Abstract
Single-nucleotide polymorphisms (SNPs) in the Nogo-A receptor gene (RTN4R) have been associated with increased risk for sporadic amyotrophic lateral sclerosis (SALS) in the French population. In the present study, we investigated the associations between RTN4R tag SNPs and SALS in a large Chinese population. Four tag SNPs (rs854971, rs887765, rs696880 and rs1567871) in the RTN4R gene with an r2 threshold of 0.8 and a minor allele frequency (MAF) greater than 0.2% were selected based on Chinese population data from HapMap. A total of 499 SALS patients and 503 healthy controls were genotyped for the SNPs by SNaPshot technology. Haplotype analysis of the four SNPs was performed using the SHEsis software platform. The results showed a significant association between the rs696880 risk allele (A) and SALS in the Han Chinese population (P = 0.009, odds ratio (OR) = 1.266 [1.06–1.51]). The allele and genotype frequencies of rs854971, rs887765 and rs1567871 were not associated with SALS. The distribution of the GAAT haplotype was different between the case and control groups (P = 0.008, OR = 1.289 [1.066–1.558]). In conclusion, our study showed an association between the RTN4R SNP rs696880 and the risk of SALS in the Han Chinese population, with the A allele increasing risk.
Keywords: Nogo-A receptor, reticulon 4 receptor gene (RTN4R), single-nucleotide polymorphisms, amyotrophic lateral sclerosis, Chinese population
Introduction
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease involving brain and spinal cord motor neurons (Wijesekera and Leigh, 2009). Although the incidence of ALS is low, the disease is fatal and has no effective treatment. The disease is considered to result from the interaction of genetics, environment and aging. Gene mutations are a well-known and significant cause of ALS. A twin study estimated that the effect of heritability on ALS is 0.61 and that of an unshared environment is 0.39 (Al-Chalabi et al., 2010). Therefore, genes play a major role in ALS. At least 25 causative genes have been described in ALS, including SOD1, DCTN1, ANG, VAPB, FUS, TARDBP, UBQLN2, PFN1, HNRNPA1, C9orf72, SQSTM1, OPTN, VCP, ATXN2, ARHGEF28, TUBA4A, MATR3, CHCHD10, TBK1, NEKI and CCNF (Brown and Al-Chalabi, 2017; Chia et al., 2018). Among them, SOD1, TARDBP and C9orf72 are the most common causative genes. Due to differences in genetic backgrounds among different ethnic groups, a difference exists in the frequency of disease-causing gene mutations in ALS patients. Zou et al. (2017) performed a meta-analysis of the mutation frequency of these four common genes and found that in the European population, the most common pathogenic gene was C9orf72 (familial ALS [FALS] 33.7%, sporadic ALS [SALS] 5.1%), followed by SOD1 (FALS 14.8%, SALS 1.2%), TARDBP (FALS 4.2%, SALS 0.8%) and FUS (FALS 2.8%, SALS 0.3%). However, in the Asian population, SOD1 (FALS 30.0%, SALS 1.5%) was the most common gene, followed by FUS (FALS 6.4%, SALS 0.9%), C9orf72 (FALS 2.3%, SALS 0.3%) and TARDBP (FALS 1.5%, SALS 0.2%).
In our previous study, the mutation rates of SOD1, FUS, TARDBP and C9orf72 in FALS patients were 34.5%, 9.5%, 4.3% and 0%, respectively (Liu R. et al., 2013). The mutation rates of SOD1, FUS, TARDBP and C9orf72 in SALS patients were 2.2%, 3.0%, 1.1% and 0.3%, respectively (He et al., 2015). The mutation rates of other genes were as follows: ATXN2, 1.6% (Liu X. et al., 2013); ARHGEF28, 0.56% (Ma et al., 2014); SQSTM1, 1.38% (Yang et al., 2015); OPTN, 0.78% (Li C. et al., 2015); TUBA4A, 0% (Li J. et al., 2015); MATR3, 0.19% (Xu et al., 2016); DCTN1, 0.39% (Liu et al., 2017); UBQLIN, 0.19% (Huang et al., 2017); and CHCHD1, 0.23% (Shen et al., 2017).
Approximately 10% of ALS cases are FALS, while approximately 90% of cases are SALS. The cause of SALS remains unknown. SALS may result from the interaction between susceptible genes and the environment; therefore, identifying SALS susceptibility genes is as important as identifying pathogenic genes. Several previous studies have identified different susceptibility genes in various ethnic cohorts, including DPP6, ITPR2, UNC13A, FGGY, ELP3, KIFAP3, 9p21.2, ZNF512B, TIMA1, SCNN1A, BDNF, C21orf2, MOBP, SCFD1 and GPX3-TNIP1 (van Es et al., 2007, 2008, 2009a,b; Cronin et al., 2008; Landers et al., 2009; Simpson et al., 2009; Iida et al., 2011; Cai et al., 2014; Chen et al., 2014; van Rheenen et al., 2016; Xu et al., 2017).
Although many ALS-related genes have been identified, approximately 44% of FALS and 91% of SALS cases in European populations and 59% of FALS and 96% of SALS cases in Asian populations still have no clear pathogenesis (Zou et al., 2017). Therefore, the genetic study of pathogenic genes in Chinese ALS patients is of great significance.
Recently, two non-coding variants (rs701427 A and rs1567871 C) in the Nogo-A receptor gene (RTN4R) were reported to confer susceptibility to SALS in the French population (Amy et al., 2015). Moreover, rs701427 showed a significant correlation with reduced gene expression. The study concluded that reduced expression of the Nogo receptor was associated with SALS. In this study, we investigated the associations between RTN4R tag single-nucleotide polymorphisms (SNPs) and SALS in a large Chinese population.
Materials and Methods
Study Population
This study consisted of 499 SALS patients and 503 healthy control subjects. All participants were of Han Chinese origin. Diagnosis of ALS was performed at Peking University Third Hospital using the E1 Escorial Word Federation criteria for definite or probable ALS (Brooks et al., 2000). This study was carried out in accordance with the recommendations of Peking University Third Hospital ethics committee with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the Peking University Third Hospital ethics committee.
SNP Selection
Based on the Chinese population data from the HapMap database (HapMap Data Rel 27 Phase II+III, Feb 2009, on NCBI B36 assembly, dbSNP126), candidate tag SNPs in RTN4R with an r2 threshold of 0.8 and a minor allele frequency (MAF) greater than 0.2% were selected.
SNP Analysis
Genomic DNA was extracted from leukocytes of venous blood using the phenol-chloroform method. The sense and antisense primers for SNPs are shown in Table 1. Primers were used to amplify the target SNPs, followed by agarose gel electrophoresis and product recovery. The product was primer-extended by one base and terminated. It was detected by an ABI sequencer at Tsingke Biotechnology Co. (Beijing, China) according to the ABI PRISM® SNaPshot™ Multiplex Kit Protocol.
Table 1.
The primes of the single-nucleotide polymorphisms (SNPs).
| SNP | Sense primers | Antisense primers |
|---|---|---|
| rs854971 | CCCACTTTCCACTCTCACCC | GGAGGAGACCCTGGAGGAAA |
| rs887765 | GGAGAATTTTGGTTGGGCCTAGA | CTGACCCTCTGTGGTGTGGG |
| rs696880 | ATCCTCAGGCCCACAGACAC | GGTCCAGTGACCTCTCACCA |
| rs1567871 | CCCAGAGAAAGGGGTTCCAC | ATGCCCAGTGGAGGACTG |
Statistical Analysis
The chi-square test was used to compare the allele frequencies and genotype differences between the case and control groups. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to assess the risk of genetic polymorphisms. Haplotype analysis was performed with the SHEsis software platform1 (Shi and He, 2005; Li et al., 2009). The original P value was multiplied by the number of comparisons to obtain a Bonferroni-corrected P value. Corrected P-values less than 0.05 were considered significant.
Functional Prediction
The functions of positive loci were analyzed using MATCH software.
Results
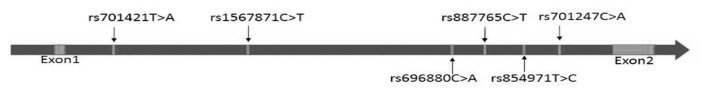
A total of four SNPs of RTN4R (rs854971, rs887765, rs696880 and rs1567871) were selected for further genotyping. All four SNPs were located in the intron between exon 1 and exon 2 (Figure 1).
Figure 1.
Genomic structure and localization of polymorphisms in the RTN4R gene.
This study included 499 SALS patients with a male-to-female ratio of 1.80 and average age of onset of 51.3 ± 11.9 years. The study also included 503 control subjects, with a male-to-female ratio of 1.30 and a mean age of 53.5 ± 12.8 years. No difference was observed between the patients and control subjects in age or sex.
The four polymorphisms (rs854971, rs887765, rs696880 and rs1567871) were in Hardy-Weinberg equilibrium in SALS patients and control subjects.
The rs696880 A allele (P = 0.009, OR = 1.266 [1.06–1.51]) and rs1567871 T allele (P = 0.045, OR = 1.204 [1.00–1.44]) were associated with SALS in terms of allele and genotype frequencies (Table 2). Since four SNPs were included in the study, the Bonferroni-corrected P value of the allele or genotype comparison was the original P value multiplied by 4. Hence, only the rs696880 allele distributions was significantly correlated with SALS after Bonferroni correction. The allele and genotype frequencies of rs854971 (P = 0.053, OR = 0.836 [0.69–1.01]) and rs887765 (P = 0.112, OR = 1.161 [0.96–1.39]]) were not associated with SALS.
Table 2.
Allele and genotype frequencies of the four SNPs within RTN4R.
| SNP | Samples | n | Allelic (frequency) | Pa(χ2) | OR (95%CI) | Genotype (frequency) | Pb(χ2) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| A | G | AA | AG | GG | ||||||
| rs854971 | Cases | 499 | 357 (0.360) | 641 (0.642) | 0.053 | 0.836 [0.69~1.01] | 66 (0.132) | 225 (0.450) | 208 (0.416) | 0.162 |
| controls | 503 | 402 (0.400) | 604 (0.600) | (3.736) | 89 (0.177) | 224 (0.445) | 190 (0.378) | (3.631) | ||
| A | G | AA | AG | GG | ||||||
| rs887765 | Cases | 499 | 357 (0.357) | 641 (0.642) | 0.112 | 1.161 [0.96~1.39] | 59 (0.123) | 239 (0.484) | 201 (0.393) | 0.067 |
| controls | 503 | 326 (0.324) | 680 (0.676) | (2.527) | 57 (0.113) | 212 (0.421) | 234 (0.465) | (5.413) | ||
| A | G | AA | AG | GG | ||||||
| rs696880 | Cases | 499 | 489 (0.490) | 509 (0.510) | 0.009 | 1.266 [1.06~1.51] | 117 (0.234) | 255 (0.511) | 127 (0.254) | 0.027 |
| controls | 503 | 434 (0.431) | 572 (0.569) | (6.917) | 95 (0.189) | 244 (0.485) | 164 (0.326) | (7.214) | ||
| C | T | CC | CT | TT | ||||||
| rs1567871 | Cases | 499 | 574 (0.575) | 424 (0.425) | 0.045 | 1.204 [1.00~1.44] | 158 (0.320) | 258 (0.506) | 83 (0.174) | 0.018 |
| controls | 503 | 585 (0.626) | 359 (0.374) | (4.002) | 202 (0.402) | 225 (0.448) | 75 (0.149) | (8.029) | ||
ad.f. = 1; bd.f. = 2. d.f, degree of freedom. P value in the table is the original P value.
Using the method of increasing sensitivity with the lowest false-negative rate, MATCH software predicted that if allele A replaced G in rs696880, one more cooperates with myogenic proteins 1 (COMP1) transcription factor binding site would be expected.
Haplotype frequencies were calculated using the SHEsis software platform. According to the sequence of rs854971-rs887765-rs696880-rs1567871, the AGGC, GAAT, GGAC, GGAT and GGGC haplotypes, with the smallest haplotype frequency >0.03, were included in the study as common haplotypes. The GAAT haplotype and non-GGAT haplotypes differed between the case group and the control group (P = 0.008). Because the GAAT haplotype was involved in five multiple comparisons, P = 0.008 × 5 = 0.04 after Bonferroni correction; therefore, this difference was still significant (Table 3).
Table 3.
Haplotype frequencies of the tag SNPs within RTN4R.
| Haplotypea | Case (freq)b | Control (freq)b | χ2 | Pearson’s P | Odds ratio[95%] |
|---|---|---|---|---|---|
| AGGC | 349.47 (0.345) | 374.75 (0.373) | 3.196 | 0.073 | 0.845 [0.703~1.016] |
| GAAT | 353.95 (0.350) | 288.21 (0.287) | 6.880 | 0.008 | 1.289 [1.066~1.558] |
| GGAC | 57.93 (0.057) | 47.34 (0.050) | 0.766 | 0.381 | 1.193 [0.804~1.770] |
| GGAT | 70.62 (0.070) | 74.84 (0.075) | 0.343 | 0.558 | 0.904 [0.645~1.268] |
| GGGC | 156.38 (0.153) | 168.84 (0.168) | 1.226 | 0.268 | 0.874 [0.689~1.109] |
aThe sequence of the SNPs (rs854971-rs887765-rs696880-rs1567871). bThe frequencies of haplotype were estimated using SHEsis platform software. Common haplotype included in this study were selected with minor frequency greater than 0.03.
Discussion
The tag SNPs in the RTN4R gene are different between the Han Chinese population and the French population. Four SNPs, rs854971, rs887765, rs696880 and rs1567871, were included in our study. Three SNPs, rs701427, rs701421 and rs1567871, were included in the French study (Amy et al., 2015). The MAFs of rs701427 and rs701421 were both lower than 0.2% in the Han Chinese population; therefore, these two SNPs were not included in our study. Only the rs1567871 SNP was included in both studies. In Amy et al.’s (2015) study, the rs1567871 C allele was associated with SALS in the French population. However, the rs1567871 C allele was not a risk allele for SALS in the Chinese population. This difference may be attributable to ethnic factors. The C9orf72 gene frequency was high in European and American ALS patients (FALS 33.7%, SALS 5.1%) but low in Asian ALS patients (FALS 2.3%, SALS 0.3%; Zou et al., 2017). The frequency of OPTN mutations was high in Japanese and Chinese populations (FALS 3.3%–3.8%, SALS 0.23%–1%), whereas these mutations occurred infrequently in Caucasian patients (Li C. et al., 2015).
Nogo-A is a myelin growth inhibitor protein that exerts an inhibitory effect by binding to the Nogo-A receptor (NgR1). NgR1 is a glycosylphosphatidylinositol (GPI)-linker that is rich in leucine-rich repeats (LRRs) and is encoded by the endoplasmic reticulum-4 receptor (RTN4R) gene. NgR1 forms a receptor complex with Lingo-1, p75NTR and Troy, binding to the growth inhibitory protein Nogo-A, oligodendrocyte myelin-associated glycoprotein (OMG), and myelin-binding glycoprotein myelin-associated glycoprotein (MAG; Schmandke et al., 2014).
Rho-A, a GTP-binding protein, can activate the signal transduction that regulates NgR1 in neurons, leading to growth inhibition and congenital atrophy (Fournier et al., 2002; GrandPré et al., 2002). Motor neuron growth inhibition leads to denervation of muscles and is an important pathological mechanism of ALS (Jokic et al., 2006; Steele and Yi, 2006). However, the NgR1 receptor may also regulate the complex role of maintaining motor neuron survival in ALS patients by blocking the p75NTR-mediated neuronal death induced by nerve growth factor (NGF; Dupuis et al., 2008). This shows that the Nogo-A/NgR1 signaling pathway and the complex role of NgR1 in ALS are closely related (Teng and Tang, 2008; Schmandke et al., 2014). Amy et al. (2015) concluded that expression of the Nogo receptor is positively associated with SALS.
Our study suggests that the rs696880 A allele in the RTN4R gene increases the risk of SALS in the Han Chinese population. However, the mechanism is not clear. The rs696880 SNP, located in an enhancer element of the RTN4R gene, may regulate RTN4R gene transcription by changing COMP1 transcription factor binding sites. As concluded by Amy et al. (2015), the A allele of rs696880 may reduce transcription of the RTN4R gene, but this requires further functional verification.
The above findings demonstrate that Nogo-A/NgR1 contributes to the neuropathology of ALS. Variation at the NgR locus is associated with schizophrenia (Hsu et al., 2007; Budel et al., 2008; Voineskos, 2009; Jitoku et al., 2011; Willi and Schwab, 2013). Hence, our data provide additional evidence for the view that a genetic overlap exists between ALS and schizophrenia (Byrne et al., 2013; Fahey et al., 2014).
To the best of our knowledge, this is the first analysis of RTN4R tag SNPs in Han Chinese patients with SALS. This case-control study may provide an association between the Nogo-A receptor gene SNP rs696880 and the risk of SALS in the Han Chinese population, with the A allele increasing risk. However, the results still need to be verified in a larger study. Larger studies in different ethnicities are also needed to confirm and extend our findings.
Author Contributions
DF conceived this study and provided financial support and was responsible for project management. LX, JL and DT performed the experiments and analyzed the data. LC and LT conducted data management and undertook data checking. LX and DF wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the National Natural Science Foundation of China.
Funding. This work was supported by the National Natural Science Foundation of China (81030019).
References
- Al-Chalabi A., Fang F., Hanby M. F., Leigh P. N., Shaw C. E., Ye W., et al. (2010). An estimate of amyotrophic lateral sclerosis heritability using twin data. J. Neurol. Neurosurg. Psychiatry 81, 1324–1326. 10.1136/jnnp.2010.207464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amy M., Staehlin O., René F., Blasco H., Marouillat S., Daoud H., et al. (2015). A common functional allele of the Nogo receptor gene, reticulon 4 receptor (RTN4R), is associated with sporadic amyotrophic lateral sclerosis in a French population. Amyotroph. Lateral Scler. Frontotemporal. Degener. 16, 490–496. 10.3109/21678421.2015.1051988 [DOI] [PubMed] [Google Scholar]
- Brooks B. R., Miller R. G., Swash M., Munsat T. L. (2000). El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Frontotemporal. Degener. 1, 293–299. 10.1080/146608200300079536 [DOI] [PubMed] [Google Scholar]
- Brown R. H., Al-Chalabi A. (2017). Amyotrophic lateral sclerosis. N. Engl. J. Med. 377, 162–172. 10.1056/NEJMra1603471 [DOI] [PubMed] [Google Scholar]
- Budel S., Padukkavidana T., Liu B. P., Feng Z., Hu F., Johnson S., et al. (2008). Genetic variants of Nogo-66 receptor with possible association to schizophrenia block myelin inhibition of axon growth. J. Neurosci. 28, 13161–13172. 10.1523/JNEUROSCI.3828-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne S., Heverin M., Elamin M., Bede P., Lynch C., Kenna K., et al. (2013). Aggregation of neurologic and neuropsychiatric disease in amyotrophic lateral sclerosis kindreds: a population-based case-control cohort study of familial and sporadic amyotrophic lateral sclerosis. Ann. Neurol. 74, 699–708. 10.1002/ana.23969 [DOI] [PubMed] [Google Scholar]
- Cai B., Tang L., Zhang N., Fan D. (2014). The single-nucleotide polymorphism rs6690993 in FGGY is not associated with amyotrophic lateral sclerosisin a large Chinese cohort. Neurobiol. Aging 35, 1512.e3–1512.e4. 10.1016/j.neurobiolaging.2013.12.018 [DOI] [PubMed] [Google Scholar]
- Chen X., Huang R., Chen Y., Zheng Z., Chen K., Song W., et al. (2014). Association analysis of four candidate genetic variants with sporadic amyotrophic lateral sclerosis in a Chinese population. Neurol. Sci. 35, 1089–1095. 10.1007/s10072-014-1656-1 [DOI] [PubMed] [Google Scholar]
- Chia R., Chiò A., Traynor B. J. (2018). Novel genes associated with amyotrophic lateral sclerosis: diagnostic and clinical implications. Lancet Neurol. 17, 94–102. 10.1016/s1474-4422(17)30401-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin S., Berger S., Ding J., Schymick J. C., Washecka N., Hernandez D. G., et al. (2008). A genome-wide association study of sporadic ALS in a homogenous Irish population. Hum. Mol. Genet. 17, 768–774. 10.1093/hmg/ddm361 [DOI] [PubMed] [Google Scholar]
- Dupuis L., Pehar M., Cassina P., Rene F., Castellanos R., Rouaux C., et al. (2008). Nogo receptor antagonizes p75NTR-dependent motor neuron death. Proc. Natl. Acad. Sci. U S A 105, 740–745. 10.1073/pnas.0703842105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey C., Byrne S., McLaughlin R., Kenna K., Shatunov A., Donohoe G., et al. (2014). Analysis of the hexanucleotide repeat expansion and founder haplotype at C9ORF72 in an Irish psychosis case-control sample. Neurobiol. Aging 35, 1510.e1–1510.e5. 10.1016/j.neurobiolaging.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Fournier A. E., Gould G. C., Liu B. P., Strittmatter S. M. (2002). Truncated soluble Nogo receptor binds Nogo-66 and blocks inhibition of axon growth by myelin. J. Neurosci. 22, 8876–8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GrandPré T., Li S., Strittmatter S. M. (2002). Nogo-66 receptor antagonist peptide promotes axonal regeneration. Nature 417, 547–551. 10.1038/417547a [DOI] [PubMed] [Google Scholar]
- He J., Tang L., Benyamin B., Shah S., Hemani G., Liu R., et al. (2015). C9orf72 hexanucleotide repeat expansions in Chinese sporadic amyotrophic lateral sclerosis. Neurobiol. Aging 36, 2660.e1–2660.e8. 10.1016/j.neurobiolaging.2015.06.0020 [DOI] [PubMed] [Google Scholar]
- Hsu R., Woodroffe A., Lai W.-S., Cook M. N., Mukai J., Dunning J. P., et al. (2007). Nogo receptor 1 (RTN4R) as a candidate gene for schizophrenia: analysis using human and mouse genetic approaches. PLoS One 2:e1234. 10.1371/journal.pone.0001234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Shen S., Fan D. (2017). No evidence for pathogenic role of UBQLN2 mutations in sporadic amyotrophic lateral sclerosis in the Mainland Chinese population. PLoS One 12:e0170943. 10.1371/journal.pone.0170943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida A., Takahashi A., Kubo M., Saito S., Hosono N., Ohnishi Y., et al. (2011). A functional variant in ZNF512B is associated with susceptibility to amyotrophic lateral sclerosis in Japanese. Hum. Mol. Genet. 20, 3684–3692. 10.1093/hmg/ddr268 [DOI] [PubMed] [Google Scholar]
- Jitoku D., Hattori E., Iwayama Y., Yamada K., Toyota T., Kikuchi M., et al. (2011). Association study of Nogo-related genes with schizophrenia in a Japanese case-control sample. Am. J. Med. Genet. B Neuropsychiatr. Genet. 156B, 581–592. 10.1002/ajmg.b.31199 [DOI] [PubMed] [Google Scholar]
- Jokic N., Gonzalez de Aguilar J.-L., Dimou L., Lin S., Fergani A., Ruegg M. A., et al. (2006). The neurite outgrowth inhibitor Nogo-A promotes denervation in an amyotrophic lateral sclerosis model. EMBO Rep. 7, 1162–1167. 10.1038/sj.embor.7400826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landers J. E., Melki J., Meininger V., Glass J. D., van den Berg L. H., van Es M. A., et al. (2009). Reduced expression of the Kinesin-Associated Protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U S A 106, 9004–9009. 10.1073/pnas.0812937106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., He J., Tang L., Chen L., Xu L., Ma Y., et al. (2015). TUBA4A may not be a significant genetic factor in Chinese ALS patients. Amyotroph. Lateral Scler. Frontotemporal. Degener. 17, 148–150. 10.3109/21678421.2015.1074705 [DOI] [PubMed] [Google Scholar]
- Li C., Ji Y., Tang L., Zhang N., He J., Ye S., et al. (2015). Optineurin mutations in patients with sporadic amyotrophic lateral sclerosis in China. Amyotroph. Lateral Scler. Frontotemporal. Degener. 16, 485–489. 10.3109/21678421.2015.1089909 [DOI] [PubMed] [Google Scholar]
- Li Z., Zhang Z., He Z., Tang W., Li T., Zeng Z., et al. (2009). A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res. 19, 519–523. 10.1038/cr.2009.33 [DOI] [PubMed] [Google Scholar]
- Liu X., Lu M., Tang L., Zhang N., Chui D., Fan D. (2013). ATXN2 CAG repeat expansions increase the risk for Chinese patients with amyotrophic lateral sclerosis. Neurobiol. Aging 34, 2236.e5–2236.e8. 10.1016/j.neurobiolaging.2013.04.009 [DOI] [PubMed] [Google Scholar]
- Liu R., Tang L., Cai B., Liu X., Ye S., Ma Y., et al. (2013). C9orf72 repeat expansions are not detected in Chinese patients with familial ALS. Amyotroph. Lateral Scler. Frontotemporal. Degener. 14, 630–631. 10.3109/21678421.2013.817588 [DOI] [PubMed] [Google Scholar]
- Liu X., Yang L., Tang L., Chen L., Liu X., Fan D. (2017). DCTN1 gene analysis in Chinese patients with sporadic amyotrophic lateral sclerosis. PLoS One 12:e0182572. 10.1371/journal.pone.0182572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Tang L., Chen L., Zhang B., Deng P., Wang J., et al. (2014). ARHGEF28 gene exon 6/intron 6 junction mutations in Chinese amyotrophic lateral sclerosis cohort. Amyotroph. Lateral Scler. Frontotemporal. Degener. 15, 309–311. 10.3109/21678421.2014.896926 [DOI] [PubMed] [Google Scholar]
- Schmandke A., Schmandke A., Schwab M. E. (2014). Nogo-A: multiple roles in CNS development, maintenance, and disease. Neuroscientist 20, 372–386. 10.1177/1073858413516800 [DOI] [PubMed] [Google Scholar]
- Shen S., He J., Tang L., Zhang N., Fan D. (2017). CHCHD10 mutations in patients with amyotrophic lateral sclerosis in Mainland China. Neurobiol. Aging 54, 214.e7–214.e10. 10.1016/j.neurobiolaging.2017.02.011 [DOI] [PubMed] [Google Scholar]
- Shi Y. Y., He L. (2005). SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction and genetic association at polymorphism loci. Cell Res. 15, 97–98. 10.1038/sj.cr.7290272 [DOI] [PubMed] [Google Scholar]
- Simpson C. L., Lemmens R., Miskiewicz K., Broom W. J., Hansen V. K., van Vught P. W. J., et al. (2009). Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum. Mol. Genet. 18, 472–481. 10.1093/hmg/ddn375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele A. D., Yi C. H. (2006). Neuromuscular denervation: bax up against the wall in amyotrophic lateral sclerosis. J. Neurosci. 26, 12849–12851. 10.1523/JNEUROSCI.4086-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F. Y. H., Tang B. L. (2008). Nogo-A and Nogo-66 receptor in amyotrophic lateral sclerosis. J. Cell. Mol. Med. 12, 1199–1204. 10.1111/j.1582-4934.2008.00351.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es M. A., van Vught P. W., Blauw H. M., Franke L., Saris C. G., Andersen P. M., et al. (2007). ITPR2 as a susceptibility gene in sporadic amyotrophic lateral sclerosis: a genome-wide association study. Lancet Neurol. 6, 869–877. 10.1016/S1474-4422(07)70222-3 [DOI] [PubMed] [Google Scholar]
- van Es M. A., van Vught P. W., Veldink J. H., Andersen P. M., Birve A., Lemmens R., et al. (2009a). Analysis of FGGY as a risk factor for sporadic amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 10, 441–447. 10.3109/17482960802673042 [DOI] [PubMed] [Google Scholar]
- van Es M. A., Veldink J. H., Saris C. G. J., Blauw H. M., van Vught P. W. J., Birve A., et al. (2009b). Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat. Genet. 41, 1083–1087. 10.1038/ng.442 [DOI] [PubMed] [Google Scholar]
- van Es M. A., van Vught P. W. J., Blauw H. M., Franke L., Saris C. G. J., van den Bosch L., et al. (2008). Genetic variation in DPP6 is associated with susceptibility to amyotrophic lateral sclerosis. Nat. Genet. 40, 29–31. 10.1038/ng.2007.52 [DOI] [PubMed] [Google Scholar]
- van Rheenen W., Shatunov A., Dekker A. M., McLaughlin R. L., Diekstra F. P., Pulit S. L., et al. (2016). Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat. Genet. 48, 1043–1048. 10.1038/ng.3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineskos A. N. (2009). Converging evidence for the Nogo-66 receptor gene in schizophrenia. J. Neurosci. 29, 5045–5047. 10.1523/JNEUROSCI.0477-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijesekera L. C., Leigh P. N. (2009). Amyotrophic lateral sclerosis. Orphanet J. Rare Dis. 4:3. 10.1186/1750-1172-4-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willi R., Schwab M. E. (2013). Nogo and Nogo receptor: relevance to schizophrenia? Neurobiol. Dis. 54, 150–157. 10.1016/j.nbd.2013.01.011 [DOI] [PubMed] [Google Scholar]
- Xu L., Li J., Tang L., Zhang N., Fan D. (2016). MATR3 mutation analysis in a Chinese cohort with sporadic amyotrophic lateral sclerosis. Neurobiol. Aging 38, 218.e3–218.e4. 10.1016/j.neurobiolaging.2015.11.023 [DOI] [PubMed] [Google Scholar]
- Xu L., Tan D., Li J., Chen L., Tang L., Zhang N., et al. (2017). The analysis of two BDNF polymorphisms G196A/C270T in Chinese sporadic amyotrophic lateral sclerosis. Front. Aging Neurosci. 9:135. 10.3389/fnagi.2017.00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Tang L., Zhang N., Pan L., Hadano S., Fan D. (2015). Six SQSTM1 mutations in a Chinese amyotrophic lateral sclerosis cohort. Amyotroph. Lateral Scler. Frontotemporal. Degener. 16, 378–384. 10.3109/21678421.2015.1009466 [DOI] [PubMed] [Google Scholar]
- Zou Z.-Y., Zhou Z.-R., Che C.-H., Liu C.-Y., He R.-L., Huang H.-P. (2017). Genetic epidemiology of amyotrophic lateral sclerosis: a systematic review and meta-analysis. J. Neurol. Neurosurg. Psychiatry 88, 540–549. 10.1136/jnnp-2016-315018 [DOI] [PubMed] [Google Scholar]