed] [Google Scholar]
- Xu Y, Yu H, Hall TC. Rice triosephosphate isomerase gene 5′ sequence directs β-glucuronidase activity in transgenic tobacco but requires an intron for expression in rice. Plant Physiol. 1994;106:459–467. doi: 10.1104/pp.106.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Kondo Y, Kato A, Tsuji H, Obokata J. Light-responsive elements of the tobacco PSI-D gene are located both upstream and within the transcribed region. Plant J. 1997;12:255–265. doi: 10.1046/j.1365-313x.1997.12020255.x. [DOI] [PubMed] [Google Scholar]
- Zhang W, McElroy D, Wu R. Analysis of rice Act1 5′ region activity in transgenic rice plants. Plant Cell. 1991;3:1155–1165. doi: 10.1105/tpc.3.11.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]