Abstract
Somatic cell differentiation is required throughout the life of a multicellular organism to maintain homeostasis. In contrast, germ cells have only one specific function; to preserve the species by conveying the parental genes to the next generation. Recent studies of the development and molecular biology of the male germ cell have identified many genes, or isoforms, that are specifically expressed in the male germ cell. In the present review, we consider the unique features of male germ cell differentiation. (Reprod Med Biol 2007; 6: 1–9)
Keywords: gene, germ cell, infertility, single nucleotide polymorphisms, spermiogenesis, testis
INTRODUCTION
ANIMALS CONSIST OF somatic cells, which comprise the organismal structure and maintain homeostasis, and germ cells, which function only to preserve the species. Germ cells differentiate into gametes (spermatozoa or ova) that convey the parental genes to the next generation through fertilization. 1 Germ cells are the only cells that undergo meiotic division, which occurs during germ cell differentiation and involves mosaic recombination of homologous chromosomes. As a result, each chromosome of a haploid germ cell is an arbitrary assemblage of parts of the two homologous parental chromosomes. Because mosaic recombination and meiosis can prevent the fixation of accumulated mutations that have arisen spontaneously on one chromosome during gene replication or other processes, sexual reproduction is the most effective mechanism for maintenance and adaptation of species. 1 Although germ cell differentiation is entirely unrelated to maintenance of homeostasis for the individual, it is one of the most important processes occurring in multicellular organisms.
As many as 15% of human couples 2 are infertile, and male infertility is associated with about half of these cases. A decrease in sperm production has recently been reported. 2 Although advances in medical technology have allowed some infertile couples to have children, more than half of all infertility is idiopathic. 2 Because unresolved environmental problems such as global pollution might be causing endocrine disruption, a thorough understanding of the basic mechanisms of germ cell differentiation is critical for development of infertility treatments. In the present review, we describe the current state of understanding of the specifics of male germ cell differentiation.
SPERMATOGENESIS
GERM CELLS DEVELOP from primordial germ cells (PGC) that are formed in the first stages of embryonic growth. 3 In mice, PGC are first observed at days 7–8 of pregnancy, in the extra‐embryonic mesoderm. They subsequently migrate through the hindgut and dorsal mesentery, undergoing repeated multiplicative divisions, and establish the gonadal primordium. At this stage, whether a particular cell will be a male or female germ cell cannot be determined. At days 12–13 of pregnancy, seminiferous cords formed by Sertoli cell precursors appear in the gonadal primordium, and whether the gonad will be a testis or ovary is determined. In the testis, PGC proliferate until days 13–14 of pregnancy, when their cell cycles are arrested, and they do not recommence proliferation and differentiation until a few days after birth. In the ovary, PGC/gonocytes enter meiosis and arrest in the prophase of meiosis I.
The male versus female germ‐cell fate of PGC is determined by the environment around the genital ridge and other factors. 4 , 5 The sex chromosome is important before meiosis, however, because the XY ovum differentiates in the ovary and the XX sperm of testicular origin cannot differentiate. 6 , 7 The systematic differentiation processes in the testis are supported by Sertoli cells and by interstitial cells of the testes (Leydig cells); functioning of these cell types is regulated by testosterone and other factors. After birth, the spermatogonia emerge from cell cycle arrest and the first wave of spermatogenesis begins.
Spermatogenesis occurs in seminiferous tubules throughout adulthood and produces huge amounts of sperm. The process requires approximately 1 month in mice and 2 months in humans and can be roughly divided into three phases. First, the spermatogonia proliferate and differentiate. Second, meiotic prophase of the spermatocytes occurs, leading to the formation of haploid round spermatids. Third, the morphogenetic events necessary for sperm formation occur in the postmeiotic phase.
Spermatogonia, the multipotent stem cells of the germ line that are reprogrammed through cell differentiation, proliferate by mitotic division. In spermatogenesis, differentiation of spermatogonia or PGC is important for subsequent meiosis and spermiogenesis; mice deficient in either Vasa or Mili, homologous genes normally expressed in PGC, exhibit imperfect meiosis and differentiated spermatids. 8 , 9 Adult germ‐line stem cells that are multipotent in culture can be isolated from spermatogonia. 10 In the leptotene phase occurring within about 2 weeks of spermatocyte formation, synapsis and crossing over are observed. Two (meiotic) cell divisions unaccompanied by DNA synthesis occur in the zygotene and pachytene.
After meiotic division (during the process of haploid germ cell differentiation, or spermiogenesis), the rounded spermatids undergo marked morphological changes to become sperm; the nucleus assumes a compact shape, the mitochondria are rearranged, the flagellum forms and the acrosome is generated. This period of differentiation lasts for about 5–6 weeks in humans 11 or 2–3 weeks in mice. As the first sperm cell is completed in the testis, the spermatogonia, spermatocytes and spermatids are systematically arranged in the seminiferous tubules; the spermatogonia are in the tubule walls, the spermatids are at the tubule centers and the spermatocytes are between them. The spermatogenic cells included in the testicular tubule section are understood to form by the combination of spermatogenic cells at a specific stage of differentiation. In the rat, the spermatogenic cycle of each tubule of the seminiferous epithelium is divided into 14 stages by this combination in stage 12 in the mouse. This stage can be crudely distinguished by the differential density of the spermiogenic cells observed by microscopy of the separated, extended tubules. 12
Stem cell proliferation and differentiation, meiosis, generation of haploid germ cells, and morphogenesis of the developing sperm require approximately 1 month in mice and 2 months in humans. The sperm that have completed morphogenesis move from the testis to the epididymis after collection in the rete testis through the center of the testicular tubule. The sperm mature in the epididymis and are finally ejaculated from the vas deferens through the caput, corpus and cauda in the epididymis. The caudal sperm are used in in vitro fertilization (IVF) procedures.
In mammals, various hormones act on specific cells to maintain homeostasis of the living organism. Many hormones, growth factors and vitamins act on the testis to regulate spermatogenesis. 13 The adrenal gland and testes secrete androgen hormones, one of which (testosterone) is required for spermatogenesis. Testosterone is secreted by the Leydig cells, which are adjacent to the seminiferous tubules. Luteinizing hormone (LH) stimulates the secretion of testosterone from the Leydig cells, and follicle‐stimulating hormone (FSH) secreted from the pituitary gland is required for proper functioning of Sertoli cells in spermatogenesis. FSH secretion from the pituitary gland is suppressed by inhibin secreted from Sertoli cells and induced by activin secreted from Sertoli, peritubular and Leydig cells. Spermatogenesis is also a temperature‐ and radiation‐sensitive process. 13
Only germ cells undergo meiosis. The multipotency of the fertilized egg is guaranteed by telomere maintenance and reprogramming. Germ cell differentiation is strictly controlled by hormones. Male germ cells are unique in morphology and function, and they have male‐specific genomic imprinting. Many specific gene products are required for spermatogenesis.
SPERMIOGENESIS‐SPECIFIC GENES
GENE DEFICIENCIES THAT cause infertility in mice have been reported. 14 The cause of the infertility might be determined by examining the efficiency of genes that are ubiquitously expressed in all tissues. However, many genes are specifically expressed during spermatogenesis, 15 and specific genes have been shown to function in spermatogenesis. If the mutation of a haploid germ cell‐specific gene is the cause of male infertility, then it will not influence somatic cell differentiation or any cell differentiation in the female.
Genetic analyses of germ cells have shown that germ cell‐specific isozymes of ubiquitously expressed genes function in testicular germ cells. 1 For example, almost all steps of glycolysis in germ cells might be catalyzed by germ cell‐specific isozymes. 1 In addition, many intronless, germ cell‐specific genes are present on germ cell chromosomes. 1 , 16
TRANSCRIPTION IN HAPLOID GERM CELLS
CHROMATIN RESTRUCTURING IS a unique characteristic of the male germ cell. Nuclear elongation and chromatin condensation occur concomitantly with modifications in the nuclear basic proteins associated with DNA. Histone displacement is accompanied by several biochemical events and a set of basic nuclear proteins appears; these proteins include tH2A, tH2B, H1t, spermatid‐specific H2B (ssH2B), haploid germ cell‐specific nuclear protein 1 (Hanp1), testis‐specific HMG (tsHMG), histone H1‐like protein in spermatids 1 (Hils 1), transition proteins (TNP), and protamines. 1 The transcription that occurs during spermatogenesis commences in the almost round spermatids and these transcripts are translated in accordance with spermatid elongation. The cAMP‐responsive element (CRE) modulator protein (CREM) has been shown to play an important regulatory role in spermiogenesis by binding to CRE sequences on the genomic DNA. 17 CREM‐deficient mice exhibit decreased testis weight and a complete lack of mature spermatozoa in the seminal fluid; histological analyses of these mice have shown that spermatogenesis is arrested at the early round spermatid stage. 17
In somatic cells, transcriptional activation by CREM requires phosphorylation of a unique regulatory site (Ser117) on CREM by the ubiquitous coactivator CBP (CREB [CRE‐binding protein]‐binding protein). 18 In the testis, CREM in testis (CREM‐τ) transcriptional activity does not require CREM phosphorylation but is, instead, controlled by the interaction between CREM‐τ and the ACT (tissue‐specific activator of CREM in testis) protein 18 (Fig. 1). TISP‐40 (the transcript induced in spermiogenesis protein‐40) is also associated with CREM‐τ. 19 ACT is regulated by its selective association with the testis‐enriched kinesin KIF17b, a kinesin highly expressed in male germ cells 20 (Fig. 1).
Figure 1.
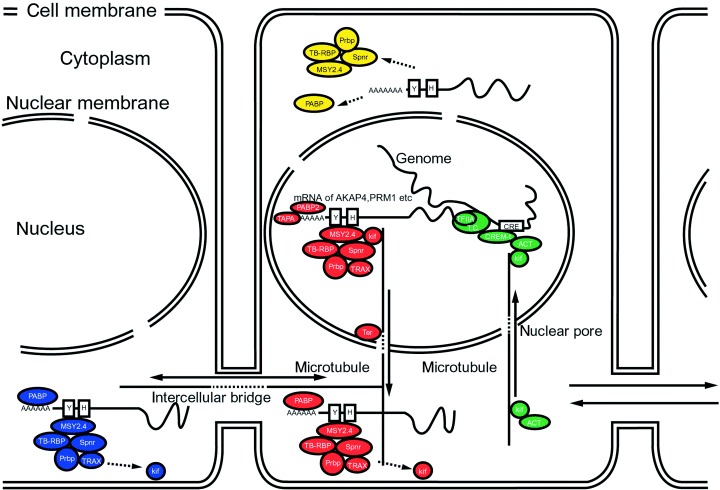
Schematic representation of the regulatory pathways affecting gene expression during mouse spermatogenesis. 1 Spermatids are connected by intercellular bridges. The pathway of transcriptional control by CREM‐τ is shown in green. RNA‐binding proteins or poly(A)‐RNA polymerases modify the translation of transcribed mRNAs, which are then transferred to the cytoplasm along microtubules by the kinesin KIF17b (red). Some mRNAs are transferred to another cell through the intercellular bridge (blue). The timing of translation is regulated by RNA‐binding proteins (yellow). Circles indicate proteins, T.C. indicates the transcriptional complex, and the boxed Y and H indicate Y‐ and H‐elements on the mRNA, respectively. Bars indicate microtubule networks, and arrows show the direction of movement of protein and protein‐mRNA complexes. Doublet boxes and circles indicate cell membranes and nuclear membranes, respectively.
Some genes that are specifically expressed in haploid germ cells have been shown to lack CRE motifs in their promoter regions, suggesting that CRE‐independent regulatory mechanisms are also present. Homeobox genes that are expressed only in specific cell lineages are of particular interest as candidates for the regulation of differentiation, but the regulatory functions of these genes in spermatogenesis are still largely unknown. Some fundamental transcription factors are expressed during spermiogenesis, such as TATA‐binding protein (TBP), which accumulates at much higher levels in early haploid germ cells than in any somatic cell type. 21
The expression of specific genes appears to be regulated by alternative splicing of transcripts. 22 The CREM gene is differentially regulated during spermatogenesis; repressors (α, β and γ) are expressed in pre‐ and early meiotic germ cells, whereas activators (τ, τ1, and τ2) are abundant in postmeiotic germ cells. 23 Low levels of CREM activator expression have also been detected in pachytene spermatocytes. Furthermore, Sertoli cells are reported to express a CREM isoform (an inducible cAMP early repressor) that is truncated as a result of transcription from an alternative promoter. 24 RNA‐binding proteins might also play roles in germ cell‐specific mRNA splicing. 25 , 26 Recent studies have shown that ejaculated human spermatozoa contain the transcription products of over 1000 genes 27 and specific RNAs function in fertilized eggs as well. 28
TRANSLATION IN HAPLOID GERM CELLS
THE CYTOPLASM OF haploid germ cells is lost during postmeiotic spermiogenesis. Many genes are translated when necessary from transcripts stored in round spermatids. An intercellular bridge connects the haploid spermatids, creating a cytoplasmic interconnection through which some mRNAs and proteins are transported. 29 RNA‐binding proteins are also important in the temporal regulation of translation; 30 they add poly(A) sequences to transcripts 31 or interact with cis‐acting elements in the 5′‐ and 3′‐untranslated regions (UTR) of certain genes 32 (Fig. 1). H‐ and Y‐elements are found in the 5′‐ and 3′‐UTR of genes expressed in spermatids. The genes for protamines 1 and 2 are first transcribed in step‐7 spermatids, but are not translated until about step 13 of spermiogenesis. 33
Many protein‐mRNA complexes have been found. Testis‐brain RNA‐binding protein (TB‐RBP/translin) and translin‐associated factor X (TRAX) bind to mRNA on protamines and to A kinase‐anchoring protein 4 (AKAP4), 34 , 35 and these protein‐mRNA complexes are transported across the interspermatid bridge. TB‐RBP/translin complexes bind to H‐ or Y‐elements on mRNA transcripts, and these protein‐mRNA complexes, together with Ter ATPase, 36 are transported by KIF17b through nuclear pores to polysomes for storage in the cytoplasm. 35
These various mechanisms of transcriptional and post‐transcriptional regulation might also act cooperatively. Some mRNA‐binding proteins, such as Prbp (Prm‐1 RNA‐binding protein), 37 MSY2 and 4, 38 Spnr (spermatid perinuclear RNA‐binding protein), 39 PABP2 (poly(A)‐binding protein 2), 40 and TPAP 31 might serve to adjust the timing of translation. The length of the poly(A) tail of mRNA expressed in the testis changes during spermiogenesis, 31 and PABP inhibit translation in vitro, 41 suggesting that the length of the poly(A) tail affects translation in spermatids.
THE Y CHROMOSOME
THE HUMAN Y chromosome is roughly one‐third the size of the X chromosome, but the number of genes encoded by the Y chromosome is less than one‐third the number of genes encoded by the X chromosome. Both telomeres of the Y chromosome include a pseudoautosomal region (PAR), which shares homology with regions of the X chromosome. The non‐recombining region of the Y (NRY) accounts for 95% of the Y chromosome, and does not undergo homologous recombination with the X chromosome. The possibility that rearrangement might occur among the many repeated regions of the Y chromosome has been raised. Because the Y chromosome has no homolog, and most areas do not participate in homologous rearrangement, the genes of the Y chromosome are passed directly from parent to child; however, as a result, genes might be maintained without mutation. The NRY is composed of euchromatin, areas of transcriptional activity, and heterochromatin. Many Y chromosome genes are multicopy genes that do not include open reading frames; however, the Y chromosome contains genes that code for approximately 30 types of proteins, and half of those genes are expressed specifically in the testis. Both the X and Y chromosomes might play roles in germ cell differentiation, and some of the genes that function in somatic cell differentiation might work together with hormones to regulate the development of the different sexes. 42 The sex‐determining region Y (SRY) gene on the short‐terminal region of the Y chromosome is also important in sex determination. 43
Deletion of the Y chromosome causes male infertility, as do microdeletions in the azoospermia factor (AZF) locus of the Y chromosome. Deletion of AZF has been reported in 5–15% of azoospermia and oligospermia cases. The long arm of the Y chromosome contains at least three distinct deletion‐sensitive regions, termed AZFa, AZFb, and AZFc. 44 AZFa has a length of 800 bp and contains the USP9Y and DDX3Y genes, which are involved in spermatogenesis. RBMY is a multicopy gene found in AZFb and deletion of this region produces azoospermia, suggesting that RBMY might be important in spermiogenesis. Human AZFc encodes seven genes and consists of inverted repeats, direct repeats and palindromic DNA sequences. Because they are exclusively expressed in the testis, deletion of these genes might cause male infertility. Indeed, deletions in the four DAZ genes in the AZFc region have been reported in about 10% of human azoospermia cases. It is difficult to understand the molecular mechanisms of the genes on the Y chromosome, because the DNA includes many repetitive sequences and species‐specific genes. Our understanding of male infertility and the evolution of genes will likely be advanced by analysis of the Y chromosome.
POLYMORPHISMS AND MUTATION OF SPERMATOGENESIS‐SPECIFIC GENES
MALE GERM CELL‐SPECIFIC genes function only in male germ cell differentiation and are not expressed in females. Various forms of partial Y chromosome deletion account for approximately 10–15% of male infertility cases. 45 The deletions that cause infertility are, by definition, not heritable. 46 The number of possible combinations of single nucleotide polymorphisms (SNP) is enormous, and mutations or combinations of SNP in genes expressed ubiquitously in somatic cells might introduce serious illness through both germ cells and somatic cells, and some rare combinations might even lead to male infertility (Table 1). Mutations and SNP in haploid germ cell‐specific genes on the autosome will, in theory, be transferred to the next generation by the female, and this mechanism might cause male infertility at a high frequency. Mutations and SNP in the PRM gene, which is not expressed in females, have been reported to cause haplo‐insufficiency in males (Table 1). Analyses of mutations and SNP germ cell‐specific genes within the autosome are therefore important in understanding the causes of male infertility. 16
Table 1.
Single nucleotide polymorphisms and mutations associated with infertility of men
THE NECESSITY OF INDEPENDENT SOMATIC AND GERM CELL DIFFERENTIATION
EACH MAMMAL REPRESENTS a collection of specially differentiated cells, such as germ cells. Germ cell differentiation is directed by a combination of germ cell‐specific gene products and gene products common to both germ cells and somatic cells. The primary differences between germ cells and somatic cells are the occurrence of meiosis with subsequent formation of male and female gametes, imprinting, and telomeric maintenance that assures cellular immortality. A PGC undergoes a differentiation process that is distinct from that of somatic cells at an extremely early stage of embryo formation. 3 PGC are distinct from the somatic cells that enter the somatic tissue that constitutes the gonadal primordium. Why don't mammalian gonadal cells simply differentiate into germ cells in many multi‐cellular organisms? This seemingly circuitous phenomenon has likely been preserved precisely because it is important.
Considering this, if a germ cell is derived from a certain type of differentiated somatic cell, then the germ cell must first differentiate into that somatic cell before dedifferentiating into a germ cell (Fig. 2, left). Alternatively, the germ cell differentiation process may be independent of somatic differentiation (Fig. 2, right). One criterion for defining the successful evolution of an organism is whether each step in its cellular differentiation allows the process to be efficiently concluded. This suggests that if germ cells differentiate after following a separate course of differentiation, cycling of germ cell and somatic differentiation could cause them to interfere with each other. As a result, it would be more difficult for evolution to act on cellular differentiation processes involving cells that share a differentiation process (somatic) than on cells that do not share a differentiation process (germ). The roles of the gonadal primordium and germ cells are very different. Considering the entire developmental process, we therefore see that evolution is facilitated by avoiding the coexistence of the two differentiation processes. An independent differentiation system allows evolution to act equivalently on various cellular differentiation processes in an individual, and contributes to a specific differentiation system for certain somatic cells (i.e. sharing the differentiation process with germ cells is avoided).
Figure 2.
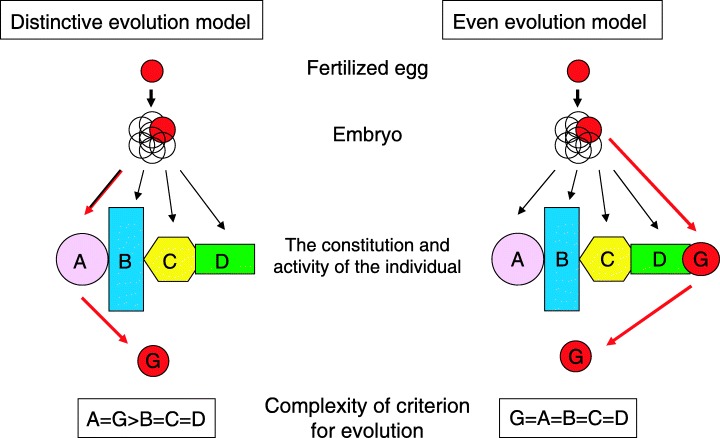
Schematic representation of the differentiation lineage of germ cells in the constitution of the individual. The left column shows the ‘Distinctive evolutionary model’ in which the gamete that carries genes to the next generation differentiates into organ A in the embryo, and next differentiates into gametes. The right column shows the ‘Equal evolution model’ in which the gamete differentiates from the embryo independent of other cells. In the left model, the differentiation of organ A is indispensable for the gamete. The criteria for the evolution of cell differentiation (A and G) in the left model are more complex than in the right model because it is necessary to serve purposes in two forms of cell differentiation (A and G). In the right model, evolution is almost equal between germ cell differentiation and other cell differentiation (the lowest boxes). The upper red circle represents a multipotent fertilized egg. The fertilized egg undergoes embryonic growth and forms an individual that consists of various differentiated organs and specific differentiated cells (A–D). The cell with the ability to differentiate into a gamete is shown in G. Red arrows indicate the lineage of germ cell differentiation.
At the gene level, germ cell differentiation is based on specific gene groups that are independent of those that support certain types of somatic cell differentiation, and this eliminates interference with the development of the individual, and allows germ cell‐specific genes to easily undergo changes that make germ cell differentiation distinct. Recent reports have described a group of isozymes distinct from somatic cell enzymes that function in the glycolytic pathway during spermiogenesis. 1 , 47 All cells in a mature organism undergo glycolysis, particularly the cells of muscles and similar somatic tissues. 48 The presence of a separate glycolytic isozyme group in germ cells therefore frees germ cells from the need to coordinate their genetic evolution with that of somatic cells. The independence of genes involved in germ cell differentiation is important for facilitating evolution, and explains why many germ cell isozymes are distinct from those of somatic cells. Many germ cell‐specific isozyme genes 49 do not have introns, 1 suggesting that they might have been duplicated in the course of evolution from ubiquitously used genes through retrotransposition, thereby establishing the genetic independence of germ cells from somatic cells. In some cases, when necessary, these isozymes are consolidated.
PERSPECTIVE
ALTHOUGH FERTILITY TREATMENT technology has improved, artificial materials in the environment are increasing, and numbers of ejaculated sperm are decreasing. A relationship between decreasing sperm numbers and an increase in infertile couples is feared, although the mechanism of action is uncertain. Therefore, a more thorough understanding of the specific mechanisms and processes of germ cell differentiation is still needed.
REFERENCES
- 1. Tanaka H, Baba T. Gene expression in spermiogenesis. Cell Mol Life Sci 2005; 62: 344–354. [DOI] [PubMed] [Google Scholar]
- 2. Skakkebaek NE, Jorgensen N, Main KM et al. Is human fecundity declining? Int J Androl 2006; 29: 2–11. [DOI] [PubMed] [Google Scholar]
- 3. McLaren A. Primordial germ cells in the mouse. Dev Biol 2003; 262: 1–15. [DOI] [PubMed] [Google Scholar]
- 4. McLaren A, Southee D. Entry of mouse embryonic germ cells into meiosis. Dev Biol 1997; 187: 107–113. [DOI] [PubMed] [Google Scholar]
- 5. Adams IR, McLaren A. Sexually dimorphic development of mouse primordial germ cells: switching from oogenesis to spermatogenesis. Development 2002; 129: 1155–1164. [DOI] [PubMed] [Google Scholar]
- 6. Amleh A, Taketo T. Live‐borns from XX but not XY oocytes in the chimeric mouse ovary composed of B6.Y (TIR) and XX cells. Biol Reprod 1998; 58: 574–582. [DOI] [PubMed] [Google Scholar]
- 7. Isotani A, Nakanishi T, Kobayashi S et al. Genomic imprinting of XX spermatogonia and XX oocytes recovered from XX↔XY chimeric testes. Proc Natl Acad Sci USA 2005; 102: 4039–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tanaka SS, Toyooka Y, Akasu R et al. The mouse homolog of Drosophila vasa is required for the development of male germ cells. Genes Dev 2000; 4: 841–853. [PMC free article] [PubMed] [Google Scholar]
- 9. Kuramochi‐Miyagawa S, Kimura T, Ijiri TW et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 2004; 131: 839–849. [DOI] [PubMed] [Google Scholar]
- 10. Guan K, Nayernia K, Maier LS et al. Pluripotency of spermatogonial stem cells from adult mouse testis. Nature 2006; 440: 1199–1203. [DOI] [PubMed] [Google Scholar]
- 11. Heller CG, Clermont Y. Spermatogenesis in man: an estimate of its duration. Science 1963; 140: 184–186. [DOI] [PubMed] [Google Scholar]
- 12. Penttila TL, Yuan L, Mali P et al. Haploid gene expression: temporal onset and storage patterns of 13 novel transcripts during rat and mouse spermiogenesis. Biol Reprod 1995; 53: 499–510. [DOI] [PubMed] [Google Scholar]
- 13. De Rooij DG. Stem cells in the testis. Int J Exp Pathol 1998; 79: 67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol 2002; 4: S41–S49. [DOI] [PubMed] [Google Scholar]
- 15. Fujii T, Tamura K, Masai K et al. Use of stepwise subtraction to comprehensively isolate mouse genes whose transcription is up‐regulated during spermiogenesis. EMBO Rep 2002; 3: 367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nishimune Y, Tanaka H. Infertility caused by polymorph‐isms or mutations in spermatogenesis‐specific genes. J Androl 2006; 27: 326–334. [DOI] [PubMed] [Google Scholar]
- 17. Nantel F, Monaco L, Foulkes NS et al. Spermiogenesis deficiency and germ‐cell apoptosis in CREM‐mutant mice. Nature 1996; 380: 159–162. [DOI] [PubMed] [Google Scholar]
- 18. Fimia GM, De Cesare D, Sassone‐Corsi P. CBP‐independent activation of CREM and CREB by the LIM‐only protein ACT. Nature 1999; 398: 165–169. [DOI] [PubMed] [Google Scholar]
- 19. Nagamori I, Yomogida K, Adams PD et al. Transcription factors, cAMP‐responsive element modulator (CREM) and Tisp40, act in concert in postmeiotic transcriptional regulation. J Biol Chem 2006; 281: 15073–15081. [DOI] [PubMed] [Google Scholar]
- 20. Macho B, Brancorsini S, Fimia GM et al. CREM‐dependent transcription in male germ cells controlled by a kinesin. Science 2002; 298: 2388–2390. [DOI] [PubMed] [Google Scholar]
- 21. Schmidt EE, Schibler U. Developmental testis‐specific regulation of mRNA levels and mRNA translational efficiencies for TATA‐binding protein mRNA isoforms. Dev Biol 1997; 184: 138–149. [DOI] [PubMed] [Google Scholar]
- 22. Walker WH, Delfino FJ, Habener JF. RNA processing and the control of spermatogenesis. Front Horm Res 1999; 25: 34–58. [DOI] [PubMed] [Google Scholar]
- 23. Blocher S, Behr R, Weinbauer GF, Bergmann M, Steger K. Different CREM‐isoform gene expression between equine and human normal and impaired spermatogenesis. Therio-genology 2003; 60: 1357–1369. [DOI] [PubMed] [Google Scholar]
- 24. Walker WH, Daniel PB, Habener JF. Inducible cAMP early repressor ICER down‐regulation of CREB gene expression in Sertoli cells. Mol Cell Endocrinol 1998; 143: 167–178. [DOI] [PubMed] [Google Scholar]
- 25. Morales CR, Leyne M, El‐Alfy M, Oko R. Molecular cloning and developmental expression of a small ribonuclear protein in the mouse testis. Mol Reprod Dev 1997; 46: 459–470. [DOI] [PubMed] [Google Scholar]
- 26. Elliott DJ, Bourgeois CF, Klink A et al. A mammalian germ cell‐specific RNA‐binding protein interacts with ubiquitously expressed proteins involved in splice site selection. Proc Natl Acad Sci USA 2000; 97: 5717–5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ostermeier GC, Dix DJ, Miller D et al. Spermatozoal RNA profiles of normal fertile men. Lancet 2002; 360: 772–777. [DOI] [PubMed] [Google Scholar]
- 28. Rassoulzadegan M, Grandjean V, Gounon P et al. RNA‐mediated non‐Mendelian inheritance of an epigenetic change in the mouse. Nature 2006; 441: 469–474. [DOI] [PubMed] [Google Scholar]
- 29. Morales CR, Wu XQ, Hecht NB. The DNA/RNA‐binding protein, TB‐RBP, moves from the nucleus to the cytoplasm and through intercellular bridges in male germ cells. Dev Biol 1998; 201: 113–123. [DOI] [PubMed] [Google Scholar]
- 30. Iguchi N, Tobias JW, Hecht NB. Expression profiling reveals meiotic male germ cell mRNAs that are translationally up‐ and down‐regulated. Proc Natl Acad Sci USA 2006; 103: 7712–7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kashiwabara S, Noguchi J, Zhuang T et al. Regulation of spermatogenesis by testis‐specific, cytoplasmic poly (A) polymerase TPAP. Science 2002; 298: 1999–2002. [DOI] [PubMed] [Google Scholar]
- 32. Han JR, Yiu GK, Hecht NB. Testis/brain RNA‐binding protein attaches translationally repressed and transported mRNAs to microtubules. Proc Natl Acad Sci USA 1995; 92: 9550–9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mali P, Kaipia A, Kangasniemi M et al. Stage‐specific expression of nucleoprotein mRNAs during rat and mouse spermiogenesis. Reprod Fertil Dev 1989; 1: 369–382. [DOI] [PubMed] [Google Scholar]
- 34. Chennathukuzhi V, Stein JM, Abel T et al. Mice deficient for testis‐brain RNA‐binding protein exhibit a coordinate loss of TRAX, reduced fertility, altered gene expression in the brain, and behavioral changes. Mol Cell Biol 2003; 23: 6419–6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chennathukuzhi V, Morales CR, El‐Alfy M, Hecht NB. The kinesin KIF17b and RNA‐binding protein TB‐RBP transport specific cAMP‐responsive element modulator‐regulated mRNAs in male germ cells. Proc Natl Acad Sci USA 2003; 100: 15566–15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morales CR, Lefrancois S, Chennathukuzhi V et al. A TB‐RBP and Ter ATPase complex accompanies specific mRNAs from nuclei through the nuclear pores and into intercellular bridges in mouse male germ cells. Dev Biol 2002; 246: 480–494. [DOI] [PubMed] [Google Scholar]
- 37. Lee K, Fajardo MA, Braun RE. A testis cytoplasmic RNA‐binding protein that has the properties of a translational repressor. Mol Cell Biol 1996; 16: 3023–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Davies HG, Giorgini F, Fajardo MA, Braun RE. A sequence‐specific RNA binding complex expressed in murine germ cells contains MSY2 and MSY4. Dev Biol 2000; 221: 87–100. [DOI] [PubMed] [Google Scholar]
- 39. Schumacher JM, Lee K, Edelhoff S, Braun RE. Spnr, a murine RNA‐binding protein that is localized to cytoplasmic microtubules. J Cell Biol 1995; 129: 1023–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kleene KC, Mulligan E, Steiger D et al. The mouse gene encoding the testis‐specific isoform of Poly (A) binding protein (Pabp2) is an expressed retroposon: intimations that gene expression in spermatogenic cells facilitates the creation of new genes. J Mol Evol 1998; 47: 275–281. [DOI] [PubMed] [Google Scholar]
- 41. Gu W, Kwon Y, Oko R et al. Poly (A) binding protein is bound to both stored and polysomal mRNAs in the mammalian testis. Mol Reprod Dev 1995; 40: 273–285. [DOI] [PubMed] [Google Scholar]
- 42. Skaletsky H, Kuroda‐Kawaguchi T et al. The male‐specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 2003; 423: 825–837. [DOI] [PubMed] [Google Scholar]
- 43. Koopman P, Munsterberg A, Capel B et al. Expression of a candidate sex‐determining gene during mouse testis differentiation. Nature 1990; 348: 450–452. [DOI] [PubMed] [Google Scholar]
- 44. Vogt PH. AZF deletions and Y chromosomal haplogroups: history and update based on sequence. Hum Reprod Update 2005; 11: 319–336. [DOI] [PubMed] [Google Scholar]
- 45. Hopps CV, Mielnik A, Goldstein M et al. Detection of sperm in men with Y chromosome microdeletions of the AZFa, AZFb and AZFc regions. Hum Reprod 2003; 18: 1660–1665. [DOI] [PubMed] [Google Scholar]
- 46. Silber SJ, Repping S. Transmission of male infertility to future generations: lessons from the Y chromosome. Hum Reprod Update 2002; 8: 217–229. [DOI] [PubMed] [Google Scholar]
- 47. Miki K, Qu W, Goulding EH et al. Glyceraldehyde‐3‐phosphate dehydrogenase‐S, a sperm‐specific glycolytic enzyme, is required for sperm motility and male fertility. Proc Natl Acad Sci USA 2004; 101: 16501–16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Piechaczyk M, Blanchard JM, Marty L et al. Post‐transcriptional regulation of glyceraldehyde‐3‐phosphate‐dehydrogenase gene expression in rat tissues. Nucl Acids Res 1984; 12: 6951–6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hecht NB. Molecular mechanisms of male germ cell differentiation. Bioessays 1998; 20: 555–561. [DOI] [PubMed] [Google Scholar]
- 50. Tanaka H, Miyagawa Y, Tsujimura A et al. Single nucleotide polymorphisms in the protamine‐1 and – 2 genes of fertile and infertile human male populations. Mol Hum Reprod 2003; 9: 69–73. [DOI] [PubMed] [Google Scholar]
- 51. Becherini L, Guarducci E. Degl’Innocenti S et al. DAZL polymorphisms and susceptibility to spermatogenic failure: an example of remarkable ethnic differences. Int J Androl 2004; 27: 375–381. [DOI] [PubMed] [Google Scholar]
- 52. Miyagawa Y, Nishimura H, Tsujimura A et al. Single‐nucleotide polymorphisms and mutation analyses of the TNP1 and TNP2 genes of fertile and infertile human male populations. J Androl 2005; 26: 779–786. [DOI] [PubMed] [Google Scholar]
- 53. Singh K, Singh SK, Sah R et al. Mutation C677T in the methylenetetrahydrofolate reductase gene is associated with male infertility in an Indian population. Int J Androl 2005; 28: 115–119. [DOI] [PubMed] [Google Scholar]
- 54. Longley MJ, Graziewicz MA, Bienstock RJ, Copeland WC. Consequences of mutations in human DNA polymerase gamma. Gene 2005; 354: 125–131. [DOI] [PubMed] [Google Scholar]
- 55. Zhoucun A, Zhang S, Yang Y et al. The common variant N372H in BRCA2 gene may be associated with idiopathic male infertility with azoospermia or severe oligozoospermia. Eur J Obstet Gynecol Reprod Biol 2006; 124: 61–64. [DOI] [PubMed] [Google Scholar]
- 56. Iguchi N, Yang S, Lamb DJ, Hecht NB. An SNP in protamine 1: a possible genetic cause of male infertility? J Med Genet 2006; 43: 382–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. A Z, Zhang S, Yang Y et al. Single nucleotide polymorphisms of the gonadotrophin‐regulated testicular helicase (GRTH) gene may be associated with the human spermatogenesis impairment. Hum Reprod 2006; 21: 755–759. [DOI] [PubMed] [Google Scholar]
- 58. Christensen GL, Wooding SP, Ivanov IP et al. Sequencing and haplotype analysis of the Activator of CREM in the Testis (ACT) gene in populations of fertile and infertile males. Mol Hum Reprod 2006; 12: 257–262. [DOI] [PubMed] [Google Scholar]


