Abstract
Identifying direct substrates targeted by protein kinases is important in understanding cellular physiology and intracellular signal transduction. Mass-spectrometry based quantitative proteomics provides a powerful tool for comprehensively characterizing the downstream substrates of protein kinases. This approach is efficiently applied to receptor kinases which can be precisely, directly, and rapidly activated by some agent, such as a growth factor. However, non-receptor tyrosine kinase Abl lacks the experimental advantage of extracellular growth factors as immediate and direct stimuli. To circumvent this limitation, we combine a chemical rescue approach with quantitative phosphoproteomics to identify targets of Abl and their phosphorylation sites with enhanced temporal resolution. Both known and novel putative substrates are identified, presenting opportunities for studying unanticipated functions of Abl under physiological and pathological conditions.
TOC image

INTRODUCTION
The proto-oncogene protein tyrosine kinase Abl (also known as Abl1) has been shown to be critical for cell signaling in health and disease1. Abl is the cellular ortholog of the transforming v-Abl oncogene from Abelson murine leukemia virus2. Genomic rearrangement of Abl resulting from a 9,22 chromosomal translocation (the Philadelphia chromosome) gives rise to the Bcr-Abl fusion protein (Fig. 1A). Bcr-Abl is a hallmark of chronic myeloid leukemia (CML) and other leukemias and a key oncogenic driver in these malignancies1. Imatinib and other tyrosine kinase inhibitors that target Bcr-Abl are used to treat CML and other cancers3. In addition to cancer, dysregulated Abl activity has been linked to neurodegenerative diseases and inflammatory/immune disorders1.
Figure 1.

Chemical rescue of FLAG-tagged Bcr-Abl R367A in HEK293 cells and abl−/−arg−/− mouse 3T3 fibroblast cells. (A) Schematic diagram of Abl and Bcr-Abl indicating the sites of Bcr, SH3, SH2, and kinase domain. (B) HEK 293 cells transiently transfected with Bcr-Abl R367A were treated with 20 mM of imidazole for 10 min. Cell lysates were immunoblotted with 4G10 anti-phosphotyrosine antibody, anti-Flag antibody, and anti-Actin antibody. The band corresponding to the size of Bcr-Abl was indicated as pBcr-Abl. (C) abl−/−arg−/− mouse embryo fibroblasts that had been immortalized using a 3T3 protocol were stably transfected with Bcr-Abl R367A. Multiple Bcr-Abl stable clones were analyzed by western blotting probing for Bcr-Abl to select clone #50 in which the expression level of Bcr-Abl is comparable to that of K562 leukemia cells. (D) Bcr-Abl R367A cells were treated with 20 mM imidazole (imi) for the indicated time (0, 2.5, 4.0, 10, 40 min). Whole cell lysates were immunoblotted with antibodies anti-FLAG, anti-phospho CRKL (Tyr207), and anti-Actin. The intensity of the band was measured by software ImageJ. The ratio relative to control was calculated using pCRKL: Actin. (E) Bcr-Abl R367A cells were incubated with serum-free medium for 17, 24, 39h prior to imidazole treatment. The ratio relative to control was calculated using pCRKL: Actin.
At the cellular level, Abl regulates the actin and microtubule cytoskeleton and has been implicated in membrane and organelle trafficking4. Moreover, Abl modulates cell proliferation and survival pathways as well as DNA damage response pathways. A number of protein substrates of Abl’s tyrosine kinase activity have been reported previously1. One of these is the protein CrkL, an SH2/SH3 domain adaptor protein, which upon tyrosine phosphorylation can stimulate multiple signaling pathways that drive cell proliferation and invasion5.
Despite many years of study, a full mechanistic understanding of Abl’s biological functions is lacking. In general, it is difficult to firmly establish cellular protein kinase phosphorylation targets since many kinases are active concurrently and may overlap in their substrate selectivity. One criterion that can connect kinases to their substrates in cells is a kinetic linkage. For receptor kinases like the epidermal growth factor receptor (EGFR) tyrosine kinase, ligands such as EGF can be used to rapidly switch on the kinase and the pattern of protein tyrosine phosphorylation can be observed within minutes6, 7. In contrast, for non-receptor kinases including Abl, which are turned on in the cell in an indirect fashion, a precise ‘on switch’ is lacking.
Previously, for the non-receptor tyrosine kinases Src, v-Src and Csk, a mutant chemical rescue approach was reported and used to obtain insights into the cellular functions of these enzymes8–10. In this technique, a conserved active site Arg is replaced with Ala which sharply reduces catalytic activity, and this reduced activity can be complemented with the small molecule imidazole11, 12. We have also shown previously that the corresponding Arg in Abl (Arg367) when replaced with Ala can also be efficiently complemented by imidazole12. In this study, we have prepared a mouse 3T3 fibroblast cell line stably expressing R367A mutant human Abl in which the native mouse Abl and Arg were genetically deleted13. We have used these R367A mutant Abl cells in combination with mass spectrometry to identify phosphorylation targets of Abl. Below, we describe these findings and their potential biological significance.
MATERIALS AND METHODS
Materials
SILAC amino acids, 13C6 15N2-Lysine and 13C6 15N4-Arginine were purchased from Cambridge Isotope Laboratories. Light amino acids, 12C6 14N2-Lysine and 12C6 14N2-Arginine, were purchased from Sigma. Sep-Pak C18 cartridge (WAT051910) was from Waters. Lipofectamine 2000, Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), arginine- and lysine-free DMEM, and phosphate-buffered saline (PBS) were from Life Technologies. Hygromycin B was from Roche. Oligonucleotides were purchased from Integrated DNA Technologies. The following antibodies were used: phospho-Hck (Tyr410) (ab40660 Abcam), Hck antibody (N-30) (SC-72), 4G10 anti-phosphotyrosine antibody (05-321 Millipore), anti-FLAG antibody (Sigma F3165), anti-Actin (A2228 Sigma), Abl antibody (24-11) (SC-23, Santa Cruz), LDLR antibody (C20) (SC-11824 Santa Cruz), phospho-CrkL (Tyr207) (3181 Cell Signaling), phospho-Abl (Tyr245) (2861 Cell Signaling), FAK antibody (3285 Cell Signaling), phospho-FAK (Tyr576/577) (3281 Cell Signaling), Paxillin antibody M107 (23510 Abcam), phosphor-Paxillin (Tyr118) (2541 Cell Signaling), and p130 Cas antibody (06-500 Millipore). P-Tyr-100 sepharose bead conjugate was from Cell Signaling. Dynabeads were from Thermo Fisher.
Plasmids
p210 Bcr-Abl in the pMSCV puro retroviral vector14 was used as a template for amplifying Abl, which was sub-cloned into mammalian expression vector pCDNA3.1/hygro(+). A R367A mutation and a C-terminal FLAG tag were introduced into Abl. Similarly, p210 Bcr-AblR367A with a C-terminal FLAG tag was cloned into pCDNA3.1/hygro(+). Note that the numbering system in the fusion protein Bcr-Abl is based on the Abl moiety.
Chemical rescue of transiently transfected Bcr-AblR367A in HEK293 cells
HEK293 cells were transfected using lipofectamine 2000 for transient expression of p210 Bcr-AblR367A. 24 h after transfection, the HEK293 cells were either treated with 20 mM imidazole, pH 7.4, for 10 min or treated with water pH 7.4. Both treated and control cells were lysed and subjected to western blotting. 4G10 antibody was used to probe for all tyrosine phosphorylated proteins in whole cell lysate.
Generation of stable mouse 3T3 fibroblast cells
abl−/−arg−/− mouse embryo fibroblasts that had been immortalized using a 3T3 protocol15 were transfected using lipofectamine 2000 for expression of AblR367A. Individual stable clones were selected with hygromycin B and verified via western blotting probing for Abl. The clone (Abl R367A) with which the expression level of Abl is similar to that of p210 Bcr-Abl in chronic myelogenous leukemia K562 cells was selected for further study. Likewise, a stable abl−/−arg−/− 3T3 fibroblast cell line expressing p210 Bcr-AblR367A was obtained.
Chemical rescue with imidazole in stable mouse 3T3 fibroblast cells
Abl R367A cells were cultured at 37 °C under 5% CO2 in DMEM supplemented with 10% FBS and 300 μg/mL of hygromycin B. When cells reached 60-70% confluence, cells were serum-starved for 17 h, followed by imidazole treatment (20 mM imidazole pH 7.4) for 10 min. Cells were then washed once with cold PBS and lysed immediately in modified RIPA buffer (50mm Tris-HCl pH7.4; 1% NP-40; 0.25% sodium deoxycholate; 150 mM NaCl; 1 mM PMSF; 1 mM Na3VO4; 1 mM NaF; plus protease cocktail inhibitor).
SILAC and peptide preparation
SILAC medium was prepared as previously reported10. In brief, arginine- and lysine-free DMEM was supplemented with either light amino acids or heavy amino acids. Abl R367A cells were adapted to SILAC medium culture after at least five cycles of cell divisions. When cells reached 60-70% confluence, they were subjected to serum starvation for 17 h. Thirty 150-mm plates of cells adapted in heavy medium then were treated with 20 mM imidazole pH 7.4 for 10 min, while the other thirty 150-mm plates of cells adapted in light medium were treated with vehicle. After rapid washes with cold PBS, both populations of cells were immediately lysed in urea lysis buffer containing 20 mM HEPES pH 8.0, 8 M urea, 1 mM sodium orthovanadate, 2.5 mM sodium pyrophosphate, and 1 mM β-glycerophosphate, followed by sonication for 3 × 15 sec. The cell lysate was centrifuged at 20,000 × g at 15 °C for 15 min. BCA assay was used to quantify the total protein concentration of the cell lysate. Twenty milligrams of cell lysate from each condition were mixed, reduced with 5 mM dithiothreitol for 30 min at 60 °C and alkylated with 20 mM iodoacetamide for 30 min in the dark at room temperature. The mixture was then diluted in 20 mM HEPES pH 8.0 to a final concentration of 1.5 M urea and incubated overnight with TPCK-treated trypsin at 25 °C with an enzyme-to-protein ratio of 1:20. The next day, the digested peptides were acidified by 1% trifluoroacetic acid (TFA) (vol/vol) and centrifuged at 2,000 × g at room temperature for 15 min to remove undigested proteins and lipids. Desalting of peptide mixtures was performed on a Sep-Pak C18 cartridge that was equilibrated with 0.1% TFA. Desalted peptides were eluted with 40% acetonitrile (v/v) containing 0.1% TFA and lyophilized for three days. Lyophilized peptides were kept at −80 °C for storage.
Immunoaffinity purification of tyrosine phosphopeptides
Immunoaffinity purification (IAP) of phosphotyrosine peptides was carried out as previously described16. Briefly, lyophilized peptides were dissolved in 1.4 ml of IAP buffer (50 mM MOPS pH 7.2, 10 mM sodium phosphate, 50 mM NaCl) and subjected to centrifugation at 2,000 × g at room temperature for 5 min. P-Tyr-100 beads were equilibrated with IAP buffer at 4 °C. The peptide solution was then incubated with P-Tyr-100 beads at 4 °C for 1 h. After washing the beads three times with IAP buffer and twice with ultrahigh pure water, phosphotyrosine peptides were eluted from the beads by incubating the beads with 0.15% TFA at room temperature for 15 min. Eluted peptides were further cleaned up by STAGE tip and the dried peptides were kept at −20°C for storage prior to LC-MS/MS experiment.
LC-MS/MS of enriched tyrosine phosphopeptides
The phosphotyrosine peptides were reconstituted in solvent A (0.1% formic acid in water) and loaded onto a trap column (75 μm × 2 cm) packed in-house with Magic C18 AQ (Michrom Bioresources, Inc.) (5 μm particle size, pore size 100Å) at a flow rate of 5 μL/min with solvent A. Peptides were resolved for 130 min on an analytical column (75 μm i.d. × 20 cm) at a flow rate of 300 nL/min using a linear gradient of 7-15% solvent B (0.1% formic acid in 90% acetonitrile) over 5 min and 15-40% solvent B over 90 min. Mass spectrometry analysis was carried out in a data-dependent manner with full scans (300-1700 m/z) acquired using an Orbitrap mass analyzer at a mass resolution of 120,000 in Elite at 400 m/z. The fifteen most intense precursor ions from a survey scan were selected for MS/MS from each duty cycle and fragment ions generated by the higher-energy collisional dissociation (HCD) method were detected at a mass resolution of 30,000 at m/z of 400 in the Orbitrap analyzer. Dynamic exclusion was set for 30 sec and the automatic gain control for full FT MS was set to 1 million ions and for FT MS/MS was set to 10,000 ions. Internal calibration was carried out using lock-mass from ambient air (m/z 445.120024).
Database searching
MS/MS data obtained from all LC-MS analysis were searched against mouse RefSeq protein database using the MaxQuant platform17. The parameters used for data analysis included trypsin as the protease with a maximum of two missed cleavages allowed. Carbamidomethylation of cysteine was specified as a fixed modification with variable modifications such as oxidation of methionine, acetylation at protein N-termini and phosphorylation at serine, threonine or tyrosine. The mass error of parent ions was set at 4.5 ppm for the main search. Proteins and phosphopeptides were identified by applying 1% false discovery rates at peptide-spectrum matches and proteins.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD007118. To access the data please visit the PRIDE website by using username: reviewer86338@ebi.ac.uk and password: O6Eyt4z5.
Immunoblotting and immunoprecipitation
Imidazole-treated or untreated Abl R367A cells were lysed using modified RIPA buffer. The total protein concentration of cell lysates was determined using Bradford protein assays (Bio-Rad). Equal amounts of cell lysate from each condition were loaded and separated by SDS-PAGE, transferred to PVDF and subjected to immunoblotting. For immunoprecipitation, equal amounts of cell lysate from each condition were incubated with antibody-conjugated Dynabeads magnetic beads for 2-4 h or overnight with rotation at 4° C. After washing the magnetic beads three times with modified RIPA buffer, bound proteins were dissociated from beads by incubating with Laemmli sample buffer at 95 °C for 5 min and then subjected to SDS-PAGE and immunoblotting. The bands were visualized using Clarity Western ECL substrate (Bio-Rad) and the intensity of each band was quantified with ImageJ software. Each experiment was performed at least twice and gave reproducible results.
RESULTS
Transiently transfected Bcr-AblR367A can be rescued by imidazole in live cells
Our previous work12 on purified isolated Abl kinase demonstrated that kinase dead AblR367A can be rescued to 70% of its wt activity level by imidazole. To investigate the feasibility of applying the same strategy in live cells, we transiently transfected kinase dead Bcr-AblR367A into HEK293 cells and compared the pTyr profiles of imidazole-treated and untreated cells. Untransfected cells were treated with imidazole under similar conditions as negative controls. As shown in Fig. 1B, a new pTyr protein was identified in imidazole-treated Bcr-AblR367A transfected cells, the size of which matches that of autophosphorylated Bcr-Abl, suggesting that kinase dead Bcr-AblR367A was rescued by imidazole in live cells, leading to its to autophosphorylation. This result indicates that, similar to Csk and Src kinases8–10, chemical rescue in combination with quantitative proteomics is a feasible approach to identify the physiological targets of Abl kinase in high confidence.
Optimization of chemical rescue in mouse fibroblast cell lines
Mouse embryonic fibroblasts that contained genetic deletions of the paralogs Abl and Arg as reported previously13 were stably transfected with R367A Abl and, for comparison, the analogous Arg to Ala mutant in Bcr-Abl. Mutant cell lines selected for further analysis showed amounts of mutant Abl and Bcr-Abl that were comparable to Bcr-Abl levels present in the K562 chronic myeloid leukemia cell line (Fig. 1C & Fig. 2A). Previous studies have identified CrkL as one well-established Bcr-Abl substrate18, therefore we used phosphorylated Tyr 207 of CrkL as a readout of Bcr-Abl activity to optimize the time of imidazole treatment and serum starvation. After comparing different conditions, we chose 17 h serum starvation and 20 mM imidazole treatment for 10 min (Fig. 1D & E). To determine whether the same conditions could be extended to Abl kinase, we assessed autophosphorylation of Tyr 245 Abl as the readout of Abl tyrosine kinase activity. Tyr 245, a key regulatory residue, is known to be autophosphorylated and this appears to be necessary to for maximal activity of Abl kinase19,20. Immunoblotting revealed that Abl R367A exhibited increased autophosphorylation after 10 min of imidazole treatment, indicating that cellular Abl R367A was rescued under conditions similar to those used for mutant Bcr-Abl (Fig. 2B). We didn’t observe an increase in pTyr 207 CrkL in Abl R367A transfected cells (Fig. 2C), consistent with the predicted greater enzymatic activity of the oncogenic Bcr-Abl vs. wt Abl protein. Nevertheless, the fact that imidazole treatment led to increased wt Abl autophosphorylation provided the basis to proceed to phosphoproteomics analysis.
Figure 2.

Chemical rescue of FLAG-tagged Abl R367A in abl−/−arg−/− mouse 3T3 fibroblast cells. (A) abl−/−arg−/− mouse 3T3 fibroblast cells were stably transfected with Abl R367A. The expression level of Abl is comparable to that of Bcr-Abl in K562 leukemia cells. (B) Abl was immunoprecipitated from Abl R367A cells treated with imidazole (imi, 10 min) or vehicle (con) and analyzed by immunoblotting with antibodies anti-phospho Abl (Tyr245) (top panel) and anti-FLAG (bottom panel). The ratio was calculated using pAbl: Abl. (C) Abl R367A cells were treated with 20 mM imidazole for the indicated time (0, 0, 10, 20 min) in the presence or absence of 1 μM Abl kinase inhibitor gleevec (imatinib). Whole cell lysates were immunoblotted with antibodies anti-phospho CRKL (Tyr 207) (top panel) and anti-Actin (bottom panel). The ratio was calculated using pCRKL: Actin.
Mass spectrometric analysis of R367A Abl cells
Fig. 3A & B illuminate the experimental strategy of chemical rescue in combination with quantitative phosphoproteomics. We used SILAC methodology21 to prepare two populations of R367A Abl cells, one of which was labeled with heavy isotope Arg and Lys and the other with standard (light) amino acids. After 5 passages, heavy atom amino acid labeled cells were treated with imidazole and the standard isotope cells with vehicle for 10 min and the same amounts of cell lysate from the two cell pools were then combined. The coomassie stained SDS-PAGE in Fig. 3C verified that the same amounts of cell lysate from treated and untreated cells were mixed, which is critical for proper SILAC analysis. After lysis, the mixtures were trypsinized and then the protein fragments subjected to phosphoTyr antibody binding to enrich for phosphoTyr-containing peptides. This enriched phosphoTyr pool was analyzed by tandem mass spectrometry and a total of 318 phosphorylated peptides that map to 300 phospho-sites on 218 proteins were detected from two biological replicates in R367 Abl cells. Of these, 245 and 228 phosphotyrosine sites were identified in each experimental replicate and 173 phosphotyrosine sites were identified in common (Fig. 4A). The relative abundance of the heavy labeled peptides derived from imidazole-treated cells versus light labeled peptides derived from vehicle treated cells was quantified to obtain a SILAC ratio for each identified peptide. We used a SILAC ratio cutoff of 1.20, a cutoff that we have found previously to be predictive of enhanced protein tyrosine phosphorylation when immunoprecipitation-western blot validation is performed9, 10. The majority of the phosphotyrosine sites showed less than a 20% change upon imidazole treatment, which is consistent with the idea that Abl influences only a subset of tyrosine phosphorylation events. A total of 82 phosphotyrosine sites in 65 proteins was found to show a SILAC ratio of 1.20 or greater (Supplementary Table 1&2; Fig. 4B). One of the pTyr peptides with the highest SILAC ratios (2.34) was derived from p130Cas (Bcar1) (Fig. 4B), a well-established SH3 domain containing signaling protein with an adaptor functionality22, 23. The SILAC ratio of the Bcar1/p130Cas pY391-peptide (RPGPGTLpYDVPR) is shown in Fig. 5A and its tandem mass spectrum indicates the exact site of phosphorylation site of the peptide (Fig. 5B). Many of the identified proteins through our study have been reported to be Abl and Bcr-Abl downstream substrates (Supplementary Table 3). Notably, a number of up-regulated proteins, such as low density lipoprotein receptor (LDLR), have not previously been identified as Abl substrates.
Figure 3.

Proteomics approach for identification of Abl substrates. (A) Schematic diagram of chemical rescue. Because of the presence of R367A mutation, Abl kinase is catalytically inactive and is highlighted in gray color (left panel). The activity of kinase-dead Abl can be rescued by small molecule imidazole (green star) in Abl R367A live cells. Upon imidazole treatment, rescued Abl (highlighted in red color, right panel) phosphorylates a variety of substrates within minutes. (B) Schematic diagram of the experimental workflow of SILAC phosphoproteomics. In order to identify the targets of Abl kinase, Abl R367A cells were treated with (experiment, cells grown in heavy isotope-containing medium) or without imidazole (control, cells grown in light isotope-containing medium) for 10 min. Whole cell lysates from two different cell populations were mixed in a ratio of 1:1, which were then subjected to trypsin digestion and pTyr-peptide enrichment, followed by LC-MS/MS for quantification and identification of pTyr-peptides. (C) 100 μg of cell lysates from two cell populations, Abl R367A cells grown in heavy isotope-containing medium and Abl R367A cells grown in light isotope-containing medium, were loaded on a 4-15% SDS-PAGE gel and were stained with Coomassie blue dye.
Figure 4.

Phosphotyrosine-containing peptides identified from LC-MS/MS analysis of Abl R367A cell lysates. (A) Venn diagram of identified phosphotyrosine peptides from two biological replicates. A total of 300 phosphotyrosine peptides were identified and 173 of them were identified in common between replicates. (B) The SILAC ratios of the 173 phosphotyrosine peptides are plotted as a scatter plot. A good correlation of SILAC ratios between replicates was observed. Several upregulated phosphotyrosine peptides are highlighted, such as pY391-containing peptide derived from Bcar1.
Figure 5.
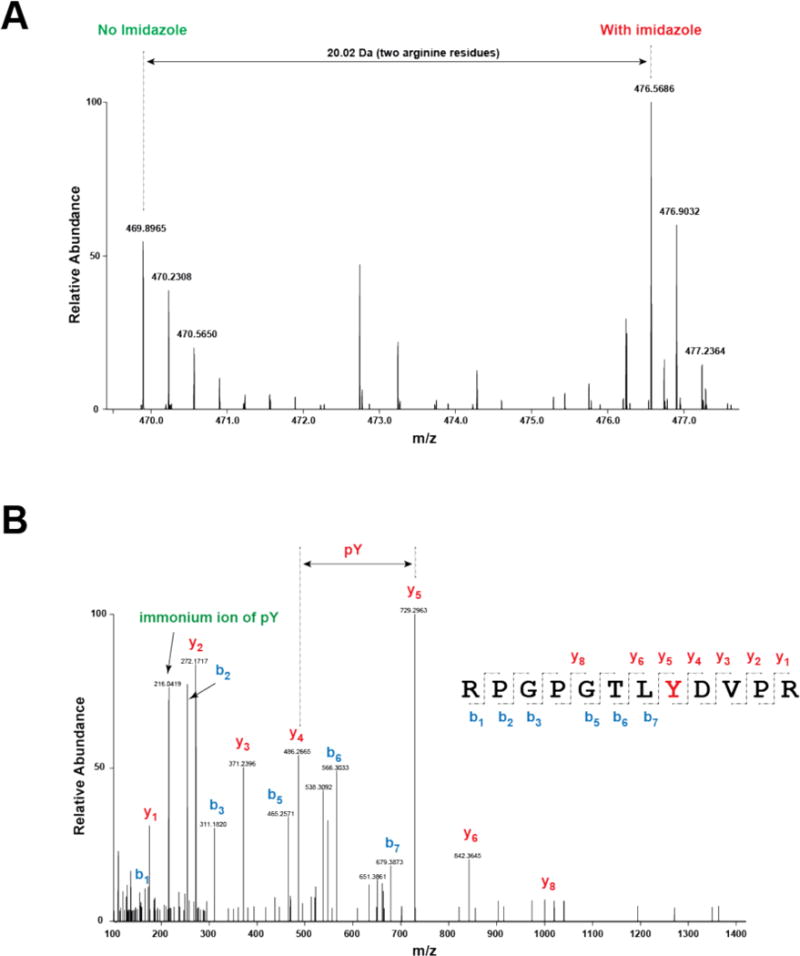
SILAC ratio and spectral annotation of the pY391-containing peptide (RPGPGTLpYDVPR) derived from Bcar1. (A) MS1 scan representing the SILAC intensities of RPGPGTLpYDVPR is shown. Two arginine residues resulted in 20.02 Da shift when SILAC-labeled peptides were observed in MS. (B) Spectral annotation of RPGPGTLpYDVPR onto the MS2 scan is shown. The m/z spacing as indicated by the arrow between y4 and y5 ions represents the exact site of phosphorylation on Tyr8 of the peptide sequence (RPGPGTLpYVPR). Immonium ion (m/z 216.04) of pY is also shown in the MS/MS spectrum, evidence of the existence of tyrosine phosphorylation on the fragmented peptide.
Gene Ontology
The tyrosine phosphorylated proteins obtained from Abl rescue were grouped in several categories using gene ontology analysis (Fig. 6A&B). The various categories that were especially well-represented among these substrates included protein kinases, cytoskeletal proteins, adaptor and scaffold proteins (SH2 and SH3 domain containing proteins), as well as proteins involved in cell adhesion, migration, and apoptosis. Concentrations of Abl targets in these clusters are consistent with extensive prior experimental studies on Abl1 and are also overlapped significantly with proteomics analysis of Src9, 10, which is considered an upstream regulator of Abl20, 24. The largest two categories populated by the Abl targets are protein kinases and cytoskeleton-related proteins.
Figure 6.

Gene ontology analysis of the tyrosine phosphorylated proteins upregulated in Abl R367A cells upon imidazole treatment. (A) Among the 300 tyrosine phosphorylated peptides identified by LC-MS/MS, 82 of them have a SILAC ratio higher than 1.20, corresponding to 65 proteins. (B) Enriched functional gene clusters for the 65 up-regulated genes.
Immunoprecipitation validation studies
We selected several proteins from the phospho-proteins identified by mass spectrometry using chemical complementation of Abl and performed immunoprecipitation and western blot analysis with antibodies against the phosphorylated site identified (when available) or anti-pTyr Ab as a method of validation. The selection of proteins for validation was in part based on the availability of high quality commercial antibodies for a given target. Of those that showed efficient and high quality immunoprecipitation and western blotting with the requisite antibodies tested, 6 proteins showed at least a 20% increase in anti-phosphotyrosine western blotting signal after cells were treated with imidazole (Fig. 7 A&B; Supplementary Fig. 1). These proteins included focal adhesion kinase (FAK), paxillin, the Src family tyrosine kinase Hck, the adaptor protein p130Cas (Bcar1), actin, and LDL receptor. We also analyzed whether these proteins were shown to have similar behavior in the context of Bcr-Abl R/A mutant cells (Fig. 8A). These proteins also appeared to be increased in the context of imidazole in cells expressing this rescuable form of activated Bcr-Abl. The abl−/−arg−/− mouse fibroblast cell line stably transfected with empty vector was used for control experiments (Fig. 8B), and the comparison revealed that the increased pTyr proteins observed in Abl and Bcr-Abl R/A mutant cells are likely induced by rescued Abl kinase activity rather than extraneous factors.
Figure 7.
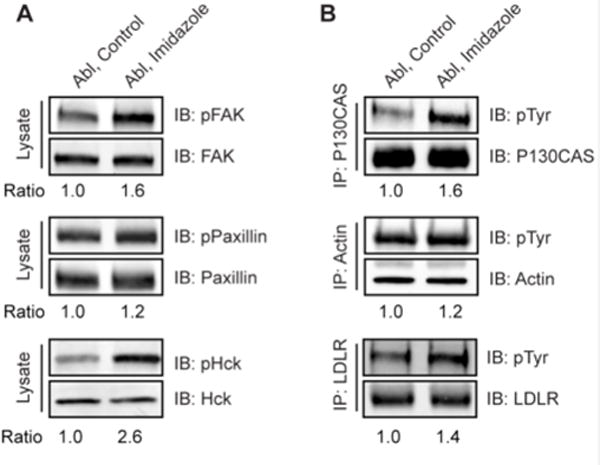
Validation of Abl substrates identified from SILAC proteomics. (A) Abl R367A cells were treated with or without 20 mM imidazole for 10 min. Whole cell lysates were immunoblotted with anti-pTyr specific antibodies phospho-FAK (Tyr576/577), phospho-Paxillin (Tyr118), and phospho-Hck (Tyr410). As a control, the total proteins were probed with antibodies recognizing total FAK, Paxillin and Hck, respectively. (B) P130CAS (BCAR1), Actin and LDLR were immunoprecipitated from Abl R367A cells treated with or without imidazole and analyzed by immunoblotting with antibodies anti-phosphotyrosine 4G10 (top panel) and anti-P130CAS, Actin, or LDLR (bottom panel). All the ratios were calculated using pTyr (top panel): loading control (bottom panel). Each experiment has been repeated at least twice and similar results were observed for each replicate.
Figure 8.

Analysis of identified Abl substrates in Bcr-Abl R367 cells. (A) Bcr-Abl R367A cells were treated with or without 20 mM imidazole for 10 min. Whole cell lysates were immunoblotted with anti-pTyr specific antibodies phospho-FAK (Tyr576/577), phosphor-Paxillin (Tyr118), and phospho-Hck (Tyr410). As a control, the total proteins were probed with antibodies recognizing FAK, Lyn, and Paxillin respectively. p130CAS was immunoprecipitated from Bcr-Abl R367A cells treated with or without imidazole and analyzed by immunoblotting with anti-phosphotyrosine 4G10 antibody (top panel) and anti-p130CAS antibody (bottom panel). (B) abl−/−arg−/− mouse 3T3 fibroblast cells stably transfected with empty vector pcDNA 3.0 (abbreviated as EV) were treated with or without 20 mM imidazole for 10 min. Whole cell lysates were immunoblotted with anti-pTyr specific antibodies. As a control, the total proteins were probed with antibodies recognizing FAK, Lyn, and Paxillin, respectively. All the ratios were calculated using pTyr (top panel): loading control (bottom panel). Each experiment has been repeated at least twice and similar results were observed for each replicate.
DISCUSSION
This study describes the application of a chemical complementation method to assess the cellular protein tyrosine phosphorylation in a rapid kinetic manner. Most alternative proteome-wide strategies that have been used previously to identify Bcr-Abl phosphorylation targets have been conducted on a time scale of hours to days, including genetic manipulation. The chemical rescue method here offers a different window for observing the phospho-signaling pattern associated with an intracellular enzyme compared with the more conventional application of even highly selective ATP-site inhibitors. Of course, many kinase inhibitors are not so specific and can hit multiple kinase enzymes. Conceptually, data from even very selective kinase inhibitors provide the perspective of protein phosphorylation events that have been reached over a chronic period in the life of the cell. While clearly relevant, such processes reach equilibrium over hours to days and may represent ‘late’ events in a signaling cascade.
The chemical rescue method is most powerful in revealing phosphorylation functions of a kinase at the outset of signaling. In this sense the chemical rescue of mutant kinases resembles signaling initiation analogous to the trigger of a ligand binding a cell surface receptor. It is accepted that nuclear-cytosolic tyrosine kinases like the Src and Abl families are activated by a range of upstream events but it has been difficult to isolate the consequences of stimulation of a single kinase rather than an entire network. We view this as the main advantage of the chemical complementation strategy.
We identified many up-regulated tyrosine phosphorylation sites on proteins involved in cytoskeletal regulation and signal transduction pathways in response to Abl activation, in agreement with previous studies on the biological functions of Abl kinase. Thus, our findings have confirmed and extended the intracellular targets of Abl and indicated clear roles for the enzyme in cellular processes. Rapid stimulation of Abl leads to many additional tyrosine and serine kinases being targeted, consistent with a key role for Abl in an extensive enzymatic phospho-amplification network. Such a kinase amplification network was perhaps underappreciated in the context of the Abl proto-oncogene which is typically viewed as quiescent in the resting cell. Our data suggests that Abl, like Src10, is quite an active cellular enzyme. However, the presence of tyrosine phosphatase activity in cells opposes this basal kinase activity, which renders it invisible. Interestingly, the tyrosine phosphorylated phospho-proteins observed by chemical rescue of R/A Abl were also detected in chemical rescue of Bcr-Abl. These findings are consistent with a hyperactivity of Bcr-Abl that has been previously reported25–27, which broadens its substrate portfolio versus Abl. In addition, our study reveals novel Abl-mediated tyrosine phosphorylation sites, including proteins involved in metabolism and transcription. The specific functions of these novel phosphorylation sites remain to be discovered. Future detailed investigation on those newly identified targets is necessary to reveal the physiological significance of such Abl-mediated tyrosine phosphorylation events, yielding new insight into network of Abl kinase.
While our manuscript was in preparation, an alternative approach was published to identify Abl substrates. Diae and co-workers engineered a split-Abl kinase that can be activated by rapamycin-induced dimerization28. They inserted FKBP and FRB to two protein fragments of Abl, which can be rapidly dimerized and activated by the presence of rapamycin in live cells. The authors conclude that the split Abl kinase provides a tool to temporally activate cytosolic kinase. They employed duplex neutron-encoded stable isotope labeling with amino acids in cell culture (NeuCode SILAC) and identified a large pool of 432 phosphotyrosine sites as a result of direct activation of Abl kinase. Both studies have identified many of the molecules involved in the same signaling pathway as potential direct Abl targets. However, only 10 of our 65 Abl phosphotyrosine targets overlapped with the split Abl kinase method, underscoring the need for multiple complementary methods to dissect the complex processes of cells.
Although we believe that the large majority of the enhanced phosphotyrosine events detected in our chemical rescue approach are likely direct substrates of activated Abl kinase because of the rapid time scale, we cannot rule out the possibility of identifying indirect substrates of Abl kinase, which are sequentially phosphorylated by Abl-activated tyrosine kinases. Ultimately, this is a general challenge emanating from the complex cellular mileu. Regardless, our methodology has the potential to study immediate post-translational changes that occur in response to activation of Abl. The chemical complementation technology described here has relied on the somewhat demanding task of somatic recombination followed by stable transfection. However, the recent development of CRISPR/Cas929 provides a more efficient and economical pathway to introducing the requisite R/A mutation in a protein tyrosine kinase and cell type of interest using a direct knock-in strategy. Such technology could greatly enhance the chemical complementation approach to address a wide array of cell signaling challenges in biomedical research and will thereby enable us to uncover global phosphorylation events induced by individual kinases.
Supplementary Material
Acknowledgments
We thank Charles L. Sawyers for providing the p210 Bcr-Abl cDNA in the pMSCV puro retroviral vector. We thank the Pandey lab and the Cole lab members for helpful discussion. The study was funded by grants from the National Institutes of Health (R01 NS089662-04 to A.J.K and R01 CA074305 to P.A.C), and the grant from the International Research & Development Program of National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (Grant number: NRF-2016K1A3A1A47921601) to M-S. Kim.
References
- 1.Khatri A, Wang J, Pendergast AM. Multifunctional Abl kinases in health and disease. J Cell Sci. 2016;129:9–16. doi: 10.1242/jcs.175521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Konopka JB, Witte ON. Activation of the abl oncogene in murine and human leukemias. Biochim Biophys Acta. 1985;823:1–17. doi: 10.1016/0304-419x(85)90012-5. [DOI] [PubMed] [Google Scholar]
- 3.Hughes T, Deininger M, Hochhaus A, Branford S, Radich J, Kaeda J, Baccarani M, Cortes J, Cross NC, Druker BJ, Gabert J, Grimwade D, Hehlmann R, Kamel-Reid S, Lipton JH, Longtine J, Martinelli G, Saglio G, Soverini S, Stock W, Goldman JM. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108:28–37. doi: 10.1182/blood-2006-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maiani E, Diederich M, Gonfloni S. DNA damage response: the emerging role of c-Abl as a regulatory switch? Biochem Pharmacol. 2011;82:1269–1276. doi: 10.1016/j.bcp.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Shi J, Meng L, Sun MZ, Guo C, Sun X, Lin Q, Liu S. CRKL knockdown promotes in vitro proliferation, migration and invasion, in vivo tumor malignancy and lymph node metastasis of murine hepatocarcinoma Hca-P cells. Biomed Pharmacother. 2015;71:84–90. doi: 10.1016/j.biopha.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 6.Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat Biotechnol. 2003;21:315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 7.Reddy RJ, Gajadhar AS, Swenson EJ, Rothenberg DA, Curran TG, White FM. Early signaling dynamics of the epidermal growth factor receptor. Proc Natl Acad Sci U S A. 2016;113:3114–3119. doi: 10.1073/pnas.1521288113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowry WE, Huang J, Ma YC, Ali S, Wang D, Williams DM, Okada M, Cole PA, Huang XY. Csk, a critical link of g protein signals to actin cytoskeletal reorganization. Dev Cell. 2002;2:733–744. doi: 10.1016/s1534-5807(02)00175-2. [DOI] [PubMed] [Google Scholar]
- 9.Qiao Y, Molina H, Pandey A, Zhang J, Cole PA. Chemical rescue of a mutant enzyme in living cells. Science. 2006;311:1293–1297. doi: 10.1126/science.1122224. [DOI] [PubMed] [Google Scholar]
- 10.Ferrando IM, Chaerkady R, Zhong J, Molina H, Jacob HK, Herbst-Robinson K, Dancy BM, Katju V, Bose R, Zhang J, Pandey A, Cole PA. Identification of targets of c-Src tyrosine kinase by chemical complementation and phosphoproteomics. Mol Cell Proteomics. 2012;11:355–369. doi: 10.1074/mcp.M111.015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Cole PA. Catalytic mechanisms and regulation of protein kinases. Methods Enzymol. 2014;548:1–21. doi: 10.1016/B978-0-12-397918-6.00001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muratore KE, Seeliger MA, Wang Z, Fomina D, Neiswinger J, Havranek JJ, Baker D, Kuriyan J, Cole PA. Comparative analysis of mutant tyrosine kinase chemical rescue. Biochemistry. 2009;48:3378–3386. doi: 10.1021/bi900057g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wetzel DM, McMahon-Pratt D, Koleske AJ. The Abl and Arg kinases mediate distinct modes of phagocytosis and are required for maximal Leishmania infection. Mol Cell Biol. 2012;32:3176–3186. doi: 10.1128/MCB.00086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burgess MR, Skaggs BJ, Shah NP, Lee FY, Sawyers CL. Comparative analysis of two clinically active BCR-ABL kinase inhibitors reveals the role of conformation-specific binding in resistance. Proc Natl Acad Sci U S A. 2005;102:3395–3400. doi: 10.1073/pnas.0409770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koleske AJ, Gifford AM, Scott ML, Nee M, Bronson RT, Miczek KA, Baltimore D. Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron. 1998;21:1259–1272. doi: 10.1016/s0896-6273(00)80646-7. [DOI] [PubMed] [Google Scholar]
- 16.Kim MS, Zhong Y, Yachida S, Rajeshkumar NV, Abel ML, Marimuthu A, Mudgal K, Hruban RH, Poling JS, Tyner JW, Maitra A, Iacobuzio-Donahue CA, Pandey A. Heterogeneity of pancreatic cancer metastases in a single patient revealed by quantitative proteomics. Mol Cell Proteomics. 2014;13:2803–2811. doi: 10.1074/mcp.M114.038547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 18.de Jong R, ten Hoeve J, Heisterkamp N, Groffen J. Tyrosine 207 in CRKL is the BCR/ABL phosphorylation site. Oncogene. 1997;14:507–513. doi: 10.1038/sj.onc.1200885. [DOI] [PubMed] [Google Scholar]
- 19.Brasher BB, Van Etten RA. c-Abl has high intrinsic tyrosine kinase activity that is stimulated by mutation of the Src homology 3 domain and by autophosphorylation at two distinct regulatory tyrosines. J Biol Chem. 2000;275:35631–35637. doi: 10.1074/jbc.M005401200. [DOI] [PubMed] [Google Scholar]
- 20.Tanis KQ, Veach D, Duewel HS, Bornmann WG, Koleske AJ. Two distinct phosphorylation pathways have additive effects on Abl family kinase activation. Mol Cell Biol. 2003;23:3884–3896. doi: 10.1128/MCB.23.11.3884-3896.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amanchy R, Kalume DE, Pandey A. Stable isotope labeling with amino acids in cell culture (SILAC) for studying dynamics of protein abundance and posttranslational modifications. Sci STKE. 2005;2005:pl2. doi: 10.1126/stke.2672005pl2. [DOI] [PubMed] [Google Scholar]
- 22.Camacho Leal Mdel P, Sciortino M, Tornillo G, Colombo S, Defilippi P, Cabodi S. p130Cas/BCAR1 scaffold protein in tissue homeostasis and pathogenesis. Gene. 2015;562:1–7. doi: 10.1016/j.gene.2015.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett A, Pellet-Many C, Zachary IC, Evans IM, Frankel P. p130Cas: a key signalling node in health and disease. Cell Signal. 2013;25:766–777. doi: 10.1016/j.cellsig.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 24.Plattner R, Irvin BJ, Guo S, Blackburn K, Kazlauskas A, Abraham RT, York JD, Pendergast AM. A new link between the c-Abl tyrosine kinase and phosphoinositide signalling through PLC-gamma1. Nat Cell Biol. 2003;5:309–319. doi: 10.1038/ncb949. [DOI] [PubMed] [Google Scholar]
- 25.Reckel S, Hamelin R, Georgeon S, Armand F, Jolliet Q, Chiappe D, Moniatte M, Hantschel O. Differential signaling networks of Bcr-Abl p210 and p190 kinases in leukemia cells defined by functional proteomics. Leukemia. 2017;31:1502–1512. doi: 10.1038/leu.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winter GE, Rix U, Carlson SM, Gleixner KV, Grebien F, Gridling M, Muller AC, Breitwieser FP, Bilban M, Colinge J, Valent P, Bennett KL, White FM, Superti-Furga G. Systems-pharmacology dissection of a drug synergy in imatinib-resistant CML. Nat Chem Biol. 2012;8:905–912. doi: 10.1038/nchembio.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breitkopf SB, Yuan M, Helenius KP, Lyssiotis CA, Asara JM. Triomics Analysis of Imatinib-Treated Myeloma Cells Connects Kinase Inhibition to RNA Processing and Decreased Lipid Biosynthesis. Anal Chem. 2015;87:10995–11006. doi: 10.1021/acs.analchem.5b03040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz JE, Morgan CW, Minogue CE, Hebert AS, Coon JJ, Wells JA. A Split-Abl Kinase for Direct Activation in Cells. Cell Chem Biol. 2017;24:1250–1258 e1254. doi: 10.1016/j.chembiol.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


