Abstract
Brain tissue from 1068 donors was analyzed for RNA quality as a function of postmortem interval (PMI) and years in storage. Approximately 83% of the cortical and cerebellar samples had an RNA integrity number (RIN) of 6 or greater, indicating their likely suitability for real-time quantitative polymerase chain reaction research. The average RIN value was independent of the PMI, up to at least 36 hours. The RNA quality for specific donated brains could not be predicted based on the PMI. Individual samples with a low PMI could have a poor RIN value, while a sample with a PMI over 36 hours may have a high RIN value. The RIN values for control brain donors, all of whom died suddenly and unexpectedly, were marginally higher than for individuals with clinical brain disorders. Polymerase chain reaction (PCR) analysis of samples confirmed that RIN values were more critical than PMI for determining suitability of tissue for molecular biological studies and samples should be matched by their RIN values rather than PMI. Importantly, PCR analysis established that tissue stored up to 23 years at −80°C yielded high-quality RNA. These results confirm that postmortem human brain tissue collected by brain and tissue banks over decades can serve as high quality material for the study of human disorders.
Keywords: : human brain, PMI, RNA quality, brain bank, long-term storage
Introduction
Research utilizing animal models, particularly genetic mouse models, provides insight into the mechanism of human diseases. Furthermore, the effects of many external interventions can only be studied in animals due to the ability to manipulate experimental conditions. However, the unique characteristics of the human intellect and human diseases require the study of the human brain because research conclusions, and identification of potential drugs developed for animal models, do not always transfer to humans.1–3 Some researchers have expressed the opinion that such criticism has been overstated, while recognizing the importance of animal research.4 Promising findings from animal studies need to be verified in humans. A significant number of studies can be performed on living individuals. However, studies at the molecular level in the human brain require either surgical or postmortem tissue.
The suitability of postmortem human brains for research has been analyzed with regard to a large number of potential variables, such as manner of death, storage conditions of the body, postmortem interval (PMI; the time between death and brain processing),5–11 and length of time in storage.12 Additional variables are different socioeconomic and nutritional status, exposure to environmental toxins, as well as the co-mingling of individuals of multiple genetic backgrounds. Nevertheless, human postmortem tissue has been extensively studied, including over 800 publications that are based on human tissue donated to the University of Maryland Brain and Tissue Bank (UMD-BTB, formerly the NICHD Brain and Tissue Bank for Developmental Disorders; www.medschool.umaryland.edu/btbank/Brain). This bank is a brain and tissue repository of the NIH NeuroBioBank (https://neurobiobank.nih.gov).
Molecular biological studies from this tissue repository have addressed many human diseases: X-linked adrenoleukodystrophy,13 autism spectrum disorder,14–16 Alexander disease,17 amyotrophic lateral sclerosis,18 Angelman syndrome,19 developing brain,20,21 diabetes,22 Down syndrome,23 epilepsy,24 Fragile X syndrome,25 Prader–Willi syndrome,26 Rett syndrome,27 schizophrenia,28 Sturge-Weber syndrome,29 and tuberous sclerosis.30 Postmortem human brain tissue has also been used for detailed gene mapping.31
Families donate tissue to further research on diseases affecting their family member; therefore, it is important to demonstrate to both families and researchers that the stored tissue retains its suitability for multiple types of research. In this report, a cohort of 1068 donors to the UMD-BTB was evaluated for RNA integrity number (RIN), pH, and gene expression as a function of PMI, agonal conditions prior and immediately after death, and years in storage. This study demonstrated that high-quality RNA can be obtained from tissue with a PMI up to 36 hours, but that the RNA quality for a specific donated brain currently cannot be predicted. In addition, frozen tissue stored for more than two decades at −80°C retained very high quality of RNA and its value to researchers who are studying human biology and diseases.
Materials and Methods
Donation of human tissue
All human tissue was collected by the UMD-BTB according to legal provisions of the medical faculty of the University of Maryland School of Medicine. The bank functions under the supervision of two Internal Review Boards (IRB): the IRB of the University of Maryland School of Medicine and the IRB of the Department of Health and Mental Hygiene of the State of Maryland. Consent was obtained from the next of kin before tissue retrieval. The majority of tissue donors resided in the United States and Canada. Exclusion criteria for donation were contagious infections, severe head trauma, and the use of ventilators during the agonal period. The standard protocol was to fix half the brain in phosphate-buffered formalin (10%) and section and freeze the other half in an isopentane-dry ice mix (www.medschool.umaryland.edu/btbank/Brain-Protocol-Methods/Brain-Sectioning---Protocol-Method-2). A variety of systemic tissue was also collected from some, but not all, donors. The formalin-fixed tissue was stored in plastic containers at room temperature. The frozen tissue was stored at −80°C in heat-sealed plastic bags. The function of each freezer was monitored continually. In case of freezer failure or loss of electricity, staff are contacted by telephone and the event is recorded. Each freezer had a backup system of liquid carbon dioxide and emergency electrical generators.
Total RNA extraction
For gene expression studies, cases were selected from control males with no known disorders. Real-time quantitative polymerase chain reaction (RT-qPCR) was performed on 12 samples for the RNA integrity (RIN) study, 15 samples for the PMI study, and 12 samples for the storage length study. Approximately 60–80 mg of frozen tissue from the prefrontal cortex and cerebellum was collected per case in prechilled (dry ice) microcentrifuge tubes and immediately stored at −80°C. To minimize RNA degradation, all materials and work surfaces were cleaned with RNaseZap (Sigma-Aldrich, St. Louis, MO). Total RNA was extracted using the Qiagen RNeasy Lipid Tissue kit (Qiagen, Hilden, Germany). Frozen tissues were disrupted using the Bullet Blender bead homogenizer (Next Advance, Averill Park, NY). QIAzol Lysis Reagent (Qiagen) and 2.0 mm zirconium oxide beads (Next Advance) were added to frozen tissue and homogenized at a speed setting of 5 for 1 minute. The supernatant was transferred to a fresh microcentrifuge tube and incubated at room temperature for 5 minutes. Chloroform (200 μL) was added and the samples were shaken vigorously for 15 seconds, then incubated for 3 minutes, and centrifuged at 12,000 RPM at room temperature to separate the homogenate into aqueous and organic phases. The upper aqueous phase containing RNA (∼600 μLl) was removed and 70% ethanol (600 μL) was added to enhance the binding conditions to the RNeasy spin column. The RNA sample (600 μL) was applied to a spin column and centrifuged at 8000 g for 15 seconds at room temperature. The flow-thru was discarded. This step was repeated for the remaining RNA sample. The sample was treated on-column with the RNase-Free DNase set (Qiagen) to eliminate any residual DNA in the sample. The column was rinsed with 500 μL Qiagen buffer RPE, centrifuged 15 seconds at 800 g, and the RNA was eluted with 100 μL of RNase-free water.
Determination of total RNA quantity, quality, integrity, and pH
The concentration of RNA was measured with a Nanodrop 2000 UV-VIS spectrophotometer (Thermo Scientific, Wilmington, DE) and expressed as nanograms per microliter. The integrity of the RNA samples was assessed by sample preparation using Agilent RNA 6000 Nano Kits (Agilent Technologies, Santa Clara, CA) and algorithmic interpretation on the Agilent 2100 Bioanalyzer© (Agilent Technologies). RNA samples were stained with an intercalating dye, separated by microcapillary electrophoresis on gel-filled microfluidic chips, and detected by laser-induced fluorescence creating an electropherogram.32 The Bioanalyzer compares ribosomal RNA fragments (18S and 28S) of the sample electropherograms to a software algorithm generating an RIN. The quality of ribosomal RNA, as reflected by an RIN value, is utilized as an alternate measurement of messenger RNA (mRNA) quality.32 The pH was determined on a 10% (wt:vol) sample (200 mg) of prefrontal cortex homogenized with a VWR pellet mixer in 2 mL Milli-Q water.
DNA extraction
The protocol from the DNeasy Blood and Tissue Kit (Qiagen) was used to extract genomic DNA from frozen cerebellum.33 Tissue lysis buffer (Qiagen) and 2.0 mm zirconium oxide beads (Next Advance) were added to the sample and homogenized at a speed of 3 for 1 minute in the Bullet Blender (Next Advance). Proteinase K (Qiagen) was added to the supernatant and incubated for 2 hours at 56°C. RNase A was added to the lysate. The lysate was added to the spin column, and DNA was bound, washed, and eluted. The concentration and the 260/280 ratio of DNA were determined with a NanoDrop 2000 Spectrophotometer (Thermo Scientific). DNA integrity was determined by gel electrophoresis and ethidium bromide staining of DNA isolated from brain tissue stored at −80°C for 0.5–1 and for 13–14 years.
Complementary DNA synthesis and qRT-qPCR
A two-step protocol was used for reverse transcription RT-qPCR. The Quantitect Reverse Transcription kit (Cat. No. 205311; Qiagen) with genomic DNA elimination buffer was used to synthesize complementary DNA (cDNA). For each case, 2 μg of total RNA was reverse transcribed in a 40 μL reaction volume. The final cDNA reaction was diluted with an equal volume of RNase-free water (Qiagen). The cDNAs were used as templates for RT-qPCR on Step-One Plus Real-Time PCR System (Applied Biosystems, Darmstadt, Germany).
Ten genes were chosen to examine the effects of RIN, PMI, and storage length on gene expression in postmortem brain tissue. The genes were selected for their RNA half-life: 4 hours (p21 protein-activated kinase 2 [PAK2], serpine mRNA binding protein 1 [SERBP1], and tubulin, alpha 4a [TUBA4A]), 8 hours (aconitase 1, soluble [ACO1], N-ethylmaleimide-sensitive factor attachment protein, alpha [NAPA], and peroxiredoxin 5 [PRDX5]), and stable >48 hours (electron-transfer flavoprotein, beta polypeptide [ETFB], glutathione S-transferase, mu 5 [GSTM5], malignant T cell amplified sequence 1 [MCTS1], and beta-actin [ACTB]).34 The half-life calculation was based on the degradation rate of existing transcripts after transcription was blocked.34 A recent study demonstrated that many gene transcriptions were increased after organism death.35 The abundance of a transcript at a given time was determined by its rates of synthesis and degradation.36 Assuming that the postmortem transcription rate in each sample was relatively constant, the degradation rate (half-life) was the major contributing factor for transcript abundance. TaqMan Gene Expression Assays (Applied Biosystems) with specific primer/probe mixtures were used for the following genes: PAK2; SERBP1; TUBA4A; ACO1; NAPA; PRDX5; ETFB; GSTM5; MCTS1; and ACTB. The amount of cDNA template was optimized for each gene. All samples tested for expression of a specific gene employed an equal amount of cDNA template (Table 3)
Table 3.
Genes Tested by Polymerase Chain Reaction
| Gene symbol | Gene name | RNA half-life | RNA (ng) | Biological function | Assay IDa |
|---|---|---|---|---|---|
| PAK2 | p21 Protein-activated kinase 2 | 4 Hours | 80 | Protein amino acid phosphorylation | Hs02559219_s1 |
| SERBP1 | Serpine mRNA binding protein 1 | 4 Hours | 40 | Regulation of mRNA stability | Hs00967385_g1 |
| TUBA4A | Tublin, alpha 4a | 4 Hours | 40 | Mitotic cell cycle | Hs01081794_g1 |
| ACO1 | Aconitase 1, soluble | 8 Hours | 60 | Tricarboxylic acid cycle | Hs00158095_m1 |
| NAPA | N-ethylmaleimide-sensitive factor attachment protein, alpha | 8 Hours | 40 | Intracellular protein function, brain development | Hs00943303_m1 |
| PRDX5 | Peroxiredoxin 5 | 8 Hours | 40 | Inflammatory response to oxidative stress | Hs00738905_g1 |
| ETFB | Electron-transfer flavoprotein, beta polypeptide | Stable | 40 | Respiratory electron transport chain | Hs01085511_m1 |
| GSTM5 | Glutathione S-transferase, mu5 | Stable | 80 | Glutathione and xenobiotic metabolism | Hs00757076_m1 |
| MCTS1 | Malignant T cell-amplified sequence 1 | Stable | 60 | Regulation of transcription | Hs01585687_m1 |
| ACTB | Beta-actin | Stable | 40 | Protein folding | Hs01060665_g1 |
| Cytoskeletal protein |
TaqMan Gene Expression Assays (FAM-dye/MBG probe) examined in this study. The RNA half-life listed in the table was determined in fibroblasts and B cells by chemically stopping degradation, which may not reflect the values in postmortem brain tissue.30 The amount of RNA listed corresponds to the quantity used to form cDNA for determination of Cq value of each analyzed gene.
Purchased from Applied Biosystems.
cDNA, complementary DNA; Cq, cycle of quantification; mRNA, messenger RNA.
The RT-PCR reactions were run in 96-well plates with a total volume of 20 μL per reaction. Each reaction contained 10 μL TaqMan Universal Master Mix II (2 × ) with uracil-N-glycosylase (UNG), 1 μL of TaqMan Gene Expression Assay (20 × ), and the corresponding amount of cDNA (1.6–3.2 μL) and PCR-grade water (Roche Diagnostics, Indianapolis, IN). RT-PCR reactions were prepared in triplicate for each of the 10 gene assays. Each plate contained nontemplate controls and a primer/probe mixture for ACTB as a methodological control. RT-PCR was run in the StepOnePlus (Applied Biosystems) using the following thermocycler parameters: 2 minutes at 50°C, 10 minutes at 95°C, and then 40 cycles of 15 seconds at 95°C and 1 minute at 60°C.
Statistical analysis
Correlations between two variables were tested by linear regressions, where p-values represent whether the correlation between variables reached significance and R2 values illustrate fit of the data to the regression line. Differences between multiple groups (comparison of multiple disease conditions to controls; Table 1) were determined by analyses of variance followed by a post hoc Bonferroni test. Differences between two groups (comparison of all conditions together to controls; Table 2) were determined by t-tests. Values were considered significant when p < 0.05. Star symbols in figures represent levels of significance, *p < 0.05; **p < 0.01; and ***p < 0.001.
Table 1.
Comparison of RNA Integrity Number Values of Control Cases Versus Disorders That Had at Least 12 Cases
| Case | n | PMI (hours) | RIN (cortex) | RIN (cerebellum) | pH |
|---|---|---|---|---|---|
| ALD | 35 | 14 ± 9.1 | 7.3 ± 1.1 | 6.9 ± 1.4 | 5.8 ± 0.3 |
| Alzheimer's disease | 74 | 12.8 ± 8.7a | 6.2 ± 1.2a | 6.5 ± 1.2a | 5.8 ± 0.6 |
| ALS | 34 | 17.1 ± 9.7 | 6.7 ± 0.9 | 6.9 ± 1.3 | 5.8 ± 0.5 |
| Autism | 60 | 21.7 ± 10.5 | 6.7 ± 1.1b | 7.1 ± 1.3 | 6.3 ± 0.5a |
| Depression | 22 | 18.9 ± 7.4 | 7.3 ± 0.9 | 7.4 ± 1 | 6.1 ± 0.5 |
| Dystonia | 20 | 20 ± 11.8 | 6.4 ± 1.5 | 6.8 ± 1.1 | 6 ± 0.5 |
| Epilepsy | 28 | 23.3 ± 8.8 | 7 ± 0.8 | 7.6 ± 0.7 | 5.6 ± 0.5 |
| Parkinson's disease | 16 | 19.2 ± 7.8 | 6.3 ± 1.4 | 6.6 ± 1.4 | 5.9 ± 0.7 |
| Prader–Willi syndrome | 12 | 19.8 ± 8 | 7.1 ± 0.9 | 7.1 ± 1.1 | 6 ± 0.5 |
| Controls | 506 | 18.7 ± 8.9 | 7.2 ± 1.2a | 7.5 ± 1.2a | 5.8 ± 0.5a |
| All conditions, other than controls | 562 | 18.4 ± 11.4 | 6.7 ± 1.3a | 6.8 ± 1.5a | 5.9 ± 0.6a |
Alzheimer's disease (PMI): af = 6, df = 9, p < 0.001, Bonferroni p < 0.001; Alzheimer's disease (RIN cortex): af = 8.5, df = 9, p < 0.001, Bonferroni p < 0.001; Alzheimer's disease (RIN cerebellum): af = 7.9, df = 9, p < 0.001, Bonferroni p < 0.001; autism (RIN cortex): bf = 8.5, df = 9, p < 0.001, Bonferroni p = 0.03; autism (pH): af = 4.5, df = 9, p < 0.001, Bonferroni p < 0.001; all conditions, other than controls (RIN cortex), at = 7.4, df = 1065, Bonferroni p < 0.001; all conditions, other than controls (RIN cerebellum): at = 9.7, df = 1057, Bonferroni p < 0.001; all conditions, other than controls (pH): at = 3.3, df = 763, Bonferroni p < 0.001.
ALD, adrenoleukodystrophy, X-linked; ALS, amyotrophic lateral sclerosis; PMI, postmortem interval; RIN, RNA integrity number.
Table 2.
Comparison of RNA Integrity Number Values in Human Tissues
| Tissue | RIN (average ± SD) | n | p |
|---|---|---|---|
| Cerebellum | 8.3 ± 0.6 | 15 | n/a |
| Prefrontal cortex | 7.8 ± 0.5 | 15 | 0.34 |
| Psoas muscle | 7.2 ± 0.9 | 15 | 0.009 |
| Heart | 7.1 ± 0.7 | 15 | 0.003 |
| Testis | 7.2 ± 0.8 | 15 | 0.007 |
| Liver | 5.4 ± 1.3 | 15 | <0.001 |
| Lung | 5.4 ± 1.6 | 15 | <0.001 |
| Kidney | 4.4 ± 1.5 | 15 | <0.001 |
| Ileum | 2.6 ± 0.2 | 5 | <0.001 |
| Spleen | 2.1 ± 0.2 | 5 | <0.001 |
RIN values were determined on RNA isolated from brain and systemic tissue from a group of 15 donors. Only five cases included ileum and spleen in addition to the other tissues. The p-values apply to the difference in RIN values between the cited tissue and cerebellum.
SD, standard deviation.
Results
RNA quality and PMI
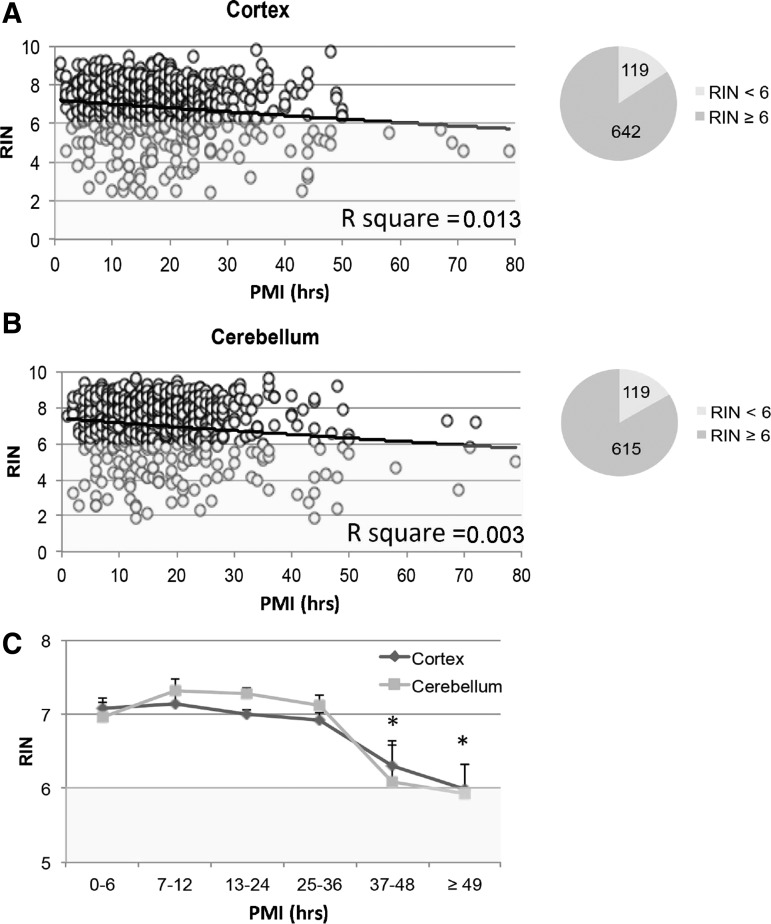
RIN values of all samples were determined in cortical and cerebellar samples from 1068 donors (Fig. 1). Average RIN values and standard deviation for the cortex and cerebellum tissue were, respectively, 6.9 ± 1.24 and 7.1 ± 1.4. RIN values for cortex and cerebellum showed a correlation of R2 = 0.46; p < 0.001. Pie charts in Figure 1A and B show that over 80% of samples had RINs ≥6, an arbitrary point used by some researchers to evaluate suitability of tissue for molecular biological studies.33 A linear regression of data in Figure 1 showed that, while there was a significant inverse correlation between variables (p < 0.001), the value of PMI to predict RIN was very low (R2 = 0.013) in cortical tissue (Fig. 1A). A similar finding was observed in the cerebellum (Fig. 1B), where a linear regression showed that there was a trend (p = 0.06) for an inversion correlation between variables and the value of PMI to predict RIN was low (R2 = 0.003). Figure 1C shows that the effect of PMI on RIN was not evident, particularly before 36 hours.
FIG. 1.
Effect of PMI and RIN in human brain cortex (A) and cerebellum (B) from 1068 donors. The number of cases per interval in (C) 0–6 hours, n = 108; 7–12 hours, n = 202; 13–24 hours, n = 523; 25–36 hours, n = 183; 37–48, n = 41; >49 hours, n = 11. Average ± SEM. PMI, postmortem interval; RIN, RNA integrity number; SEM, standard error of the mean.
RNA quality as a function of time in storage, age of donor, and pH
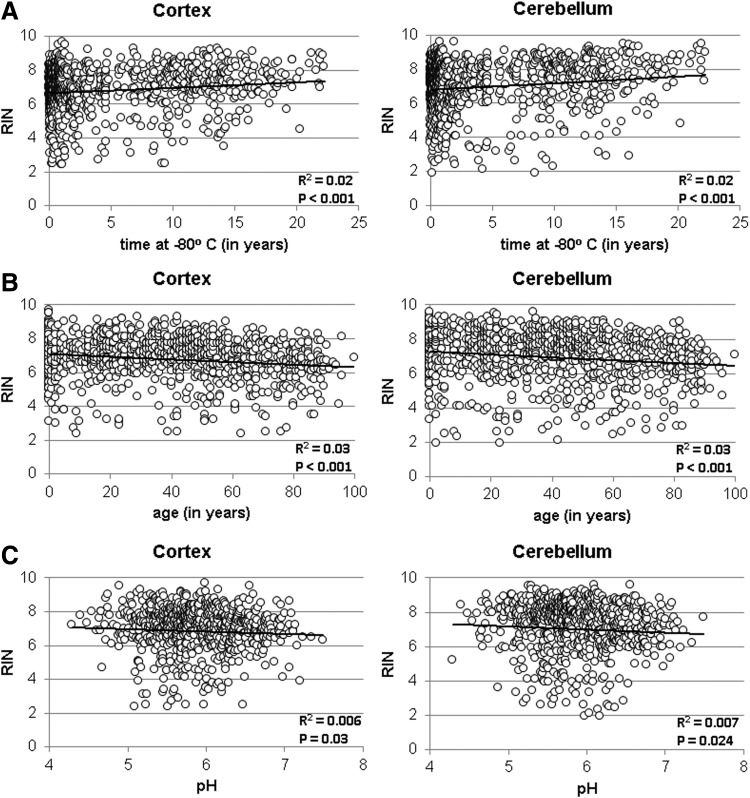
The data for Figure 1 were analyzed with respect to the number of years that the tissue had been stored at −80°C before determination of RIN values, the age of donors, and the pH of the brain tissue at the time of evaluation of RIN values (Fig. 2).
FIG. 2.
Effect of storage time at −80°C, age of donor, and pH on RIN values. The n = 1068 for (A, B) and 789 for (C).
Only a fraction of the data was explained by the trendlines for Figure 2A–C due to variability of the results (R2 = 0.01–0.03). Unexpectedly, RINs are significantly positively correlated with years in storage for the cortical tissue and the cerebellum. It is likely that this represents variables such as the nature of the donors collected or site of origin of the tissues.
RIN and agonal state
The agonal state, as defined by parameters prior and immediately after death, has been implicated in the quality of brain samples.5–11,34,37 These factors are not possible to control, although the UMD-BTB excluded cases where the donor had a respiratory infection or was on a ventilator. Limited data on agonal state are known for cases collected at sites other than the State of Maryland medical examiner. The donors consented at the Office of the Chief Medical Examiner (OCME) in Baltimore all died suddenly and were placed in refrigeration when the body arrived at the OCME. This allows a comparison of donors from the former with donors with disorders who died under medical or hospice care.
The cortical and cerebellar RIN values for Alzheimer's disease patients were consistently lower than the average for all patient donors, even though the PMI for the same patients was significantly shorter than for controls (Table 1). A slightly higher average tissue pH for autism cases was unexpected since the RIN values were not statistically different. The average RIN values and pH of all cases with a disorder were significantly different from controls. However, the actual values did not vary by more than 0.7 RIN units.
RIN values of brain and systemic tissue from the same donor
RIN values were analyzed relative to cerebellum using multivariate regression (Table 2). Substantial differences in RNA stability were observed depending on the tissue studied. The stability of RNA in brain is greater than in other body tissue. Psoas muscle, heart, and testis had RIN values ∼1 unit less than brain. The RNA extracted from liver, lung, and kidney had reduced RIN values, while ileum and spleen had significant RNA degradation.
Gene expression as a function of RIN values
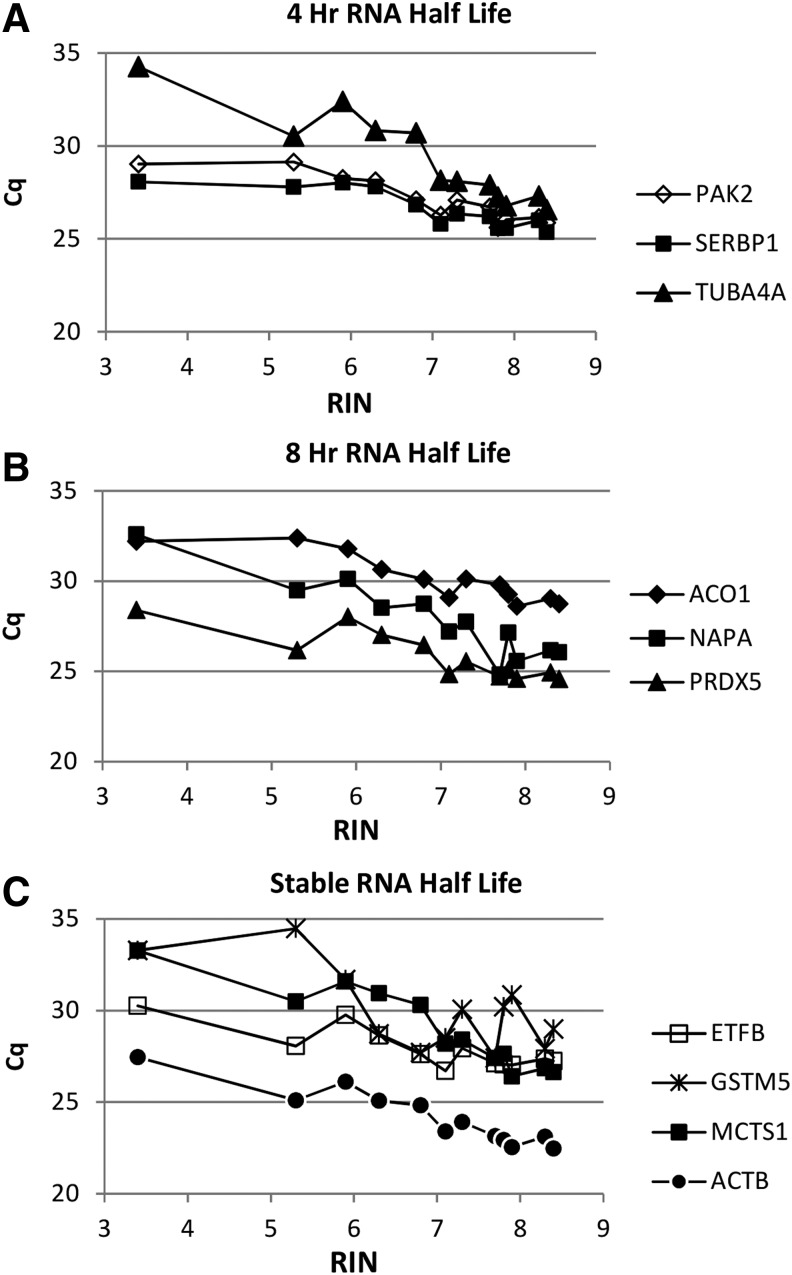
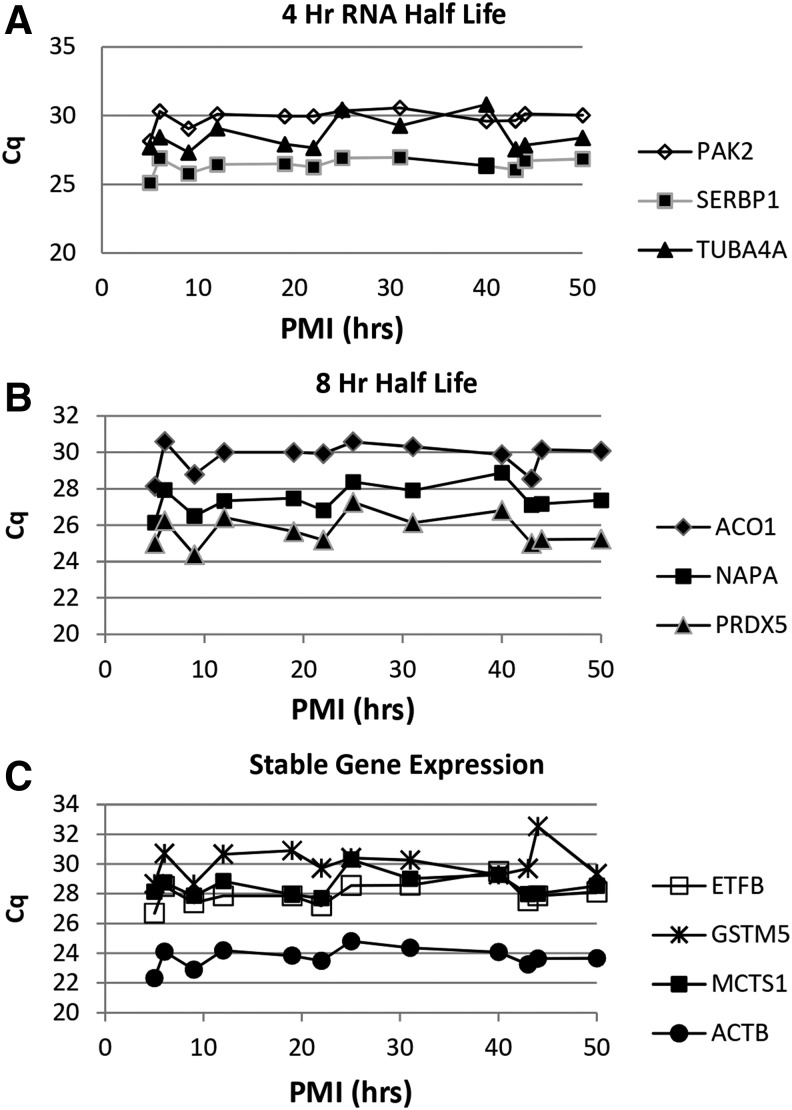
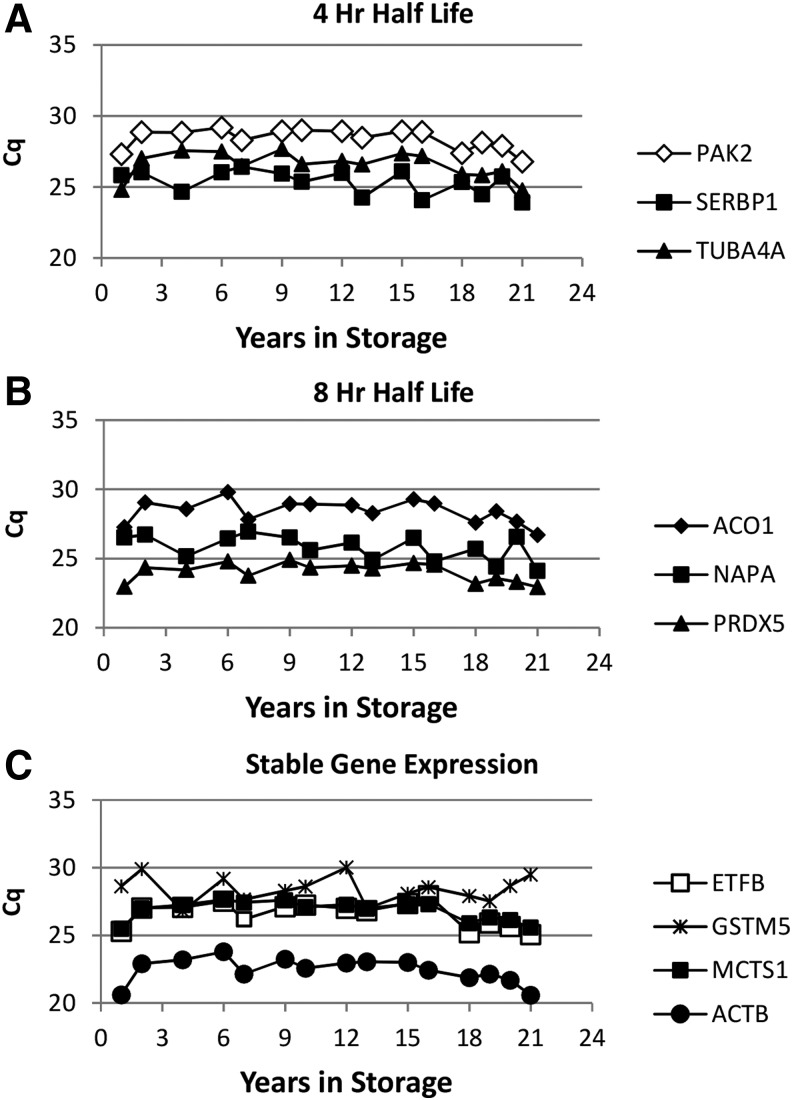
The expression of 10 genes was studied by qPCR to determine the effect of RIN, PMI, and years in storage. The effect of mRNA turnover rate was assessed by inclusion of three genes whose mRNA has a half-life of ∼4 hours, three genes whose mRNA has a half-life of ∼8 hours, and four genes that are considered stable (Table 3).34 The data are expressed as the cycle of quantification (Cq), which indicates the cycle number at which a significant number of gene amplicons has been generated above the baseline. The lower the Cq value, the greater the number of gene copies in the sample. The Cq value of samples was an inverse function for RIN values below 7 (Fig. 3), such that samples with a low Cq value have a higher RIN value. Although the individual values show scatter, the curves level off as the RIN values approached 7. This pattern is independent of the mRNA half-life. Linear regressions showed significant correlations between RIN and Cq value in all genes analyzed (p < 0.05). To determine if the PMI had an effect on the Cq value, samples were selected in which the RIN values were held constant at 7.5. The Cq value remained relatively constant for all 10 genes studied and no significant correlations between PMI and Cq value were observed for all genes analyzed (Fig. 4). These data suggest that if tissue yielded RNA with a high RIN value, the PMI had no effect. The same was found for samples with a constant RIN value of 8.5 stored at −80°C for up to 23 years (Fig. 5). With the exception of NAPA, there were no significant correlations between years in storage and Cq value. Curiously, for unknown reasons, the association between storage time and NAPA was positive, showing higher expression (lower Cq value) in samples with increased storage time. The sample points at <1 year were consistently lower for nearly all the genes tested and may reflect an unrecognized characteristic of that donor rather than an effect of shorter storage time. These results indicate that the PMI and the number of years in storage have no effect on the gene expression if the RIN is acceptable for a given study.
FIG. 3.
Cq values as a function of RIN values for genes with different turnover rates. Each point represents one sample with the indicated RIN values measured in triplicate for gene expression. Cq values were determined for genes with reported different half lives based on transcription inhibition studies34, (A) 4 hours, (B) 8 hours, (C) stable. Cq, cycle of quantification.
FIG. 4.
Cq values as a function of PMI values for genes with different turnover rates. Gene expression as a function of PMI when the RIN value was kept constant at 7.5 for all samples. Each point represents one sample with the indicated RIN values measured in triplicate for gene expression. Cq values were determined for genes with reported different half lives based on transcription inhibition studies34, (A) 4 hours, (B) 8 hours, (C) stable.
FIG. 5.
Cq values as a function of years at −80°C values for genes with different turnover rates. Gene expression was determined with samples with RIN values kept constant at 8.5 for all samples. This RIN value was selected because samples with an RIN of 7.5 could not be found at each of the years in storage. Each point represents one sample with the indicated RIN values measured in triplicate for gene expression. Cq values were determined for genes with reported different half lives based on transcription inhibition studies34, (A) 4 hours, (B) 8 hours, (C) stable.
DNA evaluation
DNA was extracted from brain sections stored either 0.5–1 or 13–14 years at −80°C (Fig. 6). Fragmentation of DNA was not observed in either set of brain tissue.
FIG. 6.
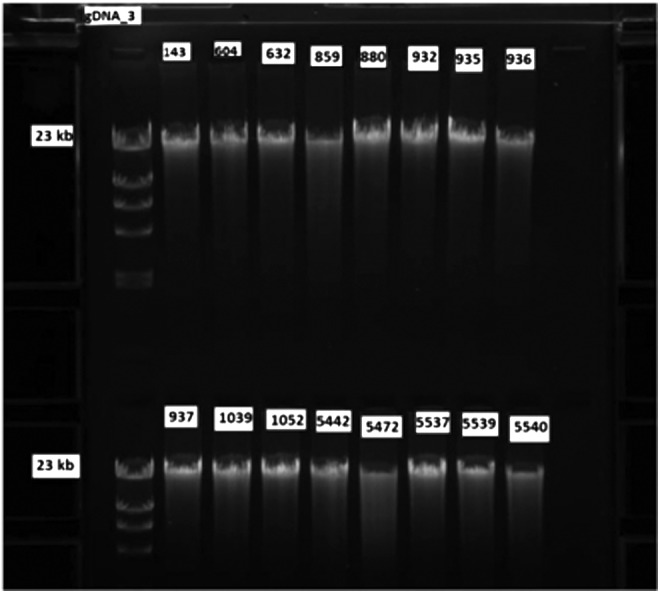
DNA gels. The isolated DNA was analyzed by gel electrophoresis and ethidium bromide staining. Samples numbered <1052 had been stored at −80°C for 13–14 years and samples numbered over 5442 and above had been stored for 0.5–1 year.
Discussion
The UMD-BTB (www.medschool.umaryland.edu/btbank/Brain) is one of six brain banks of the NIH NeuroBioBank (https://neurobiobank.nih.gov) that serve as a source of human tissue for the research community. The six banks use standardized procedures for tissue processing, quality assessment and storage. The University of Maryland bank has in its inventory postmortem brain and systemic tissue from over 3500 donors, representing several hundred clinical disorders with special emphasis on developmental disorders. This bank has shipped both formalin-fixed and frozen tissue to more than 1000 researchers worldwide, who have published about 800 scientific articles based on tissue received. The intent of this study is to provide an assessment of factors affecting RNA quality, including PMI, agonal state, types of clinical disorders, and length of time the specimens are housed in ultralow freeze storage at −80°C.
A major variable in tissue collection is the time between death and tissue recovery. Factors affecting the PMI are the notification of the UMD-BTB by the donor families, grieving time required by families, arrangement for tissue recovery if prior arrangements had not been made, time between death and refrigeration of the body, and the time required for legal processes to occur, such as performance of autopsies by medical examiners for cases under their jurisdictions. The combination of these factors and medical conditions of the patient in the hours to days before death, referred to as the agonal state, are conditions beyond the control of the tissue bank and studies have shown that these factors affected the integrity of the RNA.3,6–11 The UMD-BTB recovers tissue throughout the United States, Canada, and several other English-speaking countries. The involvement of many different pathologists makes it often difficult to control these factors. It is extremely rare that brain and systemic tissues can be recovered in less than 6 hours after death. Although the goal has been to collect tissue with a PMI of 24 hours or less, this arrangement is not always possible. Furthermore, some of the cases are so infrequent that declining a case based on extended intervals would deprive the scientific community of access to tissue from these rare disorders. The approach the UMD-BTB adopted is to collect the tissue with longer PMIs and evaluate the tissue on a case by case basis for tissue integrity, specifically in regard to RNA quality.
The data in Figure 1 indicate that there is significant variability of RIN values at all PMI periods. As reported by others, shorter PMI values do not correlate with a high RIN value.6,8,10,38 Indeed, neural tissue with a longer PMI may have high RIN values, indicating the overall tissue integrity even with longer death to freezing intervals. The data in this article support the conclusion that with PMI values up to 36 hours, and perhaps even longer, the majority of donor specimens banked at brain autopsy retain high-quality RNA. However, it is important that the tissue for each case be evaluated to determine RNA integrity. It has been reported that brain samples with an RIN value >4 or 5 are suitable for RT-qPCR studies.39,40 Eighty-three percent of the UMD-BTB samples tested had an RIN value >6. Although RIN values have been found to be useful in determining RNA quality, RIN reflects ribosomal RNA integrity, which may not reflect the integrity of specific mRNAs. Other methods have been proposed that produce a more direct measure of mRNA quality.41
The establishment of a cohort of cases for a rare disorder may take decades; therefore, the effect of storage time could be a significant factor. In Figure 2A, the RIN value was determined for tissue stored 23 years at −80°C. The RIN values did not indicate deterioration of tissue integrity with time. The slight upward slope of the regression line is assumed to reflect a higher RIN value of the tissue collected at that earlier time point. Analysis of 16 additional brain samples stored for 29–31 years, originally at −60°C and then at −80°C, had an average RIN value of 6.2 ± 0.7, range = 4.7–7.2 (data not shown). There is a small, but significant, decrease of RIN values with the age of donor (Fig. 2B). However, an R2 = 0.03 value would suggest that age is not a good predictor of RIN. The pH of a sample has been used as a marker of tissue integrity.6,33–36,38,42,43 Brain pH has a significant impact on human postmortem hippocampal gene expression profiles.6 The data in Figure 2C do not indicate a direct relationship between RIN and pH. Although unexpected, it may be either due to the wide range of values observed for both RIN and pH or due to variation in methodology such as excluding donors expected to experience hypoxia before death.
Donors who serve as controls and do not have a clinical brain disorder may experience a sudden event leading to their death, in contrast to donors with a clinical brain disorder, who may experience a more variable and protracted terminal course. Data in Table 1 indicate that for the listed disorders, there is a minimum, but significant, decrease in RIN, which could affect interpretation of experimental results. A difference in RIN values was observed between controls and Alzheimer's disease and Autism Spectrum Disorder donors, and between controls and all disorders combined. These results suggest that the disease conditions or differences in agonal states contribute to the significant differences in RIN values, which could affect experimental interpretations. Decreased RIN values for Alzheimer's disease brain tissue have been reported.37
In addition to brain donations, the UMD-BTB also collects systemic tissue and establishes fibroblast cultures from a portion of the donors. Data in Table 2 indicate that the RIN values vary greatly between brain and systemic tissues. RIN values were determined for 15 cases that had a range of tissues collected from the same donor (a smaller proportion of cases also had ileum and spleen samples). Tissue from the brain had the highest RIN values followed by psoas muscle, heart, and testis. Liver, lung, and kidney had intermediate RIN values. Ileum and spleen had lower RIN values, presumably due to release of a greater quantity of hydrolases. In an earlier study, high-quality RNA was recovered from muscle, brain, and heart, and RNA with a lower RIN from liver, kidney, and spleen.44 The data in Table 2 suggest that the collection procedures currently used by the Bank are not optimized for obtaining samples from the ileum and spleen with high RNA integrity.
We examined ten genes with different half-lives calculated from transcription inhibition studies.34 However, transcription does not cease after death, but continues after death.35 Therefore, Cq value numbers for all the genes obtained from postmortem brain likely reflects the sum of the transcription rate and the degradation rate (half-life). A given Cq value number could theoretically apply at two different time points postmortem: once as gene expression increases and then again as the mRNA degrades for these genes. The transcription rate for a given gene at a given time is relatively constant in each sample from the same brain region. The degradation rate, in other words, half-life, is a key contributor to the abundance of the transcript. Thus, the selection of genes in a range of half-lives ensures our analysis covers the spectrum of RNA turnover. In addition, most of the qPCR analyses in this study were done beyond the transcription activation stage after death.35
The Cq values of the ten genes tested were an inverse function of the RIN value (Fig. 3). A similar relationship between Cq value and RIN has been reported using samples with a lower RIN range.45 Although the individual values show scatter in this study, the curves level off as the RIN value approached 7, indicating that the number of gene copies in the extracted RNA had reached a constant level at an RIN value of 7 and higher. This pattern was independent of the mRNA half-life of the gene studied. When RNA was denatured in vitro to obtain RNA samples with RIN values from 7.5 to 1.5, the denatured samples had a direct correlation between RIN and RT-qPCR results.46 Amplification of transcripts can occur despite variable degradation of RNA.45
When RNA was extracted from samples with increasing PMI, but with a constant RIN value of 7.5, the Cq values remained relatively constant for all 10 genes studied and no significant correlations were observed (Fig. 4). These studies indicated that RIN values were more important than PMI. Consequently, researchers requesting tissue from a tissue repository for RNA research should utilize data on RNA quality rather than PMI in making their decision as to the cases they request. The question regarding the stability of RNA in brain samples that had been stored for a long time was investigated in brain samples with a constant RIN value of 8.0 and stored at −80°C for up to 23 years (Fig. 5). The RT-qPCR results indicated that high-quality mRNA can be obtained from tissue stored for many years at −80°C if the RIN value indicates that the ribosomal RN is intact. It was not feasible to determine if the results also pertained to samples with low RIN values, which had been stored over this same time period, since such samples were not available in the bank. DNA from brain sections stored between 6 months and 14 years analyzed by gel electrophoresis and ethidium bromide staining showed identical patterns (Fig. 6).
In conclusion, extensive efforts and funding have been expended on establishing brain and tissue banks for the retention of and research on postmortem human tissue. These results for a large population of donated tissue, specifically brain, indicate that RNA isolated from tissue obtained by the UMD-BTB within the period of 4 to at least 36 hours postmortem have RIN values >6 and are suitable for RT-qPCR studies.39,40 The quality of RNA was unchanged after 23 years of storage at −80°C. Although it is important to try to minimize the PMI, it often is not possible when a bank has to arrange for tissue collection at sites distal from the brain bank. The authors suggest that researchers who obtain tissue from a brain repository request tissue of similar RIN values rather than matching PMI, and that the brain repositories obtain RIN values for tissue in their collection. These results provide support for outreach to families with rare disorders with the recognition that proper storage of patient tissue with detailed medical information in a brain and tissue bank provides the best assurance that the tissue is available to be studied when a sufficient number of cases have been collected or when new methodologies or research questions arise.
Acknowledgments
The generosity of the families of the tissue donors makes possible the wide range of basic medical studies that the UMD-BTB has supported with the hope of curing the many diseases affecting humanity. The Bank is supported by NIH contract No. HHSN275200900011C, Ref. No. NO1-HD-9-0011 with additional support from the Autism Research Institute and the Blazeman Foundation for ALS.
Author Disclosure Statement
No conflicting financial interests exist.
References
- 1.Seok J, Warren HS, Cuenca AG, et al. . Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A 2013;110:3507–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGonigle P, Ruggeri B. Animal models of human disease: Challenges in enabling translation. Biochem Pharm 2014;87:162–171 [DOI] [PubMed] [Google Scholar]
- 3.Van Dam D, De Deyn PP. Animal models in the drug discovery pipeline for Azheimer's disease. Br J Pharmacol 2011;164:1285–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osuchowski MF, Remick DG, Lederer JA, et al. . Abandon the mouse research ship? Not just yet! Shock 2014;41:463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton AJL, Pearson RCA, Najlerahim A, et al. . Pre- and postmortem influences on brain RNA. J Neurochem 1993;61:1–11 [DOI] [PubMed] [Google Scholar]
- 6.Stan AD., Ghose S, Gao X-M, et al. . Human postmortem tissue: What quality makers matter? Brain Res 2006;1123:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomita H, Vawter MP, Walsh DM, et al. . Effect of agonal and postmortem factors on gene expression profile: Quality control in microarray analyses of postmortem human brain. Biol Psychiatry 2004;55:346–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trabzuni D, Ryten M, Walker R, et al. . Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem 2011;119:275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li JZ, Vawter MP, Walsh DM, et al. . Systematic changes in gene expression in postmortem human brains associated with tissue pH and terminal medical conditions. Hum Mol Gen 2004;13:609–616 [DOI] [PubMed] [Google Scholar]
- 10.Durrenberger PF, Fernando S, Kashefi SN, et al. . Effects of antemortem and postmortem variables on human brain mRNA quality: A BrainNet Europe study. J Neuropath Exp Neuorol 2010;69:70–81 [DOI] [PubMed] [Google Scholar]
- 11.Lipska BK, Deep-Soboslay A, Weickert CS, et al. . Critical factors in gene expression in postmortem human brain: Focus on studies in schizophrenia. Biol Psychiatry 2006;60:650–658 [DOI] [PubMed] [Google Scholar]
- 12.Ervin JF, Heinzen EL, Cronin KD, et al. . Postmortem delay has minimal effect on brain RNA integrity. J Neuropathol Exp Neurol 2007;66:1093–1099 [DOI] [PubMed] [Google Scholar]
- 13.Asheuer M, Bieche I, Laurendeau I, et al. . Decreased expression of ABCD4 and BG1 genes early in the pathogenesis of X-linked adrenoleukodystrophy. Hum Mol Genet 2005;14:1293–1303 [DOI] [PubMed] [Google Scholar]
- 14.Alarcón M, Abrahams BS, Stone JL, et al. . Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet 2008;82:150–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroeder DI, Lott P, Korf I, et al. . Large-scale methylation domains mark a functional subset of neuronally expressed genes. Genome Res 2011;21:1583–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoner R, Chow ML, Boyle MP, et al. . Patches of disorganization in the neocortex of children with autism. N Engl J Med 2014;370:1209–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brenner M, Johnson AB, Boespflug-Tanguy O, et al. . Mutations in GFAP, encoding glial fibrillary acidic protein, are associated with Alexander disease. Nat Genet 2001;27:117–120 [DOI] [PubMed] [Google Scholar]
- 18.Henkel JS, Beers DR, Wen S, et al. . Decreased mRNA expression of tight junction proteins in lumbar spinal cord of ALS patients. Neurology 2009;72:1614–1616 [DOI] [PubMed] [Google Scholar]
- 19.Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet 1997;17:14–15 [DOI] [PubMed] [Google Scholar]
- 20.Jen JC, Chan WM, Bosley TM, et al. . Mutations in a human ROBO gene disrupt hindbrain axon pathway crossing and morphogenesis. Science 2004;304:1509–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evrony GD, Cai X, Lee E, et al. . Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell 2012;151:483–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pugliese A, Zeller M, Fernandez A Jr., et al. . The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INSVNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat Genet 1997;15:293–297 [DOI] [PubMed] [Google Scholar]
- 23.Kuhn DE, Nuovo GJ, Terry AV Jr., et al. . Chromosome 21-derived microRNAs provide an etiological basis for aberrant protein expression in human down syndrome brains. J Biol Chem 2010;285:1529–1543 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Dibbens LM, Tarpey PS, Hynes K, et al. . X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat Genet 2008;40:776–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bittel DC, Kibiryeva N, Butler MG. Whole genome microarray analysis of gene expression in subjects with fragile X syndrome. Genet Med 2007;9:464–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bittel DC, Kibiryeva N, Sell SM, et al. . Whole genome microarray analysis of gene expression in Prader-Willi syndrome. Am J Med Genet A 2007;143A:430–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makedonski K, Abuhatzira L, Kaufman Y, et al. . MeCP2 deficiency in Rett syndrome causes epigenetic aberrations at the PWS/AS imprinting center that affect UBE3A expression. Hum Mol Genet 2005;14:1049–1058 [DOI] [PubMed] [Google Scholar]
- 28.Tao R, Li C, Newburn EN, et al. . Transcript-specific associations of SLC12A5 (KCC2) in human prefrontal cortex with development, schizophrenia, and affective disorders. J Neurosci 2012;32:5216–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shirley MD, Tang H, Gallione CJ, et al. . Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med 2013;368:1971–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin W, Chan JA, Vinters HV, et al. . Analysis of TSC cortical tubers by deep sequencing of TSC1, TSC2 and KRAS demonstrates that small second-hit mutations in these genes are rare events. Brain Path 2010;20:1096–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, et al. . An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012;489:391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroeder A, Mueller O, Stocker S, et al. . The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 2006;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolova YS, Koenen KC, Galea S, et al. . Beyond genotyping: Serotonin transporter epigenetic modification predicts human brain function. Nat Neurosci 2014;17:1153–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedel CC, Dölken L, Ruzsics Z, et al. . Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res 2009;37:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pozhitkov AE, Neme R, Domazet-Lošo T, et al. . Tracing the dynamics of gene transcripts after organismal death. Open Biol 2017;7:160267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neymotin B, Athanasiadou R, Gresham D. Determination of in vivo RNA kinetics using RATE-seq. RNA 2017;20:1645–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Preece P, Cairns NJ. Quantifying mRNA in postmortem human brain: Influence of gender, age at death, postmortem interval, brain pH, agonal state and inter-lobe mRNA variance. Mol Brain Res 2003;118:60–71 [DOI] [PubMed] [Google Scholar]
- 38.Gonzàlez-Herrera L, Valensuela A, Marchal JA, et al. . Studies on RNA integrity and gene expression in human myocardial tissue, pericardial fluid and blood, and its postmortem stability. Forensic Sci Int 2013;232:218–228 [DOI] [PubMed] [Google Scholar]
- 39.Weis S, Llenos IC, Dulay JR, et al. . Quality control for microarray analysis of human brain samples: The impact of postmortem factors, RNA characteristics, and histopathology. J Neurosci Methods 2007;165:198–209 [DOI] [PubMed] [Google Scholar]
- 40.Fleige S, Walf V, Huch S, et al. . Comparison of relative mRNA quantitation models and the impact of RNA integrity in quantitative real time RT-PCR. Biotechnol Lett 2006;28:1601–1613 [DOI] [PubMed] [Google Scholar]
- 41.Björkmam , Švec JD, Lott E, et al. . Differential amplicons (ΔAmp)-a new molecular method to assess RNA integrity. Biomol Detect Quantif 2016;6:4–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kingsbury AE, Foster OJF, Nisbet AP, et al. . Tissue pH as an indicator of mRNA preservation in human post-mortem brain. Mol Brain Res 1995;28:311–318 [DOI] [PubMed] [Google Scholar]
- 43.Mexal C, Berger R, Adams CE, et al. . Brain pH has a significant impact on human postmortem hippocampal gene expression profiles. Brain Res 2006;1106:1–11 [DOI] [PubMed] [Google Scholar]
- 44.Heinrich M, Matt K, Lutz-Bonengel S, et al. . Successful RNA extraction from various human postmortem tissues. Int J Legal Med 2007;121:136–142 [DOI] [PubMed] [Google Scholar]
- 45.Sonntag K-C, Tejada G, Subburaju S, et al. . Limited predictability of postmortem human brain tissue quality by RNA integrity numbers. J Neurochem 2016;138:53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koppelkamm J, Vennemann B, Lutz-Bonengel S, et al. . RNA integrity in post-mortem samples: Influencing parameters and implications on RT-qPCR assays. Int J Legal Med 2011;125:573–580 [DOI] [PubMed] [Google Scholar]