Abstract
Arabidopsis Rubisco was activated in vitro at rates 2- to 3-fold greater by recombinant Arabidopsis 43-kD Rubisco activase with the amino acid replacements Q111E and Q111D in a phosphate-binding loop, G-G-K-G-Q-G-K-S. However, these two mutant enzymes had only slightly greater rates of ATP hydrolysis. Activities of the Q111D enzyme were much less sensitive and those of Q111E were somewhat less sensitive to inhibition by ADP. Both mutant enzymes exhibited higher Rubisco activation activities over the physiological range of ADP to ATP ratios. Enzymes with non-polar, polar, and basic residues substituted at position Gln-111 exhibited rates of Rubisco activation less than the wild-type enzyme. Estimates of the relative affinity of the wild type and the Q111D, Q111E, and Q111S enzymes for adenosine nucleotides by a variety of methods revealed that the nucleotide affinities were the most diminished in the Q111D enzyme. The temperature stability of the Q111D and Q111E enzymes did not differ markedly from that of the 43-kD recombinant wild-type enzyme, which is somewhat thermolabile. The Q111D and Q111E enzymes, expressed in planta, may provide a means to better define the role of the ADP to ATP ratio in the regulation of Rubisco activation and photosynthesis rate.
The light-dependent activation of Rubisco (EC 4.1.1.39) in plants is dependent on the activity of the stromal protein, Rubisco activase, which usually consists of two isoforms slightly different in molecular mass (Salvucci and Ogren, 1996). Rubisco activase uses the hydrolysis of ATP to facilitate the dissociation of ribulose 1,5-bisphosphate (RuBP) bound as an inhibitor at the active site of uncarbamylated and, therefore, inactive Rubisco (Wang and Portis, 1992). Rubisco activase also facilitates the dissociation of carboxyarabinitol 1-phosphate, bound to the carbamylated form, in plants that contain this inhibitor (Robinson and Portis, 1988; Hammond et al., 1998). The activity of Rubisco activase is light regulated by the ratio of ADP to ATP (Robinson and Portis, 1989), which increases from dark to light conditions in the stroma (Stitt et al., 1982). More recently it was shown that the activity may be regulated by redox changes mediated by thioredoxin-f (Zhang and Portis, 1999), which alters the response of the larger isoform to the ratio of ADP to ATP.
Two distinct cDNAs coding for 41- and 45-kD isoforms of spinach Rubisco activase were isolated from a cDNA expression library and subsequently shown to arise by alternative splicing (Werneke et al., 1989). Sequence analysis revealed the presence of a “P-loop” or “Walker A” (Walker et al., 1982) triphosphate-binding loop consensus sequence, G105GKGQGKS112, for nucleotide binding, consistent with its ATPase activity. Site-directed mutagenesis of recombinant 41-kD spinach Rubisco activase substituted with either, Arg, Ile, or Thr at the invariant position, Lys-111, had no detectable ATPase or Rubisco activase activity (Shen et al., 1991), as expected from the numerous mutagenesis studies of this region in other proteins (Traut, 1994). Substitutions were subsequently made at the non-conserved positions (K107A, K107M, K107R, Q109E, Q109K, and S112P; Shen and Ogren, 1992). Of these, only the K107M and Q109E mutants retained activity. Most surprisingly, the 41-kD Q109E protein exhibited a higher Rubisco activation activity and a slightly reduced ATPase activity when compared to the wild-type 41-kD recombinant protein. The dependence of both activities on the concentration of ATP was not greatly altered, but Shen and Ogren (1992) did not investigate whether inhibition by ADP was affected.
Complementation of the Arabidopsis mutant, rca−, in which Rubisco activase is not present (Salvucci et al., 1985), with mutant forms of Rubisco activase provides a powerful approach to analyze Rubisco regulation. Rubisco activase mutants that exhibit altered responses to ATP or ADP would be especially informative because they might alter the normal regulation of Rubisco. Rather than continuing studies with the spinach recombinant isoforms in anticipation of using them to complement the rca− mutant, we decided to switch to the Arabidopsis protein for two reasons. First, it seemed important to establish that the unexpected results observed with Q109E in spinach were not unique and could be observed with an identical substitution, Q111E, at the same position in the loop of Arabidopsis activase. Identical amino acid substitutions in the highly conserved Rubisco large subunit of different species have produced different biochemical effects (Spreitzer, 1999). Second, expressing otherwise native forms of the protein in the rca− mutant would eliminate possible complications due to altering the specificity between Rubisco and Rubisco activase (Wang et al., 1992) or other unknown components of the regulation system.
In addition, we sought to extend the previous mutagenesis experiments with the spinach protein in two important ways before attempting transformation. Because only two replacements had been made in the P-loop at the Gln position, it was desirable to determine whether any other replacements at this position would increase either ATPase or Rubisco activation activity, or both. Shen and Ogren (1992) suggested that the increased activity of Q109E was due to the acidic nature of the substitution. If so, only a replacement of the Gln with Asp would be expected to produce an effect similar to the Q109E replacement. Also, the increased activation activity of the Q109E enzyme might not be realized under physiological conditions in planta because of the potent inhibition by ADP. Therefore, we examined the response of potentially interesting mutant enzymes to both nucleotides.
RESULTS
Rubisco Activation and ATPase Activities of the Mutant Proteins
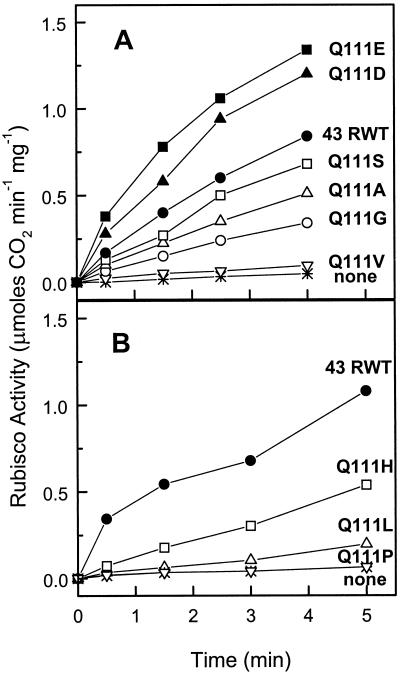
Shen and Ogren (1992) reported that the Q109E substitution in the 41-kD isoform of spinach Rubisco activase resulted in higher rates of Rubisco activation compared to the recombinant wild-type (RWT) protein. We were interested in determining the Rubisco activation and ATPase activities resulting from the same Gln to Glu substitution at the analogous site (position 111) in Arabidopsis Rubisco activase. In addition we wanted to examine the activity of proteins with additional substitutions at the 111 position to see if any other mutations would increase the Rubisco activation and/or ATPase activities. Figure 1 shows that the engineered recombinant proteins varied widely in their rate of activation of Rubisco with respect to the 43-kD RWT protein. Recombinant proteins Q111E and Q111D (Fig. 1A) exhibited similar rates of Rubisco activation, both appreciably greater than the 43-kD RWT. Q111S activated Rubisco at a rate slightly less the 43-kD RWT, whereas the Q111A, Q111G, and Q111H recombinant proteins all exhibited significantly less Rubisco activation activity. The Q111V, Q111L, and Q111P mutants retained little activation activity (Fig. 1).
Figure 1.
Activation of the inactive Rubisco-RuBP complex by the 43-kD RWT enzyme and various P-loop mutant enzymes. Rubisco activity in the absence of ADP was measured by a two-step radiometric assay. A and B are separate experiments.
The significance of the increased Rubisco activation activity of the Q111D and Q111E enzymes compared to the activity of the 43-kD RWT might be questioned because separate isolations are required. Whereas some variation (20%–30%) in the activation activities could be observed between various isolations of the same mutant enzyme made months apart, we routinely observed a 2- to 3-fold higher activation activity with the Q111D and Q111E enzymes compared to the 43-kD RWT preparations (data not shown).
Shen and Ogren (1992) also reported that the Q109E mutation in the 41-kD isoform of spinach Rubisco activase had an ATPase activity nearly equivalent to that of the 43-kD RWT, resulting in a higher efficiency because of the greater Rubisco activation activity. To determine whether this was also true for the Arabidopsis proteins, aliquots from the same protein preparation were measured spectrophotometrically on the same day for both Rubisco activation activity and ATPase activity. Table I shows that the specific activation rates for the Q111D and Q111E mutant enzymes were respectively 1.9- and 2.4-fold greater than the 43-kD RWT in this experiment. However, the ATPase activities of the Q111D and Q111E enzymes were only 1.4- and 1.2- fold greater, respectively, than the 43-kD RWT. Therefore, the activation efficiency (ratio of Rubisco activation and ATPase specific activities) was the highest for the Q111E enzyme (1.9-fold), followed by Q111D (1.4-fold), and Q111S (0.8-fold), which was less than the 43-kD RWT.
Table I.
Comparison of Rubisco activation and ATPase activities of recombinant Arabidopsis Rubisco activase
| Enzyme | Rubisco Activation
|
ATPase Activity
|
Rubisco Activation: ATPase
|
|||
|---|---|---|---|---|---|---|
| Activitya | RWT | Activityb | RWT | Ratio | RWT | |
| % | % | % | ||||
| 43-kD RWT | 4.2 | – | 0.55 | – | 7.8 | – |
| Q111D | 8.2 | 190 | 0.78 | 140 | 10.6 | 140 |
| Q111E | 10.0 | 240 | 0.68 | 120 | 14.7 | 190 |
| Q111S | 2.3 | 55 | 0.35 | 64 | 6.5 | 83 |
Rubisco activation was determined by following phosphoglyceric acid production in a coupled spectrophotometric assay. ATPase activity in the absence of ADP was measured by a spectrophotometric assay modified from Robinson and Portis (1989). Rates represent steady-state values averaged over a 2-min time span. The Rubisco activation to ATPase ratio is a measure of the coupling efficiency. The results are the average of duplicate assays.
Units of activity are μmol CO2 min−1 mg−1 Rubisco min−1 mg−1 protein.
Units of activity are μmol ADP min−1 mg protein−1.
Effect of ADP on Rubisco Activase and ATPase Activities
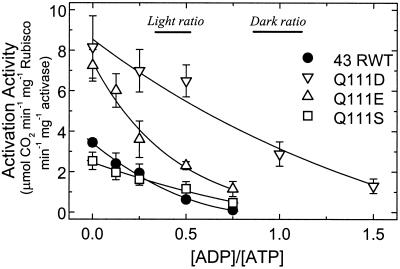
Robinson and Portis (1989) found that ADP is a potent inhibitor of Rubisco activase. A re-analysis of their data and more recent data (not shown), which cover a range (0.5–3 mm) of ATP and ADP concentrations, indicates that the enzyme's activity depends simply on the ratio of ADP to ATP. As shown below, a relationship between enzyme activity and the ratio of ADP to ATP concentrations can be derived from the equation describing competitive inhibition. Whereas Shen and Ogren (1992) showed that the spinach 41-kD Q109E Rubisco activase exhibited greater Rubisco activation activity, but had a similar dependence on ATP concentration as the 41-kD RWT enzyme, they did not examine the effects of ADP on their mutant enzymes. In the chloroplast stroma, both ADP and ATP are present and will compete for the nucleotide-binding site on Rubisco activase. Therefore, it was important to determine the effects of ADP on Rubisco activation and ATPase activity of the Q111D, Q111E, and Q111S enzymes, as well as the RWT protein at physiological ADP to ATP ratios. This ratio ranges from about 1:1 in the dark to 1:3 in the light under normal conditions (Stitt et al., 1982). Figure 2 shows that, in agreement with our previous results, the Q111D and Q111E proteins had a much higher specific activity at zero ADP:ATP than the 43-kD RWT protein. The higher activities of these proteins were maintained across a wide range of ADP to ATP ratios. At an ADP:ATP ratio of 1:3, equivalent to a typical daytime stromal ratio, the estimated values of the Q111D and Q111E enzymes are 4.3- and 2.3-fold greater, respectively, than the 43-kD RWT and Q111S enzymes, which had similar activities at this ratio. At an ADP:ATP ratio of 1:1, typical of the stromal conditions in the dark, the activity of Q111D remained nearly as high as the activity of the 43-kD RWT enzyme in the absence of ADP. However, the Q111E, Q111S, and 43-kD RWT proteins all retained minimal activity as the ADP:ATP ratio approached 1:1.
Figure 2.
Dependence of Rubisco activation activity on the ratio of ADP to ATP for the 43-kD RWT and Q111D, Q111E, and Q111S mutant enzymes. Activation activity was measured by a single step Rubisco activity assay at a constant total nucleotide concentration of 4 mm.
The equation describing the rate (v) for a reaction under conditions of competitive inhibition between a substrate (S) and inhibitor (I) can be rearranged into the following form: v = Vm/[(Km/Ki).(I/S) + 1 + Km/S]. At substrate concentrations (millimolar ATP in our experiments) much greater than the Km (μm ATP for activase), the Km/S term is very small, and plots of activity versus I/S over a range of inhibitor and substrate concentrations will have the same inverse hyperbolic form. Furthermore, the ratio (R0.5) of ADP to ATP (I/S) at which activity is reduced to one-half of the activity in the absence of ADP, which can be obtained from this plot, is nearly equal to the ratio Ki (ADP) to Km (ATP).
The R0.5 (ADP to ATP) values for the recombinant proteins are 0.79 for Q111D, 0.29 for Q111E, 0.39 for Q111S, and 0.20 for 43-kD RWT, which were calculated from the data shown in Figure 2. Therefore, the Ki(ADP) to Km(ATP) ratio is greater for all the mutant enzymes. However, the greatest change (4-fold) in the response to ADP to ATP ratio occurred with the Q111D enzyme.
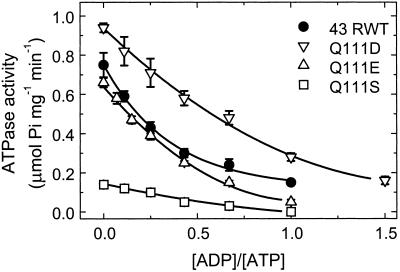
The ATPase activities of the various recombinant Rubisco activase proteins also were measured under varying ADP to ATP ratios. Figure 3 shows that all of the enzymes also exhibited decreased ATPase activity with increasing ADP to ATP ratios. Similar to its Rubisco activation response, the Q111D enzyme exhibited the highest ATPase activity in the absence of ADP and its activity at an ADP:ATP ratio of 1:1 was nearly equal to the activity of the 43-kD RWT enzyme at the lower ratio of 1:3. However, unlike the Rubisco activation results, the ATPase activity of the Q111E enzyme was more similar to the activity of the 43-kD RWT enzyme at all ADP to ATP ratios. The R0.5 (ADP to ATP) calculated from the data shown in Figure 3 are: RWT, 0.26; Q111D, 0.65; Q111E, 0.26; and Q111S, 0.37. These values are similar to those obtained from the Rubisco activation response except that with the ATPase activity, there was no difference in the values for the Q111E and 43-kD RWT proteins.
Figure 3.
Dependence of the ATPase activity on the ratio of ADP to ATP for the 43-kD RWT and Q111D, Q111S, and Q111E mutant enzymes. ATPase activity was measured by a single-step assay measuring the rate of inorganic phosphate formation from ATP. The experiments were conducted at a constant total nucleotide concentration of 4 mm.
Alteration of Nucleotide Binding Measured by Fluorescence and ATPase Activity
Because the mutant proteins exhibited altered responses to ADP and possibly ATP, as measured by the changes in the R0.5 values, we were interested in determining the extent to which the amino acid substitutions in the ATP-binding region (P-loop) had altered the individual affinities for ATP or ADP. A variety of fluorescent methods have been used to analyze nucleotide binding in Rubisco activase (Wang and Portis, 1991; Wang et al., 1993). Values for the apparent dissociation constants (Kd) for ADP and ATP were determined using 1-anilinonapthalene-8-sulfonic acid (ANS) fluorescence quenching as a function of nucleotide concentration (Table II). All of the engineered proteins had an ADP Kd much greater than that of the 43-kD RWT enzyme: 5.2-fold for Q111D, 3.4-fold for Q111E, and 5.8-fold for Q111S. However, all the mutant proteins also had an ATP Kd greater than that of the 43-kD RWT enzyme: 33-fold for Q111D, 7.3-fold for Q111E, and 1.7-fold for Q111S. The ratio of the ADP and ATP Kd values determined by this method do not agree with the values determined by the activity assays (data not shown), particularly in the case of the Q111D enzyme that exhibited a very large increase in the ATP Kd.
Table II.
Apparent nucleotide Kd for recombinant Rubisco activase as measured by various methods
| Enzyme | ANSa
|
Intrinsicb
|
TNP-ATPc
|
ATPased
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Kd ADP | RWT | Kd ATP | RWT | Kd ATP | RWT | Kd | RWT | S0.5 | RWT | |
| μm | % | μm | % | μm | % | μm | % | μm | % | |
| 43-kD RWT | 0.77 | – | 8.8 | – | 10 | – | 0.07 | – | 13 | – |
| Q111D | 4.0 | 520 | 293.0 | 3,300 | 25 | 250 | 0.82 | 1,170 | 43 | 330 |
| Q111E | 2.6 | 340 | 64 | 730 | 4.5 | 45 | 0.10 | 140 | 11 | 80 |
| Q111S | 4.5 | 580 | 15 | 580 | 13 | 130 | 0.08 | 110 | 7 | 50 |
Assay conditions for the fluorescence methods are described in “Materials and Methods.” ATPase activity was measured by the coupled spectrophotometric method.
Calculated errors for all Kd values were less than ±10% except for the Q111D protein, which had an error of ±33%.
Calculated errors for all Kd values were ±10% except for the Q111D protein which had an error of 16%.
Calculated errors for the RWT, Q111D, Q111E, and Q111S proteins were ±: 0.01, 0.68, 0.09, and 0.03, respectively.
Calculated errors for RWT, Q111D, Q111E, and Q111S proteins were ±: 16%, 30%, 23%, and 12%, respectively.
To determine relative affinities for ATP with a different method, we measured changes in intrinsic fluorescence of the various proteins in response to added ATP. Wang et al. (1993) concluded that the change in intrinsic fluorescence is indicative of a change in the aggregation state of Rubisco activase induced by the binding of this nucleotide. Table II shows that the ATP Kd as measured by intrinsic fluorescence was only 2.5-fold greater in the Q111D mutant protein compared with the 43-kD RWT protein. However, the value for the Q111E protein was 0.5-fold less, indicating a greater affinity, and the value for Q111S was 1.3-fold greater than the 43-kD RWT protein. Even excluding the value for Q111D, the relative ATP affinities using this method clearly do not agree well with those obtained with ANS.
The third measurement of ATP affinity for the mutant proteins employed an ATP analog, 2′,3′-O-(2,4,6-trinitrophenyl)-ATP (TNP-ATP), which probes hydrophobic regions in adenine nucleotide-requiring enzymes (Hiratsuka, 1982). TNP-ATP has been used to characterize the nucleotide-binding sites of such proteins as the cystic fibrosis transmembrane regulator (Thomas et al., 1991), the P-glycoprotein multidrug transporter (Liu and Sharom, 1997), and the Escherichia coli F1 ATPase (Weber and Senior, 1997). We used TNP-ATP to measure the relative affinities of the various forms of Rubisco activase for the probe. Table II shows that the Q111D protein has a TNP-ATP Kd nearly 12-fold greater than that of the 43-kD RWT protein, whereas the Q111E:43-kD RWT ratio was 1.4 and the Q111S:43-kD RWT ratio was close to 1.
The relative affinities for ATP can also be estimated from the concentration dependence of the ATPase activity. However, this method is also indirect because the ATPase activity can be influenced by the ability of the protein to aggregate (Wang et al., 1993). ATP hydrolysis has been suggested to arise from an actin-like mechanism involving a dynamic equilibrium between monomers and oligomers, with activation of Rubisco dependent an oligomeric form (Lilley and Portis, 1997). Table II shows the concentration of ATP required for one-half-maximal ATPase activity (S0.5). In this case, the value for the Q111D protein was 3.3-fold higher than that of the 43-kD RWT protein. The Q111E and Q111S proteins had a slightly higher affinity (S0.5 ratios of 0.8 and 0.5, respectively) than the 43-kD RWT protein.
The various fluorescent techniques resulted in widely varying Kd values for the nucleotides with each form of the activase protein. This variation suggests that the intrinsic binding affinities are being obscured by other factors unique to each method. However, the Q111D isoform consistently exhibited the least affinity for the respective nucleotides.
Thermal Stability of the Mutant Proteins
Site-directed mutagenesis of recombinant proteins can result in altered thermal stability as shown by Natarajan and Sierks (1996), Slaby et al. (1996), Bogin et al. (1998), and others. The spinach wild-type protein (Robinson and Portis, 1989) and both of the recombinant spinach Rubisco activase isoforms are thermolabile, particularly in the absence of nucleotides (Crafts-Brandner et al., 1997). Because the mutant Q111D and Q111E proteins exhibited altered nucleotide-binding properties compared with the RWT 43-kD isoform, we examined their thermal stability to ensure that they would be suitable candidates for transformation experiments. We also examined the thermal stability of both the Arabidopsis 43- and 46-kD isoforms because of the differences found between the spinach isoforms (Crafts-Brandner et al., 1997). Table III shows that the Q111D and Q111E mutations did not greatly alter (<2°C) the thermal stability of activase compared with the RWT 43-kD isoform when heat-treated in the absence of nucleotide. Inclusion of either ADP or the ATP analog, adenosine-5′-O-(3-thiotriphosphate) (ATP-γ-S), increased the thermal stability of all recombinant proteins by at least 10°C. However, the Q111D protein, when preincubated at various temperatures in the presence of 2 mm ADP, exhibited a temperature for 50% reduction in activity (T50) that was 3.5°C and 4.5°C less than the Q111E and 43-kD RWT proteins, respectively. The greater thermolability concurs with our previous observations indicating that the Q111D protein does not bind ADP as tightly as the Q111E or 43-kD RWT proteins. Pre-incubation of the proteins at various temperatures in the presence of ATP-γ-S also increased the thermal stability of all the recombinant proteins by at least 10°C (Table III). In this case, both the Q111D and Q111E proteins exhibited slightly greater (2.5°C) stability than the 43-kD RWT protein. However, neither ADP nor ATP-γ-S conferred significantly greater thermal stability to the 46-kD RWT protein as compared with the 43-kD RWT protein. This observation is in marked contrast to the report by Crafts-Brandner et al. (1997), in which ATP-γ-S conferred significant thermal stability to the 45-kD isoform of spinach Rubisco activase, relative to the 41-kD form.
Table III.
Effect of nucleotide on the temperature sensitivity of the ATPase activity of recombinant Rubisco activase
| Enzyme | T50
|
||
|---|---|---|---|
| No nucleotide | 2 mm ADP | 2 mm ATP-γ-S | |
| 43-kD RWT | 31 | 43.5 | 43.5 |
| Q111D | 29.5 | 39 | 46 |
| Q111E | 30 | 42.5 | 46 |
| 46-kD RWT | 32.5 | 44.5 | 43.5 |
Temperature treatments in the presence or absence of nucleotide and the spectrophotometric assay of ATPase activity are described in “Materials and Methods.” The 43- and 46-kD RWT enzymes are the small and large recombinant Arabidopsis isoforms. Q111D and Q111E are the recombinant 43-kD isoforms with Glu and Asp substitutions, respectively, at position 111. T50 is the calculated temperature in °C for 50% reduction in activity after treatment.
DISCUSSION
Rubisco Activation with Engineered Proteins
Our data show that replacement of Gln-111 with Glu in the phosphate-binding loop increases the Rubisco activation activity of recombinant 43-kD Arabidopsis Rubisco activase, confirming the results obtained by Shen and Ogren (1992) with the corresponding isoform of spinach. Replacement with Asp also increased the activation activity, which indicates that an acidic residue at this position is required for the increase in the ability to activate Rubisco. All the other replacements reduced the activation activity, with Ser causing the least reduction.
The P-loop is a ubiquitous structural domain (Kinochita et al., 1999) for binding phosphate-containing substrates. In the many cases where purine nucleotides are involved, nucleotide hydrolysis transmits conformation changes that are central to the function of the protein (Shen et al., 1994) and such a process would be applicable to Rubisco activase. The available three-dimensional structures of nucleotide-binding P-loop proteins consistently show that the two conserved Gly residues (G-X-X-X-X-G-K-S/T) allow backbone hydrogen bonds between the adjacent amino acids and the β- and γ-phosphate groups. The side-chains face away from the nucleotide pocket and interact with the remainder of the protein (Smith and Rayment, 1996). The critical Lys interacts with the γ-phosphate, whereas the hydroxyl of the following Ser or Thr ligates to the divalent cation associated with the bound nucleotide. A wide variety of residues are found in the other positions, and many functional replacements have been found in some proteins (Shen et al., 1994). However, the occurrence of a Gln residue adjacent to the critical GKS/T sequence as found in Rubisco activase is exceptional. A small hydrophobic (Gly, Ala, or Val) or a small polar (Ser or Thr) residue usually occupies this position. Therefore, it is likely that the replacement of the Gln residue with an acidic amino acid in the P-loop results in a major change in the interaction of the loop with the remainder of the protein.
A change in this interaction may explain both the small increase in ATPase activity and the much greater coupling of that activity into a greater ability to activate Rubisco, as seen in the Q111D and Q111E mutant enzymes. Other substitutions at position 111 cause a decrease in Rubisco activation (Fig. 1). The Pro and Arg (data not shown) replacements resulted in minimal activity. Pro may be too inflexible, which inhibits loop movement. Arg, being both large and basic, could disrupt the loop conformation. The bulkier, non-polar residues Leu and Val also caused a substantial decrease in Rubisco activase activity, perhaps due to steric hindrance of P-loop function. The Ser replacement is similar to Gln, being an uncharged polar residue, and the Q111S protein exhibited Rubisco activation activity most similar to the 43-kD RWT protein. Proteins with the smaller, non-polar Ala and Gly substitutions retained significant Rubisco activase activity, but these residues may not provide the sufficient polar or negative charge that seems to be required for wild-type or greater activity.
Shen and Ogren (1992) found that the Q111E substitution in the larger, 45-kD isoform of spinach activase did not increase its activation activity, but its ATPase activity was reduced. The larger isoform is much more sensitive to inhibition by ADP (Zhang and Portis, 1999), and the reduction of a disulfide in the longer carboxy-terminal region relieves the inhibition, suggesting that the oxidized region interacts with the nucleotide-binding site in a manner different from the reduced form. This interaction may dominate the effects of amino acid changes at Q111, but we have not examined the effects of similar changes in the larger isoform in Arabidopsis. Because there is no three-dimensional structure for Rubisco activase, it is difficult to make more specific conclusions or interpretations about why the Q111E and Q111D enzymes have altered activity based on the structure of the nucleotide-binding site.
The Q111E enzyme has a naturally occurring counterpart in the form of the P-loop sequence reported for a bacterial Rubisco activase from Anabaena (Li et al., 1993). A functional activase seems to be physiologically important, but it is unclear whether or not activase plays a regulatory role in this organism. (Li et al., 1999).
Rubisco Activation at Various ADP to ATP Ratios
The increased activation activity of the Q111D and Q111E proteins was maintained at ADP to ATP ratios estimated to occur in the stroma in the light. However, the R0.5 (ADP to ATP) of the Q111E protein was not very different from that of the RWT enzyme. Thus, this protein might still be effectively down-regulated at ADP to ATP ratios observed in the stroma in the dark. In contrast, the greatly altered ADP to ATP response of the Q111D protein resulted in the maintenance of very high activity even at the dark stromal ratio. The maintenance of a significant ATPase activity in the dark wastes energy when Rubisco activation is not needed, and this dark activity could be detrimental in planta. The Rubisco activation activity of the Q111S protein was slightly reduced in the absence of ADP, but was less inhibited by ADP and thus was the most similar to the 43-kD RWT protein at physiological ADP to ATP ratios.
Shen and Ogren (1992) suggested that altering the stoichiometry of Rubisco activase to ATPase activities may result in a more energy-efficient regulation of Rubisco activity. We report that the Q111E substitution in Arabidopsis Rubisco activase produces increased activation efficiency compared to the 43-kD RWT enzyme. Shen and Ogren (1992) have reported similar results for the same mutation in spinach. These authors did not make the Q109D mutation in spinach. Our Q111D enzyme was 1.4-fold more efficient than the 43-kD RWT enzyme, but less efficient than the Q111E enzyme (1.9-fold increase). However, these efficiencies are not calculated at physiological nucleotide ratios, which may be more important in terms of finding mutants that may lead to more efficient activation in planta. Comparing Figures 1 and 2, the activation:ATPase ratios of both the Q111D and Q111E enzymes are both about 2.5-fold greater than the 43-kD RWT enzyme at 1:3 and 1:1 ratios of ADP:ATP. The Q111D mutant enzyme exhibited a high level of ATPase activity under dark levels of ADP to ATP compared with all other isoforms tested. The Q111E enzyme, on the other hand, exhibited both increased activation efficiency and less ATPase activity at the 1:1 ADP:ATP ratio compared with the RWT enzyme.
We can only speculate at this point that the increased activation activity of these mutants may have some benefit in planta because antisense Arabidopsis plants with 30% to 40% of wild-type levels of Rubisco activase had significant reductions in growth under ideal conditions (Eckardt et al., 1997). An increased activation efficiency may also have some impact in planta, and the altered ADP to ATP regulation of the Q111D enzyme would seem to have considerable potential to alter the phenotype of mutant plants in some manner. The recently discovered role of the 46-kD isoform in the regulation of activase (Zhang and Portis, 1999) also needs to be explored in vivo. In any case, it is clear that the contrasting activation activities and responses to ADP to ATP ratio of these proteins provide an excellent opportunity to investigate the role and significance of ADP to ATP regulation in the regulation of Rubisco by placing these proteins in the rca− Arabidopsis mutant.
Altered Apparent Dissociation Constants for ADP and ATP
The greatly altered ADP to ATP response of the Q111D enzyme indicates that the relative affinity for ADP is reduced more than it is for ATP. We expected that by using ANS fluorescence, it would be possible to resolve the observed R0.5 (ADP to ATP) for each protein to the alteration in the binding of each nucleotide. However, the ANS method indicated that the affinity for ATP is more diminished than that for ADP in the Q111D and Q111E proteins, which clearly disagrees with the observed ADP to ATP responses. Some improvement may have occurred if additional experiments were performed in the presence of Mg2+ (Wang and Portis, 1991). However, the 33-fold increase in the ATP Kd of the Q111D mutant enzyme compared with that of the RWT enzyme, suggests that the method may not provide valid comparisons for the various mutants. In hindsight, this is reasonable because the method actually measures alteration in the binding of ANS to hydrophobic sites accessible to the probe as a result of nucleotide binding. The alterations in the P-loop, which may be communicating the status of the nucleotide-binding site to remote areas of the protein, and the unknown relationships between aggregation of the activase and ANS binding, clearly complicate the application of this method.
The relative Kd indicated by intrinsic fluorescence, TNP-ATP, and the ATPase activity were generally more consistent for each enzyme. The intrinsic fluorescence and ATPase methods indicated a 2.5- to 3.3-fold increase for the Q111D enzyme and a 0.45- to 0.8-fold increase for the Q111E enzyme, but with the Q111S enzyme, one method indicated an increased affinity and the other a decreased affinity. A much larger decrease in the affinity of the nucleotide-binding site in the Q111D mutant enzyme was also indicated with TNP-ATP, even though this analog binds much tighter than either ATP or ADP. We attribute the inconsistencies with these methods to the fact that they do not directly measure nucleotide binding, and to the possibility that these mutant enzymes not only differ in affinity for nucleotides, but also in the transmission of binding to other parts of the protein. Thus, the degree to which changes in the binding of each nucleotide contributes to the altered responses to ADP to ATP ratio may be difficult to resolve unless methods to directly measure the binding of ATP and ADP during the activation of Rubisco are employed in future studies.
MATERIALS AND METHODS
PCR Site-Directed Mutagenesis
Templates used for PCR were cDNA sequences of the 43-kD isoform of Arabidopsis Rubisco activase (Werneke et al., 1989). PCRs using four bottom-strand PCR primers, 5′-TGACGAGCTCACACTGGAAGGATTTACC(GNA or GNG or ANC or GNT) ACCTTTGCCTCCCC-3′ (where N was either, A, G, C, or T), were used to modify the codon that codes for the amino acid at position 111 (italicized trimers) and to generate a SacI site (underlined hexamer) for linking the two fragments down stream of the P-loop. An NcoI site for initiation of translation was created at the 5′ end of the coding region of the cDNA. An additional PCR using 3′ and 5′ mutagenic primers created a BamHI site at the 3′ end of the cDNA for directed cloning and a SstI site to link the front part of the complete clone. The large size (45-mer) of the PCR primers resulted in a sufficiently high melting temperature to permit a two-step PCR reaction. The time and temperature for the two steps were 1 min at 72°C and 1 min at 92°C for 35 cycles. The reaction conditions were 10 mm KCl, 10 mm (NH4)2SO4, 20 mm Tris [Tris(hydroxymethyl)-aminomethane]-HCl (pH 8.8 at 25°C), 2 mm MgSO4, 200 μm each dNTP, 100 ng of DNA template, and 100 pmol of primer, and 1 unit of Vent Polymerase3 (New England Biolabs, Beverly, MA) in a final volume of 100 μL.
Cloning and Identification of PCR Site-Directed Mutagenesis Products
PCR products were identified on 1% (w/v) agarose by size comparison to a 1-kb ladder (Bethesda Research Labs, Gaithersburg, MD) and diagnostic restriction digest of the engineered SacI site. The PCR products were excised from the gel, purified, and ligated into expression vector pTrc99a (Pharmacia Biotech, Piscataway, NJ) for expression and cloning. Products of the ligation reaction were transformed into Escherichia coli cloning vector JM101 (Stratagene, La Jolla, CA) and selected on 200 mg L−1 carbenicillin Luria Bertani plates. Single colonies containing putative cloned PCR site-directed mutagenesis products were grown overnight in liquid Luria Bertani containing 200 mg L−1 carbenicillin. Plasmid DNA was extracted from the overnight cultures by the alkaline lysis method (Maniatis et al., 1982) and the randomly introduced nucleotide changes were identified by dideoxy sequencing (Sanger et al., 1977). Clones were analyzed until lines containing either acidic, basic, aromatic, polar, or sterically large or small amino acids were identified.
Expression and Purification of Recombinant Rubisco Activase
The pTrc99a expression vectors containing the engineered Arabidopsis 43-kD Rubisco activase cDNA were transformed into BL21(DE3) (Novagen, Madison, WI). Transformed cells were grown at 37°C to an A600 of 0.8, at which time Rubisco activase expression was induced with 1 mm isopropyl β-d-thiogalactoside. Cells were incubated for an additional 4 h, chilled on ice, and centrifuged. The pellet was resuspended in 10 to 15 mL of 20 mm 1,3-bis [Tris(hydroxymethyl) methylamino] propane (pH 7.2), containing 1 mg mL−1 lysozyme and 1 mm phenylmethylsulfonyl fluoride, and incubated with moderate stirring for 30 min at 40°C. The resulting viscous mixture was sonicated on ice for 1 min with a Branson Sonifier 450 (Branson Ultrasonics, Danbury, CT). The lysed cells were centrifuged at 27,000g for 45 min at 4°C. The supernatant was decanted into a chilled graduated cylinder and solid (NH4)2SO4 was added to 35% saturation and stirred for 1 h at 4°C. The mixture was centrifuged for 20 min at 17,000g at 4°C. The subsequent pellet was gently resuspended in 1 to 2 mL of 20 mm 1,3-bis [Tris(hydroxymethyl) methylamino] propane (pH 7.2) and 0.2 mm ATP, quickly frozen in liquid nitrogen, and stored at −80°C. Prior to further purification by ion-exchange chromatography, the sample was thawed and passed over a Sephadex G-25 desalting column (Sigma, St. Louis). The collected sample was loaded onto a 25-mL Q-Sepharose column and eluted with a 0 to 400 mm NaCl gradient at a flow rate of 1 mL min−1. Eluate fractions between 220 and 300 mm NaCl were analyzed for recombinant Rubisco activase by immunoblot analysis and PAGE. Fractions containing Rubisco activase that were at least 90% pure were concentrated to 3 to 5 mg mL−1 in a Centriprep-10 concentrator (Amicon, Beverly, MA), frozen in liquid nitrogen, and stored at −80°C.
Biochemical Assays of Rubisco Activase Activity
In the absence of ADP, Rubisco activation by the various recombinant Rubisco activase proteins was measured with the two-step radiometric assay described by Shen et al. (1991). The reaction mixture contained 1 mm ATP, 0.5 mg mL−1 of Arabidopsis Rubisco-RuBP complex, and 0.1 mg mL−1 recombinant Arabidopsis Rubisco activase. Phosphoenolpyruvate and pyruvate kinase were replaced by 2 mm phosphocreatine and 20 units mL−1 phosphocreatine kinase. In the presence of ADP, the single step Rubisco activity assay described by Zhang and Portis (1999) was used. Activity was determined at the various ADP to ATP ratios indicated while maintaining a total nucleotide concentration of 4 mm.
The ATPase activity of Rubisco activase at various ADP to ATP ratios was assayed by measuring the formation of inorganic phosphate from ATP as described by Zhang and Portis (1999). The total nucleotide concentration was maintained at 4 mm and the protein concentration was 50 μg mL−1.
In some experiments, activation of the inactive Rubisco-RuBP complex by recombinant Rubisco activase was measured by following 3-phosphoglyceric acid production in a coupled spectrophotometric assay as described by Larson et al. (1997) with the following modifications: The assay mixture contained 100 mm Tricine (N-tris[hydroxymethyl]-methyl-Gly; pH 8.0), 10 mm MgCl2, 3 mm ATP, 0.3 mm NADH, 4 mm RuBP, 10 mm sodium bicarbonate, 125 μg mL−1 Arabidopsis Rubisco-RuBP complex, 1 mm phosphoenolpyruvate, 30 units mL−1 pyruvate kinase, 20 units mL−1 glycerate 3-phosphate kinase, 15 units mL−1 glyceraldehyde 3-phosphate dehydrogenase, 150 units mL−1 triose phosphate isomerase, and 15 units mL−1 glycerol phosphate dehydrogenase. The reaction was initiated by the addition of 25 μg mL−1 recombinant Rubisco activase. The specific activation activity of Rubisco activase is the rate of increase of Rubisco activity per milligram of activase protein. These rates are expressed as micromoles CO2 fixed per minute per milligram Rubisco per minute per milligram activase as used by Shen and Ogren (1992). Alternatively, the activation rate can be expressed in terms of the carbamylation of Rubisco (Larson et al., 1997) by dividing this unit by the maximal specific activity of the Rubisco used in the experiment, which averaged 2 μmol CO2 min−1 mg−1. The specific activation activity of Rubisco activase expressed in this manner has units of moles carbamylation CO2 per mole Rubisco site per minute per milligram activase.
The ATPase activity of recombinant Rubisco activase in the absence of ADP was measured by a spectrophotometric assay modified from Robinson and Portis (1989). The assay mixture contained 50 mm Tricine (pH 8.0), 20 mm KCl, 10 mm MgCl2, 0 to 2 mm ATP, 1 mm phosphoenolpyruvate, 0.3 mm NADH, 40 units mL−1 pyruvate kinase, and 40 units mL−1 lactate dehydrogenase in a total volume of 0.5 mL. The reaction was initiated by the addition of Rubisco activase to a final concentration of 25 μg mL−1.
Fluorescence Measurements
Displacement of ANS bound to Rubisco activase by the addition of ADP or ATP was determined according to Wang and Portis (1991) to obtain an estimate of nucleotide affinity. The assay buffer consisted of 50 mm Tricine (pH 8.0), 0.1 mm EDTA, 2 mm dithiothreitol (DTT), 40 μm ANS, and 25 μg mL−1 Rubisco activase. Samples were equilibrated at 4°C for 1 to 2 h. ADP or ATP was added and the change in fluorescence (ΔF) measured. Binding constants (Kd) were determined by non-linear regression analysis with the equation ΔF = ΔFmax − Kd(ΔF/[L]) where L is nucleotide concentration (Fersht, 1977) using the Origin scientific graphing software package (MicroCal, Northampton, MA).
Intrinsic fluorescence was measured according to Wang et al. (1993). The assay buffer contained 50 mm Tricine-NaOH (pH 8.0), 5 mm MgCl2, 20 mm KCl, 40 μg mL−1 Rubisco activase, and 1 mm phosphoenolpyruvate, and 10 units mL−1 pyruvate kinase. ATP (0–200 μm) was added 5 min after the addition of all other assay components. Kd were determined by non-linear regression fitting to the Hill equation as described by Wang et al. (1993).
The binding of TNP-ATP was measured by the change in its fluorescence as follows. The concentration of TNP-ATP was determined from the extinction coefficient of 26,400 m−1 cm−1 at 408 nm as described by Hiratsuka and Uchida (1973). The excitation and emission wavelengths were 408 and 540 nm, respectively. The excitation and emission optical bandwidths were 3 and 5 nm, respectively. The assay buffer contained 50 mm Tricine-NaOH (pH 8.0), 0.1 mm EDTA, 2 mm DTT, and 25 μg mL−1 Rubisco activase. TNP-ATP (Molecular Probes, Eugene, OR) was added 5 min after the addition of all other assay components. Background fluorescence due to the probe (all assay components present except Rubisco activase) was subtracted from the change in the probe's fluorescence caused by the addition of Rubisco activase. Kd were determined by non-linear regression analysis using the equation y = [−b − (b2 − 4ac)1/2]/2a, where a = Em (enzyme concentration), b = −Fm(x + Em + Kd), c = (Fm)2x, x = the total probe concentration, y = measured fluorescence, and Fm = maximal fluorescence. This equation accounts for the binding of the probe to the enzyme when the enzyme concentration is not insignificant relative to that of the probe.
Temperature Stability of Rubisco Activase
The thermal stability of Rubisco activase was determined following the procedure described by Crafts-Brandner et al. (1997). The recombinant Rubisco activase proteins (1 mg mL−1) were preincubated for 15 min in 0.2 mL 50 mm Tricine (pH 8.0), 5 mm MgCl2, and either 2 mm ADP, 2 mm ATP-γ-S, or no nucleotide at a given temperature. ATP hydrolysis was initiated by adding 35 μL of the heat-treated Rubisco activase to 0.7 mL of 50 mm Tricine (pH 8.0), 5 mm MgCl2, 2 mm DTT, 0.3 mm NADH, 20 mm KCl, 2 mm ATP, and 1 mm phosphoenolpuruvate. ATPase activity was followed by measuring the oxidation of NADH at 340 nm in an 8452A Diode Array spectrophotometer (Hewlett Packard, Palo Alto, CA). The amount of either ADP or ATP-γ-S carried over from the pre-incubation buffer to the assay buffer was 0.10 mm nucleotide. Temperature for 50% reduction in activity (T50) was determined by interpolation of the linear portion of the temperature response data immediately above and below 50% inhibition.
ACKNOWLEDGMENT
R.P.K. thanks Dr. W.L. Ogren for support and advice throughout a large portion of his thesis research, part of which is included in this report.
Footnotes
This work was supported in part by the Triagency Program for Collaborative Research in Plant Biology (grant no. DOE 92ER20095) and by the U.S. Department of Energy (grant no. 97ER20268).
Names are necessary to report factually on available data; however, the U.S. Department of Agriculture neither guarantees nor warrants the standard of the product, and the use of the name by U.S. Department of Agriculture implies no approval of the product to the exclusion of others that may also be suitable.
LITERATURE CITED
- Bogin O, Peretz M, Hacham Y, Korkhin Y, Frolow F, Kalb(Gilboa) AJ, Burstein Y. Enhanced thermal stability of Clostridium beijerinckii alcohol dehydrogenase after strategic substitution of amino acid residues with prolines from the homologous thermophilic Thermoanaerobacter brockii alcohol dehydrogenase. Protein Sci. 1998;7:1156–1163. doi: 10.1002/pro.5560070509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crafts-Brandner SJ, van de Loo FJ, Salvucci ME. The two forms of ribulose-1,5-bisphosphate carboxylase-oxygenase activase differ in sensitivity to elevated temperature. Plant Physiol. 1997;114:439–444. doi: 10.1104/pp.114.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt NA, Snyder GW, Portis AR, Jr, Ogren WL. Growth and photosynthesis under high and low irradiance of Arabidopsis thaliana antisense mutants with reduced ribulose-1,5-bisphosphate carboxylase/oxygenase activase content. Plant Physiol. 1997;113:575–586. doi: 10.1104/pp.113.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. Enzyme Structure and Mechanism. W.H. New York: Freeman; 1977. [Google Scholar]
- Hammond ET, Andrews TJ, Woodrow IE. Regulation of ribulose-1,5-bisphosphate carboxylase/oxygenase by carbamylation and 2-carboxyarabinitol 1-phosphate in tobacco: insights from studies of antisense plants containing reduced amounts of Rubisco activase. Plant Physiol. 1998;118:1463–1471. doi: 10.1104/pp.118.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka T. Biological activities and spectroscopic properties of chromophoric and fluorescent analogs of adenine nucleoside and nucleotides, 2′,3′-O-(2,4,6-trinitrocyclohexadienylidene) adenosine derivatives. Biochim Biophys Acta. 1982;719:509–517. doi: 10.1016/0304-4165(82)90240-9. [DOI] [PubMed] [Google Scholar]
- Hiratsuka T, Uchida K. Preparation and properties of 2′(or 3′)-O-(2,4,6-trinitrophenyl) adenosine 5′-triphosphate, an analog of adenesine triphosphate. Biochem Biophys Acta. 1973;320:635–647. doi: 10.1016/0304-4165(73)90143-8. [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Sadanami K, Kidera A, Go N. Structural motif of phosphate-binding site common to various protein superfamilies: all-against-all structural comparison of protein-mononucleotide complexes. Protein Eng. 1999;12:11–14. doi: 10.1093/protein/12.1.11. [DOI] [PubMed] [Google Scholar]
- Larson EM, O'Brien CM, Zhu G, Spreitzer RJ, Portis AR., Jr Specificity for activase is changed by a Pro-89 to Arg substitution in the large subunit of ribulose-1,5-bisphosphate carboxylase-oxygenase. J Biol Chem. 1997;272:17033–17037. doi: 10.1074/jbc.272.27.17033. [DOI] [PubMed] [Google Scholar]
- Li LA, Gibson JL, Tabita FR. The Rubisco activase (rca) gene is located downstream from rbcS in Anabaena sp. strain CA and is detected in other Anabaena/Nostoc strains. Plant Mol Biol. 1993;21:753–764. doi: 10.1007/BF00027109. [DOI] [PubMed] [Google Scholar]
- Li LA, Zianni MR, Tabita FR. Inactivation of the monocistronic rca gene in Anabaena variabilis suggests a physiological ribulose bisphosphate carboxylase oxygenase activase-like function in heterocystous cyanobacteria. Plant Mol Biol. 1999;40:467–478. doi: 10.1023/a:1006251808625. [DOI] [PubMed] [Google Scholar]
- Lilley RMcC, Portis AR., Jr ATP hydrolysis activity and polymerization state of ribulose-1,5-bisphosphate carboxylase oxygenase activase. Plant Physiol. 1997;114:605–613. doi: 10.1104/pp.114.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Sharom FJ. Fluorescence studies on the nucleotide binding domains of the P-glycoprotein multidrug transporter. Biochemistry. 1997;36:2836–2843. doi: 10.1021/bi9627119. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- Natarajan S, Sierks MR. Functional and structural roles of highly conserved Trp120 loop region of glucoamylase from Aspergillus awamori. Biochemistry. 1996;35:3050–3058. doi: 10.1021/bi952458x. [DOI] [PubMed] [Google Scholar]
- Robinson SP, Portis AR., Jr Release of the nocturnal inhibitor, carboxyarabinitol-1-phosphate from ribulose bisphosphate carboxylase/oxygenase by Rubisco activase. FEBS Lett. 1988;233:413–416. [Google Scholar]
- Robinson SP, Portis AR., Jr Adenosine triphosphate hydrolysis by purified Rubisco activase. Arch Biochem Biophys. 1989;268:93–99. doi: 10.1016/0003-9861(89)90568-7. [DOI] [PubMed] [Google Scholar]
- Salvucci ME, Ogren WL. The mechanism of Rubisco activase: insights from studies of the properties and structure of the enzyme. Photosynth Res. 1996;47:1–11. doi: 10.1007/BF00017748. [DOI] [PubMed] [Google Scholar]
- Salvucci ME, Portis AR, Jr, Ogren WL. A soluble chloroplast protein catalyzes ribulosebisphosphate carboxylase/oxygenase activation in vivo. Photosynth Res. 1985;7:193–201. doi: 10.1007/BF00037012. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;80:3666–3670. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Yai B, Mueller DM. Primary structural constraints of P-loop mitochondrial F1-ATPase from yeast. J Biol Chem. 1994;269:9424–9428. [PubMed] [Google Scholar]
- Shen JB, Ogren WL. Alteration of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase activase activities by site-directed mutagenesis. Plant Physiol. 1992;99:1201–1207. doi: 10.1104/pp.99.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JB, Orozco EM, Jr, Ogren WL. Expression of the two isoforms of spinach ribulose-1,5-bisphosphate carboxylase activase and essentiality of the conserved lysine in the consensus nucleotide-binding domain. J Biol Chem. 1991;266:8963–8968. [PubMed] [Google Scholar]
- Slaby I, Cerna V, Jeng M-F, Dyson HJ, Holmgren A. Replacement of Trp28 in Escherichia coli thioredoxin by site-directed mutagenesis affects thermodynamic stability but not function. J Biol Chem. 1996;271:3091–3096. doi: 10.1074/jbc.271.6.3091. [DOI] [PubMed] [Google Scholar]
- Smith CA, Rayment I. Active site comparisons highlight structural similarities between myosin and other P-loop proteins. Biophys J. 1996;70:1590–1602. doi: 10.1016/S0006-3495(96)79745-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer RJ. Questions about the complexity of chloroplast ribulose-1,5-bisphosphate carboxylase/oxygenase. Photosynth Res. 1999;60:29–42. [Google Scholar]
- Stitt M, Lilley RM, Heldt HW. Adenine nucleotide levels in the cytosol, chloroplasts, and mitochondria of wheat leaf protoplasts. Plant Physiol. 1982;70:971–977. doi: 10.1104/pp.70.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PJ, Shenbagamurthi P, Ysern X, Pedersen PL. Cystic fibrosis transmembrane conductance regulator: nucleotide binding to a synthetic peptide. Science. 1991;251:555–557. doi: 10.1126/science.1703660. [DOI] [PubMed] [Google Scholar]
- Traut TW. The functions and consensus motifs of nine types of peptide segments that form different types of nucleotide-binding sites. Eur J Biochem. 1994;222:9–19. doi: 10.1111/j.1432-1033.1994.tb18835.x. [DOI] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases, and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Portis AR., Jr A fluorometric study with 1-anilinonaphthalene-8-sulfonic acid (ANS) of the interactions of ATP and ADP with Rubisco activase. Biochim Biophys Acta. 1991;1079:263–267. doi: 10.1016/0167-4838(91)90067-a. [DOI] [PubMed] [Google Scholar]
- Wang Z-Y, Portis AR., Jr Dissociation of ribulose-1,5-bisphosphate bound to ribulose-1,5-bisphosphate carboxylase/oxygenase and its enhancement by ribulose-1,5-bisphosphate carboxylase/oxygenase activase-mediated hydrolysis of ATP. Plant Physiol. 1992;99:1348–1353. doi: 10.1104/pp.99.4.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Ramage RT, Portis AR., Jr Mg2+ and ATP or adenosine-5′-[γ-thio]-triphosphate (ATPγS) enhances intrinsic fluorescence and induces aggregation, which increases the activity of spinach Rubisco activase. Biochim Biophys Acta. 1993;1202:247–255. doi: 10.1016/0167-4838(93)90061-u. [DOI] [PubMed] [Google Scholar]
- Wang Z-Y, Snyder GW, Esau BD, Portis AR, Jr, Ogren WL. Species-dependent variation in the interaction of substrate-bound ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) and Rubisco activase. Plant Physiol. 1992;100:1858–1862. doi: 10.1104/pp.100.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J, Senior AE. Binding and hydrolysis of TNP-ATP by Escherichia coli F1-ATPase. J Biol Chem. 1997;271:3474–3477. doi: 10.1074/jbc.271.7.3474. [DOI] [PubMed] [Google Scholar]
- Werneke JM, Chatfield JM, Ogren WL. Alternative mRNA splicing generates the two ribulosebisphosphate carboxylase/oxygenase activase polypeptides in spinach and Arabidopsis. Plant Cell. 1989;1:815–826. doi: 10.1105/tpc.1.8.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Portis AR., Jr Mechanism of light regulation of Rubisco: a specific role for the larger Rubisco activase isoform involving reductive activation by thioredoxin-f. Proc Natl Acad Sci USA. 1999;96:9438–9443. doi: 10.1073/pnas.96.16.9438. [DOI] [PMC free article] [PubMed] [Google Scholar]