Abstract
NMDA receptors are critical for neuronal communication. Dysfunction in NMDA receptors has been implicated in neuropsychiatric diseases. While it is well recognized that the composition of NMDA receptors undergoes a GluN2B-to-GluN2A switch in early postnatal life, the mechanism regulating this switch remains unclear. Using transcriptomic and functional analyses in brain tissues from male and female Hipk2+/+ and Hipk2−/− mice, we showed that the HIPK2-JNK–c-Jun pathway is important in suppressing the transcription of Grin2a and Grin2c, which encodes the GluN2A and GluN2C subunits of the NMDA receptors, respectively. Loss of HIPK2 leads to a significant decrease in JNK–c-Jun signaling, which in turn derepresses the transcription of Grin2a and Grin2c mRNA and upregulates GluN2A and GluN2C protein levels. These changes result in a significant increase of GluN2A/GluN2B ratio in synapse and mitochondria, a persistent activation of the ERK-CREB pathway and the upregulation of synaptic activity-regulated genes, which collectively contribute to the resistance of Hipk2−/− neurons to cell death induced by mitochondrial toxins.
SIGNIFICANCE STATEMENT We identify HIPK2-JNK–c-Jun signaling as a key mechanism that regulates the transcription of NMDA receptor subunits GluN2A and GluN2C in vivo. Our results provide insights into a previously unrecognized molecular mechanism that control the switch of NMDA receptor subunits in early postnatal brain development. Furthermore, we provide evidence that changes in the ratio of NMDA subunits GluN2A/GluN2B can also be detected in the synapse and mitochondria, which contributes to a persistent activation of the prosurvival ERK-CREB pathway and its downstream target genes. Collectively, these changes protect HIPK2 deficient neurons from mitochondrial toxins.
Keywords: HIPK2, JNK, NMDA receptor, subunit, survival, transcription
Introduction
Homeodomain interacting protein kinase 2 (HIPK2) regulates a wide range of biological processes, including tumorigenesis, vasculogenesis, tissue fibrosis, epithelial-mesenchymal transition, and neural development (Hofmann et al., 2013; Fan et al., 2014; Blaquiere and Verheyen, 2017). There are several salient features in HIPK2 that enable it to regulate such diverse biological functions. First, HIPK2 has a versatile protein–protein interacting domain, which provides a flexible interphase to interact with a number of transcription factors and modulate gene expression in a context-dependent manner. Second, the intrinsic kinase activity in HIPK2 provides an additional mechanism that regulates the activity of HIPK2 and its downstream targets. Finally, the broad expression pattern of HIPK2 during embryogenesis and in adult animals also allows it to regulate cell growth and differentiation either as a transcriptional coactivator or corepressor depending on the signal transduction pathway upstream of HIPK2 and the downstream transcription factors with which it interacts (Blaquiere and Verheyen, 2017).
In the mouse sensory neurons, HIPK2 interacts with POU homeodomain transcription factor Brn3a and suppresses the expression of prosurvival genes trkA (Ntrk1) and bcl-xl (bcl2l1) (Wiggins et al., 2004). HIPK2 can also interact with Smad transcription factors to promote the prosurvival signals downstream of TGF-β and BMP in ventral midbrain dopaminergic (DA) neurons and enteric neurons (Zhang et al., 2007; Chalazonitis et al., 2011). Given these cell type-specific functions, loss of HIPK2 results in more sensory neurons and a significant reduction in the number of DA neurons and enteric neurons during embryonic development and in early postnatal life. In addition to the important role of HIPK2 under physiological conditions, several studies show that HIPK2 can be activated by stress conditions to promote cell death via the ataxia telangiectasia mutated or c-Jun N-terminal kinase (JNK) pathway in tumor cells and neurons (Hofmann et al., 2003; Choi et al., 2013; Lee et al., 2016). In particular, endoplasmic reticulum (ER) stress, induced either pharmacologically by tunicamycin or by the accumulation of misfolded SOD1G93A proteins, activates a series of protein kinases downstream of inositol-requiring enzyme 1α, including apoptosis signal-regulating kinase 1, HIPK2, and JNK, to promote cell death in spinal motor neurons (Lee et al., 2016). Inhibition of HIPK2 kinase activity or loss of HIPK2 protects neurons from ER stress-induced cell death and delays disease onset and prolongs survival in SOD1G93A mice (Lee et al., 2016). Together, these results highlight the role of HIPK2 in diverse pathophysiological conditions and the need to investigate the mechanisms of HIPK2 in a context-dependent fashion.
Given the role of HIPK2 in transducing the prosurvival signal of TGF-β in embryonic DA neurons (Zhang et al., 2007; Luo and Huang, 2016), we ask how loss of HIPK2 might impact on the maintenance of ventral midbrain DA neurons in the postnatal brain. By comparing the transcriptomes of ventral midbrain in adult Hipk2+/+ and Hipk2−/− mice, we uncover a previously unappreciated role of HIPK2 in regulating the subunit composition of NDMA receptors in the substantia nigra as well as several different regions in the postnatal brain. Bioinformatics data show that the majority of HIPK2 target genes contain an overrepresented number of binding motifs for AP-1 transcription factors in the promoter regions. Consistent with this idea, HIPK2 suppresses c-Jun-mediated transcription of mouse Grin2a and Grin2c genes, which encode the GluN2A and GluN2C subunits of the NMDA receptors (Collingridge et al., 2009). Loss of HIPK2 leads to an upregulation of GluN2A and GluN2C and increases the GluN2A/GluN2B ratio and the ERK-CREB signaling pathway in early postnatal brain. These changes promote the upregulation of several activity-regulated inhibitors of death (AID) genes and enhance the survival of Hipk2−/− DA neurons in mitochondrial toxin-induced cell death. Together, these results support a novel role of HIPK2 in the transcriptional regulation of NMDA receptor subunits via the JNK–c-Jun signaling pathway.
Materials and Methods
Animals.
Hipk2−/− mice (Hipk2tm1Ejh/Hipk2tm1Ejh, RRID:MGI:5008273 and RRID:MGI:3510466) have been described previously (Wiggins et al., 2004; Zhang et al., 2007). Hipk2+/+ and Hipk2−/− mice in the mixed C67BL/6 and 129 background were used at postnatal day 0 (P0), P14, P28, 2 months, and 6 months. TH-IRES-Cre mice (Thtm1(cre)Te, MGI catalog #3056580) were previously described (Lindeberg et al., 2004; Tang et al., 2009). R26RHIPK2 mice were generated as described in Fig. 8-1. Mice of both genders were selected and assigned to each age or treatment group randomly. Animal care was approved by the Institutional of Animal Care and Use Committee at the University of California–San Francisco and followed the National Institutes of Health guidelines.
Cell cultures, transfection, and luciferase reporter assays.
HEK293 and COS-7 cells were obtained from ATCC. Hipk2+/+ and Hipk2−/− mouse embryonic fibroblasts (MEFs) (Wei et al., 2007; Shang et al., 2013) were maintained in DMEM (Thermo Scientific) supplemented with 10% FBS (Invitrogen). All cell lines were authenticated and verified to be free of mycoplasma contamination. To investigate the transcriptional control of Grin2a and Grin2c, 2 kb sequences of the mouse Grin2a or 2.9 kb sequences of the mouse Grin2c promoter were amplified from mouse genomic DNA using the PCR and inserted into pGL4.10 vector to generate the Grin2a-Luc and Grin2c-Luc reporters. A series of deletions in Grin2a-Luc and Grin2c-Luc were performed using QuikChange Site-Directed Mutagenesis kit (Stratagene), and all constructs were confirmed by DNA sequencing. The luciferase constructs, c-Jun or/and HIPK2 expression plasmids, along with the Renilla firefly internal control constructs were transfected into COS-7 cells. Cells were harvested for luciferase activity measurement (dual luciferase, Promega) or for Western blot analyses. The luciferase reporter activity was measured using the dual-luciferase system on a luminometer (Turner Designs). Relative luciferase activity was reported as a ratio of firefly over Renilla luciferase activities.
RNA isolation, microarray, and data analysis.
Total RNA was extracted from the substantia nigra of 2-month-old Hipk2+/+ and Hipk2−/− mouse brain (n = 3 for each genotype) using PicoPureTM RNA Isolation Kit (Arcturus) and used as a template for reverse transcriptase with MessageAmpTM II-Biotin enhanced Kit (Ambion). Microarray analysis was performed using CodeLink Mouse Whole Genome Bioarray (Applied Microarrays). The microarray data are being deposited in Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/); the accession number will be provided once available. Resulting data were log transformed and uploaded into the Genesifter program (a web-based expression analysis software with statistical and visual tools, www.genesifter.com). Differences in gene expression were identified by using a minimal threshold value of a twofold change with no maximal threshold value. Based on the Genesifter analyses, the genes that show differential expression by microarray were uploaded on to DAVID bioinformatics resources. The functional annotation chart and clustering analysis modules were used to determine gene-term enrichment scores. To find the different transcriptional factor binding motif of HIPK2 targets, we analyzed 2 kb promoter sequence of HIPK2 target genes that were within the three clusters: membrane, channel, and transporter and cell–cell junction with online software Cluster-buster (http://zlab.bu.edu/cluster-buster/). The RNA from HEK293T cells, MEF cells, mouse cortex, or midbrain was isolated by Trizol reagent (Invitrogen) and used as a template for reverse transcriptase with random hexamer primers (Invitrogen). Primer sequences for specific genes are given in Table 1-1.
List of primer sequences (5’ to 3’). Download Table 1-1, DOCX file (100.8KB, docx)
Chromatin immunoprecipitation (ChIP) assays.
ChIP assays were performed as described previously (Shang et al., 2013). Briefly, 2-month-old mice were perfused and fixed with 4% PFA and treated with SDS lysis buffer. After shearing with a sonicator and centrifugation, the supernatant of cell lysates was used for immunoprecipitation with different antibodies. The DNA protein–antibody complexes were isolated using antibodies for HIPK2 (c-15, sc-110294), c-Jun (H-179, SC-1694) (Santa Cruz Biotechnology), and Smad2/3 (#3102, Cell Signaling Technology). The complexes were washed with buffers, and the DNA was eluted and purified. qRT-PCR was used to analyze the DNA abundance in protein-DNA precipitates. qRT-PCR primer sequences are given in Table 1-1.
Primary cortical and dopaminergic (DA) neuron cultures.
Primary cortical neurons were prepared from the cerebral cortex of E17.5 mouse embryos and placed in MEM supplemented with 10% FBS, 1× penicillin/streptomycin, and 2 mm glutamine (Invitrogen). On in vitro day 2 (DIV2), 5 mm 5-fluoro-2′-deoxyuridine (Sigma) was added to the cultures. From DIV3, the primary cortical neurons were maintained in Neurobasal medium with B-27 supplement (Invitrogen). On DIV14, the cells were fixed with cold methanol for 15 min, followed by immunofluorescent staining with GluN2A or GluN2B antibody (NR2a, Code #GluRe1C-Rb-Af542; NR2b, Code #GluRe2N-Rb-Af660, Frontier Institute). Primary DA neurons were prepared according to published procedures (Zhang et al., 2007). Briefly, E13.5 Hipk2+/+ and Hipk2−/− mouse embryos were collected from time-pregnant Hipk2+/− females. The ventral mesencephalon was dissected, dissociated after treatment with trypsin, and cultured in DMEM-F12 medium (Invitrogen #11765–054) supplemented with 10% FBS, 1× penicillin/streptomycin, and 2 mm glutamine (Invitrogen) on cover slides coated with poly-dl-ornithine hydrobromide (Sigma-Aldrich, catalog #P8638) and laminin (Sigma-Aldrich, catalog #L2020) overnight. On DIV2, the medium was replaced with DMEM-F12 medium supplemented with 10% FBS, 20 ng/ml FGF2, 100 ng/ml FGF8, and 1× penicillin/streptomycin for 2 d. The cells were either treated with 0.5 nm carbonyl cyanide m-chlorophenyl hydrazone (CCCP) directly or pretreated with GluN2A inhibitor NVP-AAM077 (Calbiochem, catalog #CAS459836-30-7) or ERK1/2 inhibitor SCH-772984 (AbMole Bioscience, catalog #M2084) for 4 h, then treated with CCCP for 20 h. The cells were fixed with 4% PFA for 10 min and stained with anti-TH antibody (Millipore Bioscience Research Reagents, catalog #AB152) or anti-Tuj1 antibody (Covance, catalog #PRB-435P) following the standard straining procedures. Confocal images of the cultured neurons were captured using the confocal microscope (TCS SP, Leica). Laser intensity (measured as the PMT levels) for each fluorophor was kept within the linear range. TH+ or NeuN+ Cell number was determined by National Institutes of Health ImageJ online software.
Immunogold electron microscopy.
Mice were deeply anesthetized with avertin (150 mg/kg) and were perfused transcardially with 2% PFA/0.2% glutaraldehyde in 0.1 m phosphate buffer at pH 7.4 with gravity. Brains were quickly removed, fixed overnight in 2% PFA at 4°C, and cut into 60-μm-thick frontal sections with vibratome. To enhance the penetration of the immunoreagents, the sections were equilibrated in a cryoprotectant solution, freeze-thawed, and stored in PBS with 0.03% sodium azide. After blocked with 4% normal goat serum in PBS, the sections were incubated with primary antibody in 1% normal goat serum blocking solution overnight (NR2a, Code #GluRe1C-Rb-Af542; NR2b, Code #GluRe2N-Rb-Af660, Frontier Institute). After washed with washing buffer (PBS with 0.2% BSAc and 0.2% fish gelatin), the samples were incubated with secondary antibody conjugated to gold particles (Electron Microscopy Sciences, catalog #25100, or GAR, Ultra Small, Code 100,011) in washing solution for 2 h, and refixed with 1% glutaraldehyde in PBS 10 min. The signal of the immunogold particles was boosted using R-gent SE-EM silver enhancement solution (Aurion, 500.033). Finally, the sections were postfixed in 0.5% osmium tetroxide for 10 min, dehydrated, and embedded in resin (Durcupan ACM, Sigma-Aldrich). Serial ultrathin sections were cut with a Reichert Ultracut S, contrasted with lead citrate. Ultrathin sections were cut at 1 μm thick with a Reichert Ultracut S, contrasted with lead citrate, and imaged in a Phillips Tecnai10 transmission electron microscope using FEI software.
Western blot analysis and synaptosomal fractionation.
Total cell lysates were prepared from cultured cells in NP-40 lysis buffer (1% NP-40, 20 mm Tris, pH 7.6, 150 mm NaCl, 10 mm NaF, 1 mm Na3VO4) supplemented with protease inhibitor mixture. RIPA buffer (0.1% SDS, 1% sodium deoxycholate, 1% NP-40, 20 mm Tris, pH 7.6, 150 mm NaCl, 10 mm NaF, 1 mm Na3VO4) supplemented with protease inhibitor mixture was used for protein extraction from tissues. Proteins in cell lysates were separated by SDS-PAGE and transferred to the PVDF membrane (Millipore Bioscience Research Reagents). The membrane was blocked in 4% BSA (for phosphor antibodies) or 5% nonfat milk for nonphosphorylated antibodies before incubation with primary antibodies overnight at 4°C. Antibodies to p-JNK (9255), p-c-Jun (9164), JNK (9252), c-Jun (9165), p-ERK1/2 (4370), ERK1/2 (9102), CREB (9197), p-CaMKII (3361), and CaMKII (3362) were from Cell Signaling Technology. p-CREB was from Millipore Bioscience Research Reagents (06-519). Anti-TH antibody was from Millipore Bioscience Research Reagents (AB152) and Tuj1 antibody from Covance (PRB-435P). Actin antibody (CP01) was from Calbiochem. HIPK2 antibody was purchased from Santa Cruz Biotechnology (sc-10294) or Abcam (ab28507). The membranes were washed with 0.1% TBST washing buffer followed by incubation with secondary antibodies conjugated with HRP. Western blots were developed by ECL chemiluminescence (Thermo Scientific).
To characterize the GluN2A, GluN2B, and GluN2C protein level in synapses, the synaptosomes were isolated from 2-month-old Hipk2+/+ and Hipk2−/− mouse brains as described previously (Carlin et al., 1980). Briefly, mouse brains without cerebellums were homogenized in buffer A (0.32 m sucrose, 1 mm NaHCO3, 1 mm MgCl2, 0.5 mm CaCl2, 5 mm NaF, and 2 mm Na3VO4, supplemented with protease inhibitor, at a ratio of 4 ml/g of brain tissue) using a Dounce tissue grinder (Kontes glass homogenizer No 20, 12 strokes). Cell debris were removed by centrifugation at 710 × g for 10 min at 4°C. The pellet was suspended with 3 strokes in solution A, and then the nuclear fraction was removed by centrifugation at 1400 × g for 10 min at 4°C. The crude membrane fraction in the supernatant was collected by centrifugation at 13,800 × g for 10 min at 4°C. The pellet (crude synaptosomal fraction) was suspended in solution B (0.32 m sucrose, 1 mm NaHCO3, 5 mm NaF, and 2 mm Na3VO4, 3.2 ml/g of starting tissue) with 6 strokes of homogenizer. The resulting supernatant was collected and separated on a discontinuous sucrose gradient (1.2 m, 1 m, and 0.8 m) by centrifugation at 82,500 × g with a Beckman SW55Ti rotor at 4°C for 2 h. Following centrifugation, cytosolic, synaptosomal, and mitochondrial fractions were collected from top to bottom. Equal aliquots from individual fractions were separated by SDS-PAGE and analyzed by immunoblotting with antibodies for PSD95, synaptophysin (SPH), GluN2A, GluN2B, GluN2C, and actin.
Chronic MPTP exposure in Hipk2+/+, Hipk2−/−, and TH-IRES-Cre;R26RHIPK2/HIPK2 mice.
MPTP (4 mg/kg, Sigma-Aldrich catalog #M103) or equivalent volume of PBS was injected intraperitoneally into 2-month-old male and female mice (littermates) by one injection per day for 10 consecutive days. The mice were monitored according to the approved Institutional of Animal Care and Use Committee protocol, and their health was scored before each injection. Seven days after the last MPTP treatment, mice were killed and perfused with 4% PFA before brain extraction. The brains were postfixed in 4% PFA overnight, followed by serial cryoprotection in 15% and 30% sucrose for 24 h each. The brains were embedded for cryosectioning and cut into 40 μm coronal sections. The sections containing the substantia nigra pars compacta were stained using anti-TH (Millipore Bioscience Research Reagents, catalog #AB152) and the staining results developed with DAB. Stereological counting was used to quantify the number of TH-positive cells in the substantia nigra pars compacta (single hemisphere) at bregma −2.8 to −4.04 mm (6 serial sections) at 60× magnification using the StereoInvestigator Software, version 9 (MBF Bioscience) according to the protocols previously reported (Zhang et al., 2007; Martens et al., 2012).
Experimental design and statistical analyses.
For both in vivo and in vitro studies, at least three biological replicates were used in each study. In experiments in which N was >3, the exact number of replicates will be indicated. Data were analyzed by two-tailed Student's t test for pairwise comparisons or two-way ANOVA for multiple comparisons using Prism (GraphPad Software). All data were expressed as mean ± SEM. For stereology counting, investigators were blinded to the genotypes.
Results
Loss of HIPK2 increases Grin2a and Grin2c genes expression
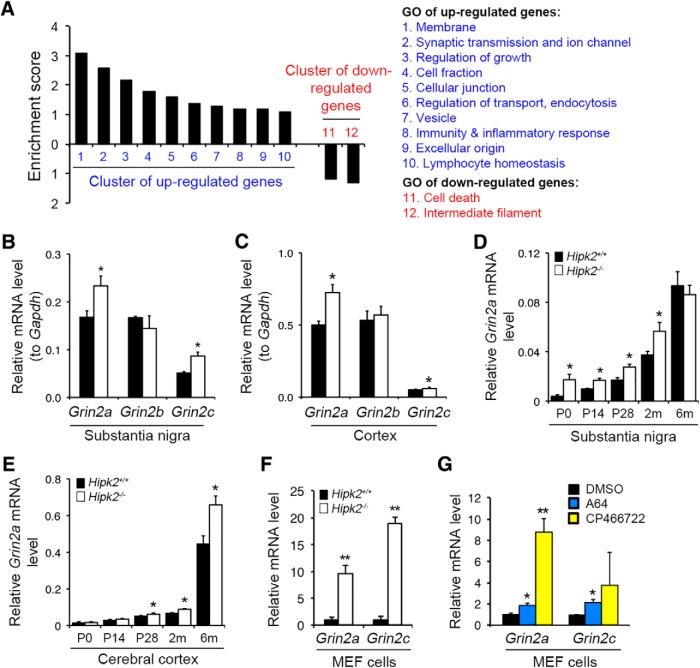
To characterize how loss of HIPK2 affects the long-term survival of DA neurons, we compared the transcriptomes of substantia nigra microdissected from 2-month-old Hipk2+/+ and Hipk2−/− mice, and found 279 upregulated and 256 downregulated genes in Hipk2−/− mice (Table 1-2. Functional annotations using DAVID Gene Ontology (GO) analyses showed that the upregulated genes belonged to 10 groups, including membrane functions, synaptic transmission and ion channel, regulation of growth, cell fraction, cellular junction, regulation of transport and endocytosis, vesicles, immunity and inflammatory response, extracellular origin, and lymphocyte homeostasis (Fig. 1A). In contrast, most of the GO clusters of the downregulated genes showed very low enrichment scores; therefore, only two functional categories were identified among these genes, including cell death and intermediate filament.
Figure 1.
Transcriptomic analyses of the substantia nigra in adult Hipk2−/− mouse brain. A, DAVID GO analyses showed the 12 groups of the upregulated and downregulated genes in the substantia nigra of 2-month-old Hipk2−/− mice. B, C, Grin2a and Grin2c mRNA levels were upregulated in the substantia nigra and cerebral cortex of 2-month-old Hipk2−/− mice, whereas the Grin2b mRNA expression was similar between Hipk2+/+ and Hipk2−/− mice. D, E, Grin2a mRNA was elevated in the substantia nigra of Hipk2−/− from postnatal day 0 (P0) until 2 months old. However, in the cerebral cortex, Grin2a mRNA started to increase from P20 to adulthood. F, G, Grin2a and Grin2c mRNA levels were also robustly increased in Hipk2−/− MEFs, and blocking of HIPK2 kinase activity by its specific inhibitors enhanced Grin2a and Grin2c expression in MEF cells. Data are mean ± SEM from at least three independent experiments. *p < 0.05 (ANOVA). **p < 0.01 (ANOVA). Not significant: p > 0.05.
DAVID gene ontology (GO) analyses of microarray data from the substantia nigra of Hipk2+/+ and Hipk2-/- mice. Download Table 1-2, DOCX file (41.8KB, docx)
Of the upregulated genes in Hipk2−/− mice, we focused on two NMDA receptor subunits, Grin2a and Grin2c, because they were the top hits in the upregulated GO categories and because both have well-established prosurvival functions (Hardingham and Bading, 2010; Paoletti et al., 2013). To validate the microarray data, we used qRT-PCR to show that both Grin2a and Grin2c were indeed upregulated in the substantia nigra and cerebral cortex of Hipk2−/− mice (Fig. 1B,C). Consistent with microarray data, there was no significant difference in Grin2b mRNA level. The upregulation of Grin2a in postnatal brain appeared to be stage-dependent. In the substantia nigra, Grin2a mRNA was elevated in Hipk2−/− mice as early as postnatal day 0 (P0) and persisted until 2 months of age. By 6 months of age, there was no difference in Grin2a mRNA levels in the substantia nigra of Hipk2+/+ and Hipk2−/− mice (Fig. 1D). In contrast, Grin2a mRNA levels in the cerebral cortex showed no difference between Hipk2+/+ and Hipk2−/− mice at P0 and P14 but were significantly higher in Hipk2−/− cortex from P28 to 6 months of age (Fig. 1E). Interestingly, Grin2a and Grin2c mRNA levels were also robustly increased in Hipk2−/− MEFs. Furthermore, treating Hipk2+/+ MEF with HIPK2 inhibitors, A64 and CP466722 (Miduturu et al., 2011; Lee et al., 2016), was sufficient to upregulate Grin2a and Grin2c mRNA levels (Fig. 1F,G). Together, these results revealed a broader role of HIPK2 in the transcription of NMDA receptor subunits in postnatal brain development and in non-neuronal cells.
HIPK2 suppresses Grin2a and Grin2c gene expression via AP-1 transcription factor c-Jun
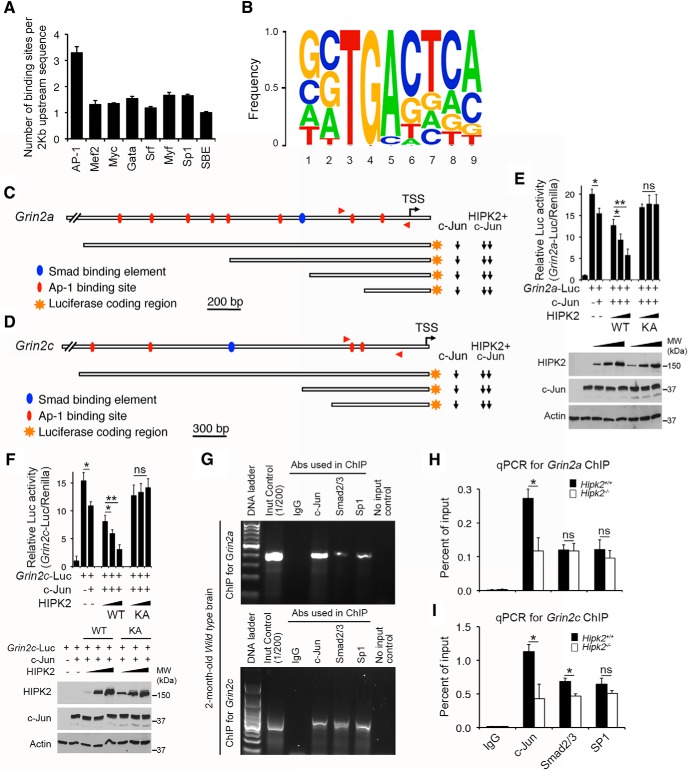
Given the previously reported role of HIPK2 as a transcriptional cosuppressor or coactivator in a variety of signaling pathways, we hypothesized that HIPK2-dependent expression of Grin2a and Grin2c is likely mediated through transcription factors that directly bind to the promoter and/or enhancer elements of Grin2a and Grin2c genes. To test this, we procured 2 kb genomic DNA sequence upstream of the transcriptional start sites in 130 HIPK2 target genes in the top three GO categories, including “cell–cell junction,” “membrane,” and “ion channel and transport” (Table 1-3, and analyzed them with the Cluster-Buster software (Frith et al., 2003). Our goal was to identify consensus transcription factor binding elements that were shared among the HIPK2 target genes. Of all the transcription factors binding elements annotated in Cluster Buster, the AP-1 binding motif was overrepresented in the 2 kb promoter/enhancer sequences of HIPK2 target genes, with 3–4 AP-1 binding motifs per gene (Fig. 2A). In contrast, the binding motifs for other transcription factors were significantly fewer, ranging from 1 or 2 per gene. Further annotations of all the AP-1 binding motif in HIPK2 target genes revealed a highly conserved TGA core sequence in positions 3–5, flanked by more flexible nucleotide sequences at both 5′ and 3′ ends (Fig. 2B).
Figure 2.
Characterizations of HIPK2 as a transcriptional repressor of c-Jun in the transcriptional control of mouse Grin2a and Grin2c genes. A, B, Compared with the DNA binding motifs for other transcription factors, the AP-1 binding motif was overrepresented and showed a highly conserved binding core sequence, in the 2 kb promoter/enhancer sequences of HIPK2 target genes. C, D, Luciferase activities of both Grin2a-Luc and Grin2c-Luc were suppressed by c-Jun, and the transcriptional suppressor activity of c-Jun in Grin2a-Luc and Grin2c-Luc reporters can be further enhanced by the addition of HIPK2 (Figure 2-1. E, F, Western blot showed the expression of the exogenous HIPK2 and c-Jun constructs, and the endogenous actin protein level. The results showed that HIPK2 kinase dead mutant, HIPK2K221A, was unable to promote the c-Jun-mediated suppression, indicating that the kinase is required for its cosuppressor function. G–I, ChIP assays showed that the endogenous c-Jun was bound to the AP-1 motif in the promoters of Grin2a and Grin2c in 2-month-old Hipk2+/+ mouse brain. In contrast, the same promoter regions in Grin2a and Grin2c showed much lower binding of transcription factors Smad2/3 and SP-1. Loss of HIPK2 decreased the binding between c-Jun and the conserved AP-1 binding motif in the promoter/enhancer sequences of Grin2a and Grin2c. Data are mean ± SEM from at least three independent experiments. *p < 0.05 (ANOVA). **p < 0.01 (ANOVA). Not significant: p > 0.05.
Genes analyzed by Cluster Buster to identify potential transcription factor binding elements in HIPK2 target genes. Download Table 1-3, DOCX file (165.5KB, docx)
HIPK2 suppresses, whereas MEF2C and HDAC7 do not affect, Grin2a-Luc and Grin2c-Luc reporter activities. Expression of HIPK2 or c-Jun alone in COS-7 cells was sufficient to suppress the basal activity of Grin2a-Luc and Grin2c-Luc reporters. Unlike c-Jun or HIPK2, transcription factor MEF2C and transcriptional cofactor HDAC7, which interact with HIPK2 and regulate vasculogenesis during embryonic development, had very modest or no detectable effects on the Grin2a-Luc or Grin2c-Luc reporter activity. Download Figure 2-1, DOCX file (25.5MB, docx)
The identification of AP-1 binding motifs in HIPK2 target genes is in agreement with our recent data that HIPK2 promotes ER stress-mediated cell death via the JNK–c-Jun signaling pathway (Lee et al., 2016). To characterize the role of AP-1 binding motifs in Grin2a and Grin2c, we generated luciferase reporters that contained the 2 kb upstream sequence of Grin2a or 2.9 kb upstream sequence of Grin2c gene. These two reporters, named Grin2a-Luc and Grin2c-Luc, respectively, were then used as surrogates to determine how HIPK2 and c-Jun regulate the transcription of Grin2a and Grin2c. Consistent with the microarray data, our results showed that c-Jun suppressed the luciferase activities of both Grin2a-Luc and Grin2c-Luc, whereas HIPK2 worked cooperatively with c-Jun to further suppress Grin2a-Luc and Grin2c-Luc (Fig. 2C,D). Interestingly, HIPK2 alone also suppressed Grin2a-Luc and Grin2c-Luc reporter activities, probably due to the high endogenous c-Jun levels in COS-7 cells (Fig. 2-1. In contrast, transcription factors and cofactors, such as MEF2C and HDAC7, which can interact with HIPK2 and regulate vasculogenesis during embryonic development (Shang et al., 2013), did not affect Grin2a-Luc and Grin2c-Luc reporter activities (Fig. 2-1.
A series of deletions in Grin2a-Luc and Grin2c-Luc showed that, of the 8 AP-1 binding sites in Grin2a promoter, the one closest to the transcriptional start site was required for c-Jun and HIPK2 to suppress Grin2a-Luc. Similarly, of the 4 AP-1 binding sites in Grin2c-Luc, the two closest to transcriptional start site were required for c-Jun and HIPK2 to suppress Grin2c-Luc. In addition, we showed that the kinase activity of HIPK2 was required for HIPK2 to promote c-Jun-mediated suppression because the kinase dead form of HIPK2 (HIPK2-K221A) was unable to suppress Grin2a-Luc or Grin2c-Luc activity, even in the presence of c-Jun (Fig. 2E,F). The cooperative effects of HIPK2 and c-Jun in suppressing Grin2a-Luc and Grin2c-Luc reporter activities were summarized using downward arrows to the right of the schematic diagrams in Figure 2C, D. To further characterize how HIPK2 and c-Jun regulate Grin2a and Grin2c genes expression, we performed ChIP assays using native chromatin extracts from 2-month-old Hipk2+/+ mouse brain, and detected binding of the endogenous c-Jun, Smad2/3, and SP1 proteins to the Grin2a and Grin2c promoters (Fig. 2G). In contrast, there was significantly less c-Jun binding to the Grin2a and Grin2c promoters in the chromatin from Hipk2−/− mouse brain (Fig. 2H,I). These results supported the role of HIPK2 as a cosuppressor of c-Jun in the transcriptional control of Grin2a and Grin2c gene expression.
Increased GluN2A and GluN2A/GluN2B ratio in the synaptosomes of Hipk2−/− mouse brain
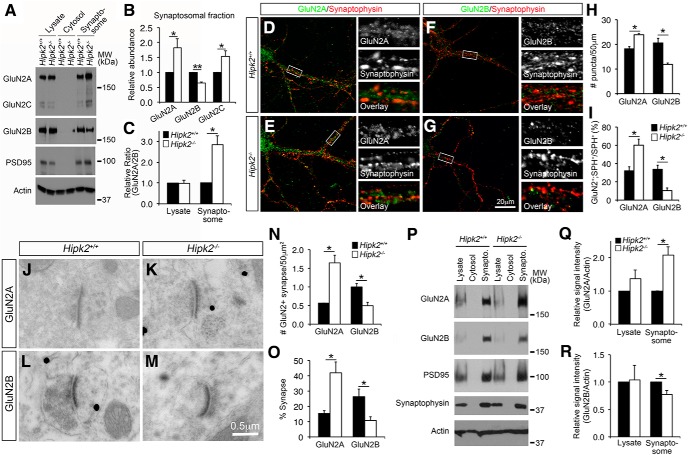
Having demonstrated the increased Grin2a and Grin2c mRNA in the substantia nigra and cerebral cortex of Hipk2−/− mice, we next asked whether there were also increases in GluN2A and GluN2C protein levels. Because NMDA receptors are enriched in synapses, we isolated synaptosomes from the cerebral cortex of 2-month-old Hipk2+/+ and Hipk2−/− mice (Carlin et al., 1980) (Fig. 3-1 and used Western blots to detect the relative abundance of GluN2A and GluN2C proteins (Fig. 3A). The results showed that GluN2A and GluN2C protein levels were indeed increased in the synaptosomes of cerebral cortex Hipk2−/− mice, whereas GluN2B protein level was decreased (Fig. 3A,B). These changes led to >2-fold increase in the ratio of GluN2A versus GluN2B in the synaptosomes of Hipk2−/− mice (Fig. 3C). To further characterize this phenotype, we prepared primary cortical neuron cultures from embryonic day 17.5 (E17.5) Hipk2+/+ and Hipk2−/− embryos. These neurons were cultured for 14 d (DIV14) and then stained with antibodies that were specific for GluN2A or GluN2B, and the presynaptic protein, SPH. Consistent with the Western blot data, confocal microscopy showed that the number of GluN2A+ puncta in Hipk2−/− dendrites were 31.2% (23.9 vs 18.2) more abundant than that in Hipk2+/+ dendrites (Fig. 3D,E,H), whereas the number of GluN2B+ puncta was decreased in Hipk2−/− dendrites (Fig. 3F–H). Interestingly, in Hipk2+/+ dendrites, the percentage of GluN2A+ and GluN2B+ puncta that were also positive for presynaptic marker SPH was 31.2% and 33.8%, respectively. In contrast, the percentage of GluN2A+;SPH+ synapse on the dendrites of Hipk2−/− neurons increased to 60.1%, whereas the percentage of GluN2B+;SPH+ synapse reduced to 10.3% (Fig. 3I). Consistent with these results, immunogold electron microscopy performed on the sensorimotor cortex using GluN2A or GluN2B antibody showed higher percentage of GluN2A+ synapse but reduced GluN2B+ synapse per unit area in Hipk2−/− brain (Fig. 3J–O). Finally, using similar approaches, we also found a significant increase in GluN2A proteins and a decrease in GluN2B proteins in the synaptosomes prepared from the substantia nigra of Hipk2−/− mouse brain (Fig. 3P–R). Together, these results support the idea that loss of HIPK2 has a broader effect on the levels of GluN2A and GluN2B in multiple areas in the mouse brain.
Figure 3.
Increased expression of GluN2A and GluN2C proteins in the synapses of Hipk2−/− mouse brain. A–C, Western blots using the total cell lysates, cytosolic fraction, and the synaptosomes from 2-month-old Hipk2+/+ and Hipk2−/− mouse brain showed that GluN2A and GluN2C protein levels were increased in the synaptosomes of cerebral cortex in Hipk2−/− mice, whereas GluN2B protein level was decreased, leading to a significant increase in the GluN2A/GluN2B ratio in the cerebral cortex in Hipk2−/− mice (Figure 3-1. The relative signal intensity of GluN2A, GluN2B, and GluN2C protein bands was quantified with National Institutes of Health ImageJ software. D–I, Confocal microscopic images of the primary cortical neuron cultures showed that the number of GluN2A+ puncta in the dendrites of Hipk2−/− neurons were 31.2% more, whereas the GluN2B+ puncta were 40% less, abundant than that in the dendrites of Hipk2+/+ neurons. Interestingly, the percentage of GluN2A+;SPH+ synapse increased to 60.1%, whereas the percentage of GluN2B+;SPH+ synapse reduced to 10.3% on the dendrites of Hipk2−/− neurons. The GluN2A+ or GluN2B+ puncta were quantified with National Institutes of Health ImageJ online software. J–O, Immunogold electron microscopy performed in the sensorimotor cortex of 2-month-old Hipk2+/+ and Hipk2−/− brains showed more GluN2A+ synaptic terminals, but fewer GluN2B+ synaptic terminals in Hipk2−/− mutants. Overall, there was a higher percentage of GluN2A+ synapse, but lower percentage of GluN2B+ synapse in the sensorimotor cortex in Hipk2−/− mutants. P–R, Western blots showed a significant increase in GluN2A proteins and a decrease in GluN2B proteins in the synaptosomes from the substantia nigra of 2-month-old Hipk2−/− mouse brain. The signal intensity of GluN2A, GluN2B, and actin was quantified with National Institutes of Health ImageJ software. Data are mean ± SEM from at least three independent experiments. *p < 0.05 (ANOVA). **p < 0.01 (ANOVA). Not significant: p > 0.05.
Schematic diagram for isolating different subcellular organelles from mouse brain. (a) Flow chart illustrating the basic steps for the preparation of cytosolic, synaptosomal, and mitochondrial extracts from 2-month-old mice brain. The procedure requires approximately 3 hour from tissue homogenization to collection of the synaptosomal fraction. (b-c) Western blot showing the purity of the synaptosomal and mitochondrial fractions. PSD95: post-synaptic marker, Synaptophysin: pre-synaptic marker, VEGFR: a membrane associated protein, Tom20: a translocase of the outer membrane in mitochondria, and Actin: ubiquitous expressed protein as loading control. Download Figure 3-1, DOCX file (25.5MB, docx)
Altered JNK and ERK-CREB signaling pathways in Hipk2−/− mouse brain
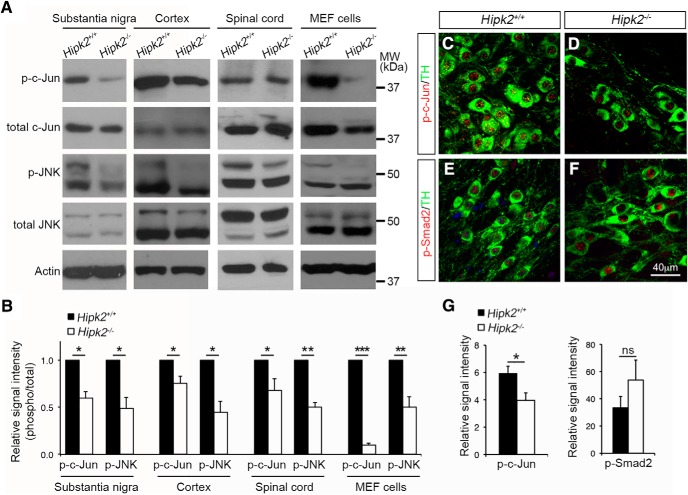
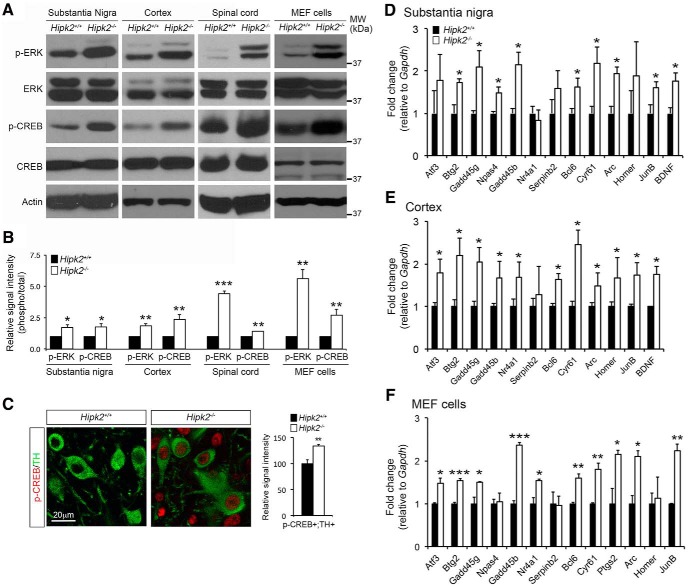
In the canonical JNK–c-Jun signaling pathway, diverse upstream signals activate JNK kinase activity, which in turn phosphorylates c-Jun to regulate the transcription of AP-1 target genes. Our recent study shows that HIPK2 can activate JNK under ER stress to promote neuronal cell death (Lee et al., 2016). In addition, results in Figure 2 indicate that HIPK2 facilitates c-Jun binding to Grin2a/2c promoter and functions as a cosuppressor to regulate Grin2a and Grin2c transcription in a kinase-dependent manner. Together, these results suggest that HIPK2-mediated JNK–c-Jun phosphorylation and activation may be required for transcriptional control of Grin2a and Grin2c. To test this, we prepared protein lysates from substantia nigra, sensorimotor cortex, spinal cord, and MEF cells, and analyzed the relative abundance of p-JNK and p-c-Jun in Hipk2+/+ and Hipk2−/− tissues using Western blot. As predicted, there was a consistent reduction in the levels of p-JNK and p-c-Jun in tissues from different regions of Hipk2−/− mouse brain, but no significant reduction in the level of total c-Jun or JNK (Fig. 4A,B). Interestingly, Hipk2−/− MEF cells also showed significant reductions in p-c-Jun and p-JNK, although the decrease in p-c-Jun was much more drastic compared with tissues from Hipk2−/− mouse brain (Fig. 4A,B). Consistent with the Western blot results, immunofluorescent microscopy showed a significant reduction of p-c-Jun staining intensity in the nuclei of Hipk2−/− dopaminergic neurons compared with that in Hipk2+/+ neurons (Fig. 4C,D,G). In contrast, no significant reduction of p-Smad2 was detected in the dopaminergic neurons of Hipk2−/− mice (Fig. 4E–G).
Figure 4.
Reduced activation of the JNK–c-Jun signaling pathway in Hipk2−/− mouse brain. A, B, Western blots using lysates from the cerebral cortex, substantia nigra, and spinal cord of 2-month-old Hipk2+/+ and Hipk2−/− mice showed a consistent reduction in the levels of p-JNK and p-c-Jun, but no changes in total JNK and c-Jun. Similar results were identified in Western blots using protein lysates from Hipk2−/− MEF cells. The signal intensity of p-c-Jun, p-JNK, c-Jun, and JNK bands was quantified with National Institutes of Health ImageJ online software. C–F, Confocal microscopy of p-c-Jun and p-Smad2 expression in DA neurons of 2-month-old Hipk2+/+ and Hipk2−/− mice. G, Quantification of p-c-Jun and p-Smad2 fluorescent signal intensity in C–F. Data are mean ± SEM from at least three independent experiments. *p < 0.05 (ANOVA). **p < 0.01 (ANOVA). ***p < 0.001 (ANOVA). Not significant: p > 0.05.
Previous studies showed that activation of extrasynaptic GluN2B triggers calcium accumulation in mitochondria that is strongly associated with mitochondrial swelling and neuronal cell death. In contrast, activation of GluN2A at the synapse provides a prosurvival mechanism by activating the ERK-CREB signaling pathway to promote the expression of prosurvival genes (Hardingham and Bading, 2010). Given the increased ratio of GluN2A/GluN2B in the synapses of Hipk2−/− neurons, we asked whether loss of HIPK2 might alter the activation of ERK and CREB. To test this, we first examined the state of CaMKII activation, which is the major downstream kinase regulated by the synaptic calcium influx, and found no difference in the level of phosphorylated CaMKII and total CaMKII between Hipk2+/+ and Hipk2−/− brain tissues or synaptosomes (Fig. 5-1. However, loss of HIPK2 resulted in an elevated level of p-ERK and p-CREB in the protein lysates from substantia nigra, sensorimotor cortex, spinal cord, and MEF cells, without altering the total level of ERK or CREB (Fig. 5A,B). Consistent with these results, confocal microscopy showed a significant increase in the relative signal intensity for p-CREB in TH+ DA neurons in the substantia nigra of Hipk2−/− mouse brain (Fig. 5C).
Figure 5.
Enhanced activation of the ERK-CREB signaling pathway in Hipk2−/− mouse brain. A, B, Western blots using protein lysates from the substantia nigra, sensorimotor cortex, and spinal cord of 2-month-old Hipk2+/+ and Hipk2−/− mice showed elevated levels of p-ERK and p-CREB in Hipk2−/− mouse brains, without altering the total level of ERK or CREB. Similar results were identified in cell lysates from Hipk2−/− MEF cells (Figure 5-1. The signal intensity of p-ERK, total ERK, p-CREB, total CREB, and actin was quantified with National Institutes of Health ImageJ software. C, Confocal microscopy showed a significant increase in the relative signal intensity for p-CREB in TH+ DA neuron in the substantia nigra of Hipk2−/− mouse brain. The signal intensity of p-CREB puncta was quantified with National Institutes of Health ImageJ software. D–F, qRT-PCR analyses using mRNA from the substantia nigra and cerebral cortex of 2-month-old Hipk2+/+ and Hipk2−/− mice showed consistent upregulation of several AID genes, and the similar AID genes could also be identified in Hipk2−/− MEF cells. Data are mean ± SEM from at least three independent experiments. *p < 0.05 (ANOVA). **p < 0.01 (ANOVA). ***p < 0.001 (ANOVA). Not significant: p > 0.05.
Increased relative abundance of GluN2A and the GluN2A/GluN2B ratio in Hipk2-/- mouse brain have no effects on CamKII phosphorylation in synaptosomes. (a) The western blot results showed that the p-CamKII and total CamKII protein levels in the whole cell lysates, or in synaptosomes isolated from cerebral cortex of Hipk2+/+ and Hipk2-/-mice. (b) The quantification results showed comparable signal intensity for p-CamKII and total CamKII in Hipk2+/+ and Hipk2-/- mouse brains. The signal intensity of p-CamKII, total CamKII, and Actin bands was quantified with NIH ImageJ software. Download Figure 5-1, DOCX file (25.5MB, docx)
As a prototypical signal-regulated transcription factor, CREB regulates a number of target genes that are critical for neuronal survival, synaptic plasticity, and learning and memory. Comprehensive whole-genome transcriptome profiling has identified a number of nuclear Ca2+-regulated genes, termed AID genes, which have been shown to provide neurons with broad-spectrum neuroprotective effects both in cultured cells and in animal models of neurodegeneration (Lau and Bading, 2009; Zhang et al., 2009). Given that some of the AID genes are potential CREB target genes, we reasoned that the elevated level of p-CREB in Hipk2−/− brain tissues might increase the expression of AID genes. In support of this idea, qRT-PCR analyses using mRNA from the substantia nigra and cerebral cortex of 2-month-old Hipk2+/+ and Hipk2−/− mice showed consistent upregulation of several AID genes, including Btg2, Gadd45 g, Gadd45b, Serpinb2, Bcl6, Cyr61, Arc, Jun B, and Bdnf (Fig. 5D,E). Upregulation of a similar set of AID genes was also identified in Hipk2−/− MEF cells (Fig. 5F). These results support the idea that loss of HIPK2 increases CREB phosphorylation and promotes the transcription of CREB target genes, including many AID genes.
Increased GluN2A in the mitochondria of Hipk2−/− neurons
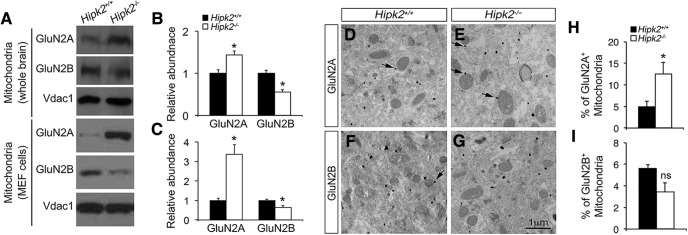
Functional NMDA receptors, including GluN1 and GluN2A, have been identified on mitochondria in neurons and play a neuroprotective role during injury (Zipfel et al., 2000; Korde and Maragos, 2012). Given the increased GluN2A protein level in the synaptosomes of Hipk2−/− brain, we asked whether there was also a similar increase of GluN2A in the mitochondria. To test this, we used sucrose density gradients to purify mitochondria from 2-month-old Hipk2+/+ and Hipk2−/− mouse brains (Fig. 3-1. Consistent with the prediction, GluN2A protein level showed a significant increase in the mitochondria isolated from Hipk2−/− mouse brain, whereas GluN2B protein level was decreased (Fig. 6A,B). Similar changes in GluN2A and GluN2B protein level were also observed in mitochondria isolated from Hipk2−/− MEF cells (Fig. 6A–C). To provide morphological evidence supporting the increase of GluN2A in mitochondria, we performed immunogold electron microscopy and showed that the majority of GluN2A+ and GluN2B+ immunogold particles were located on the outer membrane of the mitochondria (Fig. 6D–G). Consistent with the Western blot results, the percentage of GluN2A+ mitochondria was indeed increased in Hipk2−/− neurons (Fig. 6H). Although the percentage of GluN2B+ mitochondria seemed decreased, the extent of reduction did not reach statistical significance.
Figure 6.
Altered GluN2A/GluN2B protein ratio in the mitochondria of Hipk2−/− mouse brain. A, Western blot results showed that the GluN2A protein level was significantly increased, whereas the GluN2B protein level was decreased, in the mitochondria isolated from the whole brain of 2-month-old Hipk2−/− mice (top) or from Hipk2−/− MEF cells (bottom). B, C, The relative signal intensity of GluN2A, GluN2B, and Vadc1 (a mitochondria-specific marker) protein bands in Hipk2+/+ and Hipk2−/− mouse brains and in Hipk2+/+ and Hipk2−/− MEF cells was quantified with National Institutes of Health ImageJ software, and reported in B and C, respectively. D–I, Immunogold electron microscopy results showed that the percentage of GluN2A+ mitochondria was indeed increased in Hipk2−/− neurons, which were mainly located on the outer membrane of the mitochondria. Arrows indicate the GluN2A- or GluN2B-positive mitochondria. Data are mean ± SEM from at least three independent experiments. *p < 0.05 (ANOVA). Not significant: p > 0.05.
Blocking GluN2A or ERK reverses Hipk2−/− DA neuron resistance to mitochondrial toxin
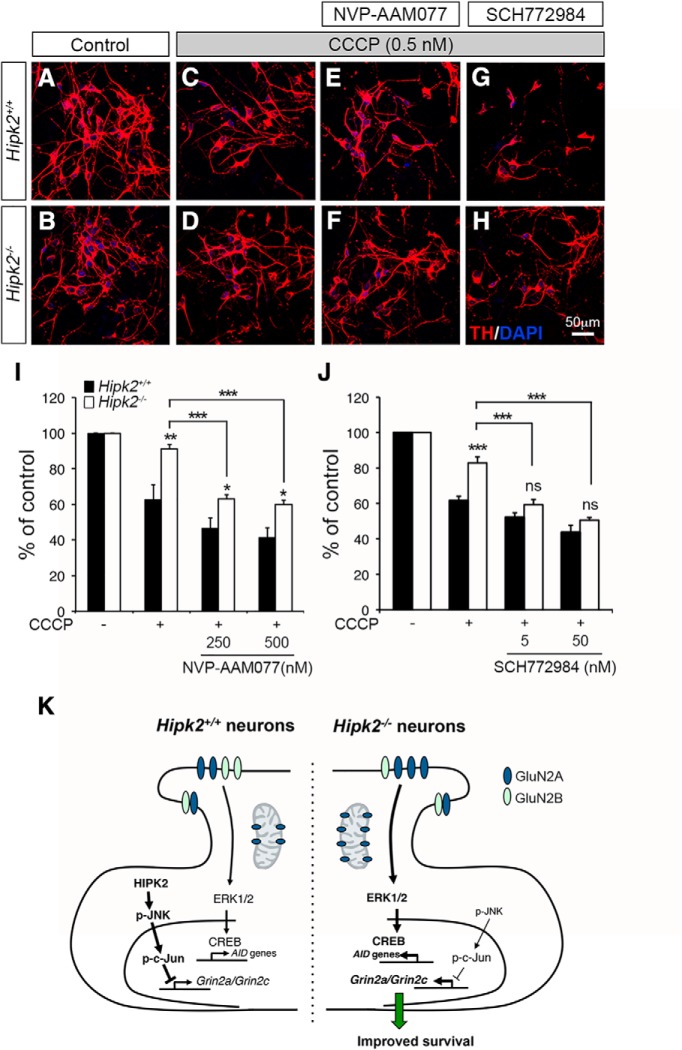
Our results thus far indicate that changes in the JNK–c-Jun and ERK-CREB pathways, upregulation of AID genes, and the increase of GluN2A in the mitochondria of Hipk2−/− neurons may render these neurons more resistant to mitochondrial toxins. In support of this idea, our results showed that primary Hipk2−/− DA neurons were more resistant to CCCP, which inhibited oxidative phosphorylation in the mitochondria by uncoupling the proton gradient (Fig. 7A–D,I,J). To characterize the role of GluN2A in the CCCP-resistant properties in Hipk2−/− neurons, we treated both Hipk2+/+ and Hipk2−/− DA neurons with 5-phosphonomethylquinoxalinedione derivative (NVP-AAM077), which is a selective antagonist of GluN2A (>100-fold over GluN2B) (Auberson et al., 2002). Consistent with our prediction, blocking GluN2A with NVP-AAM077 increased the sensitivity of Hipk2−/− DA neurons to CCCP-mediated toxicity. Interestingly, under NVP-AAM077 treatment, Hipk2−/− DA neurons were still more resistant to CCCP toxicity than Hipk2+/+ neurons (Fig. 7E,F,I). These results suggested that, in addition to the increase in GluN2A protein levels in the synaptosomes and mitochondria, additional unknown mechanism(s), which could upregulate the ERK-CREB signaling, might also contribute to the resistance of Hipk2−/− DA neurons to CCCP. To test this, we treated Hipk2+/+ and Hipk2−/− DA neurons with SCH772984, which has nanomolar potency in blocking ERK kinase activity in cancer cells (Morris et al., 2013). Similar to GluN2A inhibitor NVP-AAM077, ERK inhibitor SCH772984 dose-dependently restored the sensitivity of Hipk2−/− DA neurons to CCCP-mediated toxicity. Interestingly, unlike NVP-AAM077, SCH772984 normalized the differences in CCCP toxicity between Hipk2+/+ and Hipk2−/− DA neurons (Fig. 7G,H,J). Together, these pharmacological data from GluN2A inhibitor NVP-AAM077 and ERK inhibitor SCH772984 supported the model that the upregulation of GluN2A and activation the ERK-CREB signaling pathway in Hipk2−/− DA neurons cooperatively contribute to the resistance of these neurons to CCCP-induced mitochondrial toxicity (Fig. 7K).
Figure 7.
Pharmacological characterizations of CCCP-induced mitochondrial toxicity in Hipk2−/− DA neurons. A–J, Primary DA neurons from Hipk2−/− mice were more resistant to mitochondrial toxicity induced by CCCP than Hipk2+/+ DA neurons. CCCP toxicity in neurons was quantified by the percentage of surviving TH+ neurons based on confocal microscopy. Although blocking GluN2A with NVP-AAM077 increased the sensitivity of Hipk2−/− DA neurons to CCCP-mediated toxicity, the treated neurons were still more resistant to CCCP toxicity than Hipk2+/+ neurons. ERK inhibitor SCH772984 completely eliminated the resistance of Hipk2−/− DA neurons to CCCP-mediated toxicity. Data are mean ± SEM from at least three independent experiments. *p < 0.05 (ANOVA). **p < 0.01 (ANOVA). ***p < 0.001 (ANOVA). Not significant: p > 0.05. K, Schematic diagram depicting the working model by which the HIPK2-JNK pathway regulates c-Jun-mediated suppression of Grin2a and Grin2c in Hipk2+/+ neurons, which maintain the GluN2A/GluN2B ratio to regulate synaptic transmission and Ca2+-mediated ERK-CREB signaling pathway. Loss of HIPK2 or inhibition HIPK2 kinase activity by its specific inhibitors reduces JNK–c-Jun signaling, which derepresses the transcription of Grin2a and Grin2c, alters the GluN2A/GluN2B ratio, and activates ERK-CREB signal pathway that promotes the expression of AID genes to promote the survival of Hipk2−/− neurons in toxicity-induced by mitochondrial toxin CCCP.
Dosage-dependent effects of HIPK2 in MPTP toxicity in DA neurons
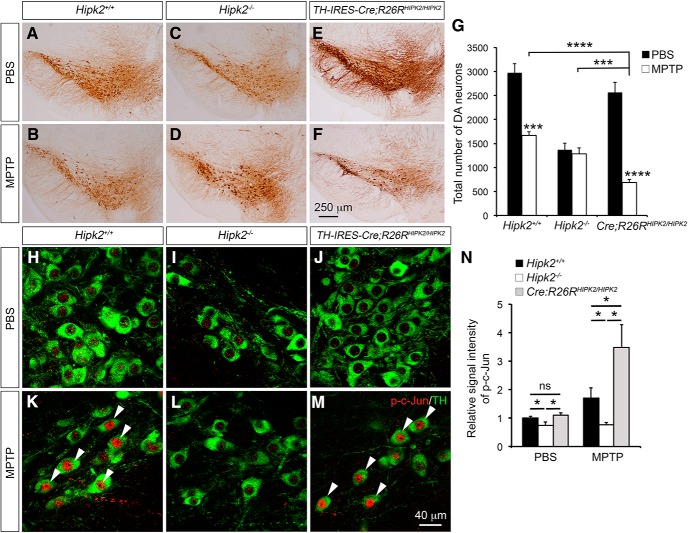
To characterize the role of HIPK2 in neuronal cell death in vivo, we generated a conditional HIPK2 “knock-in” allele by inserting EGFP-HIPK2 cDNA into the ROSA26 (R26R) locus (Fig. 8-1. This allele, named R26RHIPK2, provides the advantage of expressing additional HIPK2 proteins in ventral midbrain DA neurons using TH-IRES-Cre (Tang et al., 2009) (Fig. 8-2. To determine how HIPK2 affects neuronal cell death induced by mitochondrial toxins in vivo, we developed a chronic treatment paradigm by intraperitoneally injecting 2-month-old Hipk2+/+, Hipk2−/−, and TH-IRES-Cre;R26RHIPK2/HIPK2 mice with MPTP (4 mg/kg) once daily for 10 d. Seven days after the last injection, mice were killed for quantification of DA neuron deficits using stereology (Dauer and Przedborski, 2003; Martens et al., 2012). In Hipk2+/+ mice, chronic MPTP treatment resulted in ∼40% reduction in the number of DA neurons in the substantia nigra (Fig. 8A,B,G). Although the number of DA neurons in the substantia nigra of Hipk2−/− mice was lower than in Hipk2+/+ littermates (Zhang et al., 2007), there was no reduction of DA neurons in Hipk2−/− mice after the same MPTP treatment, supporting the idea that the majority of Hipk2−/− DA neurons were resistant to MPTP-induced toxicity (Fig. 8C,D,G). Interestingly, expressing additional HIPK2 in DA neurons in TH-IRES-Cre;R26RHIPK2/HIPK2 mice increased their vulnerability to MPTP toxicity, leading to a much more severe loss of DA neurons in the substantia nigra than that in Hipk2+/+ mice (Fig. 8E–G). Finally, to characterize the effect of MPTP in the JNK–c-Jun signaling, we quantified the signal intensity of p-c-Jun in the DA neurons using confocal microscopy. In PBS-injected mice, there was a slight reduction of p-c-Jun in the DA neurons of Hipk2−/− mice, whereas the levels of p-c-Jun in the DA neurons were similar between Hipk2+/+ and TH-IRES-Cre;R26RHIPK2/HIPK2 mice (Fig. 8H–J,N). Consistent with the essential role of HIPK2 in JNK–c-Jun activation (Fig. 4) (Lee et al., 2016), MPTP treatment significantly increased the p-c-Jun signal intensity in the DA neurons of Hipk2+/+ and even more so in TH-IRES-Cre;R26RHIPK2/HIPK2 mice, but no detectable increase in the DA neurons in Hipk2−/− mice (Fig. 8K–N).
Figure 8.
Essential role of HIPK2 in mitochondrial toxin-mediated cell death in DA neurons. A–F, TH immunohistochemical stain highlights the ventral midbrain DA neurons in 2-month-old Hipk2+/+, Hipk2−/−, and TH-IRES-Cre;R26RHIPK2/HIPK2 mice that received intraperitoneal injection of PBS or MPTP (Figure 8-1. G, Stereology counting of DA neuron number shows that MPTP causes ∼41% reduction in DA neurons in the substantia nigra of Hipk2+/+ mice, no reduction in the DA neurons in Hipk2−/− mice, and a much more severe reduction in DA neurons in TH-IRES-Cre;R26RHIPK2/HIPK2 mice (Figure 8-2. H–M, Immunofluorescent confocal microscopy shows the staining of p-Jun in TH+ DA neurons in the substantia nigra of 2-month-old Hipk2+/+, Hipk2−/−, and TH-IRES-Cre;R26RHIPK2/HIPK2 mice treated with either PBS or MPTP. K, M, Arrowheads indicate the TH+ neurons in Hipk2+/+ and TH-IRES-Cre;R26RHIPK2/HIPK2 mice show more intense p-c-Jun staining. N, Relative signal intensity of p-c-Jun in TH+ DA neurons is quantified with National Institutes of Health ImageJ software. Results are from at least 3 mice in each condition and genotype. Data are mean ± SEM from at least three independent experiments. *p < 0.05 (paired Student's t test). ***p < 0.001 (paired Student's t test). ****p < 0.0005. Not significant: p > 0.05.
Generation of the conditional “knock-in” allele that expresses EGFP-HIPK2 in a Cre-dependent manner. (a) Step-wise construction of the R26RHIPK2 targeting construct by inserting the loxP-PGKneo-STOP-loxP-EGFP-HIPK2 cassette into the R26R locus using homologous recombination. Cre-mediated recombination removes the loxP-PGKneo-STOP-loxP sequence, leading to the expression of EGFP-HIPK2 in a cell type-specific manner. (b) Genomic DNA from mouse embryonic stem (ES) cells are analyzed using Southern blots and probes that specifically detect the recombined R26REGFP-HIPK2/+ allele after restriction digestion using the EcoRV enzyme. (c) Polymerase chain reaction (PCR) using genomic DNA from ES cells to confirm the genotype of R26REGFP-HIPK2/+ allele. Download Figure 8-1, DOCX file (25.5MB, docx)
Generation of TH-IRES-Cre;R26RHIPK2/HIPK2 mice that express additional HIPK2 in DA neurons in the substantia nigra. (a) A schematic diagram showing the mating scheme to generate conditional knock-in mice that express EGFP-HIPK2 in DA neurons in the substantia nigra by crossing TH-IRES-Cre with R26RHIPK2 mice. (b) Quantitative RT-PCR (QRT-PCR) to detect the relative abundance of Hipk2 mRNA in microdissected substantia nigra tissues from 2 months old wild type (Hipk2+/+), Hipk2-/- and TH-IRES-Cre;R26RHIPK2/HIPK2 mice. The results show that the substantia nigra of TH-IRES-Cre;R26RHIPK2/HIPK2 mice contain about 50% increase of Hipk2 mRNA, whereas there is no Hipk2 mRNA detected in the substantia nigra of Hipk2-/- mice. (c) Western blots using tissues dissected from the substantia nigra of wild type (Hipk2+/+), Hipk2-/-, R26RHIPK2/+, R26RHIPK2/HIPK2, TH-IRES-Cre;R26RHIPK2/+ and TH-IRES-Cre;R26RHIPK2/HIPK2 mice. Anti-GFP antibody can detect low level of EGFP-tagged HIPK2 fusion proteins in the substantia nigra of TH-IRES-Cre;R26RHIPK2/+ and TH-IRES-Cre;R26RHIPK2/HIPK2 mice. (d-e) Immunofluorescent confocal microscopy using anti-tyrosine hydroxylase (TH) and anti-GFP antibodies in the substantia nigra of R26RHIPK2/HIPK2 and TH-IRES-Cre;R26RHIPK2/HIPK2 mice. Scale bar in panel d is 40 μm and in panel d’ 8 μm. Download Figure 8-2, DOCX file (25.5MB, docx)
Discussion
One important feature of NMDA receptors is its complex and dynamic changes in the subunit composition, which contributes to synaptic maturation, plasticity, and diversity (Paoletti et al., 2013). Although it is well recognized that the composition of NMDA receptors undergoes a switch from predominantly GluN2B subunit during embryonic brain development to GluN2A subunit in early postnatal life (the GluN2B-to-GluN2A switch), the molecular mechanism regulating this switch remains poorly understood. Using brain region-specific transcriptomic analyses, we have uncovered a previously unappreciated role of the HIPK2-JNK–c-Jun pathway in suppressing the transcription of the Grin2a and Grin2c genes, which encode NMDA receptor subunits GluN2A and GluN2C, respectively, in early postnatal brain development in mice. First, using bioinformatics, luciferase reporters, and biochemical and genetic approaches, we showed that HIPK2 is a transcriptional corepressor to suppress the expression of Grin2a and Grin2c. Loss of HIPK2 leads to a significant decrease in the JNK–c-Jun signaling pathway, increases in Grin2a and Grin2c mRNA due to reduced transcriptional repression by c-Jun, and an upregulation of GluN2A and GluN2C protein levels in DA neurons in the substantia nigra and neurons in the cerebral cortex. Second, in agreement with these findings, ultrastructural and biochemical analyses show that the increase in GluN2A results in a significant increase of GluN2A/GluN2B ratio in synapse and mitochondria, a persistent activation of the ERK-CREB pathway, and the upregulation of several prosurvival Ca2+-regulated genes (Fig. 7K). Finally, pharmacological approaches showed that Hipk2−/− DA neurons were more resistant to neurotoxin CCCP, and that this resistance can be reversed by GluN2A inhibitor NVP-AAM077 or ERK1/2 inhibitor SCH-772984. Together, these results provide strong evidence supporting the resistance of Hipk2−/− neurons to cell death induced by mitochondrial toxins.
The GluN2B-to-GluN2A switch in postnatal brain development is highly evolutionarily conserved from amphibians to mammals and is considered an important mechanism that has profound impacts from synapse maturation to associative learning (Dumas, 2005). In addition, this switch contributes to changes in the biophysical and pharmacological properties of NMDA receptors. Our results provide both in vitro and in vivo evidence that the HIPK2-JNK–c-Jun signaling pathway regulates the GluN2B-to-GluN2A switch in postnatal brain development via transcriptional control of Grin2a and Grin2c. These findings are complementary to the previously reported role of transcriptional repressor element-1 silencing transcription factor, which uses an epigenetic remodeling mechanism to suppress the transcription of Grin2b in the hippocampal synapses during postnatal development in rats (Rodenas-Ruano et al., 2012). Although our study does not directly investigate the effect of HIPK2 on Grin2a and Grin2c expression in the hippocampal synapses, given their ubiquitous expression in brain, it is plausible that HIPK2 and repressor element-1 silencing transcription factor may be critical components of an integral mechanism that cooperatively regulates the GluN2A-to-GluN2B switch during synapse maturation in postnatal brain development. With the inherent differences in the biophysical properties of GluN2A and GluN2B, it is conceivable that changes in GluN2A/GluN2B ratio in Hipk2−/− mouse brain may alter the GluN1/GluN2A diheteromic and GluN1/GluN2A/GluN2B triheteromeric composition in synapses in substantia nigra and cerebral cortex.
The identification of HIPK2-JNK–c-Jun pathway as a key mechanism that regulates the GluN2B-to-GluN2A switch has important functional implications because perturbations to this switch have been implicated in many neuropsychiatric disorders. For instance, the Rett syndrome gene MECP2 has cell type-specific effects in regulating the NMDA receptor subunit composition in visual cortex during postnatal development (Durand et al., 2012). In mouse models, loss of Mecp2 leads to a premature NMDA receptor maturation with more abundant GluN2A on the parvalbumin+ interneurons and regression in vision (Mierau et al., 2016). Interestingly, reducing GluN2A expression in Mecp2−/− mice alleviates the decline in visual function, suggesting that targeting NMDA subunit composition might provide feasible therapeutic targets for Rett syndrome. Interestingly, a recent forward genetic screen in human kinases and phosphatases for druggable regulators of MeCP2 stability identifies HIPK2 and protein phosphatase PP2A as strong candidates that can stabilize MeCP2 protein levels in vivo (Lombardi et al., 2017). These results further reinforce the important role of HIPK2 in regulating the GluN2B-to-GluN2A switch in the context of neurodevelopmental diseases, such as Rett syndrome. In addition, we have recently shown that the autocrine TGF-β signaling in ventral midbrain DA neurons is critical to maintain a balanced excitation-inhibition synaptic input. Loss of TGF-β Type II receptor in DA neurons reduces excitatory input and alters the phasic firing pattern in these neurons. As a consequence, mice lacking TGF-β Type II receptor in DA neurons (DAT-iCre;TbRIIfl/fl) exhibit hyperactivity and reward learning deficits (Luo et al., 2016). Given the important role of HIPK2 in TGF-β/BMP signaling pathway (Zhang et al., 2007; Chalazonitis et al., 2011), it is very likely that reduced HIPK2-mediated regulation of NMDA receptor subunit expression may contribute to the synaptic and behavioral changes in DAT-iCre;TbRIIfl/fl mice.
Aside from its critical roles in synaptic transmission, neurotoxicity mediated by NMDA receptors has been implicated in the pathogenesis of neurodegenerative diseases. In Alzheimer's disease model, GluN2B has been shown to mediate amyloid β-mediated perturbation of LTP and synaptic loss (Hu et al., 2009; Li et al., 2011; Rönicke et al., 2011). In patients with Parkinson's disease, the degeneration of DA neurons in the substantia nigra is associated with an overactivation of the glutaminergic projection to the striatum and basal ganglia output nuclei (Sgambato-Faure and Cenci, 2012). Several plausible mechanisms have been proposed to explain the potential contributions of NMDA receptor in these disease settings. First, excessive Ca2+ influx through NMDA receptors causing Ca2+ overload can be deleterious to neurons. Second, activation of the synaptic and extrasynaptic NMDA receptors can promote distinct signaling pathways that contribute to neuronal survival and cell death, respectively. Finally, activation of GluN2A-containing NMDA receptors can provide prosurvival effects in cultured neurons and in stroke models via CREB signaling pathway (Liu et al., 2007; von Engelhardt et al., 2007; Chen et al., 2008; Terasaki et al., 2010). Our results show that the reduced JNK–c-Jun signaling in the substantia nigra and cerebral cortex in Hipk2−/− mice is accompanied by a robust increase in the relative abundance of GluN2A and an increase in GluN2A/GluN2B ratio. In addition, the same brain regions in Hipk2−/− mice show a robust increase in ERK-CREB signaling and upregulation of several AID genes. These results are consistent with previous reports (Liu et al., 2007; von Engelhardt et al., 2007; Chen et al., 2008; Terasaki et al., 2010) and support the idea that the increase in GluN2A subunit, CREB signaling, and the expression of AID genes collectively contribute to the resistance of Hipk2−/− DA neurons to CCCP- and MPTP-induced neuronal toxicity. This idea is further supported by the pharmacological inhibition of GluN2A and ERK1/2, which reverses the resistance of Hipk2−/− DA neurons to CCCP.
Recent studies show that NMDA agonists can also increase the Ca2+ level in isolated brain mitochondria, which attenuates ROS-induced cytochrome c release and cell death induced by exposure to excessive glutamate (Korde and Maragos, 2012, 2016). Given the beneficial effects of GluN2A in mitochondria, it is tempting to speculate that the increase of GluN2A and reduced GluN2B in the mitochondria of Hipk2−/− brains might also contribute to the resistance of Hipk2−/− DA neurons to mitochondrial toxins CCCP and MPTP. However, it will require more in-depth analyses of mitochondrial physiology in Hipk2−/− neurons to determine (1) how an increase in GluN2A alters mitochondrial membrane potentials under resting condition or when exposed to CCCP, and (2) whether GluN2A-independent mechanism(s) might contribute to the resistance of Hipk2−/− neurons to mitochondrial toxins.
In conclusion, our investigation on the transcriptomes of the substantia nigra in adult Hipk2−/− mice reveals an unexpected mechanism for HIPK2 in the transcriptional control of NMDA receptor subunits GluN2A and GluN2C. Together with our previous study (Zhang et al., 2007), these results support the idea that HIPK2 is a transcriptional cofactor that has stage- and context-dependent role in regulating TGF-β-dependent survival of DA neurons during embryonic development and maturation of NMDA receptors in the same neurons in early postnatal life. This mechanism appears to have a broader role in neurons in different brain regions, including the substantia nigra, cerebral cortex, and spinal cord. These results are in agreement with our recent study that ER stress, induced by pharmacological agents or by the accumulation of misfolded proteins, activates a cascade of kinases downstream of inositol-requiring enzyme 1α, including apoptosis signal-regulating kinase 1, HIPK2, and JNK, to promote cell death in neurons, MEFs, and HEK293 cells (Lee et al., 2016). Genetically removal of Hipk2 or blocking HIPK2 kinase activation using HIPK2 kinase inhibitors protects ER stress-induced neuronal cell death. Based on the results from our current study, it is possible that, during ER stress, the activation of HIPK2-JNK–c-Jun signaling might suppress the transcription of Grin2a and Grin2c, which can alter the GluN2A-to-GluN2B ratio and potentially contribute to synaptic maturation in early postnatal brain development, as well as neuronal cell death under stress conditions.
Footnotes
This work was supported by National Institutes of Health Grant NS098516 and Department of the Veterans Affairs Grant BX001108. The contents do not represent the views of the U.S. Department of Veterans Affairs or the U.S. Government. We thank Dr. Phil Soriano for providing the R26R constructs; Dr. Guangwei Wei for help with the generation of the R26RHIPK2 allele; Ivy Hsieh for assistance with immunogold EM; and members of the E.J.H. laboratory for feedback during the course of this study.
The authors declare no competing financial interests.
References
- Auberson YP, Allgeier H, Bischoff S, Lingenhoehl K, Moretti R, Schmutz M (2002) 5-Phosphonomethylquinoxalinediones as competitive NMDA receptor antagonists with a preference for the human 1A/2A, rather than 1A/2B receptor composition. Bioorg Med Chem Lett 12:1099–1102. 10.1016/S0960-894X(02)00074-4 [DOI] [PubMed] [Google Scholar]
- Blaquiere JA, Verheyen EM (2017) Homeodomain-interacting protein kinases: diverse and complex roles in development and disease. Curr Top Dev Biol 123:73–103. 10.1016/bs.ctdb.2016.10.002 [DOI] [PubMed] [Google Scholar]
- Carlin RK, Grab DJ, Cohen RS, Siekevitz P (1980) Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J Cell Biol 86:831–845. 10.1083/jcb.86.3.831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalazonitis A, Tang AA, Shang Y, Pham TD, Hsieh I, Setlik W, Gershon MD, Huang EJ (2011) Homeodomain interacting protein kinase 2 regulates postnatal development of enteric dopaminergic neurons and glia via BMP signaling. J Neurosci 31:13746–13757. 10.1523/JNEUROSCI.1078-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Lu TJ, Chen XJ, Zhou Y, Chen Q, Feng XY, Xu L, Duan WH, Xiong ZQ (2008) Differential roles of NMDA receptor subtypes in ischemic neuronal cell death and ischemic tolerance. Stroke 39:3042–3048. 10.1161/STROKEAHA.108.521898 [DOI] [PubMed] [Google Scholar]
- Choi DW, Na W, Kabir MH, Yi E, Kwon S, Yeom J, Ahn JW, Choi HH, Lee Y, Seo KW, Shin MK, Park SH, Yoo HY, Isono K, Koseki H, Kim ST, Lee C, Kwon YK, Choi CY (2013) WIP1, a homeostatic regulator of the DNA damage response, is targeted by HIPK2 for phosphorylation and degradation. Mol Cell 51:374–385. 10.1016/j.molcel.2013.06.010 [DOI] [PubMed] [Google Scholar]
- Collingridge GL, Olsen RW, Peters J, Spedding M (2009) A nomenclature for ligand-gated ion channels. Neuropharmacology 56:2–5. 10.1016/j.neuropharm.2008.06.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer W, Przedborski S (2003) Parkinson's disease: mechanisms and models. Neuron 39:889–909. 10.1016/S0896-6273(03)00568-3 [DOI] [PubMed] [Google Scholar]
- Dumas TC. (2005) Developmental regulation of cognitive abilities: modified composition of a molecular switch turns on associative learning. Prog Neurobiol 76:189–211. 10.1016/j.pneurobio.2005.08.002 [DOI] [PubMed] [Google Scholar]
- Durand S, Patrizi A, Quast KB, Hachigian L, Pavlyuk R, Saxena A, Carninci P, Hensch TK, Fagiolini M (2012) NMDA receptor regulation prevents regression of visual cortical function in the absence of Mecp2. Neuron 76:1078–1090. 10.1016/j.neuron.2012.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Wang N, Chuang P, He JC (2014) Role of HIPK2 in kidney fibrosis. Kidney Int Suppl 4:97–101. 10.1038/kisup.2014.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Li MC, Weng Z (2003) Cluster-buster: finding dense clusters of motifs in DNA sequences. Nucleic Acids Res 31:3666–3668. 10.1093/nar/gkg540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham GE, Bading H (2010) Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat Rev Neurosci 11:682–696. 10.1038/nrn2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann TG, Stollberg N, Schmitz ML, Will H (2003) HIPK2 regulates transforming growth factor-beta-induced c-jun NH(2)-terminal kinase activation and apoptosis in human hepatoma cells. Cancer Res 63:8271–8277. [PubMed] [Google Scholar]
- Hofmann TG, Glas C, Bitomsky N (2013) HIPK2: a tumour suppressor that controls DNA damage-induced cell fate and cytokinesis. Bioessays 35:55–64. 10.1002/bies.201200060 [DOI] [PubMed] [Google Scholar]
- Hu NW, Klyubin I, Anwy R, Rowan MJ (2009) GluN2B subunit-containing NMDA receptor antagonists prevent abeta-mediated synaptic plasticity disruption in vivo. Proc Natl Acad Sci U S A 106:20504–20509. 10.1073/pnas.0908083106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korde AS, Maragos WF (2012) Identification of an N-methyl-D-aspartate receptor in isolated nervous system mitochondria. J Biol Chem 287:35192–35200. 10.1074/jbc.M111.322032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korde AS, Maragos WF (2016) Direct exposure to N-methyl-D-aspartate alters mitochondrial function. Neurosci Lett 623:47–51. 10.1016/j.neulet.2016.04.054 [DOI] [PubMed] [Google Scholar]
- Lau D, Bading H (2009) Synaptic activity-mediated suppression of p53 and induction of nuclear calcium-regulated neuroprotective genes promote survival through inhibition of mitochondrial permeability transition. J Neurosci 29:4420–4429. 10.1523/JNEUROSCI.0802-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Shang Y, Redmond SA, Urisman A, Tang AA, Li KH, Burlingame AL, Pak RA, Jovičić A, Gitler AD, Wang J, Gray NS, Seeley WW, Siddique T, Bigio EH, Lee VM, Trojanowski JQ, Chan JR, Huang EJ (2016) Activation of HIPK2 promotes ER stress-mediated neurodegeneration in amyotrophic lateral sclerosis. Neuron 91:41–55. 10.1016/j.neuron.2016.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Jin M, Koeglsperger T, Shepardson NE, Shankar GM, Selkoe DJ (2011) Soluble abeta oligomers inhibit long-term potentiation through a mechanism involving excessive activation of extrasynaptic NR2B-containing NMDA receptors. J Neurosci 31:6627–6638. 10.1523/JNEUROSCI.0203-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg J, Usoskin D, Bengtsson H, Gustafsson A, Kylberg A, Söderstrom S, Ebendal T (2004) Transgenic expression of cre recombinase from the tyrosine hydroxylase locus. Genesis 40:67–73. 10.1002/gene.20065 [DOI] [PubMed] [Google Scholar]
- Liu Y, Wong TP, Aarts M, Rooyakkers A, Liu L, Lai TW, Wu DC, Lu J, Tymianski M, Craig AM, Wang YT (2007) NMDA receptor subunits have differential roles in mediating excitotoxic neuronal death both in vitro and in vivo. J Neurosci 27:2846–2857. 10.1523/JNEUROSCI.0116-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi LM, Zaghlula M, Sztainberg Y, Baker SA, Klisch TJ, Tang AA, Huang EJ, Zoghbi HY (2017) An RNA interference screen identifies druggable regulators of MeCP2 stability. Sci Transl Med 9:eaaf7588. 10.1126/scitranslmed.aaf7588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo SX, Huang EJ (2016) Dopaminergic neurons and brain reward pathways: from neurogenesis to circuit assembly. Am J Pathol 186:478–488. 10.1016/j.ajpath.2015.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo SX, Timbang L, Kim JI, Shang Y, Sandoval K, Tang AA, Whistler JL, Ding JB, Huang EJ (2016) TGF-beta signaling in dopaminergic neurons regulates dendritic growth, excitatory-inhibitory synaptic balance, and reversal learning. Cell Rep 17:3233–3245. 10.1016/j.celrep.2016.11.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens LH, Zhang J, Barmada SJ, Zhou P, Kamiya S, Sun B, Min SW, Gan L, Finkbeiner S, Huang EJ, Farese RV Jr (2012) Progranulin deficiency promotes neuroinflammation and neuron loss following toxin-induced injury. J Clin Invest 122:3955–3959. 10.1172/JCI63113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miduturu CV, Deng X, Kwiatkowski N, Yang W, Brault L, Filippakopoulos P, Chung E, Yang Q, Schwaller J, Knapp S, King RW, Lee JD, Herrgard S, Zarrinkar P, Gray NS (2011) High-throughput kinase profiling: a more efficient approach toward the discovery of new kinase inhibitors. Chem Biol 18:868–879. 10.1016/j.chembiol.2011.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierau SB, Patrizi A, Hensch TK, Fagiolini M (2016) Cell-specific regulation of N-methyl-D-aspartate receptor maturation by Mecp2 in cortical circuits. Biol Psychiatry 79:746–754. 10.1016/j.biopsych.2015.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A, Carr D, Deng Y, Jin W, Black S, Long B, Liu J, Dinunzio E, Windsor W, Zhang R, Zhao S, Angagaw MH, Pinheiro EM, Desai J, Xiao L, et al. (2013) Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov 3:742–750. 10.1158/2159-8290.CD-13-0070 [DOI] [PubMed] [Google Scholar]
- Paoletti P, Bellone C, Zhou Q (2013) NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci 14:383–400. 10.1038/nrn3504 [DOI] [PubMed] [Google Scholar]
- Rodenas-Ruano A, Chávez AE, Cossio MJ, Castillo PE, Zukin RS (2012) REST-dependent epigenetic remodeling promotes the developmental switch in synaptic NMDA receptors. Nat Neurosci 15:1382–1390. 10.1038/nn.3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönicke R, Mikhaylova M, Rönicke S, Meinhardt J, Schröder UH, Fändrich M, Reiser G, Kreutz MR, Reymann KG (2011) Early neuronal dysfunction by amyloid beta oligomers depends on activation of NR2B-containing NMDA receptors. Neurobiol Aging 32:2219–2228. 10.1016/j.neurobiolaging.2010.01.011 [DOI] [PubMed] [Google Scholar]
- Sgambato-Faure V, Cenci MA (2012) Glutamatergic mechanisms in the dyskinesias induced by pharmacological dopamine replacement and deep brain stimulation for the treatment of Parkinson's disease. Prog Neurobiol 96:69–86. 10.1016/j.pneurobio.2011.10.005 [DOI] [PubMed] [Google Scholar]
- Shang Y, Doan CN, Arnold TD, Lee S, Tang AA, Reichardt LF, Huang EJ (2013) Transcriptional corepressors HIPK1 and HIPK2 control angiogenesis via TGF-beta-TAK1-dependent mechanism. PLoS Biol 11:e1001527. 10.1371/journal.pbio.1001527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Miyamoto Y, Huang EJ (2009) Multiple roles of beta-catenin in controlling the neurogenic niche for midbrain dopamine neurons. Development 136:2027–2038. 10.1242/dev.034330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki Y, Sasaki T, Yagita Y, Okazaki S, Sugiyama Y, Oyama N, Omura-Matsuoka E, Sakoda S, Kitagawa K (2010) Activation of NR2A receptors induces ischemic tolerance through CREB signaling. J Cereb Blood Flow Metab 30:1441–1449. 10.1038/jcbfm.2010.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Engelhardt J, Coserea I, Pawlak V, Fuchs EC, Köhr G, Seeburg PH, Monyer H (2007) Excitotoxicity in vitro by NR2A- and NR2B-containing NMDA receptors. Neuropharmacology 53:10–17. 10.1016/j.neuropharm.2007.04.015 [DOI] [PubMed] [Google Scholar]
- Wei G, Ku S, Ma GK, Saito S, Tang AA, Zhang J, Mao JH, Appella E, Balmain A, Huang EJ (2007) HIPK2 represses beta-catenin-mediated transcription, epidermal stem cell expansion, and skin tumorigenesis. Proc Natl Acad Sci U S A 104:13040–13045. 10.1073/pnas.0703213104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins AK, Wei G, Doxakis E, Wong C, Tang AA, Zang K, Luo EJ, Neve RL, Reichardt LF, Huang EJ (2004) Interaction of Brn3a and HIPK2 mediates transcriptional repression of sensory neuron survival. J Cell Biol 167:257–267. 10.1083/jcb.200406131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Pho V, Bonasera SJ, Holtzman J, Tang AT, Hellmuth J, Tang S, Janak PH, Tecott LH, Huang EJ (2007) Essential function of HIPK2 in TGFbeta-dependent survival of midbrain dopamine neurons. Nat Neurosci 10:77–86. 10.1038/nn1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Zou M, Lu L, Lau D, Ditzel DA, Delucinge-Vivier C, Aso Y, Descombes P, Bading H (2009) Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet 5:e1000604. 10.1371/journal.pgen.1000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel GJ, Babcock DJ, Lee JM, Choi DW (2000) Neuronal apoptosis after CNS injury: the roles of glutamate and calcium. J Neurotrauma 17:857–869. 10.1089/neu.2000.17.857 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primer sequences (5’ to 3’). Download Table 1-1, DOCX file (100.8KB, docx)
DAVID gene ontology (GO) analyses of microarray data from the substantia nigra of Hipk2+/+ and Hipk2-/- mice. Download Table 1-2, DOCX file (41.8KB, docx)
Genes analyzed by Cluster Buster to identify potential transcription factor binding elements in HIPK2 target genes. Download Table 1-3, DOCX file (165.5KB, docx)
HIPK2 suppresses, whereas MEF2C and HDAC7 do not affect, Grin2a-Luc and Grin2c-Luc reporter activities. Expression of HIPK2 or c-Jun alone in COS-7 cells was sufficient to suppress the basal activity of Grin2a-Luc and Grin2c-Luc reporters. Unlike c-Jun or HIPK2, transcription factor MEF2C and transcriptional cofactor HDAC7, which interact with HIPK2 and regulate vasculogenesis during embryonic development, had very modest or no detectable effects on the Grin2a-Luc or Grin2c-Luc reporter activity. Download Figure 2-1, DOCX file (25.5MB, docx)
Schematic diagram for isolating different subcellular organelles from mouse brain. (a) Flow chart illustrating the basic steps for the preparation of cytosolic, synaptosomal, and mitochondrial extracts from 2-month-old mice brain. The procedure requires approximately 3 hour from tissue homogenization to collection of the synaptosomal fraction. (b-c) Western blot showing the purity of the synaptosomal and mitochondrial fractions. PSD95: post-synaptic marker, Synaptophysin: pre-synaptic marker, VEGFR: a membrane associated protein, Tom20: a translocase of the outer membrane in mitochondria, and Actin: ubiquitous expressed protein as loading control. Download Figure 3-1, DOCX file (25.5MB, docx)
Increased relative abundance of GluN2A and the GluN2A/GluN2B ratio in Hipk2-/- mouse brain have no effects on CamKII phosphorylation in synaptosomes. (a) The western blot results showed that the p-CamKII and total CamKII protein levels in the whole cell lysates, or in synaptosomes isolated from cerebral cortex of Hipk2+/+ and Hipk2-/-mice. (b) The quantification results showed comparable signal intensity for p-CamKII and total CamKII in Hipk2+/+ and Hipk2-/- mouse brains. The signal intensity of p-CamKII, total CamKII, and Actin bands was quantified with NIH ImageJ software. Download Figure 5-1, DOCX file (25.5MB, docx)
Generation of the conditional “knock-in” allele that expresses EGFP-HIPK2 in a Cre-dependent manner. (a) Step-wise construction of the R26RHIPK2 targeting construct by inserting the loxP-PGKneo-STOP-loxP-EGFP-HIPK2 cassette into the R26R locus using homologous recombination. Cre-mediated recombination removes the loxP-PGKneo-STOP-loxP sequence, leading to the expression of EGFP-HIPK2 in a cell type-specific manner. (b) Genomic DNA from mouse embryonic stem (ES) cells are analyzed using Southern blots and probes that specifically detect the recombined R26REGFP-HIPK2/+ allele after restriction digestion using the EcoRV enzyme. (c) Polymerase chain reaction (PCR) using genomic DNA from ES cells to confirm the genotype of R26REGFP-HIPK2/+ allele. Download Figure 8-1, DOCX file (25.5MB, docx)
Generation of TH-IRES-Cre;R26RHIPK2/HIPK2 mice that express additional HIPK2 in DA neurons in the substantia nigra. (a) A schematic diagram showing the mating scheme to generate conditional knock-in mice that express EGFP-HIPK2 in DA neurons in the substantia nigra by crossing TH-IRES-Cre with R26RHIPK2 mice. (b) Quantitative RT-PCR (QRT-PCR) to detect the relative abundance of Hipk2 mRNA in microdissected substantia nigra tissues from 2 months old wild type (Hipk2+/+), Hipk2-/- and TH-IRES-Cre;R26RHIPK2/HIPK2 mice. The results show that the substantia nigra of TH-IRES-Cre;R26RHIPK2/HIPK2 mice contain about 50% increase of Hipk2 mRNA, whereas there is no Hipk2 mRNA detected in the substantia nigra of Hipk2-/- mice. (c) Western blots using tissues dissected from the substantia nigra of wild type (Hipk2+/+), Hipk2-/-, R26RHIPK2/+, R26RHIPK2/HIPK2, TH-IRES-Cre;R26RHIPK2/+ and TH-IRES-Cre;R26RHIPK2/HIPK2 mice. Anti-GFP antibody can detect low level of EGFP-tagged HIPK2 fusion proteins in the substantia nigra of TH-IRES-Cre;R26RHIPK2/+ and TH-IRES-Cre;R26RHIPK2/HIPK2 mice. (d-e) Immunofluorescent confocal microscopy using anti-tyrosine hydroxylase (TH) and anti-GFP antibodies in the substantia nigra of R26RHIPK2/HIPK2 and TH-IRES-Cre;R26RHIPK2/HIPK2 mice. Scale bar in panel d is 40 μm and in panel d’ 8 μm. Download Figure 8-2, DOCX file (25.5MB, docx)