Abstract
The difficulty of long-term management has produced a high rate of failure for obesity patients. Therefore, improving the efficacy of current obesity treatment is a significant goal. We hypothesized that combining a probiotic Lactobacillus mali APS1 intervention with dieting could improve the efficacy of obesity and hepatic steatosis treatment compared to dieting alone. Mice were fed a high-fat diet for 6 weeks and then treated with: saline + normal diet and APS1 + normal diet (NDAPS1) for 3 weeks. NDAPS1 accelerated body weight loss and reduced caloric intake and fat accumulation. The fecal microbiome showed that accelerating weight loss by NDAPS1 resulted in restoring intestinal microbiota toward a pre-obese state, with alteration of specific changes in the obesity-associated bacteria. APS1 manipulated the gut microbiome’s obesity-associated metabolites, followed by regulation of lipid metabolism, enhancement of energy expenditure and inhibition of appetite. The specific hepatic metabolites induced by the APS1-manipulated gut microbiome also contributed to the amelioration of hepatic steatosis. Our results highlighted a possible microbiome and metabolome that contributed to accelerating weight loss following treatment with a combination of APS1 and dieting and suggested that probiotics could serve as a potential therapy for modulating physiological function and downstream of the microbiota.
Introduction
An alarming increase in the prevalence of overweight and obesity worldwide has occurred in the past decade1. Obesity usually accompanies a high risk of metabolic disorder and several complications, such as multiple disabilities, diabetes, heart disease and cancers. Physical exercise and dieting have been considered the first-line therapeutic treatment of obesity and obesity-associated metabolic disorders2. Recent studies have focused on dietary modifications, such as calorie restriction3 or antioxidant intervention4, to limit and protect against high fat diet (HFD)-induced weight gain and lipogenesis in vivo. Nevertheless, on a spectrum of medical and scientific supports, long-term management intending to attenuate or reverse the prevalence of obesity still fails to fulfill expectations. Therefore, it is urgent to investigate effective treatment that could enhance the effects of current management protocols for obesity.
The gut microbiome plays a critical role affecting metabolic homeostasis and risk of developing obesity with its metabolic complications5,6. The compositional and functional microbiome alterations, such as proportions of the two predominant bacteria, Firmicutes and Bacteroidetes7, and intestinal endotoxin-producing bacteria8, have been suggested to contribute to the pathogenesis of obesity in both animal models and humans. Fecal microbiota transplantation from obese mice into germ-free mice led to an increase in body weight gain and energy absorption9, also indicating that dysbiosis of the gut microbiome is associated with pathological activities in obesity. Moreover, accumulating evidence has demonstrated that changes in the gut microbiome are linked to a variety of factors related to host metabolism, such as the obesity-related metabolome10, regulation of appetitive gut hormones11, fermentation of intestinal short chain fatty acids (SCFAs)12, and progress of nonalcoholic fatty liver disease5 in obesity.
Dietary changes have been demonstrated to be central drivers of microbiome composition and function. Probiotics are generally recognized as safe for human use13 and have been demonstrated to have beneficial effects on improvement of obesity through manipulating gut microbiota composition14. Specific probiotic strain interventions also showed protective abilities against hepatic steatosis by inhibiting fatty acid synthesis and suppressing inflammation in vivo15,16. However, most probiotic studies were focused on the protective effect on obesity. There have been few studies reporting the efficacy of probiotics in combination with dietary modifications on the improvement of obesity. The mechanisms underlying probiotic-induced weight-loss with dieting remain to be determined.
In our previous study, we demonstrated that administration of Lactobacillus mali APS1 (APS1), which was isolated from sugary kefir grain, reduced body weight gain and inflammatory activity and changed body composition in HFD-induced obese mice17. Thus, based on our previsions findings, we hypothesized that combining APS1 intervention and dieting could accelerate body weight loss and improve obesity-related pathologies comparing with dieting along. To test the hypothesis, we evaluated the impact of a combination of APS1 and dieting on body weight control and obesity-related metabolome and structure of the gut microbiota in vivo and gained an understanding of the mechanisms of obesity treatment. The mechanisms underlying the probiotic-induced weight-loss with dieting were also evaluated through analysis of the microbiome and metabolome. Our findings indicated that the combination of probiotic APS1 and dieting might be a potential alternative to enhance the efficacy of obesity management.
Results
APS1 accelerated body weight loss, reduced caloric intake, and lowered fat mass in obese mice on a diet
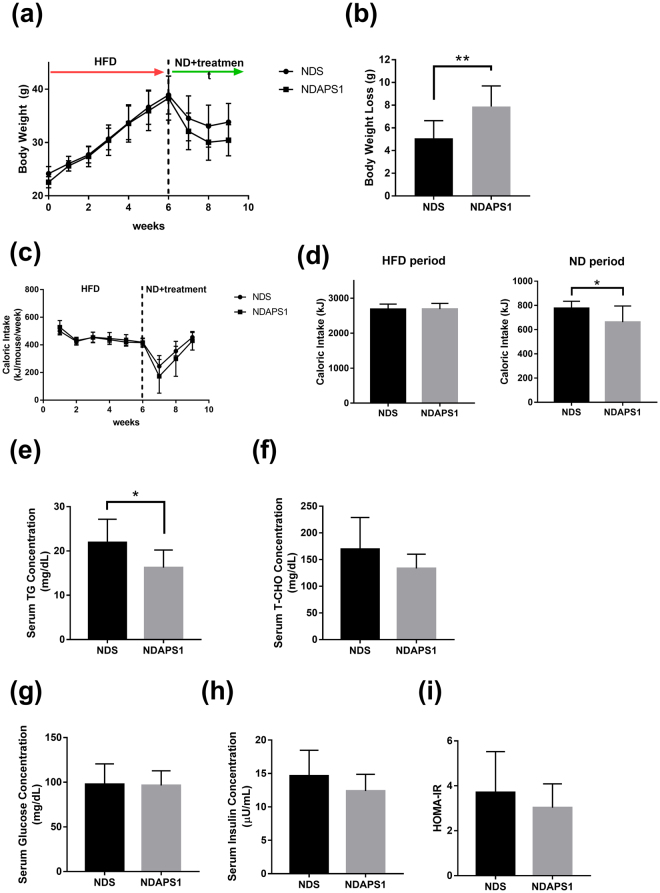
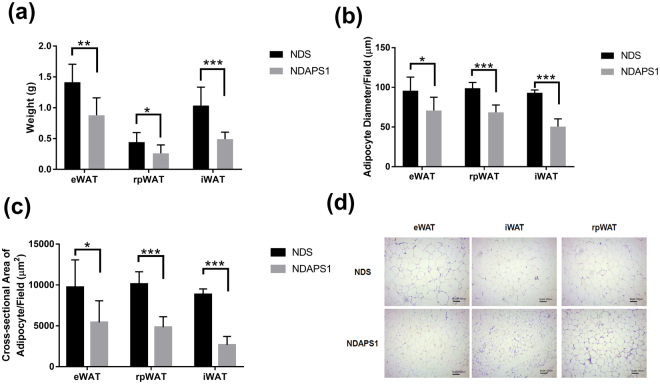
To examine the effect of APS1 intervention on physiological features of preexisting obesity after dieting, test mice were exposed to an HFD first and then subsequently fed an ND. Our results indicated that body weight gain and metabolic disorder syndromes developed during primary exposure to an HFD followed by weight reduction during exposure to an ND in all test mice (Fig. 1a). APS1 intervention (NDAPS1 group) produced significantly higher weight loss (7.89 ± 1.69 g) when compared with saline treatment (NDS) (5.09 ± 1.45 g) after exposure to an ND (P < 0.01) (Fig. 1b). Acceleration of weight loss during the dieting stage in the NDAPS1 group was associated with a decrease in cumulative caloric intake. The cumulative caloric intake of the NDAPS1 group was significantly reduced compared to that of the NDS group (P < 0.05) (Fig. 1c,d). Moreover, compared to the NDS group, the NDAPS1 group showed decreases in the serum TG concentration (by 25.83%, P < 0.05) (Fig. 1e), weights of eWAT (by 37.93%, P < 0.01), rpWAT (by 41.24%, P < 0.05) and iWAT (by 52.53%, P < 0.001) (Fig. 2a), adipocyte diameters of eWAT (by 26.07%, P < 0.05), rpWAT (by 30.70%, P < 0.001) and iWAT (by 45.71%, P < 0.001) (Fig. 2b), and cross-sectional areas of adipocytes in eWAT (by 44.24%, P < 0.05), rpWAT (by 52.21%, P < 0.001) and iWAT (by 69.77%, P < 0.001) (Fig. 2c,d), but not the serum total cholesterol concentration (P > 0.05) (Fig. 1f). The level of fasting glucose showed no difference between the NDS and NDAPS1 groups (P > 0.05) (Fig. 1g). A tendency to decrease the levels of fasting insulin and HOMA-IR in the NDAPS1 group was observed in this study but there was no significant difference between two groups (P > 0.05) (Fig. 1h,i). Overall, these results indicated that APS1 intervention accelerated body weight loss and reduced fat mass in preexisting obese mice on a diet.
Figure 1.
NDAPS1 accelerated body weight loss compared to NDS in preexisting obese mice after dieting. Effect of NDAPS1 on (a) body weight, (b) body weight loss, (c,d) caloric intake, (e) serum triglyceride, (f) total cholesterol, (g) glucose, (h) insulin levels and (i) HOMA-IR in preexisting obese mice are shown. n = 8 per group. The data are expressed as the mean ± SD. Statistical analysis was conducted by using unpaired two-tailed student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
Figure 2.
NDAPS1 reduced fat accumulation in preexisting obese mice after dieting. NDAPS1 reduced (a) WAT weight, (b) diameter and (c) cross-sectional area of adipocytes. (d) Representative histological sections of WAT from mice treated with NDS and NDAPS1 were stained with H&E staining. Scale bar indicates 100 μm. n = 8 per group. The data are expressed as the mean ± SD. Statistical analysis was conducted by using unpaired two-tailed student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
APS1 ameliorated hepatic steatosis in preexisting obese mice after dieting
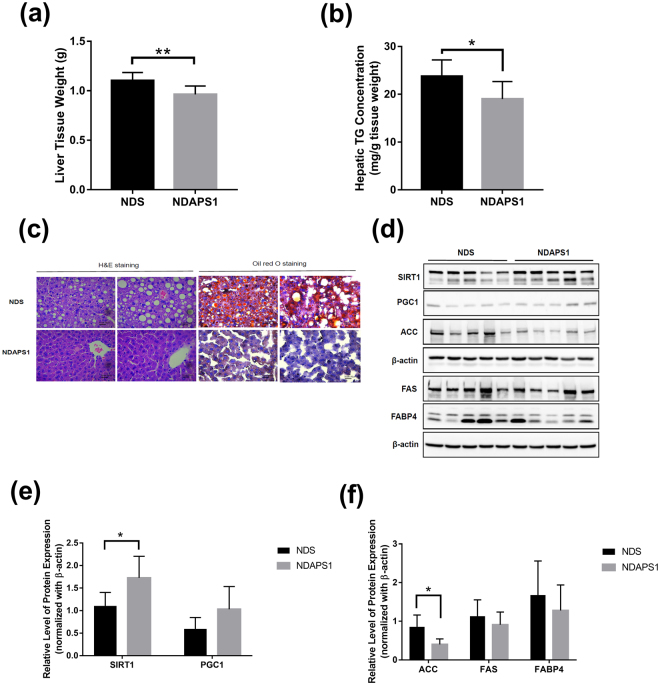
We further evaluated the effect of APS1 intervention on the improvement of hepatic steatosis in preexisting obese mice after dieting. The NDAPS1 group had significantly lower liver tissue weight (P < 0.01) (Fig. 3a) and liver TG level (P < 0.05) (Fig. 3b) compared with the NDS group. Reducing the incidence of hepatocyte vacuolation and suppressing accumulation of lipid droplets in liver tissue was also observed in the NDAPS1 group by histological H&E staining and Oil Red O staining (Fig. 3c).
Figure 3.
NDAPS1 reduced hepatic steatosis in preexisting obese mice after dieting. (a) NDAPS1 reduced liver weight. (b) NDAPS1 reduced hepatic TG levels. (c) Representative histological sections of liver tissues from mice treated with NDS and NDAPS1 were stained with H&E and Oil Red O staining. Scale bar indicates 25 μm. (d) Representative Western blot analysis for expression of SIRT1, PGC1, ACC, FAS, FABP4 and β-actin proteins in liver tissues of NDS and NDAPS1-treated mice. The cropped blots are displayed in the figure, the cropping line is delineated with the black frame line. Un-cropped full length blots are presented in Supplementary Figure S4. (e,f) Quantitative Western blot analysis by ImageJ software. n = 8 per group. The data are expressed as the mean ± SD. Statistical analysis was conducted by using unpaired two-tailed student’s t-test. *P < 0.05, **P < 0.01.
The molecular mechanism involving APS1-ameliorated hepatic steatosis including sirtuin (SIRT) 1, peroxisome proliferator-activated receptor gamma coactivator 1 (PGC1), lipid synthesis factors (ACC, FAS) and fatty acid transporter FABP4 was investigated by immunoblot assay (Fig. 3d–f). The results indicated that the NDAPS1 group showed significantly higher expression of SIRT1 (P < 0.05) and lower ACC (P < 0.05) in the liver tissue of mice compared with the NDS group. The mice that received the APS1 intervention also showed a trend of increasing expression of PGC1 and decreasing expression of FAS and FABP4 in liver tissue. These results suggested that the preexisting obese mice in the APS1 intervention not only showed accelerated weight loss after dieting but also showed enhanced amelioration of hepatic steatosis through regulation of expression of lipogenesis molecules.
APS1 manipulated obesity-associated gut microbiota in preexisting obese mice after dieting
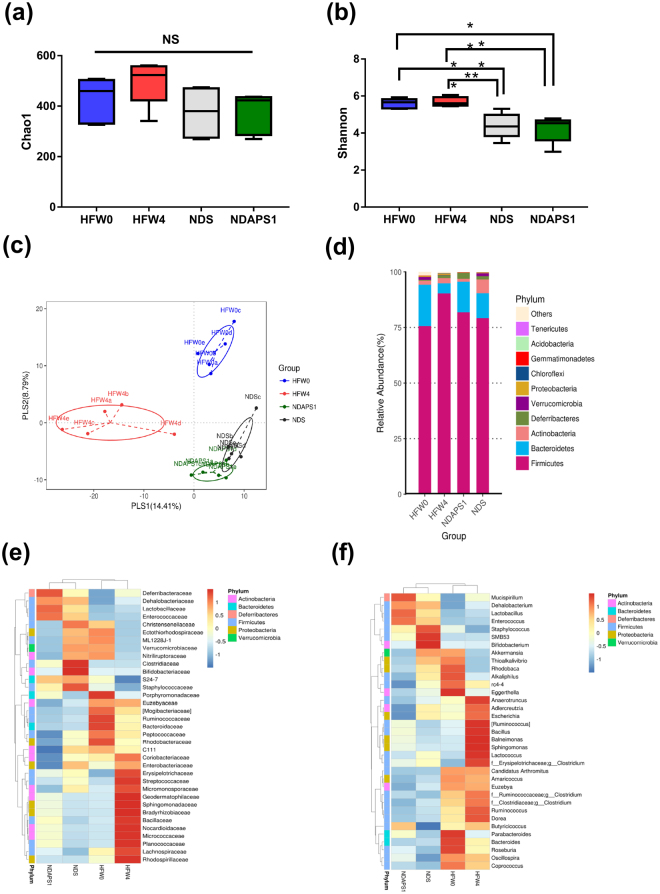
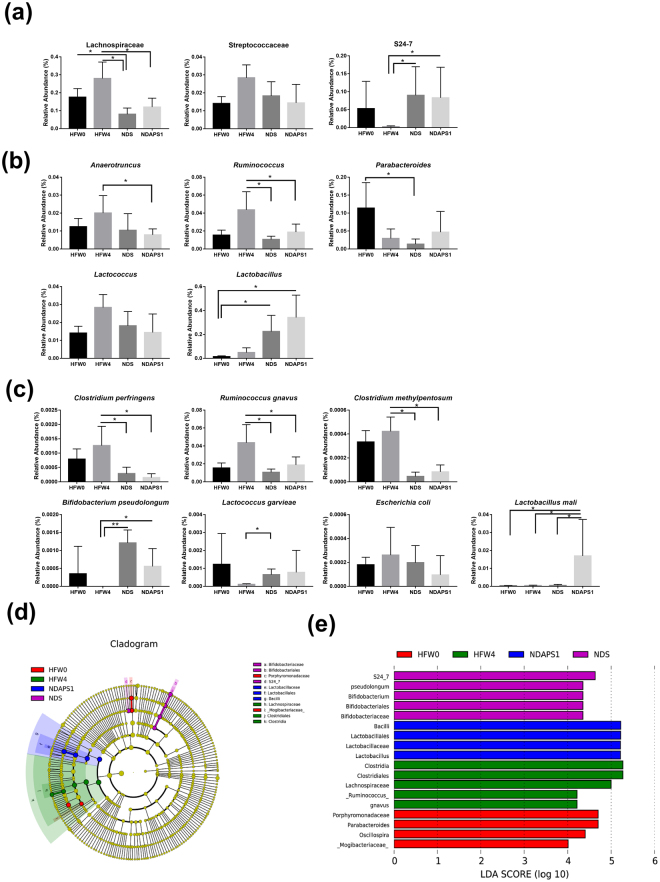
We next investigated whether APS1 intervention affected the composition of the gut microbiota in the feces of the test mice, as the gut microbiota might be involved in the metabolic complications of obesity and associated with the acceleration of weight loss after dieting. The chao1 index (Fig. 4a) showed that the bacterial richness was no difference among the groups. Whereas, Shannon’s diversity index (Fig. 4b), indicating the bacterial α-diversity, showed a tendency to increase in the HFW4 group compared with the HFW0 group. Both NDS and NDAPS1 groups also have a significantly lower Shannon diversity than the HFW0 and HFW4 groups.
Figure 4.
NDAPS1 manipulated gut microbiota in preexisting obese mice after dieting. (a) The chao1 richness estimator, (b) Shannon’s diversity index and (c) PLS-DA plots represented changes between samples collected from HFW0, HFW4, NDS and NDAPS1. NDAPS1 manipulated the abundance of the gut microbiota at the (d) phylum, (e) family and (f) genus levels. n = 5 per group. The nonparametric Wilcoxon signed rank test for paired data and Mann-Whitney U test for unpaired data (NDS vs. NDAPS1) were used. *P < 0.05, **P < 0.01, ***P < 0.001. NS: not significant.
The partial least squares discriminant analysis (PLS-DA) was performed based on OTUs level to evaluate the variant of gut microbiota composition among the groups (Fig. 4c). PLS-DA plot showed that PLS 1 and PLS 2 explained 14.41% and 8.79% of variation of gut microbiota composition, respectively. A significant separation in PLS 1 and PLS2 between each group was observed (P < 0.05) (Supplemental Fig. S1). These findings indicated that the composition of gut microbiota, which has been assumed to be in a dysbiotic state during the obesity phase, could be altered to an intermediate configuration between dysbiotic and normal states.
To further investigate the variability of gut microbiota between NDS and NDAPS1, a Venn diagram showing the overlaps between the groups was performed. The results indicated that 409 of the total richness of 751 OTUs were shared. Among shared OTUs, 74 OTUs and 127 OTUs were upregulated and downregulated, respectively, more than two folds in NDAPS1 when compared with NDS (Supplemental Fig. S2). Thus, we also analyzed the abundance of specific bacterial taxa to clarify the role of APS1 in alteration of intestinal microbiota. The results of the histogram of phyla (Fig. 4d) and heat maps of family (Fig. 4e) and genus (Fig. 4f) demonstrated very diverse patterns among treatments. After further taxonomic analysis, we observed that the mice in the diet period (NDS and NDAPS1 groups) had significantly reduced abundance of the Lachnospiraceae family, Ruminococcus genus and Clostridium perfringens, Ruminococcus gnavus, and Clostridium methylpentosum species (P < 0.05) and enhanced abundance of the S24_7 family and Lactococcus garvieae, and Bifidobacterium pseudolongum species when compared with obese mice (HFW4) (P < 0.05). Only APS1 intervention significantly increased the abundance of Lactobacillus mali and decreased the abundance of the Streptococcaceae family and Anaerotruncus genus (P < 0.05) (Fig. 5a–c).
Figure 5.
NDAPS1 manipulated gut microbiota in preexisting obese mice after dieting. NDAPS1 manipulated the abundance of specific bacteria at the (a) family, (b) genus and (c) species level. (d,e) LEfSe comparison of gut microbiota between HFW0, HFW4, NDS and NDAPS1 groups. n = 5 per group. The data are expressed as the mean ± SD. The nonparametric Wilcoxon signed rank test for paired data and Mann-Whitney U test for unpaired data (NDS vs. NDAPS1) were used. *P < 0.05, **P < 0.01, ***P < 0.001.
Linear discriminant analysis effect size (LEfSe) was also calculated to further identify the specific bacterial taxa that were predominant in the 4 treatment groups. A total of 18 differentially abundant taxonomic clades (α = 0.05) were observed (Fig. 5d,e). The family Mogibacteriaceae (within the phylum Firmicutes) and genera Parabacteroides (within the phylum Bacteroidetes) and Oscillospira (within the phylum Firmicutes) were initially abundant in the HFW0 group. After HFD feeding for 4 weeks, the species Ruminococcus gnavus (within the phylum Firmicutes) became abundant. When on a diet, the most abundant taxa were the family S24_7 (within the phylum Bacteroidetes) and species Bifidobacterium pseudolongum (within the phylum Actinobacteria) in the NDS group. APS1 intervention specifically enriched the abundance of the Lactobacillus genus (within the phylum Firmicutes) in test mice.
We further used green fluorescent protein (GFP) to trace APS1 in the gastrointestinal tract of test mice (Supplemental Fig. S3a). After gavage for three hours, the results of immunofluorescence staining of cryostat sections confirmed that GFP-expressing APS1 was detectable in the terminal of the ileum of the mice, which is the region that Lactobacillus species frequently colonize18 (Supplemental Fig. S3b). These finding might indicate that accelerating weight loss by APS1 intervention in mice results in manipulating the intestinal microbiota with alteration of specific bacterial taxa.
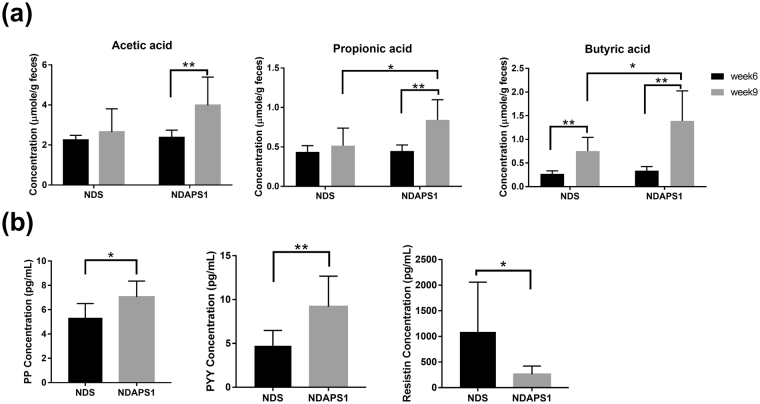
APS1 affected fecal SCFAs excretion and expression of satiety hormones in preexisting obese mice after dieting
Given the above causal connection between microbiota configuration and weight loss after dieting, we asked whether APS1 intervention could affect fecal SCFAs through alteration of gut microorganisms and further regulate the expression of satiety hormones to accelerate weight loss and ameliorate hepatic steatosis. The results indicated that the mice in the APS1 intervention significantly excreted higher fecal propionic acid and butyric acid levels compared to the NDS group at week 9 (P < 0.05) (Fig. 6a). We then investigated satiety hormones, PP, PYY and resistin, which are associated with appetite regulation. The results showed that in the mice that received the APS1 intervention, the production of serum PP (P < 0.05) and PYY (P < 0.01) were significantly induced and the production of resistin was significantly decreased (P < 0.05) compared with the NDS group after dieting (Fig. 6b).
Figure 6.
NDAPS1 increased fecal SCFA extraction and serum satiety hormone production in preexisting obese mice after dieting. (a) NDAPS1 increased SCFA extraction in feces of test mice at 6 weeks and 9 weeks. (b) NDAPS1 regulated satiety hormone production in the serum of test mice n = 8 per group. The data are expressed as the mean ± SD. Statistical analysis was conducted by using paired (week 6 vs. week 9) and unpaired (NDS vs. NDAPS1) two-tailed student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
APS1-regulating metabolites contributed to accelerated weight loss in preexisting obese mice after dieting
To further gain insight into how the APS1 intervention accelerated weight loss and ameliorated hepatic steatosis, we compared the serum metabolomics profiles of the NDS and NDAPS1 groups. The PCA plot showed that PC 1 and PC2 explained 26.78% and 19.54% of variation, respectively, with a significant difference in PC2 between NDS and NDAPS1 (P < 0.001) (Fig. 7a). Among the metabolites, 11 compounds showed significant differences between the NDS and NDAPS1 groups (P < 0.05) (Table 1). These metabolites were carnitine, hexadecanoyl-L-carnitine, L-gulono-1,4-lactone, L-acetylcarnitine, D-glucurono-6,3-lactone, hydroxykynurenine, pyruvic acid, taurocholate, D-glutamine, β-aminoisobutyric acid and glycerol 3-phosphate. After analyzing the possible metabolic pathways analysis using the KEGG database (Table 1), the APS1 intervention increased metabolites predominantly related to fatty acid degradation, ascorbate biosynthesis, conjugated bile acid biosynthesis and branched-chain amino acid degradation and reduced the triacylglycerol biosynthesis-associated metabolites in preexisting obese mice.
Figure 7.
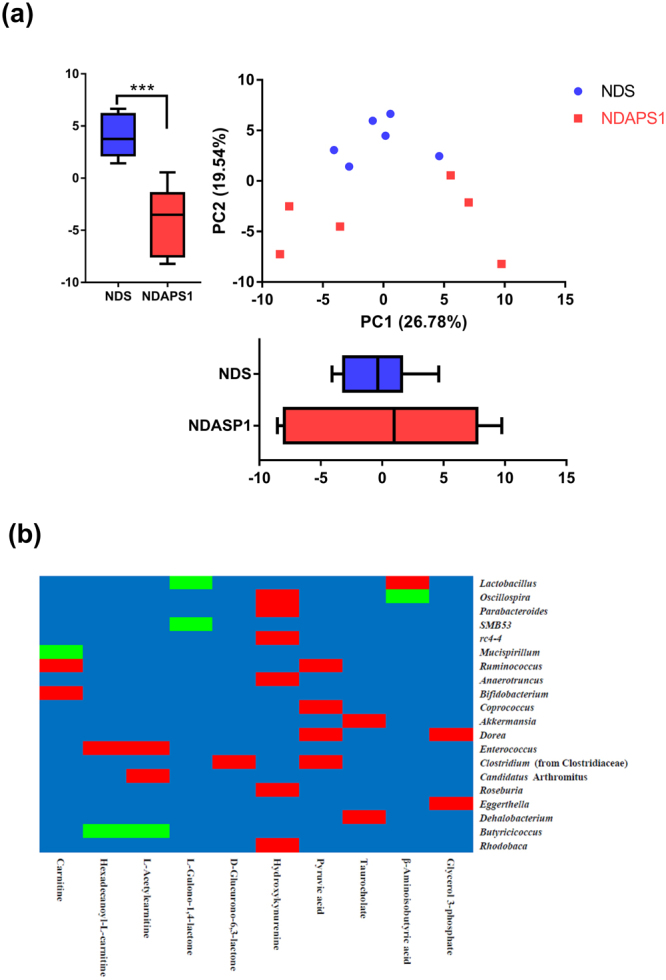
Two-dimensional PCA of metabolomics profile and Pearson’s correlation tests between predominant genera-level microbiota and metabolites. (a) PCA was based on PC1 and PC2 from mean intensity values of total detected metabolites in serum samples obtained from the NDS (solid blue circle) and NDAPS1 (solid orange square) groups. n = 6 per group. The data are expressed as the mean ± SD. Statistical analysis was conducted by using unpaired two-tailed student’s t-test. ***P < 0.001. (b) The relative abundance of bacterial taxa was significantly correlated with specific metabolites. Each cell was colored corresponding to Pearson’s correlation results. The significant positive correlation (P < 0.05) is represented as a red cell, and the significant negative correlation (P < 0.05) is represented as a green cell. The blue cells indicate the correlations were not significant (P > 0.05).
Table 1.
Detected differential metabolites between NDS and NDAPS1 groups by LC-MS.
| Metabolites | Ion(m/z) | Mean intensity | Fold change | p-value | Physiological function | Metabolic pathway | |
|---|---|---|---|---|---|---|---|
| NDS | NDAPS1 | (NDAPS1/NDS) | |||||
| Carnitine | 162.112 | 267596.07 | 441492.50 | 1.65 | 0.009 | Fatty acid degradation | Lipid metabolism |
| Hexadecanoyl-L-carnitine | 422.324 | 134.10 | 1892.88 | 14.12 | 0.017 | Fatty acid degradation | Lipid metabolism |
| L-Gulono-1,4-lactone | 177.040 | 199.88 | 1880.93 | 9.41 | 0.017 | Ascorbate biosynthesis | Ascorbate and aldarate metabolism |
| L-Acetylcarnitine | 204.123 | 845970.59 | 1018787.57 | 1.20 | 0.026 | Insulin resistance | Lipid metabolism |
| D-Glucurono-6,3-lactone | 175.024 | 12205.64 | 2117.76 | 0.17 | 0.026 | Ascorbic acid synthesis | Ascorbate and aldarate metabolism |
| Hydroxykynurenine | 225.086 | 1663.02 | 283.58 | 0.17 | 0.026 | Amino acid biosynthesis | Tryptophan metabolism |
| Pyruvic acid | 87.008 | 4556.93 | 332.37 | 0.07 | 0.026 | Energy source | Citrate cycle |
| Taurocholate | 514.284 | 1666.34 | 89307.51 | 53.60 | 0.041 | Conjugated bile acid biosynthesis | Taurine and hypotaurine metabolism |
| D-Glutamine | 169.058 | 6640.24 | 8602.06 | 1.30 | 0.041 | Insulin and glucose modulation | D-Glutamine and D-glutamate metabolism |
| β-Aminoisobutyric acid | 104.070 | 1459.53 | 3631.36 | 2.49 | 0.044 | Branched-chain amino acid degradation | Amino acid metabolism |
| Glycerol 3-phosphate | 171.006 | 1163.82 | 420.82 | 0.36 | 0.044 | Triacylglycerol biosynthesis | Lipid metabolism |
1 n = 6 per group.
2 Fold change was calculated by dividing the mean of the peak intensity of each metabolite from NDS and NDAPS1 groups.
3 Statistical analysis was conducted by using unpaired two-tailed student’s t-test.
Moreover, to explore the functional correlation between the NDAPS1-mediated gut microbiome and metabolites, the correlations among 20 predominant bacterial taxa at the genus level and 10 metabolites were calculated using Pearson’s correlation tests in Fig. 7b. Ruminococcus, Bifidobacterium, Enterococcus and Candidatus Arthromitus were positively correlated with fatty acid oxidation-related metabolites (including carnitine, hexadecanoyl-L-carnitine and L-acetylcarnitine), whereas Mucispirillum and Butyricicoccus were negatively correlated. Both Lactobacillus and SMB53 were negatively correlated with L-gulono-1,4-lactone. Clostridium (from Clostridiaceae) was positively correlated with D-glucurono-6,3-lactone. Oscillospira, Parabacteroides, rc4–4, Anaerotruncus, Roseburia and Rhodobaca were positively correlated with hydroxykynurenine. Ruminococcus, Coprococcus, Dorea and Clostridium (from Clostridiaceae) were positively correlated with pyruvic acid. Akkermansia and Dehalobacterium were positively correlated with taurocholate. β-Aminoisobutyric acid was positively correlated with Lactobacillus and was negatively correlated with Oscillospira. Glycerol 3-phosphate was positively correlated with Dorea and Eggerthella.
Discussion
In the present study, we demonstrated that administration of Lb. mali APS1, isolated from sugary kefir grain, could accelerate weight loss, improve the features of obesity and ameliorate hepatic steatosis in preexisting obese mice after dieting. Few similar papers have shown the effects of different probiotic interventions on weight loss during the dieting stage in rodent models and human pilot studies19–23. Among these studies, most results did not show significant effects, and only one study using Lb. plantarum showed a positive impact on weight loss20.
To further evaluate the possible mechanisms, we hypothesized that alteration of the microbiota configuration by APS1 intervention contributed to enhanced metabolic homeostasis through metabolite-mediated effects on host metabolism. The mechanism results highlighted three interdependent effects of the gut microbiome on acceleration of weight loss, metabolic modulation and hepatic lipid metabolism through APS1 intervention in combination with dieting.
First, the APS1 intervention induced specific changes in the diversity and abundance of obesity-associated bacteria in intestinal microbiota. A tendency to increase Shannon diversity index in HFW4 as compared with HFW0 was observed. Both NDS and NDAPS1 groups also showed a significantly lower Shannon diversity than HFW0 and HFW4. These findings differed from most studies on the obese individual showing the decrease in gut microbial diversity24,25. However, certain studies reported an increase in microbial diversity in HFD-fed rats26 and the diabetic subjects with high BMI27, which were parallel with our findings. The occasion of this inconsistency is unknown and remains to re-evaluate, but the differences in small sample size, diet, caloric intake or other conditions may influence the observation28,29. Moreover, the distribution of bacterial abundance is also a key factor affecting the bacterial diversity. The NDS and NDAPS1 groups demonstrated more than 10 and 25 folds in abundance ratios of Lactobacillus / Clostridium than HFW4, respectively, resulting in reduction of Shannon diversity index. This finding might suggest that both APS1 intervention and weight loss could promote the beneficial bacteria on competitive exclusion of specific deleterious bacteria.
The abundance of obesity-associated bacteria in intestinal microbiota was also affected by the APS1 intervention. The decreased abundances of Lachnospiraceae, Ruminococcus and Clostridium were found in both dieting groups (NDS and NDAPS1). The Lachnospiraceae family has been shown to have a positive correlation with inflammation markers in white adipose tissues and body weight gain in diet-induced obese mice30. Ruminococcus and Clostridium were butyrate producers, which were related to reduce dietary energy31,32. Uniquely, APS1 intervention regulated the levels of Streptococcaceae, Anaerotruncus and Lactococcus. Increasing the abundance of the Streptococcaceae family was associated with obesity, but it had also been shown that some strains in Streptococcaceae could also induce mild inflammation in humans, leading to the uneven morbidity related to chronic wounds among diabetic subjects33. Anaerotruncus was reported to be linked to hepatic cirrhosis with Holdemania and Dorea and type 1 diabetes, but not specific to insulin resistance, which was normally associated with HFD34,35. Interestingly, we found that the abundance of Lactobacillus mali was significantly increased in NDAPS1 group. This result reflected that the APS1 strain may survive in the intestine and associated with increased proportion of Lactobacillus.
Second, the APS1-manipulated gut microbiome influenced obesity-associated metabolites, regulated lipid metabolism, enhanced energy expenditure and inhibited appetite. Carnitine, hexadecanoyl-L-carnitine, L-acetylcarnitine, β-aminoisobutyric acid and glycerol 3-phosphate have been reported to be involved in the lipid/fatty acid metabolic pathway. Carnitine aids in a shuttle process that makes long-chain fatty-acid coenzyme A derivatives available for β-oxidation, leading to increasing ketogenesis and lipolysis36, whereas other long-chain acyl-carnitines could be metabolic intermediates of the fatty acid incorporation pathway into erythrocyte membrane phosphatidylcholine, and phosphatidylethanolamine37. The secretion of β-aminoisobutyric acid can be triggered by increasing PGC1α protein expression in skeleton myocytes after exercise, resulting in activation of β-oxidation in hepatocytes and induction of brown adipocyte-specific gene expression in white adipocytes in vitro and in vivo38. Additionally, plasma β-aminoisobutyric acid could be increased by the degradation of branched-chain amino acids, which are associated with insulin resistance in obesity39. L-Glycerol 3-phosphate is a recognized precursor of glyceride glycerol. A rise in the concentration of L-glycerol 3-phosphate contributes to an increase in fatty acid esterification and triacylglycerol biosynthesis40. Glutamine, one of the most effective substrates for gluconeogenesis, is the major gluconeogenic precursor in the kidney41. The glucose-lowering effect of glutamine could be linked to the known effect of glutamine on fat metabolism such as inhibition of fatty acid oxidation and lipolysis42. A previous study reported that the reduction in serum glutamine concentration in obese individuals was associated with an increase in certain bacteria, such as Ruminococcus spp. and Dorea longicatena, which produce glutamate from glutamine43. An increase in serum taurocholate was associated with weight loss and energy expenditure by cholic acid treatment in vivo44. Taurocholate stimulating secretion of glucagon-like peptide-1 and PYY modulated insulin resistance and glucose metabolism in obese patients45,46.
We further revealed that several bacterial taxa were positively or negatively correlated with APS1-mediated specific metabolites in test mice. For instance, Akkermansia was found to be positively correlated with taurocholate, which is synthesized via oxidation of cholesterol in the liver. This finding was similar to one indicating that Akkermansia muciniphila was negatively associated with serum cholesterol and low-density lipoprotein cholesterol levels in postmenopausal women with obesity47. Dysbiosis of Oscillospira leading to obesity and metabolic disorder in vivo48 might positively and negatively correlate with hydroxykynurenine and β-aminoisobutyric acid, respectively. Based on our results, we propose that the association between specific gut microbiota and metabolites might be used for the prediction of metabolic disease.
In addition to serum metabolites, intestinal metabolites, propionic acid and butyric acid, were also modulated by APS1 supplement and then contributed to increased energy expenditure and suppressed appetite. The intestinal SCFAs, especially butyrate, could increase energy expenditure through increasing mitochondrial respiration in mice49. SCFAs also play crucial roles in the gut-brain axis in the host50. They could stimulate the secretion of satiety hormones PYY and PP, which can regulate anorexigenic signaling in appetite51. Besides, higher SCFA excretion and increased PYY and PP expression in the NDAPS1 group than the NDS group might be due to an increase in the populations of Lactobacillus spp.52,53.
Third, APS1-manipulated gut microbiome influenced hepatic-associated metabolites, followed by the amelioration of hepatic steatosis through the gut-liver axis. D-glucurono-6,3-lactone, glycerol-3-phosphate, and pyruvic acid, which are associated with fatty liver disease54, have been identified as intermediates of TG synthesis in the liver tissues of mice on an HFD55. It is plausible that the ameliorating hepatic steatosis effect of APS1 might be partially due to the lower serum levels of certain fatty liver-associated metabolites. Additionally, butyrate plays important roles in the gut-liver axis in the host50. Butyrate could be transferred from the intestine to the liver via the portal vein and activate SIRT1 and induce SIRT1-dependent PGC1 deacetylation through AMPK activation56. SIRT1 has been shown to modulate lipid metabolism via activating PGC1 and upregulation of fatty acid oxidation genes57.The activation of SIRT1/PGC1 could further suppress the expression of lipid synthesis factors, FAS and ACC, and fatty acid transporter FABP4, and thus limited the accumulation of fatty acids58. The results of protein expression including upregulation of SIRT1 and PGC1 and downregulation of FAS and FABP4 in liver tissue of the NDS group provided evidences to support amelioration of hepatic steatosis of APS1 might through the gut-liver axis. Therefore, suppressing in hepatic steatosis may attribute to APS1-induced butyrate expression triggering hepatic protective functions of SIRT1.
Finally, our study provides an example of how probiotics could serve as a potential therapy for modulating physiological function downstream of the microbiota. We found that APS1-induced modulation of SCFAs and circulating metabolites enhanced susceptibility to accelerated weight loss and ameliorated hepatic steatosis, potentially through improvement of energy expenditure and inhibition of appetite. Future studies are warranted that examine the potential clinical use of APS1 as a novel therapeutic in the quest for effective long-term weight management solutions.
Material and Methods
Bacterial strain
Lactobacillus mali APS1 was isolated and identified from sugary kefir59. The preparation of APS1 followed the procedure previously described17.
Animal experimental protocol
Male C57BL/6 J mice at 7 weeks of age were used (BioLASCO Taiwan Co., Ltd, Yi-Lan, Taiwan). Test mice were housed in cages (one mouse per cage) at a controlled room temperature and under a 12 h light–dark cycle and provided food and filtered water ad libitum. After one week for acclimatization, all test mice were fed 60% calorie fat HFD (D12492, Research Diets, Inc., NJ, USA) for 6 weeks, at which point mice showed obesity phenotype characteristics60. After HFD feeding, test mice were then treated for the following three weeks and were randomized into two groups: control group (NDS) received 10% calorie fat normal diet (D12450B, Research Diets, Inc.) administered with saline by gavage daily, and the APS1 intervention group (NDAPS1) received a normal diet and APS1 at 5 × 108 CFU in saline by gavage daily. The body weight and food intake of the test mice were recorded once a week. The cumulative caloric intake was calculated by the total gram weight of diet taken multiplied by the caloric content of the diet (ND: 16.11 kJ/g; HFD2:1.92 kJ/g). At the end of the experiment, all test mice were fasted for 8 hours, anesthetized with isoflurane and sacrificed for serum and tissue collection. All experiments were performed in accordance with relevant guidelines, the animal experimental protocol was approved by the Institutional Animal Care and Use Committee of the National Taiwan University (Approval No: NTU103-EL-00096).
Analysis of serum biochemistry and satiety hormones
The serum samples were collected from test mice and stored at −80 °C until analysis. Serum glucose, triglyceride (TG) and total cholesterol (T-Chol) levels were measured using a Fuji DRI-CHEM Clinical Chemistry Analyzer FDC 3500 (Fujifilm, Tokyo, Japan) at the National Laboratory Animal Center (Taipei, Taiwan). Serum insulin level was measured using a mouse insulin assay kit following the manufacturer’s protocol (Mercodia, Uppsala, Sweden). Homeostasis model assessment of insulin resistance (HOMA-IR) index was calculated according to the formula: HOMA-IR = insulin (μU/mL) × glucose (mg/dL)/405. Serum peptide YY (PYY), pancreatic polypeptide (PP) and resistin levels were measured using the Milliplex® MAP Kit and Mouse Metabolic Hormone 96-Well Plate Assay (Merck Millipore, MA, USA).
Analysis of fecal SCFAs
Fecal samples were collected from test mice at 6 weeks and 9 weeks. SCFAs in feces were extracted with 70% ethanol solution and analyzed using published methods61. Briefly, the samples were processed to fatty acid derivatization and then extracted twice with ether. Each ether layer was collected and concentrated under nitrogen gas. The obtained fatty acid hydrazides were analyzed by HPLC.
Histological analysis
Liver and white adipose tissues (epididymal, retroperitoneal and inguinal white adipose tissue; namely, eWAT, rpWAT and iWAT) were obtained from test mice and fixed with 4% formalin overnight and then embedded within paraffin or optimal cutting temperature (OCT) compound for frozen section procedures. Tissue sections (5-μm thickness) were stained with hematoxylin and eosin (H&E) or Oil Red O (O0625, Sigma-Aldrich, MO, USA) staining. Histological images were obtained under a microscope (AxioObserver Z1, Göttingen, Germany). The mean adipocyte size was measured using ImageJ software (NIH, MD, USA).
Western blot analysis
Total protein from the liver tissues of mice was extracted with RIPA buffer containing protease and phosphatase inhibitors. The procedures for electrophoresis and blotting were conducted following described methods62 with some modifications. The primary antibodies, including anti-SIRT1 (D1D7, Cell Signaling, MA, USA), anti-acetyl-CoA carboxylase (ACC) (C83B10, Cell Signaling), anti- peroxisome proliferator-activated receptor gamma coactivator 1 (PGC1) (ab54481, Abcam, Cambridge, UK), anti-fatty acid synthase (FAS) (C20G5, Cell Signaling), anti-fatty acid binding protein 4 (FABP4) (D25B3, Cell Signaling) and anti-β-actin (13E5, Cell Signaling) antibodies, were used for identification of each protein. Quantification of protein levels in luminescent bands was performed using ImageJ software.
Analysis of hepatic triglyceride
Hepatic triglyceride levels were measured using a triglyceride colorimetric assay kit (Cayman Chemical, MI, USA). Briefly, weighed liver tissues were homogenized in cold buffer on ice. The homogenates were centrifuged at 10,000 g for 15 minutes at 4 °C. Supernatants were harvested and stored at −80 °C for further assay following the manufacturer’s protocol.
DNA extraction and sequencing of gut microbiota
Fecal samples were collected from test mice at 0 weeks (HFW0) and 4 weeks (HFW4) during HFD feeding, and at 3 weeks after switching to an ND (NDS and NDAPS1, respectively). Bacteria total genomic DNA from fecal samples was extracted using published methods63 with modifications. The next-generation sequencing results of bacterial 16 S ribosomal DNA genes were then analyzed following procedures described previously64. Although, different variable regions of 16 s rRNA have been targeted for distinguishing the bacteria65,66, the V3-V4 region has been identified as the most useful for distinguishing among species of the intestinal bacteria67,68. This region is generally used for intestinal microbiome studies with accumulation of information and database. Therefore, the V3–V4 regions of 16S rRNA genes were amplified using a specific primer with a barcode. Barcoded PCR amplicons were sequenced on an Illumina HiSeq. 2500 platform. Operational taxonomic unit (OTU) clustering and species annotation were performed from representative sequences using UPARSE software (Version 7.0.1001) and the Greengenes Database based on Ribosomal Database Project classifier (Version 2.2), respectively. For reads assembly and quality control, paired-end reads was assigned and based on their unique barcode and truncated by cutting off the barcode and primer sequence. Paired-end reads were merged using FLASH (V1.2.7)69. Quality filtering on the raw tags was performed under specific filtering conditions according to the Qiime (V1.7.0)70,71. The tags were compared with the reference database (Gold database) using UCHIME algorithm72 to detect and remove chimera sequences73. For OTU cluster and species annotation, sequences analysis were performed by Uparse software (Uparse v7.0.1001)74. OTUs were assigned at sequences ≥97% similarity threshold. The GreenGene Database75 was used based on RDP classifier (Version 2.2)76 algorithm to annotate taxonomic information. OTUs abundance information was normalized using a standard of sequence number corresponding to the sample with the least sequences. Prior to various analyses, each sample was rarefied to an even sequence depth of 31,315 to control the various sequencing depth. Subsequent analysis of alpha diversity and beta diversity were all performed according to this output normalized data. The alpha diversity (Chao 1 and Shannon index) and PLS-DA were calculated with QIIME (Version 1.7.0) using R software (Version 2.15.3).
Analysis of serum metabolomic profile
The metabolomic profile experiments were conducted by the commission service of the Metabolomics Core Laboratory at Center of Genomic Medicine, National Taiwan University (Taipei, Taiwan). Briefly, serum samples were collected from test mice and stored at −80 °C until analysis. Then, 100 μL of each serum sample was extracted for 2 minutes in 400 μL of ice-cold methanol, followed by vaporization. The vaporized serum residues were reconstituted with 200 μL of 50% methanol and then subjected to UHPLC-QTOF-MS analysis. The untargeted metabolic profile was analyzed using an Agilent 1290 UHPLC system (Agilent Technologies, CA, USA) coupled with an Agilent 6540 quadrupole-time of flight mass spectrometer (Agilent Technologies, CA, US) with electrospray ionization. The Acquity HSS T3 column (2.1 × 100 mm, 1.8 μm, MA, USA) was used and maintained at 40 °C. For analysis of metabolomics mass spectra, the mass spectra data were analyzed using TIPick77, XCMS278 and Batch Normalizer79 methods.
Analysis of the correlation between the gut microbiome and specific metabolites
The correlation relationships between the relative abundance of the gut microbiota at the genus level and 10 specific metabolites were evaluated by Pearson’s correlation test using SAS version 9.4 software (SAS Institute Inc., Cary, NC, USA) following a previously described method with modifications80. All metabolite data were mean-centered and scaled to unit variance prior to the test.
Statistical analysis
All values are given as the mean ± standard deviation (SD). The data were proved to be normally distribution by using a Shapiro-Wilk test for normality (P > 0.05). Statistical analysis for animal experiments comparing two groups used paired and unpaired, two-tailed Student’s t-test. P < 0.05 was indicative of a significant difference. To perform and compare the statistical significant difference between two groups without a normal distribution assumption in NGS analysis, NGS data were normalized and the nonparametric Wilcoxon signed rank test for paired data and Mann-Whitney U test for unpaired data were used. LEfSe analysis with LDA scores greater than 4 and significance at α < 0.05 was determined by Kruskal–Wallis test. For All statistical analyses were performed using Statistical Analysis System software 9.4.
Electronic supplementary material
Acknowledgements
This project was supported by grant (MOST 104-2313-B-002-043-MY3) from Ministry of Science and Technology in Taiwan. We appreciated Dr. Bor-Rung Ou at TungHai University in Taiwan for kindly supporting plasmid DNA. We appreciated Dr. Yu-Lun Kuo at BIOTOOLS Co., Ltd in Taiwan for kindly supporting analysis of NGS data.
Author Contributions
Y.T.C. and N.S.Y. drafted and prepared the article. Y.T.C. and Y.C.L. conceived and designed the study. S.T.H. conducted the animal experiment. K.Y.L. performed statistical analyses. J.S.L., N.S.Y., J.R.L. and M.J.C. were involved in the planning and design of the study. M.J.C. contributed to writing and editing of the article. All authors approved the final version of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Yung-Tsung Chen and Ning-Sun Yang contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-23844-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stevens J, Oakkar EE, Cui Z, Cai J, Truesdale KP. US adults recommended for weight reduction by 1998 and 2013 obesity guidelines, NHANES 2007-2012. Obesity. 2015;23:527–531. doi: 10.1002/oby.20985. [DOI] [PubMed] [Google Scholar]
- 2.Hruby A, Hu FB. The epidemiology of obesity: A big picture. PharmacoEconomics. 2015;33:673–689. doi: 10.1007/s40273-014-0243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lombardo GE, et al. Normocaloric diet restores weight gain and insulin sensitivity in obese mice. Front. Endocrinol. 2016;7:49. doi: 10.3389/fendo.2016.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brestoff JR, et al. Manganese [III] tetrakis [5,10,15,20]-benzoic acid porphyrin reduces adiposity and improves insulin action in mice with pre-existing obesity. PLOS ONE. 2015;10:e0137388. doi: 10.1371/journal.pone.0137388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He X, Ji G, Jia W, Li H. Gut microbiota and nonalcoholic fatty liver disease: Insights on mechanism and application of metabolomics. Int. J. Mol. Sci. 2016;17:300. doi: 10.3390/ijms17030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao L. The gut microbiota and obesity: from correlation to causality. Nat. Rev. Micro. 2013;11:639–647. doi: 10.1038/nrmicro3089. [DOI] [PubMed] [Google Scholar]
- 7.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 8.Fei N, Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2013;7:880–884. doi: 10.1038/ismej.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 10.Pedersen HK, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 11.Breton J, et al. Gut commensal E.coli proteins activate host satiety pathways following nutrient-induced bacterial growth. Cell Metab. 2016;23:324–334. doi: 10.1016/j.cmet.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 12.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders ME, et al. Safety assessment of probiotics for human use. Gut Microbes. 2010;1:164–185. doi: 10.4161/gmic.1.3.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobyliak N, et al. Probiotics in prevention and treatment of obesity: a critical view. Nutr. Metab. 2016;13:14. doi: 10.1186/s12986-016-0067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okubo H, et al. Lactobacillus casei strain Shirota protects against nonalcoholic steatohepatitis development in a rodent model. Am. J. Physiol. Gastrointest. Liver Physiol. 2013;305:G911–8. doi: 10.1152/ajpgi.00225.2013. [DOI] [PubMed] [Google Scholar]
- 16.Yoo SR, et al. Probiotics L. plantarum and L. curvatus in combination alter hepatic lipid metabolism and suppress diet-induced obesity. Obesity. 2013;21:2571–2578. doi: 10.1002/oby.20428. [DOI] [PubMed] [Google Scholar]
- 17.Lin YC, Chen YT, Hsieh HH, Chen MJ. Effect of Lactobacillus mali APS1 and L. kefiranofaciens M1 on obesity and glucose homeostasis in diet-induced obese mice. J. Funct. Foods. 2016;23:580–589. doi: 10.1016/j.jff.2016.03.015. [DOI] [Google Scholar]
- 18.Nishiyama K, Sugiyama M, Mukai T. Adhesion properties of lactic acid bacteria on intestinal mucin. Microorganisms. 2016;4:34. doi: 10.3390/microorganisms4030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez M, et al. Effect of Lactobacillus rhamnosus CGMCC1.3724 supplementation on weight loss and maintenance in obese men and women. Br. J. Nutr. 2014;111:1507–1519. doi: 10.1017/S0007114513003875. [DOI] [PubMed] [Google Scholar]
- 20.Sharafedtinov KK, et al. Hypocaloric diet supplemented with probiotic cheese improves body mass index and blood pressure indices of obese hypertensive patients - a randomized double-blind placebo-controlled pilot study. Nutr. J. 2013;12:138. doi: 10.1186/1475-2891-12-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omar JM, Chan YM, Jones ML, Prakash S, Jones PJH. Lactobacillus fermentum and Lactobacillus amylovorus as probiotics alter body adiposity and gut microflora in healthy persons. J. Funct. Foods. 2013;5:116–123. doi: 10.1016/j.jff.2012.09.001. [DOI] [Google Scholar]
- 22.Takahashi S, et al. Effect of Bifidobacterium animalis ssp. lactis GCL2505 on visceral fat accumulation in healthy Japanese adults: a randomized controlled trial. Biosci. Microbiota. Food Health. 2016;35:163–171. doi: 10.12938/bmfh.2016-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Remely M, et al. Increased gut microbiota diversity and abundance of Faecalibacterium prausnitzii and Akkermansia after fasting: a pilot study. Wien. Klin. Wochenschr. 2015;127:394–398. doi: 10.1007/s00508-015-0755-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Chatelier E, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 25.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lecomte V, et al. Changes in gut microbiota in rats fed a high fat diet correlate with obesity-associated metabolic parameters. PLOS ONE. 2015;10:e0126931. doi: 10.1371/journal.pone.0126931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsen N, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLOS ONE. 2010;5:e90855. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finucane MM, Sharpton TJ, Laurent TJ, Pollard KS. A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter. PLOS ONE. 2014;9:e84689. doi: 10.1371/journal.pone.0084689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dicksved J, et al. Molecular fingerprinting of the fecal microbiota of children raised according to different lifestyles. Appl. Environ. Microbiol. 2007;73:2284–2289. doi: 10.1128/AEM.02223-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravussin Y, et al. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity. 2012;20:738–747. doi: 10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patrone V, et al. Postoperative changes in fecal bacterial communities and fermentation products in obese patients undergoing bilio-intestinal bypass. Front. Microbiol. 2016;7:200. doi: 10.3389/fmicb.2016.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwiertz A, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 33.Price LB, et al. Community analysis of chronic wound bacteria using 16 S rRNA gene-based pyrosequencing: Impact of diabetes and antibiotics on chronic wound microbiota. PLOS ONE. 2009;4:e6462. doi: 10.1371/journal.pone.0006462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma W, et al. An analysis of human microbe–disease associations. Brief. Bioinform. 2017;18:85–97. doi: 10.1093/bib/bbw005. [DOI] [PubMed] [Google Scholar]
- 35.Togo AH, Valero R, Delerce J, Raoult D, Million M. “Anaerotruncus massiliensis,” a new species identified from human stool from an obese patient after bariatric surgery. New Microbes. New Infect. 2016;14:56–57. doi: 10.1016/j.nmni.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoppel C. The role of carnitine in normal and altered fatty acid metabolism. Am. J. Kidney Dis. 2003;41:S4–S12. doi: 10.1016/S0272-6386(03)00112-4. [DOI] [PubMed] [Google Scholar]
- 37.Arduini A, Mancinelli G, Ramsay RR. Palmitoyl-L-carnitine, a metabolic intermediate of the fatty acid incorporation pathway in erythrocyte membrane phospholipids. Biochem. Biophys. Res. Commun. 1990;173:212–217. doi: 10.1016/S0006-291X(05)81043-5. [DOI] [PubMed] [Google Scholar]
- 38.Roberts LD, et al. β-aminoisobutyric acid induces browning of white fat and hepatic β-oxidation and is inversely correlated with cardiometabolic risk factors. Cell Metab. 2014;19:96–108. doi: 10.1016/j.cmet.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newgard CB, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Denton RM, Halperin ML. The control of fatty acid and triglyceride synthesis in rat epididymal adipose tissue. Roles of coenzyme A derivatives, citrate and L-glycerol 3-phosphate. Biochem. J. 1968;110:27–38. doi: 10.1042/bj1100027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nurjhan N, et al. Glutamine: a major gluconeogenic precursor and vehicle for interorgan carbon transport in man. J. Clin. Invest. 1995;95:272–277. doi: 10.1172/JCI117651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roth E. Nonnutritive Effects of Glutamine. J. Nutr. 2008;138:2025S–2031S. doi: 10.1093/jn/138.10.2025S. [DOI] [PubMed] [Google Scholar]
- 43.Liu R, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 2017;23:859–868. doi: 10.1038/nm.4358. [DOI] [PubMed] [Google Scholar]
- 44.Watanabe M, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 45.Liu, S. et al. Agut–brain axis regulating glucose metabolism mediated by bile acids and competitive fibroblast growth factor actions at the hypothalamus. Mol. Metab, https://doi.org/10.1016/j.molmet.2017.12.003 (2017). [DOI] [PMC free article] [PubMed]
- 46.Adrian TE, et al. Rectal taurocholate increases L cell and insulin secretion, and decreases blood glucose and food intake in obese type 2 diabetic volunteers. Diabetologia. 2012;55:2343–2347. doi: 10.1007/s00125-012-2593-2. [DOI] [PubMed] [Google Scholar]
- 47.Brahe LK, et al. Specific gut microbiota features and metabolic markers in postmenopausal women with obesity. Nutr. Diabetes. 2015;5:e159. doi: 10.1038/nutd.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamilton MK, Boudry G, Lemay DG, Raybould HE. Changes in intestinal barrier function and gut microbiota in high-fat diet-fed rats are dynamic and region dependent. Am. J. Physiol. Gastrointest. Liver Physiol. 2015;308:G840–G851. doi: 10.1152/ajpgi.00029.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao Z, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58:1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Endo H, Niioka M, Kobayashi N, Tanaka M, Watanabe T. Butyrate-producing probiotics reduce nonalcoholic fatty liver disease progression in rats: New insight into the probiotics for the gut-liver axis. PLOS ONE. 2013;8:e63388. doi: 10.1371/journal.pone.0063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Byrne CS, Chambers ES, Morrison DJ, Frost G. The role of short chain fatty acids in appetite regulation and energy homeostasis. Int. J. Obes. 2015;39:1331–1338. doi: 10.1038/ijo.2015.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meimandipour A, et al. Gastrointestinal tract morphological alteration by unpleasant physical treatment and modulating role of Lactobacillus in broilers. Br. Poult. Sci. 2010;51:52–59. doi: 10.1080/00071660903394455. [DOI] [PubMed] [Google Scholar]
- 53.Sivieri K, et al. Lactobacillus acidophilus CRL 1014 improved “gut health” in the SHIME®reactor. BMC Gastroenterology. 2013;13:100. doi: 10.1186/1471-230X-13-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu C-T, et al. Metabolomics of ginger essential oil against alcoholic fatty liver in mice. J. Agric. Food Chem. 2013;61:11231–11240. doi: 10.1021/jf403523g. [DOI] [PubMed] [Google Scholar]
- 55.Lai Y-S, et al. Mass-spectrometry-based serum metabolomics of a C57BL/6J mouse model of high-fat-diet-induced non-alcoholic fatty liver disease development. J. Agric. Food Chem. 2015;63:7873–7884. doi: 10.1021/acs.jafc.5b02830. [DOI] [PubMed] [Google Scholar]
- 56.Wang, R. H. et al. Hepatic Sirt1 deficiency in mice impairs mTorc2/Akt signaling and results in hyperglycemia, oxidative damage, and insulin resistance. J. Clin. Invest. 121, 4477–4490 (2011). [DOI] [PMC free article] [PubMed]
- 57.Cantó, C. & Auwerx, J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr. Opin. Lipidol. 20, 98–105 (2009). [DOI] [PMC free article] [PubMed]
- 58.Simmons, G. E., Pruitt, W. M. & Pruitt, K. Diverse Roles of SIRT1 in cancer biology and lipid metabolism. Int. J. Mol. Sci. 16, 950–965 (2015). [DOI] [PMC free article] [PubMed]
- 59.Hsieh HH, Wang SY, Chen TL, Huang YL, Chen MJ. Effects of cow’s and goat’s milk as fermentation media on the microbial ecology of sugary kefir grains. Int. J. Food Microbiol. 2012;157:73–81. doi: 10.1016/j.ijfoodmicro.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 60.Krishna S, et al. Time-dependent behavioral, neurochemical, and metabolic dysregulation in female C57BL/6 mice caused by chronic high-fat diet intake. Physiol. Behav. 2016;157:196–208. doi: 10.1016/j.physbeh.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Torii T, et al. Measurement of short-chain fatty acids in human faeces using high-performance liquid chromatography: specimen stability. Ann. Clin. Biochem. 2010;47:447–452. doi: 10.1258/acb.2010.010047. [DOI] [PubMed] [Google Scholar]
- 62.Yang Y, et al. Alpha-lipoic acid improves high-fat diet-induced hepatic steatosis by modulating the transcription factors SREBP-1, FoxO1 and Nrf2 via the SIRT1/LKB1/AMPK pathway. J. Nutr. Biochem. 2014;25:1207–1217. doi: 10.1016/j.jnutbio.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 63.Henderson G, et al. Effect of DNA extraction methods and sampling techniques on the apparent structure of cow and sheep rumen microbial communities. PLOS ONE. 2013;8:e74787. doi: 10.1371/journal.pone.0074787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruan JW, et al. Dual-specificity phosphatase 6 deficiency regulates gut microbiome and transcriptome response against diet-induced obesity in mice. Nat. Microbiol. 2016;2:16220. doi: 10.1038/nmicrobiol.2016.220. [DOI] [PubMed] [Google Scholar]
- 65.Lane DJ, et al. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Claesson MJ, et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids. Res. 2010;38:e200–e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Q, et al. Accelerated dysbiosis of gut microbiota during aggravation of DSS-induced colitis by a butyrate-producing bacterium. Sci. Rep. 2016;6:27572. doi: 10.1038/srep27572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sánchez-Tapia M, et al. Nopal (Opuntia ficus indica) protects from metabolic endotoxemia by modifying gut microbiota in obese rats fed high fat/sucrose diet. Sci. Rep. 2017;7:4716. doi: 10.1038/s41598-017-05096-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bokulich NA, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods. 2012;10:57. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haas BJ, et al. Chimeric 16 S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome. Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 75.DeSantis TZ, et al. Greengenes, a chimera-checked 16 S rRNA Gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ho TJ, Kuo CH, Wang SY, Chen GY, Tseng YJ. True ion pick (TIPick): a denoising and peak picking algorithm to extract ion signals from liquid chromatography/mass spectrometry data. J. Mass Spectrom. 2013;48:234–242. doi: 10.1002/jms.3154. [DOI] [PubMed] [Google Scholar]
- 78.Benton HP, Wong DM, Trauger SA, Siuzdak G. XCMS2: Processing tandem mass spectrometry data for metabolite identification and structural characterization. Anal. Chem. 2008;80:6382–6389. doi: 10.1021/ac800795f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang SY, Kuo CH, Tseng YJ. Batch Normalizer: A fast total abundance regression calibration method to simultaneously adjust batch and injection order effects in liquid chromatography/time-of-flight mass spectrometry-based metabolomics data and comparison with current calibration methods. Anal. Chem. 2013;85:1037–1046. doi: 10.1021/ac302877x. [DOI] [PubMed] [Google Scholar]
- 80.Mao SY, Huo WJ, Zhu WY. Microbiome–metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ. Microbiol. 2016;18:525–541. doi: 10.1111/1462-2920.12724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.