Abstract
Acinetobacter baumannii is one of the major causes of hard to treat multidrug-resistant hospital infections. A. baumannii features contributing to its spread and persistence in clinical environment are only beginning to be explored. Bacterial toxin-antitoxin (TA) systems are genetic loci shown to be involved in plasmid maintenance and proposed to function as components of stress response networks. Here we present a thorough characterization of type II system of A. baumannii, which is the most ubiquitous TA module present in A. baumannii plasmids. higBA of A. baumannii is a reverse TA (the toxin gene is the first in the operon) and shows little homology to other TA systems of RelE superfamily. It is represented by two variants, which both are functional albeit exhibit strong difference in sequence conservation. The higBA2 operon is found on ubiquitous 11 Kb pAB120 plasmid, conferring carbapenem resistance to clinical A. baumannii isolates and represents a higBA variant that can be found with multiple sequence variations. We show here that higBA2 is capable to confer maintenance of unstable plasmid in Acinetobacter species. HigB2 toxin functions as a ribonuclease and its activity is neutralized by HigA2 antitoxin through formation of an unusually large heterooligomeric complex. Based on the in vivo expression analysis of gfp reporter gene we propose that HigA2 antitoxin and HigBA2 protein complex bind the higBA2 promoter region to downregulate its transcription. We also demonstrate that higBA2 is a stress responsive locus, whose transcription changes in conditions encountered by A. baumannii in clinical environment and within the host. We show elevated expression of higBA2 during stationary phase, under iron deficiency and downregulated expression after antibiotic (rifampicin) treatment.
Keywords: toxin-antitoxin, HigBA, plasmid maintenance, Acinetobacter baumannii, protein complex
Introduction
Acinetobacter baumannii is an emerging Gram-negative opportunistic pathogen, causing serious hospital-acquired infections (Antunes et al., 2014). These bacteria are well adapted to survive in hospital environment such as intensive care units, burn wards, and field hospitals (Rosa et al., 2014). During the last decade, several highly successful multidrug-resistant A. baumannii clonal lineages have spread in clinical settings worldwide causing difficult to treat hospital outbreaks (Howard et al., 2012). A. baumannii is known for its ability to withstand harsh environmental conditions such as prolonged periods of dryness, disinfectants, and antibiotic treatment (Jawad et al., 1998; Cardoso et al., 2010; Nwugo et al., 2012).
Bacterial type II toxin-antitoxin (TA) systems are the most ubiquitous among six types of prokaryotic toxin-antitoxin systems (TAs), known to date (Chan et al., 2016; Page and Peti, 2016; Rocker and Meinhart, 2016). They are encoded on the low copy plasmids or chromosomes and code for two proteins, one of which (toxin) is toxic to the cell, whereas the other (antitoxin) neutralizes its toxicity by forming strong protein–protein complex, which is non-harmful. Upon release from the complex, the toxin acts within the cell by interfering with essential processes, such as protein (Díaz-Orejas et al., 2010; Goeders and Van Melderen, 2014) or DNA synthesis (Harms et al., 2015), cell wall synthesis (Mutschler et al., 2011), and cell division (Masuda et al., 2012). The toxin action results in a rapid cell growth arrest or even leads to cell death (Page and Peti, 2016). The majority of toxins from type II systems, characterized to date, are endoribonucleases (Cook et al., 2013), which, if not neutralized by its cognate antitoxin protein, cleave mRNAs at specific sequences either within or outside the ribosome and inhibit translation (Chan et al., 2016). The plasmid-borne type II TAs often function as plasmid stabilization elements by allowing growth of the cells that inherit plasmid with the TA system upon segregation, whereas cells that have lost plasmid are killed by more stable toxin after the more labile antitoxin is degraded by proteases (Engelberg-Kulka and Glaser, 1999; Hernández-Arriaga et al., 2015). The biological role of chromosomally encoded type II systems is not clearly elucidated yet. The proposed functions of type II TAs range from viewing them as selfish DNA, anti-addiction elements to stress-responsive genes, which can regulate bacterial growth and survival adapting to various environmental changes (Magnuson, 2007; Van Melderen and Saavedra De Bast, 2009; Ramisetty and Santhosh, 2017). The TA systems can adjust the metabolic processes at a large scale, such as shutting down protein synthesis and switching to a dormant cellular state (Kędzierska and Hayes, 2016; Lee and Lee, 2016).
Genome analysis has shown a wide variety of TA modules in pathogenic species (Makarova et al., 2009; Leplae et al., 2011). The role of TAs in the life of bacterial pathogens is now beginning to be explored (Fernández-García et al., 2016; Kędzierska and Hayes, 2016; Lee and Lee, 2016; Lobato-Márquez et al., 2016a). Recent reports have demonstrated the significance of TAs in the stabilization of virulence plasmids in Shigella (McVicker and Tang, 2016) and Salmonella enterica (Lobato-Márquez et al., 2016b), superintegron in Vibrio cholerae (Iqbal et al., 2015), also in the promoting of S. enterica persister formation (Cheverton et al., 2016; Jaiswal et al., 2016) and in mediating the transcriptional response to environmental cues in Helicobacter pylori and Brucella abortus (Heaton et al., 2012; Cárdenas-Mondragón et al., 2016).
We have recently shown that type II HigBA TA system is one of the most prevalent plasmid-borne TA systems in A. baumannii isolates of clinical origin. It is encoded by the higBAAb operon, where higBAb toxin gene (357 bp) precedes higAAb antitoxin (303 bp) (Jurenaite et al., 2013; Sužiedėlienė et al., 2016). higBAAb locus was also found to be encoded by the newly observed ubiquitous A. baumannii 11 kb plasmid pAB120. Plasmid carries two copies of blaOXA-72 genes, conferring resistance to carbapenems, a broad spectrum β-lactam antibiotics class, which is used to treat A. baumannii infections (Povilonis et al., 2013; Supplementary Figure S1). Here we report characterization of A. baumannii type II TA system, by demonstrating that higBA locus is represented in A. baumannii by two functional variants, named higBA1Ab and higBA2Ab. The higBA2Ab is encoded on pAB120 plasmid and was further thoroughly characterized. We demonstrate that HigB2Ab toxin acts as a ribonuclease and forms an unusually large complex with the antitoxin. Both HigA2Ab and the HigBA2Ab protein complex transcriptionally autoregulate their own operon. We show that higBA2Ab represents a stress responsive TA locus, which also possesses plasmid stabilization ability.
Materials and Methods
Bacterial Strains and Growth Conditions
Escherichia coli and Acinetobacter strains were grown in LB at 37°C with appropriate antibiotics added (ampicillin 100 μg/mL, meropenem 8 μg/mL, gentamicin 10 μg/mL, kanamycin 50 μg/mL, and chloramphenicol 30 μg/mL), unless otherwise indicated. The strains and plasmids used in the study are described in Supplementary Table S1. A. baumannii was transformed with plasmid pAB120 variants by electroporation, and selected on LB containing meropenem. Minimal inhibitory concentration (MIC) values were detected as described (Wiegand et al., 2008).
Plasmid Construction
Gene cloning procedures were performed using high-fidelity Phusion polymerase (Thermo Fisher Scientific), cleavage with restriction enzymes and ligation was performed according manufacturers recommendations (New England Biolabs, Thermo Fisher Scientific). All final constructs were verified by sequencing. pAB120ΔhigBA: to introduce higBA2Ab deletion to pAB120, primers described in Supplementary Table S2 were used for inverse PCR (Imai et al., 1991). The resulting PCR product was cleaved with NcoI and ligated. The deletion of higBA2Ab was confirmed by PCR. pAcORI∗higBA: a derivative of the plasmid pWH1266 (Hunger et al., 1990; Supplementary Table S1), exhibiting faulty inheritance in Acinetobacter sp. was constructed. pAcORI∗ plasmid contained pUC19 backbone with introduced gentamicin resistance gene and a defective Acinetobacter calcoaceticus ORI from pWH1266, amplified using primers listed in Supplementary Table S2. higBA2Ab TA system from pAB120, together with predicted promoter region, was then cloned to pAcORI∗ using primers in Supplementary Table S2. BACTH constructs: for T18 and T25 fusions plasmids pUT18 and pKNT25 (Euromedex BACTH System Kit) were used. higB2Ab, higA2Ab, relBAb, relEAb genes were PCR-amplified using primer pairs described in Supplementary Table S2. pAB120 plasmid was used as a template for higBA2Ab gene amplification, while for relBEAb A. baumannii clinical (35, ECII) strain was used (Jurenaite et al., 2013). Both toxins and antitoxins were N-terminally fused to appropriate Cya domains. Protein expression plasmids: for protein purification, higBA2Ab operon was cloned to pET28b protein expression vector, fusing the antitoxin (C-terminally) or the toxin (N-terminally) with his-tag (Supplementary Table S2). To construct plasmids containing TEV cleavage sites, the thrombin recognition site in the plasmid was replaced to TEV site by inverse PCR using primers listed in Supplementary Table S2 to generate pET-His6-TEV-HigB-HigA plasmid. For pET-HigB-His6-TEV-HigA, TEV site and His-tag was first removed from previous plasmid and then added to the N-terminus of higA2Ab. Constructs for promoter repression assay: pPROBE’-gfp vectors were constructed by inserting PCR amplified predicted promoter DNA sequences (200 bp in length upstream of higB2Ab or higA2Ab genes; primers indicated in Supplementary Table S2) upstream of gfp gene. higBA2Ab and higA2Ab genes were cloned to pBAD24 under arabinose inducible promoter (primers indicated in Supplementary Table S2). pBAD-higBpAB120 and pUHEcat-higApAB120 for kill-rescue assay: the plasmids were constructed as described elsewhere (Jurenaite et al., 2013).
Bacterial Adenylate Cyclase Two Hybrid System (BACTH) Assay
Five independent clones, with compatible toxin and antitoxin containing pKNT25 and pUT18 vectors were grown in LB for 16 h, then the cultures of the five clones were mixed and 5 μL of the mix was spotted on LB agar plates containing appropriate antibiotics, 100 μg/mL IPTG and 100 μg/mL X-Gal. The plates were incubated for 24 h at 30°C.
Purification of HigBA2Ab Protein Complex
For purification of His-HigBA2 and HigBA2-His protein complexes, plasmids pET-His-HigBA or pET-HigBA-His, were introduced into E. coli strain BL21 (DE3) (Supplementary Table S1) and the expression of protein complex induced by incubation with 1 mM IPTG for 4 h during mid-logarithmic phase. Cells were collected by centrifugation for 10 min at 4°C 5500 g, bacterial pellets were resuspended in lysis buffer (20 mM NaH2PO4 pH 7.4, 500 mM NaCl, 20 mM imidazole) and lysed by sonication. Lysate was centrifuged for 10 min at 4°C 13000 g to remove cell debris. The protein complexes were purified from soluble fraction by affinity chromatography, using 1 mL HisTrapHPTM nickel-Sepharose column (GE Healthcare), equilibrated with 20 mM NaH2PO4 pH 7.4, 500 mM NaCl, 20 mM imidazole buffer. After loading the protein lysate, the column was washed with the same buffer for column volumes followed by 10 volumes of wash buffer (20 mM NaH2PO4 pH 7.4, 500 mM NaCl, 50 mM imidazole) to remove impurities. Proteins were eluted by linear gradient using buffer 20 mM NaH2PO4 pH 7.4, 500 mM NaCl, 500 mM imidazole. The eluted fractions were desalted using Sephadex G–25 (GE Healthcare) column, exchanging the buffer to 20 mM NaH2PO4 pH 7.2, 300 mM NaCl. Eluted proteins were analyzed by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE), stained with Coomassie Brilliant Blue. To generate tag-less complex, His-TEV-HigBA2 proteins were incubated with TEV protease at a molar ratio of 1 to 100 overnight at room temperature.
Protein Complex Size Determination by Size-Exclusion Chromatography
Purified His-HigBA2 and HigBA2-His complexes were analyzed by FPLC gel filtration chromatography using Superose 12 10/300 GL column (GE Healthcare). Column was washed with 20 mM NaH2PO4 pH 7.2, 300 mM NaCl buffer. Gel filtration flow rate was 0.5 mL/min. The relative quantities of the proteins present in the fractions were determined after SDS–PAGE using Image Lab software (Bio-Rad). The tag-less HigBA2 complex was analyzed by Superdex 200 16/60 column (GE Healthcare), using 50 mM Tris, pH 8.0, 500 mM NaCl buffer. The protein molecular mass standards were thyroglobulin (670 kDa); γ-globulin (158 kDa); bovine serum albumin (66 kDa); ovalbumin (43 kDa); myoglobin (17 kDa); vitamin B12 (1.35 kDa) (Bio-Rad Gel filtration standard 151–1901) and Blue Dextran (GE Healthcare).
Toxin Purification From Protein Complex
To separate HigB2 toxin from His-HigBA2 complex, denaturant-induced dissociation of the toxin–antitoxin complex on-column method was used (Sterckx et al., 2015). Briefly, the protein complex was denatured on the affinity column using guanidine HCl buffer (5 volumes of 50 mM Tris, pH 8.0, 500 mM NaCl, 5 M guanidine HCl), followed by renaturing on the resin (5 volumes of 25 mM Tris pH 8.0, 250 mM NaCl, 5% glycerol, followed by 5 volumes of 25 mM Tris pH 8.0, 250 mM NaCl, 1% glycerol). The toxin was eluted with 50 mM Tris pH 8.0, 500 mM NaCl, 200 mM imidazole, then immediately dialyzed to 50 mM Tris–HCl pH 8.0.
RNA Cleavage Analysis in Vitro
Total A. baumannii RNA, 5S rRNA and E. coli total tRNA were used as substrates for in vitro RNA cleavage analysis by HigB2 toxin. The mixtures (20 μL) contained 1.5 μg total RNA or 3 μg of 5S or tRNA in 25 mM Tris–HCl pH 7.8, and 0–1 μM final concentration of His-HigB2 protein. As a control, 1 μM of His-HigBA2 protein complex was added. The mixes were incubated for 30 min at 37°C and fractionated using 2% agarose electrophoresis.
Promoter Activity and Repression Measurements in Vivo
For promoter activity assays DJ624Δara cells were transformed with pPROBE’-gfp vectors with or without inserted predicted promoter sequences upstream from gfp. Together, pBAD24, pBAD24-HigBA or pBAD24-HigA vectors were co-transformed. Overnight cultures of resulting strains were diluted to optical density at 600 nm (OD600) = 0.02 in minimal M9 media with casamino acids, 50 μg/mL kanamycin, 100 μg/mL ampicillin and 0.2% glucose. 1 mL of cultures was grown in glass bottom black 24 well plates (Greiner) in Spectramax microplate reader at 37°C with constant shaking. OD600 and fluorescence (excitation 485 nm emission 520 nm) were registered every 15 min. After 3 h of growth 0.2% arabinose was added to induce the protein expression from pBAD24 vectors and measurements were continued for 10 more hours.
Analysis of Gene Expression by qPCR
Acinetobacter baumannii clinical strain K60 (Povilonis et al., 2013), containing pAB120 plasmid, was grown in LB, to exponential phase (OD600 = 1) and to stationary phase (48 h, OD600 = 4.5). For stress conditions, the cells were grown in LB with 200 mM 2,20-bipyridine (Eijkelkamp et al., 2011) or in LB with 1 or 2% ethanol to OD600 = 1. For antibiotic stress, 1/10 of MIC values for gentamicin and rifampicin (2 and 0.3 μg/mL, respectively) and 1/2 MIC for meropenem (16 μg/mL) was added to exponentially growing cells and incubated for 1 h to a final OD600 = 1. Total RNA was isolated, DNA removed and cDNA synthesized as recommended by the kit supplier (Thermo Fisher Scientific). qPCR was performed using primer pairs listed in Supplementary Table S2 (all primers exhibited 100–103% amplification efficiency at selected concentrations). The changes in gene expression were calculated as ΔΔCt, using rpoB as house-keeping gene. At least three biological replicas were performed.
Plasmid Stability Assay
Bacteria containing plasmid of interest were grown in LB containing appropriate antibiotic as an overnight culture (16 h), and then diluted 500-fold to a fresh media without antibiotic. The culture was restarted to a new batch of LB without antibiotic every 12 h. After each inoculation the colony forming units (CFU) of antibiotic resistant (retaining the plasmid encoding resistance) and sensitive bacteria (plasmid lost) was calculated by serial dilutions and plating on LB agar with or without appropriate antibiotic, and plasmid retention was calculated as a percentage of resistant bacteria.
Plasmid Copy Number (PCN) Calculation
Plasmid copy number (PCN) was calculated as described elsewhere (Providenti et al., 2006). Briefly, the bacteria were grown for 24 h without antibiotic pressure, the cells were collected and their lysates used for qPCR, with primers detecting rep gene of the pAB120 plasmid and rpoB as genome encoded control. Copy number was calculated as PCN = EΔCt (E = 2, if the product amplifies at 100% efficiency, which was the case for the primers used). The experiment was independently repeated 10 times.
Kill-Rescue Assay
The assay was performed as previously described (Jurenaite et al., 2013). Briefly, E. coli BW25113 F‘ pUHEcat-“antitoxin” pBAD-“toxin” strains were grown in LB medium until the early logarithmic phase (optical density at 600 nm (OD600) = 0.1). The culture was then induced with arabinose (0 or 0.02%) and/or IPTG (0, 0.1, and 1 mM). The growth was observed in Tecan Infinite M200 Pro plate reader at 37°C with shaking.
In Silico Analysis
Cluster of GP49-domain toxin family was prepared as neighbor joining tree (Saitou and Nei, 1987) using BLOSUM62 matrix, average distance tree of two major lineages of HigB toxins based on sequence identity as well as alignment of HigBAb toxins obtained using ClustalW and Jalview 2.10.0b1.
Results
higBAAb System in A. baumannii Is Represented by Two Variants
Acinetobacter baumannii higBA module has been classified on the basis of low but significant homology of its predicted GP49-like domain toxin HigB to the RelE/ParE superfamily toxins (Jurenaite et al., 2013). Further homology search has shown that the closest GP49-domain homologues of HigBAb mentioned in the literature are Mycobacterium tuberculosis Rv2022c-Rv2021c (also called higBA2) and Rv3182-Rv3183 (higBA3) TA systems, whose toxins share more than 40% protein sequence identity with HigBAb (Figure 1A). Based on the BLAST search, close homologues of HigBAb (more than 50% sequence identity) are present in other gammaproteobacteria closely related to Acinetobacter, such as Psychrobacter, as well as various members of Enterobacteriaceae family, such as Klebsiella (Supplementary Figure S2).
FIGURE 1.
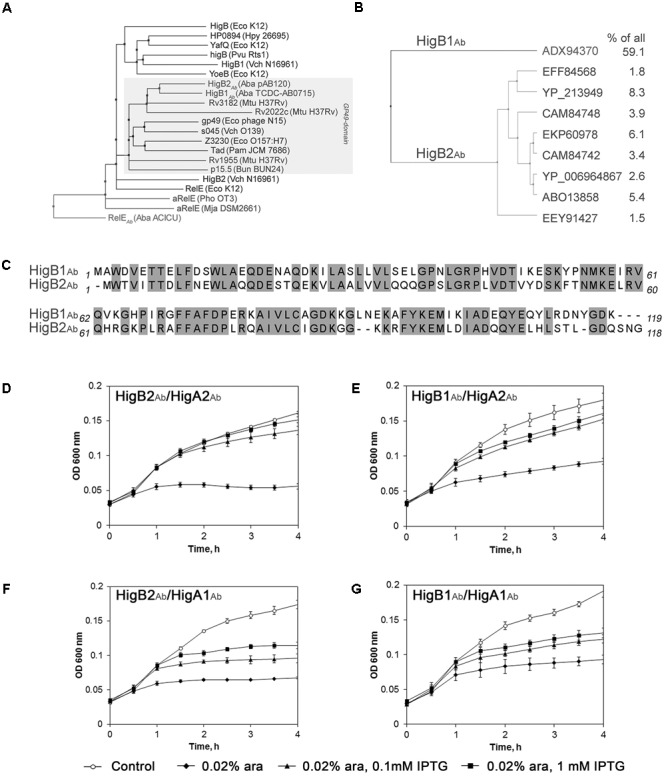
Phylogenetic and homology analysis of Acinetobacter baumannii HigBAb. (A) Cluster of GP49-domain toxin family within RelE superfamily, represented as neighbor joining tree. Gray box covers all GP49-domain proteins. The full organism names are provided in the Supplementary Material (Supplementary Table S3); (B) Two major lineages of HigBAb toxins, represented as average distance tree. All proteins that have more than 1% identical hits in all deposited sequences are shown and their frequency in percentage is provided; (C) Alignment of pAB120-borne HigB2Ab (Povilonis et al., 2013) and HigB1Ab (Jurenaite et al., 2013) toxin sequences. Identical amino acid residues are outlined in gray boxes. All alignments were prepared as described in Section “Materials and Methods”; (D–G) Growth of Escherichia coli BW25113F′ harboring components of higBAAb and their combinations. The E. coli strains with pUHE-“antitoxin” and pBAD30-“toxin” plasmids were grown until OD600 = 0.1 and induced with 0.02% arabinose and/or with 0.1–1 mM IPTG as described in Section “Materials and Methods.” The data from three independent experiments are represented, the error bars indicate standard deviation.
Strikingly, BLAST search revealed a variety of amino acid sequences for HigBAb toxin in A. baumannii species. Therefore, we have aligned all A. baumannii HigBAb sequences existing to date and found that they are, in fact, represented by two distinct versions of HigBAb modules showing around 60% sequence identity (Figures 1B,C). The most prevalent version is largely conserved (called HigB1Ab further), while the other version of HigBAb is represented by many sequence variations ranging from 85 to 99% identity (HigB2Ab) (Figure 1B). The higBAAb module found in plasmid pAB120 (Povilonis et al., 2013), which we characterize in this study, represents the less conservative version higBA2Ab, while the conserved version higBA1Ab has been characterized previously as a representative higBAAb module carried by strain 35 in a pilot study of A. baumannii TA systems (Jurenaite et al., 2013). We were interested whether the two versions of TA systems constitute stand-alone TA modules. Expression of both versions of HigBAb toxin resulted in an inhibition of E. coli growth, and interestingly, co-expression of HigAAb antitoxin counteracted toxin-mediated growth inhibition regardless the HigAAb variant used for growth rescue (Figures 1D–G). This experiment has shown that the components of two TA variants are still able to interact despite 42 and 45% amino acid sequence difference between the toxins and antitoxins of the two groups, respectively. Structure models of both versions of HigB and HigA (Supplementary Figure S3) also predicted high similarity of folds, despite some differences at the termini of the proteins. Notably, the pAB120-borne HigBA2Ab pair exhibited more pronounced kill-rescue effect compared to the more conserved HigBA1Ab version (Figures 1D vs. G). These results are in line with the proposed anti-addiction hypothesis (Saavedra De Bast et al., 2008), where the conserved but less active HigBA1Ab module could provide evolutionary pressure for the HigBA2Ab lineage.
We then searched for Acinetobacter sp. strains having both higBAAb versions in the GenBank sequence database. We were not able to find two distinct versions of higBAAb on the same plasmid or in the same strain. This phenomenon has been reported previously by analysis of identity of bacterial and chromosomal TAs (Ramisetty and Santhosh, 2016). However, we were able to find two sequenced Acinetobacter sp. isolates with copies of higBAAb on several plasmids [isolates DUT-2 (GenBank accessions CP014652, CP014654, CP014655) and VE-C3 (GenBank accessions NC_010310, NZ_ALIG01000010)], and five plasmids containing two copies of same version of higBAAb (GenBank accessions CM008888, CP010369, CM001803, NC_010404, NC_025173) (Supplementary Table S3). Interestingly, all of them had the less conserved higBA2Ab versions. In two cases [plasmid “unnamed2” from A. baumannii strain ZQ1 (Supplementary Table S3) and DUT-2 (plasmids CP014652, CP014654)], both copies of higBAAb were identical, indicating a recent event of duplication. The other cases with two distinct higBA2Ab variants had 86 to 88% DNA sequence identity. The presence of two diverse copies of higBAAb in the same bacteria or on a single plasmid could indicate the event of ongoing evolution of the system and its separation into two different TA systems.
HigB2Ab and HigA2Ab Proteins Form Complex
pAB120 plasmid-borne HigBA2Ab and its toxin HigB2Ab shares only low similarity to RelE/ParE type TA toxins. Therefore we asked whether this TA system is and acts as a canonical one. Type II TA systems are known to employ different mechanisms of action and their proteins may form different oligomeric complexes (Chan et al., 2016; Rocker and Meinhart, 2016). We asked whether HigB2Ab and HigA2Ab form a physical complex which neutralizes toxicity in a typical way to type II TA systems. For this purpose we employed bacterial adenylate cyclase two hybrid system (BACTH) (Karimova et al., 1998) and verified that HigB2Ab toxin and HigA2Ab antitoxin directly interact in vivo by restoring adenylate cyclase activity (Figure 2A). Moreover, the A. baumannii RelEAb and RelBAb proteins [homologues of well-described RelBE TA system of E. coli (Jurenaite et al., 2013)], which we used as a control, showed even weaker interaction in comparison to that of HigB2Ab and HigA2Ab.
FIGURE 2.

Analysis of HigB2Ab–HigA2Ab protein interaction. (A) Two-hybrid analysis of interaction of toxin-antitoxin (TA) components. E. coli BTH101 with two plasmids, one encoding Cya-T18 fused to A. baumanii antitoxins HigA2Ab or RelBAb, and another encoding Cya-T25 fused to toxins HigB2Ab or RelEAb. The ability of the proteins to interact and reconstitute functional Cya from T25 and T18 was observed as blue colony formation when grown on LB agar with IPTG and X-gal for 24 h; (B) Size-exclusion chromatography and SDS–PAGE analysis of His-HigBA2 and HigBA2-His protein complexes. The proteins used for molecular mass standard curve are indicated as black diamonds, empty diamonds indicate the positions of His-HigBA2 and HigBA2-His protein complexes and single His-HigB2 protein and their calculated size; inside box – 15% SDS–PAGE gel analysis lane 1 – His-HigB2, lane 2 – His-HigBA2, lane 3 – HigBA2-His proteins and complexes after gel filtration; (C,D) E. coli BL21(DE3) strain, containing pET-HigBA-His (C) and pET-His-HigBA (D) plasmids were grown to mid-exponential phase and protein expression was induced with 1 mM IPTG for 4 h. Cells were disrupted by sonication and the His-tag containing proteins were purified by affinity chromatography from the soluble fraction as described in Section “Materials and Methods”. Proteins were visualized by 15% SDS–PAGE stained with Coomassie Brilliant Blue. M – protein molecular mass markers (band sizes in kDa are shown on the Left), –/+ cell lysate before and after induction with IPTG, S – soluble protein fraction after cell disruption, F – protein purification flow-through fraction, W – protein purification wash fraction, Elution – protein purification elution fractions.
To further assess the complex forming ability between A. baumannii HigB2Ab and HigA2Ab proteins, we have constructed two versions of the expression vectors encoding higBA2Ab operon with HigB2Ab toxin N-terminal His-tag fusion (pET-His-HigBA) and with antitoxin HigA2Ab C-terminal His-tag fusion (pET-HigBA-His). Proteins were expressed in E. coli and purified by affinity chromatography as described in Section “Materials and Methods.” In both cases HigB2Ab and HigA2Ab proteins co-purified, indicating that they form a strong complex (Figures 2C,D). To assess the size of complexes we have analyzed them by size-exclusion chromatography. The elution profiles showed single peaks (not shown) corresponding to the entities with estimated molecular masses of approximately 99.3 kDa and 53.7 kDa for His-HigBA2Ab and HigBA2Ab-His complexes, respectively (Figure 2B). Protein analysis of pooled fractions that corresponded eluted peaks by 15% SDS–PAGE and subsequent gel densitometric analysis showed two bands present in ratio (toxin:antitoxin) of approximately 1.5:1 and 1:1 (Figure 2B). As the differences in observed complex sizes could have been due to the interference of His-tag, we aimed to confirm the complex size with the tag-less protein complex. To eliminate the probable His-tag effect, we constructed plasmids with cleavable His-tags situated at the N-terminus of HigB2Ab (plasmid pET-His6-TEV-HigB-HigA) or HigA2Ab (plasmid pET-HigB-His6-TEV-HigA). However, the cleavage of His-tag was only efficient for the protein complex with N-terminal His-tag fusion of HigA2Ab. Upon removal of His-tag from HigBA2Ab complex, size exclusion chromatography resulted in an entity of approximately 109 kDa (Supplementary Figure S4), thus confirming that HigBA2Ab is able to form unusually large complex.
HigB2Ab Toxin Is a Ribonuclease
We have demonstrated previously, that HigB1Ab toxin from higBA1Ab system (named as higBAAb in the previous research) when expressed in E. coli caused degradation of cellular RNAs and inhibited translation by impairing the incorporation of protein synthesis precursor, therefore suggesting that it might act as a ribonuclease (Jurenaite et al., 2013). To investigate whether HigB2Ab possesses a ribonuclease activity in vitro, we have purified recombinant His-HigB2 toxin from His-HigBA2Ab complex and tested whether it is able to degrade A. baumannii total RNA as well as RNA with known secondary structure such as E. coli 5S rRNA and tRNA. HigB2Ab toxin efficiently degraded total A. baumannii RNA in a dose dependent manner, whereas in a complex with HigA2Ab protein its ribonuclease activity was blocked (Figure 3A). The degrading activity against E. coli 5S rRNA was only detected using high concentrations of HigB2Ab toxin (Figure 3B), whereas effect against tRNA at the same conditions was negligible, indicating that HigB2Ab toxin more likely targets unstructured RNAs.
FIGURE 3.
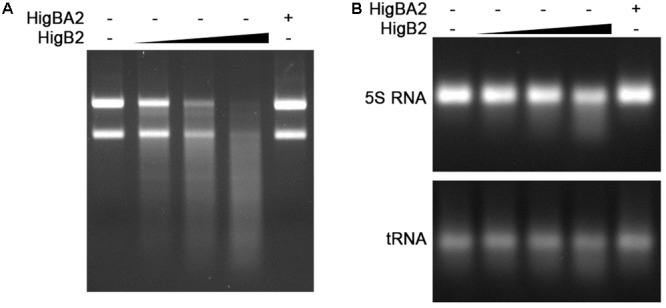
HigB2Ab toxin acts as a ribonuclease. His-HigBA2 protein complex at the concentration of 1 μM and His-HigB2 toxin at the concentrations of 0, 0.25, 0.5, and 1 μM, were incubated with 1.5 μg of total A. baumannii RNA (A) and with 3 μg of E. coli 5S rRNA (B) in 10 μl reactions for 30 min at 37°C. The samples were visualized in 1% agarose gel.
HigA2Ab Antitoxin Represses Transcription From higBA2Ab Promoter Whereas HigB2Ab Toxin Acts as a Corepressor
Type II TA complexes or single antitoxin proteins have been shown to bind to the operator DNA of their own promoter and negatively autoregulate operon transcription (Kedzierska et al., 2007). HigA2Ab has a HTH domain and could bind to DNA alone or in complex with HigB2Ab. It is not known how reverse TA systems, where the toxin is the first gene in the operon, regulate their toxin:antitoxin ratio. We looked if an additional promoter could be found for higAAb, indicating additional regulation for antitoxin expression. BPROM tool (Solovyev and Salamov, 2011) predicted promoter elements (-10 and -35) upstream the higBA2Ab operon and higA2Ab coding sequence (Figure 4A), indicating that besides promoter for the operon, higA2Ab might also have an additional promoter. We have tested the activity of these promoters and their regulation by HigBA2Ab complex and HigA2Ab protein. 200 bp DNA fragments containing higBA2Ab and higA2Ab promoters were cloned upstream of the gfp gene (plasmids pPROBE-PhigBA-gfp and pPROBE-PhigA-gfp). Promoter activities were measured by following GFP fluorescence upon induction of HigA2Ab antitoxin and HigBA2Ab complex (plasmids pBAD24-HigA, pBAD24-HigBA) in E. coli. Promoterless gfp (plasmid pPROBE-gfp) was used to assess autofluorescence level of the cells. Induction of HigA2Ab expression strongly repressed the activity of higBA2Ab promoter, and the induction of HigBA2Ab complex showed even more pronounced inhibitory effect (Figure 4C). Putative higA2Ab promoter located within the coding sequence of higB2Ab gene did not display any activity (Figures 4B,D). Our results clearly show that higBA2Ab operon is transcriptionally autorepressed by its cognate antitoxin HigA2Ab, and HigB2Ab toxin acts as a co-repressor. The predicted higA2Ab promoter was not functional.
FIGURE 4.
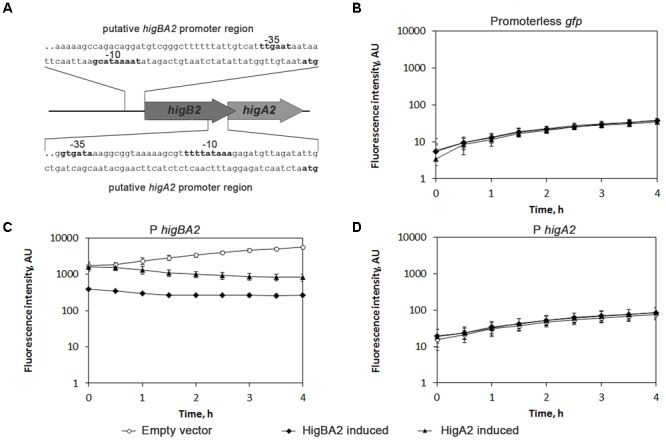
Analysis of HigBA2Ab interaction with own promoter region. (A) Schematic representation of A. baumannii higBA2Ab locus. A part of pAB120 plasmid containing higBA2Ab operon with predicted promoter is presented (not to scale). Predicted promoter DNA sequences (100 bp out of 200 bp cloned to vectors) used for promoter activity assay are shown, predicted –35 and –10 regions (BPROM) and ATG are indicated in bold; (B–D) The assessment of activity of predicted higBA2Ab operon promoters. Predicted promoter regions of higBA2Ab operon and higA2Ab gene were cloned into pPROBE vector, placing them upstream the gfp gene as described in Section “Materials and Methods”. Then pPROBE with respective promoter sequence or lacking any promoter (Promoterless gfp) was co-transformed into E. coli DJ624Δara harboring pBAD24 plasmids, with cloned higBA2Ab operon or higAAb antitoxin genes under arabinose inducible promoter. After addition of 0.2% arabinose, GFP fluorescence was measured every 30 min, reflecting the effect of induced proteins on the activity of measured promoters. Three independent experiments were performed, error bars indicate standard deviation. AU, arbitrary units.
Expression of A. baumannii higB2Ab and higA2Ab Genes in Stress Conditions
We have demonstrated that HigB2Ab and HigA2Ab form a strong complex which causes repression of its own operon. We next asked how higBA2Ab locus is expressed in A. baumannii K60 (Povilonis et al., 2013) under various conditions. We have chosen to investigate growth and stress conditions relevant to those found in clinical environment and within the host: the stationary phase, conditions mimicking the iron deficiency, growth with ethanol and sub-lethal amounts of antibiotics. The expression changes in both higB2Ab and higA2Ab were evaluated separately by qPCR to see if there are any differences in expression levels which would indicate the presence of separate transcripts. We have found that transcripts of higB2Ab and higA2Ab genes were more abundant in stationary phase (more than fivefold increase) comparing to the exponential growth conditions (Figure 5). Interestingly, the iron deficiency caused by the addition of iron chelator 2,2′-bipyridine (Eijkelkamp et al., 2011) to the LB medium resulted in the up to threefold induction of the higB2Ab and higA2Ab genes (Figure 5). The presence of ethanol in the media did not have pronounced effect on the gene expression (Figure 5). We further tested sub-lethal concentrations of antibiotics. As A. baumannii strain K60 was highly resistant to several classes of antibiotics [aminoglycosides, β-lactams, fluoroquinolones, (Supplementary Table S4)], we chose antibiotics which exhibited moderate to low MIC (gentamicin MIC 20 μg/mL, rifampicin MIC 3.125 μg/mL). The antibiotics were added at 1/10 of the MIC, as higher concentrations resulted in severe growth impairment. Effect of meropenem was also tested (1/2 of MIC), due to the presence blaOXA-72 gene in the proximity to higBA2Ab in pAB120 plasmid. No significant changes in higBA2Ab expression were observed for gentamicin or meropenem, but the addition of rifampicin caused a decrease of expression of both genes (Figure 5). We therefore conclude that higBA2Ab module could be expressed during stress conditions linked to stationary phase and iron deficiency stress, as well as it could play a role during RNA synthesis inhibition or yet in other unknown conditions. Additionally, since all expression changes were similar for both higB2Ab and higA2Ab, we can expect both genes to be expressed from a single transcript of the whole operon.
FIGURE 5.
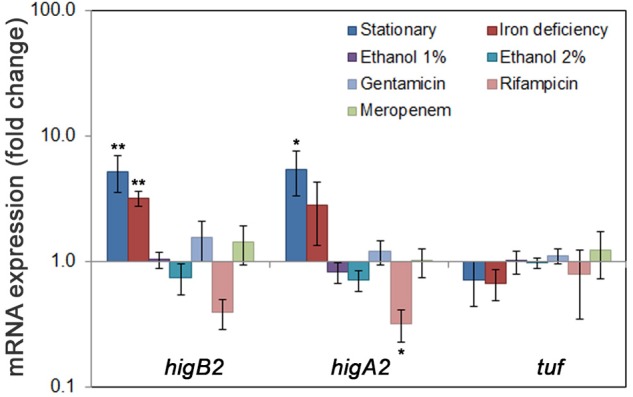
Expression of A. baumannii higBA2Ab locus during stress conditions. A. baumannii clinical strain K60 was grown in LB to exponential phase (OD600 = 1) and to stationary phase (blue bars); in conditions mimicking iron deficiency (LB with or without 2,2′-bipyridine) (red bars), in LB with 1 or 2% ethanol added (purple and cyan bars, respectively) and LB with antibiotics (gentamicin, rifampicin, meropenem, in light blue, light red, and light green, respectively) added. The bacteria were collected at OD600 = 1, except for stationary phase, where the bacteria were grown for 48 h reaching OD600 of 4.5. Total RNA was isolated and RT-qPCR performed to detect the expression of TA genes as described in Section “Materials and Methods”. The differences in transcript amounts were evaluated by comparative Ct method (ΔΔCt) using rpoB as a house-keeping gene. At least three biological replicas were performed, error bars indicate standard deviation. Statistically significant difference from tuf expression, used as a control, is indicated by one asterisk (t-test, p < 0.05), or two asterisks (t-test, p < 0.005).
The Role of higBA2Ab TA System in Stabilization of Acinetobacter Plasmids
Toxin-antitoxin modules are known plasmid stabilization factors in bacteria (Engelberg-Kulka and Glaser, 1999; Hernández-Arriaga et al., 2015) and their presence on plasmids conferring resistance to clinically important antibiotics may contribute to the persistence and spread of antibiotic resistant strains. We have previously shown that higBA2Ab module is widely spread among A. baumannii plasmids (Povilonis et al., 2013; Sužiedėlienė et al., 2016), therefore we were interested whether it provides stabilization function for cognate plasmid carrying resistance genes, found in clinical A. baumannii isolates. For this purpose plasmid pAB120, which originally carries two copies of blaOXA-72 gene conferring resistance to carbapenems (meropenem and imipenem) was purified from clinical A. baumannii strain K60 and higBA2Ab locus was deleted (Supplementary Figure S1). Given that pAB120 plasmid is equipped with another type II TA module, splTA (Jurenaite et al., 2013; Povilonis et al., 2013), we sought if the latter could be sufficient to supply plasmid stabilization even when higBA2Ab was deleted. Therefore, plasmid with deletion of both TA modules was also constructed (Supplementary Figure S1). The pAB120ΔhigBAAb and pAB120ΔhigBAAbΔsplTAAb plasmids were transformed into non-pathogenic Acinetobacter baylyi strain ADP1, which is known not to contain any plasmids or higBA homologs (Supplementary Table S1). Unexpectedly, neither the deletion of higBA2Ab locus, nor the elimination of both higBA2Ab and splTAAb modules caused any loss of pAB120 variants in A. baylyi (not shown). To eliminate the possibility of different plasmid maintenance effects in different Acinetobacter species, A. baumannii clinical strain K53 was then used as a host (the strain also does not contain any plasmids of known A. baumannii replication groups and higBA modules (Jurenaite et al., 2013; Povilonis et al., 2013; Supplementary Table S1). pAB120 variants did not show decrease in plasmid retention in A. baumannii as well (Figure 6A). We then analyzed whether higBA2Ab locus has an impact on the copy number of pAB120 plasmid. A. baumannii strains K53 bearing pAB120 with or without higBA2Ab were grown without antibiotic pressure for 24 h and PCN was calculated by qPCR. A slight reduction in PCN was observed, which was statistically significant (Figure 6A). These results indicate that while higBA2Ab and splTAAb are not the main players in stabilization of pAB120 under tested conditions, the deletion of higBA2Ab has an impact on its copy number, and could influence plasmid retention in the long run or in different conditions.
FIGURE 6.
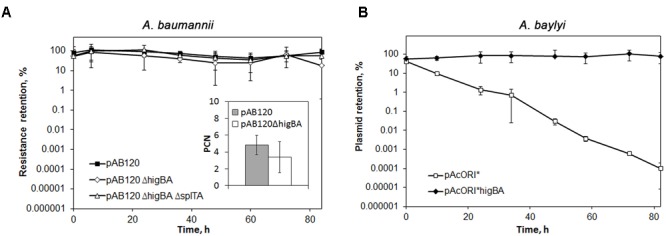
The role of A. baumannii higBA2Ab locus in plasmid stabilization. Bacteria containing plasmids were grown in LB without antibiotic pressure, restarting the culture every 12 h. The colony forming units (CFU) of total bacteria and antibiotic resistant bacteria was assessed by serial dilution and plating. At least three independent experiments were performed, error bars indicate standard deviation. (A) The effect of higBA2Ab deletion on A. baumannii pAB120 plasmid maintenance and copy number. Bacteria containing pAB120 variants with or without higBA2Ab or both higBA2Ab and splTAAb TA system were selected on media containing meropenem. Inside the box: plasmid copy number (PCN) of pAB120 variants with and without higBA2Ab was calculated after 24 h growth without antibiotic pressure. PCN was calculated as described in Section “Materials and Methods”. The experiment was independently repeated 10 times, the difference of PCN between the two strains was statistically significant (t-test, p < 0.05); (B) Effect of higBA2Ab locus in stabilization of unstable plasmid pAcORI∗. Acinetobacter baylyi containing variants pAcORI∗ with and without higBA2Ab were selected on gentamicin (pAcORI∗ marker).
To be sure if higBA2Ab is able to confer stability when introduced into unstable plasmid lacking any stabilizing determinants, we used higBA2Ab from pAB120 in additional plasmid stabilization experiments. The higBA2Ab locus with its own promoter region was cloned into unstable Acinetobacter plasmid pAcORI∗, conferring resistance to gentamicin (Supplementary Table S1). A. baylyi ADP1 strain containing plasmid pAcORI∗ with higBA2Ab locus and plasmid without the TA system were grown without antibiotic pressure and plasmid retention was calculated. Strikingly, in this background the presence of higBA2Ab locus on the pAcORI∗ plasmid ensured its stability when bacteria were grown for over 80 h (Figure 6B). This result together with the previous observations of the effect on PCN confirms that higBA2Ab module can play a role in plasmid maintenance.
Discussion
The TA modules which are grouped into higBA family (host inhibition of growth) encode a RelE-like toxin and antitoxin that contains a HTH Xre-domain (Gerdes et al., 2005; Makarova et al., 2009). Toxins of HigB family belong to a large RelE/ParE superfamily consisting of mRNAses such as RelE toxin as well as gyrase poisons such as ParE toxin (Anantharaman and Aravind, 2003). Within this superfamily, GP49-domain toxins show low but significant homology to RelE (Makarova et al., 2009). TA modules containing GP49-domain toxin in all studied cases constitute the reverse type of TA operons where toxin is encoded first and is followed by an antitoxin (Dziewit et al., 2007; Ramage et al., 2009; Jurenaite et al., 2013; Sala et al., 2014). Despite being classified as HigB family toxins, the GP49-domain toxins show little similarity to well-studied HigB toxins from E. coli, Vibrio cholerae, Proteus vulgaris (Tian et al., 1996; Christensen-Dalsgaard and Gerdes, 2006; Christensen-Dalsgaard et al., 2010) as well as to toxins of other TA families of RelE superfamily. HigBAAb shows only ∼20% sequence identity to other validated TA modules that have GP49-domain toxins and were named as Tad toxins (Dziewit et al., 2007).
pAB120 plasmid-borne HigBA2Ab proteins form strong complex with molecular mass of approximately 100 kDa suggesting that up to eight protein molecules might be present within the oligomer (the predicted molecular masses of HigB2Ab and HigA2Ab proteins are 13.5 and 11.2 kDa, respectively). According to the currently available structural data, the RelE toxin-based TA superfamily TA complexes are known to form heterotrimers (E. coli YefM-YeoB, PDB accession: 2A6Q), and most commonly, heterotetramers (E. coli DinJ-YafQ, PDB accession: 4Q2U; MqsR-MqsA, PDB accession: 3HI2, E. coli RelB-RelE, PDB accession: 4FXE; Brucella abortus BrnT-BrnA (Heaton et al., 2012), Proteus vulgaris HigBA (Schureck et al., 2014)]. In some cases complexes crystallize as more complex oligomeric structures: a so far unique hetero-hexadecameric complex has been observed for ParE2-PaaA2 TA system (Sterckx et al., 2016). ParE-family toxins inhibit DNA replication and despite sharing common fold they are functionally different from RelE-family toxins. Therefore A. baumannii HigBA2Ab complex of 100 kDa, the size strongly exceeding that of a heterotetramer, observed in this study might represent one of the most complex oligomeric structures known for RelE family TAs.
Taking into account the organization of A. baumannii higBA2Ab module, where toxin gene is located upstream the antitoxin gene, a feature known for a limited number of TAs (Christensen-Dalsgaard and Gerdes, 2006), the expression of equal amounts of both proteins must be ensured in the cell to avoid a harmful effect of the toxin. According to our observations, equal levels of higB2Ab and higA2Ab transcripts are produced at the standard growth conditions in A. baumannii indicating that TA balance at least at the transcription level is properly preserved. The transcript of the whole operon (spanning both higB2Ab and higA2Ab genes) could also be detected by qPCR (Armalytė, Unpublished data), whereas an additional promoter for the higA2Ab gene was not, indicating that most likely higBA2Ab is transcribed as a single transcript. HigA2Ab antitoxin and HigBA2Ab complex autorepressed their own promoter, indicating the transcriptional regulatory mode common to type II TA operons in other bacteria (Chan et al., 2016). HigA2Ab antitoxin harbors HTH Xre-like domain, which is present in some antitoxins of type II TAs such as HigA from Proteus vulgaris, and has been shown to bind operator sequences which overlap with the promoter region (Schureck et al., 2014). The more pronounced inhibition by HigBA2Ab complex than by single antitoxin indicates HigB2Ab toxin also takes part in transcriptional autoregulation of the system.
Despite well studied transcriptional regulation for TA systems, the data on their expression in vivo in conditions relevant to bacterial lifestyle, in particular to that of pathogenic species, is only beginning to emerge. A baumannii represents interesting example to study as it is well adapted to colonize the abiotic (glass, plastic, and metal) and biotic (human skin and mucous membranes) surfaces, withstand prolonged periods of dryness, treatment by disinfectants, antibiotics and nutrient restriction (Giannouli et al., 2013). In the host, A. baumannii encounters responses mediated by complement and professional and non-professional phagocytic cells. Among stress factors playing essential role in the host defense is the nutritional immunity exerted by the iron limitation (McConnell et al., 2013). Our data suggest that stationary growth and iron depletion causes the induction of both toxin and antitoxin genes of higBA2Ab module at a similar level, therefore, expression of both components could play a role for A. baumannii stress response. This is supported by recent observation that closest homologues of A. baumannii higBA in M. tuberculosis, Rv2022c-Rv2021c (higBA2) and Rv3182-Rv3183 (higBA3) are among the strongest stress responsive modules of this pathogen, activated in response to multiple stressors including antibiotics, starvation and low pH (Gupta et al., 2017). In our study, we found that only rifampicin influenced the expression of higBA2Ab by reducing it, which was the opposite effect from that observed for M. tuberculosis higBAs. Sub-inhibitory concentrations of antibiotics are known to induce variable gene expression outcomes in bacteria. SOS response is commonly induced, which includes upregulation of stress proteases such as Lon and Clp. Generally they target antitoxins and relieve transcriptional repression of TAs (Muthuramalingam et al., 2016). However, the observations that particular TAs are downregulated after antibiotic exposure indicate that specific inhibition of the activities of the TA promoters could be involved. Type II loci of other human pathogens have been recently shown to transcriptionally respond under stress conditions. H. pylori hp0893-hp0892 module, which is the most prevalent TA system among H. pylori clinical isolates was induced during stationary growth, in the low concentration of iron and nickel as well as in a high concentration of urea, conditions mimicking the host environment, suggesting that this TA gene pair might represent a novel H. pylori stress responsive virulence factor (Cárdenas-Mondragón et al., 2016). Intracellular pathogen B. abortus increased the expression of type II brnTA system in the presence of chloramphenicol, H2O2 and low pH stress (Heaton et al., 2012). In S. enterica type II sehAB module responded to minimal medium and within macrophages (De la Cruz et al., 2013), whereas different expression of some toxins of type II systems has been observed inside fibroblasts and epithelial cells (Lobato-Márquez et al., 2015).
The induction of bacterial TAs in stress conditions has been attributed to the enhanced degradation of antitoxin by stress peptidases or decreased translation under stress conditions (Ramisetty et al., 2016). This, in turn, might result in the derepression of transcription of TA operon to a different level depending on the stress influence on antitoxin proteolysis and translation rates. Therefore, the decrease in antitoxin concentration might result in various different physiological outcomes: the toxin, freed from the complex, could quickly come into action leading to growth reduction, or an increased transcriptional response of TA genes could be induced, led by derepression of the operon. Such a range in TA-mediated outcomes would be beneficial to A. baumannii in contributing to its persistence in clinical environment and in sensing the host.
The higBAAb locus, with a few exceptions, is found so far on the A. baumannii plasmids, which frequently harbor antibiotic resistance genes coding for carbapenemases OXA-72, OXA-23, and OXA-58 (Sužiedėlienė et al., 2016). We found that 67 out of 307 (21.8%) Acinetobacter sp. plasmids sequenced to date (GenBank accessed 2018/01/11) had higBAAb TA systems. The location of higBAAb could not be attributed to a certain plasmid replication group or plasmid size (plasmids sizes varied from 7.4 to 121 kb). The high prevalence of these TA systems indicates their easy spread and stable maintenance on the plasmids. The mechanisms responsible for plasmid stabilization in A. baumannii are largely unknown (Lean and Yeo, 2017) and this study is the first attempt to assess the role of TA systems in A. baumannii. Surprisingly, we have shown that despite the ability of higBA2Ab module to function as a bone fide addiction system by supporting maintenance of unstable plasmid in Acinetobacter, it did not play that role for pAB120 plasmid from clinical A. baumannii strain, as did not another type II module, carried by the same plasmid. This data suggest that other elements than plasmid-borne post-segregational killing pathway components are responsible for pAB120 stabilization. Among the possible candidates are XerC/XerD sites, which are known to participate in recombinase-based proper resolution of plasmid copies (Sengupta and Austin, 2011) and are present in pAB120 plasmid in multiple copies as well as in series of A. baumannii plasmids of the same replicon type (Lean and Yeo, 2017).
Another unexpected observation was the detection of several higBAAb copies in plasmids and A. baumannii isolates in the sequence databases. Interestingly, only the less conserved higBA2Ab group was detected in that setting, and we were unable to find both groups higBAAb in one isolate or plasmid in the pool of Acinetobacter sp. sequences, available to this day. In agreement with this observation we have demonstrated that both groups’ antitoxins are able to counteract the toxins. Such counter-activity has been reported for other TA systems (Zhu et al., 2010). In case of HigBAAb, the two versions might work as counter-addiction modules for each other. The observed duplication of higBA2Ab could indicate the ongoing evolution and divergence of the novel TA species.
Author Contributions
JA, DJ, RK, and ES designed the experiments. JA, DJ, RK, and AČ performed the experiments. JA, DJ, and ES analyzed the data and wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank K. Montrimas, M. Bendorius, and S. Šimaitytė for their excellent technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00732/full#supplementary-material
References
- Anantharaman V., Aravind L. (2003). Application of comparative genomics in the identification and analysis of novel families of membrane-associated receptors in bacteria. BMC Genomics 4:34. 10.1186/1471-2164-4-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes L. C., Visca P., Towner K. J. (2014). Acinetobacter baumannii: evolution of a global pathogen. Pathog. Dis. 71 292–301. 10.1111/2049-632X.12125 [DOI] [PubMed] [Google Scholar]
- Cárdenas-Mondragón M. G., Ares M. A., Panunzi L. G., Pacheco S., Camorlinga-Ponce M., Girón J. A., et al. (2016). Transcriptional profiling of type II toxin-antitoxin genes of Helicobacter pylori under different environmental conditions: identification of HP0967-HP0968 system. Front. Microbiol. 7:1872. 10.3389/fmicb.2016.01872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso K., Gandra R. F., Wisniewski E. S., Osaku C. A., Kadowaki M. K., Felipach-Neto V., et al. (2010). DnaK and GroEL are induced in response to antibiotic and heat shock in Acinetobacter baumannii. J. Med. Microbiol. 59 1061–1068. 10.1099/jmm.0.020339-0 [DOI] [PubMed] [Google Scholar]
- Chan W. T., Espinosa M., Yeo C. C. (2016). Keeping the wolves at bay: antitoxins of prokaryotic type II toxin-antitoxin systems. Front. Mol. Biosci. 3:9. 10.3389/fmolb.2016.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverton A. M., Gollan B., Przydacz M., Wong C. T., Mylona A., Hare S. A., et al. (2016). A Salmonella toxin promotes persister formation through acetylation of tRNA. Mol. Cell 63 86–96. 10.1016/j.molcel.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen-Dalsgaard M., Gerdes K. (2006). Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol. Microbiol. 62 397–411. 10.1111/j.1365-2958.2006.05385.x [DOI] [PubMed] [Google Scholar]
- Christensen-Dalsgaard M., Jørgensen M. G., Gerdes K. (2010). Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 75 333–348. 10.1111/j.1365-2958.2009.06969.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook G. M., Robson J. R., Frampton R. A., McKenzie J., Przybilski R., Fineran P. C., et al. (2013). Ribonucleases in bacterial toxin-antitoxin systems. Biochim. Biophys. Acta 1829 523–531. 10.1016/j.bbagrm.2013.02.007 [DOI] [PubMed] [Google Scholar]
- De la Cruz M. A., Zhao W., Farenc C., Gimenez G., Raoult D., Cambillau C., et al. (2013). A toxin-antitoxin module of Salmonella promotes virulence in mice. PLoS Pathog. 9:e1003827. 10.1371/journal.ppat.1003827 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Díaz-Orejas R., Diago-Navarro E., Arriaga A. M. H., López-Villarejo J., Lemonnier M., Moreno-Córdoba I., et al. (2010). Bacterial toxin-antitoxin systems targeting translation. J. Appl. Biomed. 8 179–188. 10.2478/v10136-009-0021-9 [DOI] [Google Scholar]
- Dziewit L., Jazurek M., Drewniak L., Baj J., Bartosik D. (2007). The SXT conjugative element and linear prophage N15 encode toxin-antitoxin-stabilizing systems homologous to the tad-ata module of the Paracoccus aminophilus plasmid pAMI2. J. Bacteriol. 189 1983–1997. 10.1128/JB.01610-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelkamp B. A., Hassan K. A., Paulsen I. T., Brown M. H. (2011). Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics 12:126. 10.1186/1471-2164-12-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg-Kulka H., Glaser G. (1999). Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53 43–70. 10.1146/annurev.micro.53.1.43 [DOI] [PubMed] [Google Scholar]
- Fernández-García L., Blasco L., Lopez M., Bou G., García-Contreras R., Wood T., et al. (2016). Toxin-antitoxin systems in clinical pathogens. Toxins 8:227. 10.3390/toxins8070227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Christensen S. K., Løbner-Olesen A. (2005). Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3 371–382. 10.1038/nrmicro1147 [DOI] [PubMed] [Google Scholar]
- Giannouli M., Antunes L. C. S., Marchetti V., Triassi M., Visca P., Zarrilli R. (2013). Virulence-related traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I-III and to the emerging genotypes ST25 and ST78. BMC Infect. Dis. 13:282. 10.1186/1471-2334-13-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeders N., Van Melderen L. (2014). Toxin-antitoxin systems as multilevel interaction systems. Toxins 6 304–324. 10.3390/toxins6010304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Venkataraman B., Vasudevan M., Gopinath Bankar K. (2017). Co-expression network analysis of toxin-antitoxin loci in Mycobacterium tuberculosis reveals key modulators of cellular stress. Sci. Rep. 7:5868. 10.1038/s41598-017-06003-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A., Stanger F. V., Scheu P. D., de Jong I. G., Goepfert A., Glatter T., et al. (2015). Adenylylation of gyrase and topo IV by FicT toxins disrupts bacterial DNA topology. Cell Rep. 12 1497–1507. 10.1016/j.celrep.2015.07.056 [DOI] [PubMed] [Google Scholar]
- Heaton B. E., Herrou J., Blackwell A. E., Wysocki V. H., Crosson S. (2012). Molecular structure and function of the novel BrnT/BrnA toxin-antitoxin system of Brucella abortus. J. Biol. Chem. 287 12098–12110. 10.1074/jbc.M111.332163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Arriaga A. M., Chan W. T., Espinosa M., Díaz-Orejas R. (2015). “Conditional activation of toxin-antitoxin systems: postsegregational killing and beyond,” in Plasmids: Biology and Impact in Biotechnology and Discovery eds Tolmasky M., Alonso J. (Washington, DC: ASM Press; ) 175–192. 10.1128/microbiolspec.PLAS-0009-2013 [DOI] [Google Scholar]
- Howard A., O’Donoghue M., Feeney A., Sleator R. D. (2012). Acinetobacter baumannii: an emerging opportunistic pathogen. Virulence 3 243–250. 10.4161/viru.19700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger M., Schmucker R., Kishan V., Hillen W. (1990). Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87 45–51. 10.1016/0378-1119(90)90494-C [DOI] [PubMed] [Google Scholar]
- Imai Y., Matsushima Y., Sugimura T., Terada M. (1991). A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 19:2785 10.1093/nar/19.10.2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal N., Guérout A.-M., Krin E., Le Roux F., Mazel D. (2015). Comprehensive functional analysis of the 18 Vibrio cholerae N16961 toxin-antitoxin systems substantiates their role in stabilizing the superintegron. J. Bacteriol. 197 2150–2159. 10.1128/JB.00108-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S., Paul P., Padhi C., Ray S., Ryan D., Dash S., et al. (2016). The Hha-TomB toxin-antitoxin system shows conditional toxicity and promotes persister cell formation by inhibiting apoptosis-like death in S. Typhimurium. Sci. Rep. 6:38204. 10.1038/srep38204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawad A., Seifert H., Snelling A. M., Heritage J., Hawkey P. M. (1998). Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J. Clin. Microbiol. 36 1938–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurenaite M., Markuckas A., Suziedeliene E. (2013). Identification and characterization of type II toxin-antitoxin systems in the opportunistic pathogen Acinetobacter baumannii. J. Bacteriol. 195 3165–3172. 10.1128/JB.00237-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G., Pidoux J., Ullmann A., Ladant D. (1998). A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95 5752–5756. 10.1073/pnas.95.10.5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kędzierska B., Hayes F. (2016). Emerging roles of toxin-antitoxin modules in bacterial pathogenesis. Molecules 21:790. 10.3390/molecules21060790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedzierska B., Lian L.-Y., Hayes F. (2007). Toxin-antitoxin regulation: bimodal interaction of YefM-YoeB with paired DNA palindromes exerts transcriptional autorepression. Nucleic Acids Res. 35 325–339. 10.1093/nar/gkl1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lean S. S., Yeo C. C. (2017). Small, enigmatic plasmids of the nosocomial pathogen, Acinetobacter baumannii: good, bad, Who Knows? Front. Microbiol. 8:1547. 10.3389/fmicb.2017.01547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.-Y., Lee B.-J. (2016). Structure, biology, and therapeutic application of toxin-antitoxin systems in pathogenic bacteria. Toxins 8:e305. 10.3390/toxins8100305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplae R., Geeraerts D., Hallez R., Guglielmini J., Drèze P., Van Melderen L. (2011). Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 39 5513–5525. 10.1093/nar/gkr131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobato-Márquez D., Díaz-Orejas R., García-Del Portillo F. (2016a). Toxin-antitoxins and bacterial virulence. FEMS Microbiol. Rev. 40 592–609. 10.1093/femsre/fuw022 [DOI] [PubMed] [Google Scholar]
- Lobato-Márquez D., Molina-García L., Moreno-Córdoba I., García-Del Portillo F., Díaz-Orejas R. (2016b). Stabilization of the virulence plasmid pSLT of Salmonella Typhimurium by three maintenance systems and its evaluation by using a new stability test. Front. Mol. Biosci. 3:66. 10.3389/fmolb.2016.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobato-Márquez D., Moreno-Córdoba I., Figueroa V., Díaz-Orejas R., García-del Portillo F. (2015). Distinct type I and type II toxin-antitoxin modules control Salmonella lifestyle inside eukaryotic cells. Sci. Rep. 5:9374. 10.1038/srep09374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson R. D. (2007). Hypothetical functions of toxin-antitoxin systems. J. Bacteriol. 189 6089–6092. 10.1128/JB.00958-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K. S., Wolf Y. I., Koonin E. V. (2009). Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct 4:19. 10.1186/1745-6150-4-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H., Tan Q., Awano N., Wu K.-P., Inouye M. (2012). YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol. Microbiol. 84 979–989. 10.1111/j.1365-2958.2012.08068.x [DOI] [PubMed] [Google Scholar]
- McConnell M. J., Actis L., Pachón J. (2013). Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol. Rev. 37 130–155. 10.1111/j.1574-6976.2012.00344.x [DOI] [PubMed] [Google Scholar]
- McVicker G., Tang C. M. (2016). Deletion of toxin-antitoxin systems in the evolution of Shigella sonnei as a host-adapted pathogen. Nat. Microbiol. 2:16204. 10.1038/nmicrobiol.2016.204 [DOI] [PubMed] [Google Scholar]
- Muthuramalingam M., White J. C., Bourne C. R. (2016). Toxin-antitoxin modules are pliable switches activated by multiple protease pathways. Toxins 8:e214. 10.3390/toxins8070214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutschler H., Gebhardt M., Shoeman R. L., Meinhart A. (2011). A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis. PLoS Biol. 9:e1001033. 10.1371/journal.pbio.1001033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwugo C. C., Arivett B. A., Zimbler D. L., Gaddy J. A., Richards A. M., Actis L. A. (2012). Effect of ethanol on differential protein production and expression of potential virulence functions in the opportunistic pathogen Acinetobacter baumannii. PLoS One 7:e51936. 10.1371/journal.pone.0051936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R., Peti W. (2016). Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 12 208–214. 10.1038/nchembio.2044 [DOI] [PubMed] [Google Scholar]
- Povilonis J., Seputiene V., Krasauskas R., Juskaite R., Miskinyte M., Suziedelis K., et al. (2013). Spread of carbapenem-resistant Acinetobacter baumannii carrying a plasmid with two genes encoding OXA-72 carbapenemase in Lithuanian hospitals. J. Antimicrob. Chemother. 68 1000–1006. 10.1093/jac/dks499 [DOI] [PubMed] [Google Scholar]
- Providenti M. A., O’Brien J. M., Ewing R. J., Paterson E. S., Smith M. L. (2006). The copy-number of plasmids and other genetic elements can be determined by SYBR-Green-based quantitative real-time PCR. J. Microbiol. Methods 65 476–487. 10.1016/j.mimet.2005.09.007 [DOI] [PubMed] [Google Scholar]
- Ramage H. R., Connolly L. E., Cox J. S. (2009). Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 5:e1000767. 10.1371/journal.pgen.1000767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramisetty B. C., Santhosh R. S. (2016). Horizontal gene transfer of chromosomal Type II toxin-antitoxin systems of Escherichia coli. FEMS Microbiol. Lett. 363:fnv238. 10.1093/femsle/fnv238 [DOI] [PubMed] [Google Scholar]
- Ramisetty B. C. M., Ghosh D., Roy Chowdhury M., Santhosh R. S. (2016). What is the link between stringent response, endoribonuclease encoding type II toxin-antitoxin systems and persistence? Front. Microbiol. 7:1882 10.3389/fmicb.2016.01882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramisetty B. C. M., Santhosh R. S. (2017). Endoribonuclease type II toxin-antitoxin systems: functional or selfish? Microbiology 163 931–939. 10.1099/mic.0.000487 [DOI] [PubMed] [Google Scholar]
- Rocker A., Meinhart A. (2016). Type II toxin: antitoxin systems. More than small selfish entities? Curr. Genet. 62 287–290. 10.1007/s00294-015-0541-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa R., Depascale D., Cleary T., Fajardo-Aquino Y., Kett D. H., Munoz-Price L. S. (2014). Differential environmental contamination with Acinetobacter baumannii based on the anatomic source of colonization. Am. J. Infect. Control 42 755–757. 10.1016/j.ajic.2014.03.016 [DOI] [PubMed] [Google Scholar]
- Saavedra De Bast M., Mine N., Van Melderen L. (2008). Chromosomal toxin-antitoxin systems may act as anti addiction modules. J. Bacteriol. 190 4603–4609. 10.1128/JB.00357-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Sala A., Bordes P., Genevaux P. (2014). Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins 6 1002–1020. 10.3390/toxins6031002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schureck M. A., Maehigashi T., Miles S. J., Marquez J., Cho S. E., Erdman R., et al. (2014). Structure of the Proteus vulgaris HigB-(HigA)2-HigB toxin-antitoxin complex. J. Biol. Chem. 289 1060–1070. 10.1074/jbc.M113.512095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta M., Austin S. (2011). Prevalence and significance of plasmid maintenance functions in the virulence plasmids of pathogenic bacteria. Infect. Immun. 79 2502–2509. 10.1128/IAI.00127-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev V. V., Salamov A. (2011). “Automatic annotation of microbial genomes and metagenomic sequences,” in Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies Genetics - Research and Issues ed. Robert L. W. (Hauppauge, NY: Nova Science Publishers; ) 61–78. [Google Scholar]
- Sterckx Y. G., De Gieter S., Zorzini V., Hadži S., Haesaerts S., Loris R., et al. (2015). An efficient method for the purification of proteins from four distinct toxin-antitoxin modules. Protein Expr. Purif. 108 30–40. 10.1016/j.pep.2015.01.001 [DOI] [PubMed] [Google Scholar]
- Sterckx Y. G., Jové T., Shkumatov A. V., Garcia-Pino A., Geerts L., De Kerpel M., et al. (2016). A unique hetero-hexadecameric architecture displayed by the Escherichia coli O157 PaaA2-ParE2 antitoxin-toxin complex. J. Mol. Biol. 428 1589–1603. 10.1016/j.jmb.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Sužiedėlienė E., Jurėnaitė M., Armalytė J. (2016). “Identification and characterization of type II toxin–antitoxin systems in the opportunistic pathogen Acinetobacter Baumannii,” in Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria ed. de Bruijn F. J. (Hoboken, NJ: John Wiley & Sons, Inc.) 454–462. 10.1002/9781119004813.ch41 [DOI] [Google Scholar]
- Tian Q. B., Ohnishi M., Tabuchi A., Terawaki Y. (1996). A new plasmid-encoded proteic killer gene system: cloning, sequencing, and analyzing hig locus of plasmid Rts1. Biochem. Biophys. Res. Commun. 220 280–284. 10.1006/bbrc.1996.0396 [DOI] [PubMed] [Google Scholar]
- Van Melderen L., Saavedra De Bast M. (2009). Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 5:e1000437. 10.1371/journal.pgen.1000437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand I., Hilpert K., Hancock R. E. W. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3 163–175. 10.1038/nprot.2007.521 [DOI] [PubMed] [Google Scholar]
- Zhu L., Sharp J. D., Kobayashi H., Woychik N. A., Inouye M. (2010). Noncognate Mycobacterium tuberculosis toxin-antitoxins can physically and functionally interact. J. Biol. Chem. 285 39732–39738. 10.1074/jbc.M110.163105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


