Abstract
Background and Aims: It has been widely accepted that sperm hyperactivation is regulated by protein phosphorylations. But, the sperm hyperactivation phosphorylation pathway is not well understood yet because several different proteins have been detected in other studies. In order to understand the phosphorylation pathway that regulates hyperactivation, we established how to extract sperm protein completely and detected proteins that were phosphorylated during hyperactivation.
Methods: Protein phosphorylation of hamster spermatozoa was detected by western blotting using antiphospho‐amino acid monoclonal antibodies or the SELDI ProteinChip system with IMAC‐Ga(III).
Results: We detected 75 protein/peptide phosphoryations using the method established in the present study. Tyrosine phosphorylations occurred during hyperactivation. Serine or threonine phosphorylations occurred for 30 min. Furthermore, four of the serine or threonine phosphorations were phosphorylated by A‐kinase. As for peptides, 15 peptides were dephosphorylated for 30 min. Other peptides were phosphorylated during hyperactivation.
Conclusions: Because most of the proteins detected in the present study have been described previously, we could detect comprehensive protein phosphorylations. Moreover, we also detected many novel phosphopeptides. Although we did not understand the role of peptide, it was likely that motility was basically regulated by serine/threonine phosphorylations and hyperactivation was mainly regulated by tyrosine phosphorylations. (Reprod Med Biol 2006; 5: 123–135)
Keywords: dephosphorylation, hyperactivation, phosphorylation, SELDI ProteinChip, spermatozoa
INTRODUCTION
EJACULATED MAMMALIAN SPERMATOZOA cannot fertilize the oocyte. After many physiological and molecular changes, spermatozoa are capacitated and hyperactivated, and are able to fertilize the oocyte. 1 Capacitated spermatozoa show the acrosome reaction. After capacitation, moreover, spermatozoa show hyperactivation. 1 Hyperactivation of motility exhibits large bend amplitude, whiplash and frenzied flagellar movements, and is regulated in vitro by the addition of albumin, bicarbonate, calcium and so on. Albumin removes cholesterol from the sperm plasma membrane. 2 Bicarbonate increases intracellular pH and stimulates adenylate cyclase. 3 , 4 , 5 Calcium is involved in intracellular signal transduction, such as regulation of adenylate cyclise, and phosphodiesterase and protein phosphorylation. 6 , 7 , 8 , 9
Recent understanding is that sperm hyperactivation is closely associated with protein phosphorylations. 1 , 6 , 7 So, it has been reported that serine, threonine and tyrosine phosphorylation are associated with capacitation and hyperactivation; for example, 175‐, 93‐, 44‐, 40‐, 38‐ and 20‐kDa proteins in boar spermatozoa, 10 120‐, 115‐, 90‐, 80‐, 62‐, 50/48‐, 40‐, 16‐ and 10‐kDa proteins in hamster spermatozoa, 11 , 12 , 13 , 14 , 15 and 190‐, 110‐, 94‐, 43–55‐, 35‐ and 18‐kDa proteins in human spermatozoa. 16 It has been accepted that tyrosine phosphorylation of A‐kinase anchoring protein (AKAP), which corresponds to tyrosine phosphorylation of 80‐kDa protein, is closely associated with sperm capacitation and hyperactivation. 11 , 13 , 17 , 18 Phosphorylation of AKAP is regulated by a cAMP‐dependent pathway, 19 calcium/calmodulin dependent pathway 17 and/or protein phosphatase. 11 Sperm motility and hyperactivation are also regulated by cAMP, calcium and protein phosphatase. 20 It is likely, therefore, that phosphorylation of AKAP closely associates with regulatory mechanisms of sperm hyperactivation. In contrast, it has been suggested that serine and threonine phosphorylation is also associated with sperm capacitation and hyperactivation. In hamster 14 , 15 , 21 and human spermatozoa, 16 it has been reported that serine and threonine phosphorylation occurs during capacitation and hyperactivation. Although there are many reports about the relationships between protein phosphorylations, capacitation and hyperactivation, the protein phosphorylation cascade that regulates sperm capacitation and hyperactivation is not well understood yet.
The authors previously investigated nine flagellar proteins, 120‐, 115‐, 90‐, 80‐, 62‐, 50/48‐, 40‐, 16‐ and 10‐kDa protein, that were phosphorylated on tyrosine residues during hyperactivation through activation on hamster spermatozoa. 12 It was shown that phosphorylation of 80‐kDa protein, which was supposed to be AKAP, was regulated by serine/threonine protein phosphatase. 11 As for serine phosphorylation, we also showed that seven flagellar proteins, 90‐, 66‐, 58‐, two 38/36‐, 32‐ and 10‐kDa protein, were phosphorylated during hyperactivation through activation. 15 , 21 When hamster spermatozoa began to move, four of the seven flagellar proteins, such as 66‐, 58‐ and two 38/36‐kDa proteins, were phosphorylated and two 38/36‐kDa proteins were phosphorylated in a cAMP dependent manner. 21 , 22 Four proteins were identified as tubulin, 23 ATP synthase F1β 24 and two types of pyruvate dehydrogenase E1β. 22 , 25 As for threonine phosphorylation, we suggested that 90‐, 65‐, 35‐ and 10‐kDa protein were phosphorylated and 70‐kDa protein was dephosphorylated during hyperactivation. 15 Furthermore, the 10‐kDa protein was multiphosphorylated on tyrosine, serine and/or threonine residues during hyperactivation through activation and had sequences like carcinustatin, which is a neuropeptide isolated from an insect. 12 , 15
Jha et al. also studied protein phosphorylation associated with sperm capacitation and hyperactivation using hamster spermatozoa. 13 , 14 They detected five tyrosine phosphorylations which were 83‐, 72‐, 56‐, 53‐ and 50‐kDa; four serine/threonine phosphorylations which were 100‐, 66‐, 63‐ and 56‐kDa; and five threonine phosphorylations which were 61‐, 53‐, 45‐, 39‐ and 32‐kDa. They showed that 83 k‐Da protein was AKAP. 26
Several differences in phosphorylation were found between Okuno and Fujinoki's studies, 8 , 11 , 12 , 15 , 21 and Shivaji's studies 13 , 14 because different methods were used in extracting protein. Although sperm proteins were dissolved in SDS buffer in many cases, it is difficult to extract stable sperm proteins because of the viscosity derived from DNA. When 7 mol urea solution was used in order to extract proteins from salmon spermatozoa, viscosity derived from DNA was not found and the sperm structure, except the head, were completely dissolved. 27 Thus, it is likely that using urea is better than using SDS buffer to extract proteins from spermatozoa.
In the present study, we tried to establish a method to completely extract hamster sperm proteins. In the next step, we comprehensively detected proteins phosphorylated during hyperactivation in order to understand the phosphorylation cascade regulated by hyperactivation.
MATERIALS AND METHODS
Reagents
ANTIPHOSPHOSERINE MONOCLONAL ANTIBODY (PSR‐45), antiphosphothreonine monoclonal antibody (PTR‐8) and antiphosphotyrosine monoclonal antibody (PT‐66) were purchased from Sigma Chemical Company (St Louis, MO, USA). Phospho‐(Ser/Thr)PKA substrate antibody was purchased from Cell Signaling Technology (Beverly, MA, USA). Polyvinylidene difluoride (PVDF) membrane was purchased from Millipore (Bedford, MA, USA). Enhanced chemiluminescence (ECL) plus kit was purchased from Amersham Biosciences (Piscataway, NJ, USA). Immobilized metal affinity capture (IMAC) chip was purchased from Ciphergen Biosystems (Fremont, CA, USA). α‐cyano‐4‐hydroxycinnamic acid (CHCA) was purchased from Aldrich Chem (Milwaukee, WI, USA). Other chemicals were of reagent grade from Wako Pure Chemical Industries (Osaka, Japan).
Animals and preparation of hyperactivated spermatozoa
Sexually mature male golden hamsters (Mesocricetus auratus) were used as experimental animals. Spermatozoa were obtained from the caudal epididymis.
Hyperactivated spermatozoa were prepared according to the method described previously 21 with modified Tyrode's albumin lactate pyruvate (mTALP) medium. An aliquot of caudal epididymal spermatozoa was placed at the bottom of a test tube. Several ml of the mTALP medium was then carefully added and overlayed with mineral oil. They were incubated for 10 min at 37°C to allow spermatozoa to swim up. The supernatant containing motile spermatozoa was collected, placed on the culture dish, covered with mineral oil and incubated for 3 h at 37°C under 5% CO2 in air to accomplish hyperactivation. After the incubation, spermatozoa were collected. Spermatozoa exhibiting 80% over motility were used in the experiment.
Motility and hyperactivation measurement of spermatozoa
Motility and hyperactivation measurement were carried out according to the method described in our previous study. 15 Hamster spermatozoa suspended in the mTALP medium were diluted to 10‐fold of the medium and placed on a glass slide coated with BSA. Sperm motility and hyperactivation were recorded on a videotape by a CCD camera (Progressive 3CCD, Sony, Tokyo, Japan) attached to a microscope (IX70, Olympus, Tokyo, Japan) with phase contrast illumination and a warm stage (MP10Pm, Kitazato Supply, Shizuoka, Japan). Each observation was carried out at 37°C, recorded for 5 min and analyzed by counting motile spermatozoa or hyperactivated spermatozoa in 30 different fields.
Preparation of sperm protein extracts
Sperm proteins were extracted using the following method. At first, spermatozoa were suspended at 30 mg/mL in the urea solution containing 7 mol/L urea and 10% 2‐ME (2‐mercaptoethanol). After pipetting, the suspension was incubated on ice for 10 min. As shown in Figure 1a, sperm structures except sperm heads and fibers were dissolved. After centrifugation at 15 000 × g for 10 min at 4°C, the supernatants were used as the urea extracts. The precipitate was resuspended in the same volume of the urea‐thiourea solution containing 5 mol/L urea, 1 mol/L thiourea, 10% 2‐ME and 2% nonidet P‐40 (NP‐40) as the urea solution. After pipetting, the resuspension was incubated on ice for 10 min. As shown in Figure 1b, fibers were dissolved. From observation under electron microscopy (Fig. 1c), only nuclei were not dissolved. After centrifugation at 15 000 × g for 10 min at 4°C to remove sperm nuclei, the supernatant was used as the urea‐thiourea extracts.
Figure 1.
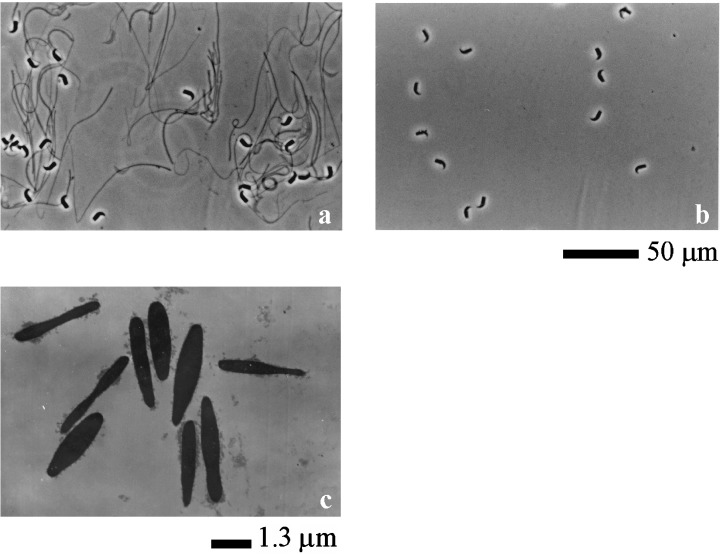
Observation of extracted spermatozoa under microscope and electron microscope. (a) Spermatozoa which were extracted by the urea solution. Sperm head and fibrous sheath were observed under microscope. (b) Spermatozoa which were extracted by the urea‐thiourea solution after urea extracting. Only sperm head was observed under microscope. Scale bar shows 50 µm. (c) Spermatozoa which were extracted by the urea‐thiourea solution after urea extracting under electron microscope. Only nucleus of sperm head was observed under electron microscope. Scale bar shows 1.3 µm.
Electron microscopy
The pellet of the urea‐thiourea extract was fixed in fixative solution containing 2% paraformaldehyde, 2.5% glutaraldehyde and 0.1 mol/L cacodylate buffer (pH 7.4) at room temperature for 2 h. The fixed pellet was washed in 0.1 mol/L cacodylalte buffer (pH 7.4) and postfixed in reduced 1% osmium tetraoxide containing 1% OsO4 and 0.8% K4Fe(CN)6 at 4°C for 1 h. The postfixed pellet was dehydrated in alcohol dehydration series and embedded in epoxy resin. After making an ultra thin section, the pellet was stained with uranyl acetate and lead. Then, the pellet was examined under the transmission electron microscope (HITACHI H‐7600, Hitachi Instruments, Tokyo, Japan).
SDS‐PAGE
SDS‐PAGE was carried out according to the method used by Laemmli. 28 The separating gel was 10% (w/v) polyacrylamide containing 0.1% (w/v) SDS.
Western blotting
Western blotting was carried out according to the method used by Towbin et al. 29 The blotted PVDF membrane was blocked with 5% (w/v) skim milk in tris buffered saline (TBS) containing 0.15 mol/L NaCl and 20 mmol/L Tris‐HCl (pH 7.4) for 1 h at 20°C, and incubated with the first antibodies (1 : 1000 dilutions) with 5% (w/v) skim milk in TBS for 1 h at 20°C. After washing with TBS, it was incubated with second antibodies conjugated to horseradish peroxidase (HRP, 1 : 5000 dilution) with 5% (w/v) skim milk in TBS. Color reaction was carried out with ECL plus kit. Reactivity of western blotting was measured by means of densitometer (GS‐800 densitometer, Bio‐Rad laboratories, Hercules, CA, USA), analyzed by Quantity One Software version 4.6.1 (Bio‐Rad laboratories) and a one‐factor anova was carried out.
Detection of phosphopeptide by surface enhanced desorption ionization proteinchip system
Detection of phosphopeptide was carried out using the surface enhanced desorption ionization (SELDI) proteinchip system (Ciphergen Biosystems) with IMAC chip SELDI proteinchip system, which is new technology used to analyze proteins and peptides using a protein chip and time of flight mass spectrometry. Because phosphorylated amino acids bind to Ga(III), 30 IMAC with Ga(III) can bind to peptides, including phosphorylated amino acids. At first, the IMAC chip was charged with Ga(III) by adding 5 µL of 100 mmol GaCl3. After 5 min incubation, the chip was quickly rinsed with water to remove unbound metal. The chip was then equilibrated with 5 µL of 50 mmol/L 2‐(N‐Morpholino) ethanesulfonic acid, monohydrate (MES) pH 4.0. After 5 min, MES was removed. Then, 5 µL of the protein extracts, diluted two fold by 50 mmol/L MES pH 4.0, were loaded on the chip, incubated for 20 min at ambient temperature and washed three times with 50 mmol/L MES pH 4.0. The washed chip was rinsed twice with pure water. After air‐drying, 1 µL of 10 mg/mL CHCA solution containing 50% acetonitrile and 0.3% tri‐fluoro acetic acid was loaded on the chip. The chip was analyzed in the Proteinchip Reader (Model PBS II, Ciphergen Biosystems) and the data were analyzed by Peak Software version 3.0.2 (Ciphergen Biosystems). All data were normalized by total ion current normalization function following the software instructions. After normalization, statistical analysis using Biomarker Wizard Software (Ciphergen Biosystems) was carried out on all data.
RESULTS
Protocol to extract of sperm proteins completely
GENERALLY, SPERM SUSPENSION makes the solution viscous due to the solubilized DNA from sperm heads when spermatozoa are incubated in a salt solution. Itoh et al. 27 showed that viscosity derived from DNA was not found, and all sperm structures except for the head were completely dissolved when 7 mol urea solution was used in order to extract proteins from salmon spermatozoa. So, we used this method described by Itoh et al. 27 to extract hamster sperm proteins completely. However, sperm heads and fibers were not dissolved (Fig. 1a). It was likely that the fibers were fibrous sheaths because they were not dissolved by high concentrations of urea solution. 31 Therefore, sperm structures, such as acrosomes, axonemes, outer dense fibers, mitochondria sheaths and membranes were dissolved by the urea solution. After centrifugation, the supernatant was used as urea extract. In the next step, the precipitate was resuspended in the urea‐thiourea solution containing 5 mol/L urea, 1 mol/L thiourea, 2% NP‐40 and 10% 2‐ME. After incubation, only sperm heads were observed (Fig. 1b). After centrifugation, the supernatant was used as urea‐thiourea extracts. The precipitate was nuclei of sperm heads (Fig. 1c). Therefore, sperm components except nuclei of heads were completely dissolved. As shown in Figure 2, different bands were detected from each extract on the SDS‐PAGE. Therefore, it was likely that different proteins were extracted by the urea solution and the urea‐thiourea solution.
Figure 2.
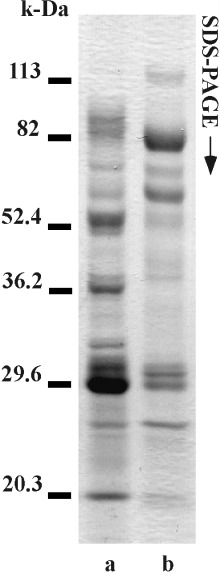
SDS‐PAGE of two sperm extracts. Lane a shows electrophoresis of the urea extracts. Lane b shows electrophoresis of the urea‐thiourea extracts. Numbers on left side show molecular weight markers.
Detection of proteins phosphorylated during sperm hyperactivation
Hamster spermatozoa were hyperactivated in vitro by incubation for 180 or 240 min. 11 As shown in Figure 3, most of motile spermatozoa were hyperactivated by incubation for 180 min. During hyperactivation, a dynamic change of sperm motility occurred in the early stage (∼30 min) and late stage (120–180 min). In the early stage, spermatozoa were activated. In the late stage, activated spermatozoa were hyperactivated.
Figure 3.
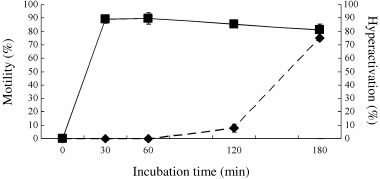
Motility and hyperactivation of hamster spermatozoa. Values are means ± SD (n = 5). (▪) Sperm motility, (◆) sperm hyperactivation.
As shown in 4, 5, 6, we detected many proteins that phosphorylated on tyrosine, serine and threonine residues during hyperactivation via activation. As for tyrosine phosphorylation, 12 major phosphoproteins, at least, were significantly detected (Fig. 4). Ten of them were detected from the urea‐extracts (Fig. 4a). Approximate molecular weights of the ten proteins were 120‐, 115‐, 90‐, 85‐, 80‐, 67.2‐, 52.4‐, 45.5‐, 40‐ and 31‐kDa, respectively. Moreover, molecular weights of two phosphoproteins that were detected from the urea‐thiourea extracts were 85‐ and 80‐kDa, respectively (Fig. 4b). Twelve phosphoproteins were designated as pY120u, pY115u, pY90u, pY85u, pY85ut pY80u, pY80ut, pY67u, pY52u, pY45u, pY40u and pY31u. As shown in Figure 4c, phosphorylations except pY31u gradationally increased during hyperactivation. In contrast, phosphorylation of pY31u increased at 30 min and was constant after 30 min.
Figure 4.
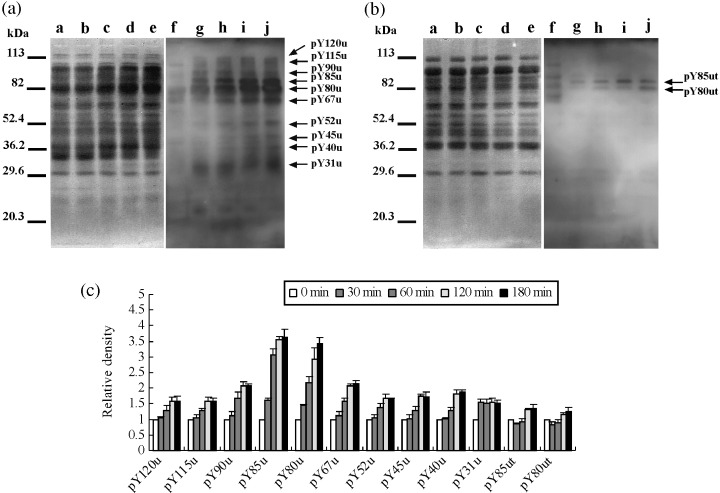
Protein tyrosine phosphorylation during hyperactivation. (a) Western blotting of the urea extracts. (b) Western blotting of the urea‐thiourea extracts. Lanes a–e and f–j showed Coomassie Brilliant Blue stain of membrane and enhanced chemiluminescence, respectively. Lanes a and f, lanes b and g, lanes c and h, lanse d and i, and lanes e and j show 0 min, 30 min, 60 min, 120 min and 180 min, respectively. Arrows show protein phosphorylations detected as a significant reaction on statistical analysis by one‐factor anova. Ten significant phosphorylations detected on (a) were designated as pY120u, pY115u, pY90u, pY85u, pY80u, pY67u, pY52u, pY45u, pY40u and pY31u. Two phosphorylations detected on (b) were designated pY85ut and pY80ut. Numbers the left side showmolecular markers. Experiment number was five. (c) Relative density of significant phosphorylations detected on (a) and (b). When reactivities of 0 min were taken, reactivities of each incubation times are shown.
Figure 5.
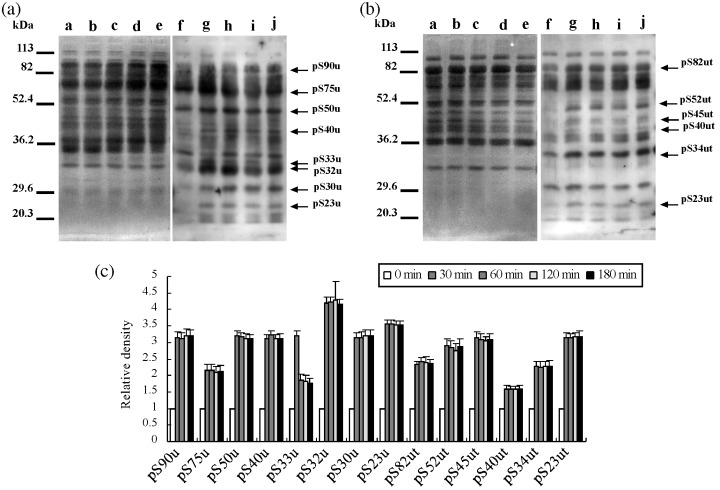
Protein serine phosphorylation during hyperactivation. (a) Western blotting of the urea extracts. (b) Western blotting of the urea‐thiourea extracts. Lanes a–e and f–j showed Coomassie Brilliant Blue stain of membrane and enhanced chemiluminescence, respectively. Lanes a and f, lanes b and g, lanes c and h, lane d and i, and lane e and j showed 0 min, 30 min, 60 min, 120 min and 180 min, respectively. Arrows show protein phosphorylations detected as a significant reaction on statistical analysis by one‐factor anova. Eight phosphoproteins detected on (a) were designated as pS90u, pS75u, pS50u, pS40u, pS33u, pS32u, pS30u and pS23u. Six phosphoproteins detected on (b) were designated as pS82ut, pS52ut, pS45ut, pS40ut, pS34ut and pS23ut. Numbers of left side show molecular markers. Experiment number was five. (c) Relative density of significant phosphorylations detected on (a) and (b). When reactivities of 0 min were taken, reactivities of each incubation time were showed.
Figure 6.
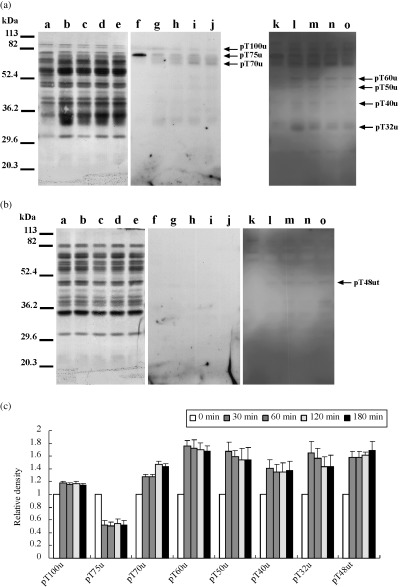
Protein threonine phosphorylation during hyperactivation. (a) Western blotting of the urea extracts. (b) Western blotting of the urea‐thiourea extracts. Lanes a–e and f–o showed Coomassie Brilliant Blue stain of membrane and enhanced chemiluminescence, respectively. Exposure time of lanes f–j and lanes k–o was 5 min and 15 min, respectively. Lanes a, f and k, lanes b, g and l, lanes c, h and m, lanes d, i and n, and lanes e, j and o showed 0 min, 30 min, 60 min, 120 min and 180 min, respectively. Arrows show protein phosphorylations detected as a significant reaction on statistical analysis by one‐factor anova. Seven phosphoproteins detected on (a) were designated as pT100u, pT75u, pT70u, pT60u, pT50u, pT40u and pT32u. One phosphoprotein detected on (b) was designated as pT48ut. Numbers of left side show molecular markers. Experiment number was five. (c) Relative density of significant phosphorylations detected on (a) and (b). When reactivities of 0 min were taken, reactivities of each incubation times are shown.
As shown in Figure 5 and 14 serine‐phosphoproteins, at least, were significantly detected. Eight proteins were detected from the urea‐extracts and approximate molecular weights of them were 90‐, 75‐, 50‐, 40‐, 33‐, 32‐, 30‐ and 23.4‐kDa, respectively (Fig. 5a). In contrast, six proteins that had molecular weights of approximately 82‐, 52‐, 45‐, 40‐, 34‐ and another 23.4‐kDa, respectively, were extracted by the urea‐thiourea solution (Fig. 5b). Fourteen phosphoproteins were designated as pS90u, pS82ut, pS75u, pS52ut, pS50u, pS45ut, pS40u, pS40ut, pS34ut, pS33u, pS32u, pS30u, pS23u and pS23ut. As shown in Fig. 5c, serine phosphorylations, except pS33u, increased at 30 min and were constant after 30 min. Phosphorylation of pS33u strongly increased at 30 min but decreased after 30 min.
As for threonine phosphorylation (Fig. 6), eight proteins, at least, were significantly detected. Seven proteins were detected from the urea solution and their molecular weights were approximately 100‐, 75‐, 70‐, 60‐, 50‐, 48‐, 40‐ and 32‐kDa, respectively (Fig. 56a). One protein was detected from the urea‐thiourea solution and its molecular weight was approximately 48‐kDa (Fig. 6b). They were designated as pT100u, pT75u, pT70u, pT60u, pT50u, pT48ut, pT40u and pT32u. Phosphorylations of six proteins, except pT75u and pT70u, increased at 30 min and were constant after 30 min (Fig. 6c). In contrast, phosphorylation of pT70u increased step‐by‐step. Moreover, pT75u was dephosphorylated at 30 min.
Phosphorylation by A‐kinase
It is widely accepted that sperm motility and hyperactivation are regulated by phosphorylation via A‐kinase 1 , 20 and we also detected phosphorylation by A‐kinase during sperm hyperactivation. As shown in Figure 7, at least four phosphoproteins were significantly detected as phosphorylation by A‐kinase. Approximate molecular weights of them were 75‐, 70‐, 60‐ and 50‐kDa, respectively. They were extracted by the urea solution (Fig. 8a) and their phosphorylations increased at 30 min and were constant after 30 min (Fig. 8c).
Figure 7.
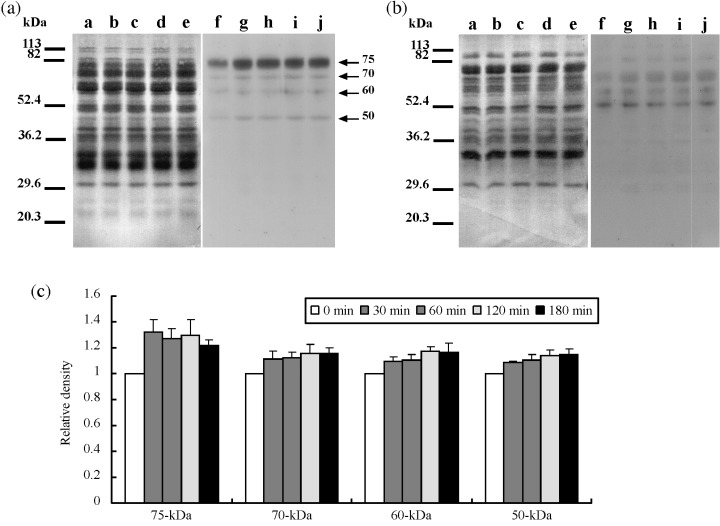
Protein phosphorylation by A‐kinase during hyperactivation. (a) Western blotting of the urea extracts. (b) Western blotting of the urea‐thiourea extracts. Lanes a–e and f–j showed Coomassie Brilliant Blue stain of membrane and enhanced chemiluminescence, respectively. Lanes a and f, lanes b and g, lanes c and h, lanes d and i, and lanes e and j showed 0 min, 30 min, 60 min, 120 min and 180 min, respectively. Arrows showed protein phosphorylations detected as a significant reaction on statistical analysis by one‐factor anova. Numbers of left side show molecular markers. Experiment number was five. (c) Relative density of significant phosphorylations detected on (a) and (b). When reactivities of 0 min were taken, reactivities of each incubation times are shown.
Figure 8.
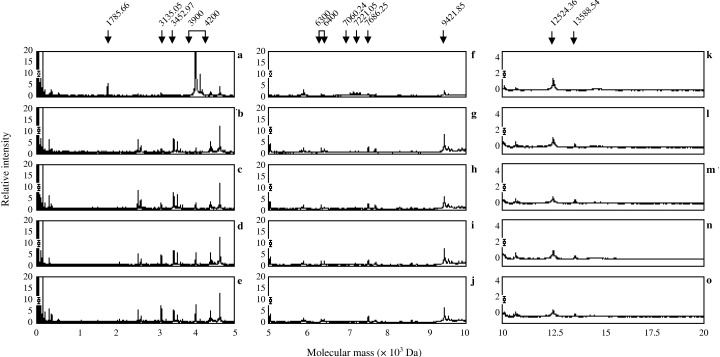
Detection of phosphopeptides by IMAC‐Ga(III) from the urea extracts. (a–e) Show mass spectrogram mass range from 0 Da to 5000 Da (f–j), show mass spectrogram mass range from 5000 Da to 10 000 Da (k–o), show mass spectrogram mass range from 10000 Da to 20000 Da and (a, f and k) show mass spectrogram of 0 min. (b, g and l) Show mass spectrogram of incubation at 30 min. (c, h and m) Show mass spectrogram of incubation at 60 min. (d, i and n) Show mass spectrogram of incubation at 120 min. (e, j and o) Show mass spectrogram of incubation at 180 min. Arrows indicate significant peaks.
Detection of phosphopeptides by SELDI proteinchip system
As shown in Figure 8, many peptides were detected from the urea extracts and 15 of them were significantly phosphorylated or dephosphorylated during sperm hyperactivation (Fig. 10a). Peptides of 1785.66‐ (Fig. 8a–e), 3960.37‐ (Fig. 8a–e), 3986.64‐ (Fig. 8a–e), 4001.37‐ (Fig. 8a–e), 4121.44‐ (Fig. 8a–e), 7060.24‐ (Fig. 8f–j) and 7221.05‐Da (Fig. 8f–j) were dephosphorylated at 30 min (Fig. 10a). In contrast, peptides of 6317.07‐ (Fig. 8f–j), 6393.81‐ (Fig. 8f–j), 7686.25‐ (Fig. 8f–j), 9421.85‐ (Fig. 8f–j) and 13588.54‐Da (Fig. 8k–o) increased at 30 min and were constant (Fig. 10a). The peptide of 3452.97‐Da (Fig. 8a–e) increased until 120 min, but it decreased at 180 min (Fig. 10a). Phosphorylation of 3135.05‐Da (Fig. 8a–e) peptide increased gradationally during hyperactivation (Fig. 10a). Conversely, phosphorylation of 12524.36‐Da (Fig. 8k–o) peptide decreased gradationally during hyperactivation (Fig. 10a).
Figure 10.
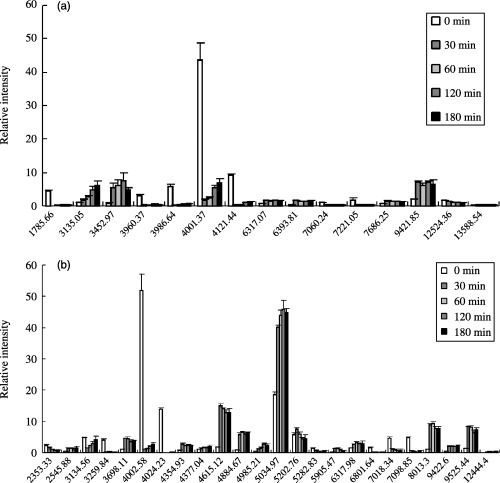
Relative intensity of significant phosphopeptides detected on 8, 9. (a) Shows relative intensity of significant peaks detected on Figure 8. (b) Shows relative intensity of significant peaks detected on Figure 9. Vertical axis indicates relative density. Cross axis indicates molecular mass.
On the urea‐thiourea extracts, many peptides were also detected (Fig. 9). Twenty‐four of them were significantly phosphorylated or dephosphorylated (Fig. 10b). Peptides of 3259.84‐ (Fig. 9a–e), 4002.58‐ (Fig. 9a–e), 4024.23‐ (Fig. 9a–e), 5282.83‐ (Fig. 9f–j), 6801.64‐ (Fig. 9f–j), 7018.34‐ (Fig. 9f–j) and 7098.52‐Da (Fig. 9f–j) were dephosphorylated at 30 min (Fig. 10b). Phosphorylation of 3134.56‐Da peptide (Fig. 9a–e) was decreased at 30 min, although it increased gradationally after 30 min (Fig. 10b). Moreover, peptides of 2545.88‐ (Fig. 9a–e), 3698.11‐ (Fig. 9a–e), 4354.93‐ (Fig. 9a–e), 4377.04‐ (Fig. 9a–e), 4615.12‐ (Fig. 9a–e), 4884.67‐ (Fig. 9a–e), 5034.97‐ (Fig. 9f–j), 6317.98‐ (Fig. 9f–j), 9422.60‐ (Fig. 9f–j), 9525.44‐ (Fig. 9f–j) and 12444.43‐Da (Fig. 8k–o) were phosphorylated at 30 min and their phosphorylations were constant (Fig. 10b). Phosphorylation of 2353.33‐Da peptide (Fig. 9a–e) was decreased gradationally during hyperactivation (Fig. 10b). Phosphorylation of 5202.76‐Da peptide (Fig. 9f–j) increased until 30 min but decreased after 30 min (Fig. 10b). Phosphorylations of 5905.47‐ (Fig. 9f–j) and 8013.3‐Da (Fig. 9f–j) peptide increased until 60 min, although they decreased gradationally after 60 min (Fig. 10b). Phosphorylation of 4985.21‐Da peptide (Fig. 9a–e) increased gradationally until 120 min, although it decreased at 180 min (Fig. 109b).
Figure 9.
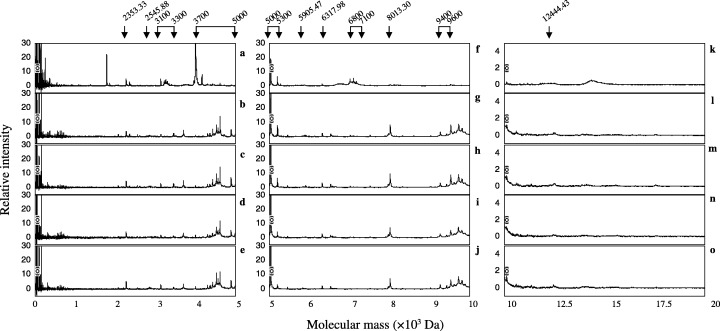
Detection of phosphopeptides by IMAC‐Ga(III) from the urea‐thiourea extracts. (a–e) Show mass spectrogram mass range from 0 Da to 5000 Da, (f–j) show mass spectrogram mass range from 5000 Da to 10 000 Da, (k–o) show mass spectrogram mass range from 10 000 Da to 20 000 Da, (a, f and k) show mass spectrogram of 0 min. (b, g and l) Show mass spectrogram of incubation at 30 min. (c, h and m) Show mass spectrogram of incubation at 60 min. (d, i and n) Show mass spectrogram of incubation at 120 min. (e, j and o) Show mass spectrogram of incubation at 180 min. Arrows indicate significant peaks.
DISCUSSION
BECAUSE IT HAS been accepted that sperm capacitation and hyperactivation is regulated thought the protein phosphorylation pathway, 1 many scientists’ research about protein phosphorylation is associated with capacitation and hyperactivations. However, the results regarding detected phosphorylation differ among reports, even if the same animals are used. One of reasons could be the instability of extracting sperm proteins. In many cases, SDS buffer for SDS‐PAGE 28 is used to solubilize sperm proteins before carrying out SDS‐PAGE. After boiling, buffer is often sticky because viscosity derived from DNA occurs. This is a weak point when SDS buffer is used. In our previous studies, 12 , 15 , 21 we used a solution containing 8M guanidine hydrochloride to dissolve sperm proteins. After dissolution, guanidine hydrochloride had to be removed from the solution before electrophoresis. The weak point was when guanidine hydrochloride was used. In a previous study, 27 viscosity derived from DNA was not found when 7 mol urea solution was used to extract sperm proteins from salmon spermatozoa. So, we used urea to extract sperm proteins. However, the solution containing only urea could not completely dissolve hamster sperm structures (Fig. 1a). As shown in ‘Materials and Methods’, we established a protocol to completely extract sperm proteins by trial and error. By the protocol, sperm structures except nuclei were dissolved (Fig. 1). In the next step, we comprehensively detected proteins phosphorylated and dephosphorylated during sperm hyperactivation. As shown in Table 1, we could detect 75 proteins from hamster spermatozoa. Most of the proteins described in our previous studies 8 , 11 , 12 , 15 , 21 and Jha et al.'s previous studies 13 , 14 were detected. Moreover, there were many novel proteins. As for high molecular weight range, 34 phosphoproteins were detected as tyrosine, serine and threonine phosphorylation. Many of them corresponded to phosphoproteins described in previous studies (Table 1). However, several of them were did not correspond at all and were novel proteins. In contrast, 37 phosphopeptides were detected for low molecular weight range. Three of them corresponded to phosphoprotein as described in our previous study (Table 1). However, many of the proteins did not corresponded at all and were novel peptides.
Table 1.
Correspondence between detected proteins and previously described proteins in hamster spermatozoa
| Band | Corresponded or included protein | References | Peak | Corresponded or included protein | References |
|---|---|---|---|---|---|
| pY120u | unknown | 12 | Urea extracts | ||
| pY115u | unknown | 12 | 1785.66 | novel | – |
| pY90u | unknown | 12 | 3135.05 | novel | – |
| pY85u | AKAP | 11, 12, 13 | 3452.97 | novel | – |
| pY80u | AKAP | 11, 12, 13 | 3960.37 | novel | – |
| pY67u | unknown | 12 | 3986.64 | novel | – |
| pY52u | unknown | 12 | 4001.37 | novel | – |
| pY45u | unknown | 12 | 4121.44 | novel | – |
| pY40u | unknown | 12 | 6393.81 | novel | – |
| pY31u | novel | – | 7060.24 | novel | – |
| pY85ut | AKAP | 11, 12, 13 | 7221.05 | novel | – |
| pY80ut | AKAP | 11, 12, 13 | 7686.25 | novel | – |
| pS90u | unknown | 15 | 9421.85 | carcinustatin like protein | 12, 15 |
| pS75u | novel or tubulin | 14, 21, 23 | 12524.36 | novel | – |
| pS50u | novel or ATP synthase E1b | 14, 21, 23 | 13588.54 | novel | – |
| pS40u | novel or Pyruvate dehydrogenase E1b | 8, 21, 22 | Urea‐Thiourea extracts | ||
| pS33u | novel | – | 2353.33 | novel | – |
| pS32u | unknown | 15 | 2545.88 | novel | – |
| pS30u | novel | – | 3134.56 | novel | – |
| pS23u | unknown | 15 | 3259.84 | novel | – |
| pS82ut | unknown | 15 | 3698.11 | novel | – |
| pS52ut | unknown | 14 | 4002.58 | novel | – |
| pS40ut | novel or Pyruvate dehydrogenase E1b | 8, 21, 22 | 4024.23 | novel | – |
| pS45ut | unknown | 14 | 4354.93 | novel | – |
| pS34ut | novel | – | 4377.04 | novel | – |
| pS23ut | novel | – | 4615.12 | novel | – |
| pT100u | unknown | 14, 15 | 4884.67 | novel | – |
| pT75u | unknown | 15 | 4985.21 | novel | – |
| pT70u | unknown | 14, 15 | 5034.97 | novel | – |
| pT60u | unknown | 14 | 5202.76 | novel | – |
| pT50u | unknown | 14 | 5282.83 | novel | – |
| pT40u | unknown | 14, 15 | 5905.47 | novel | – |
| pT32u | unknown | 14 | 6317.98 | novel | – |
| pT48ut | unknown | 14 | 6801.64 | novel | – |
| 75‐kDa | novel | – | 7098.85 | novel | – |
| 70‐kDa | novel | – | 8013.3 | novel | – |
| 60‐kDa | novel | – | 9422.6 | carcinustatin like protein | 12, 15 |
| 50‐kDa | novel | – | 9525.44 | carcinustatin like protein | 12, 15 |
| 12444.4 | novel | – |
In the present study, many novel phosphopeptides were detected (8, 10, 9, and Table 1). In previous studies on intracellular regulation of sperm motility or capacitation, peptides were not discussed. We do not know the reasons why studies about peptides associated with intracellular regulation of sperm motility or capacitation have not been popular, but analysis of peptides has always been difficult. About 10 years ago, new technology was developed to more easily research protein/peptides, such as the SELDI proteinchip system. 32 The SELDI proteinchip system is a form of MALDI TOF Mass spectrometry and only MALDI TOF Mass can be used to quantitatively analyze protein/peptide. Because we used the SELDI ProteinChip system in the present study, we could analyze phosphopeptides associated with sperm motility or hyperactivation. As a result, the present study is first report on spermatozoa analyzed using the SELDI ProteinChip system. Therefore, most of the peptides detected in the present study were novel. However, we have identified three problems that occurred as a result of analysis using SELDI proteinchip system. The first problem was that all peptides detected in the present study were not always phosphopeptide because acidic amino acid is able to bind to IMAC‐Ga. If protein/peptides include acidic amino acid, such as aspartic acid and glutamic acid, thus, it can bind to IMAC‐Ga. The second problem was that the peaks of the peptides detected in the present study were multicharge peaks. The third problem was that peptides that were detected in the present study were reduced products. These problems will be solved in future studies by identification and characterization of each peptide detected in the present study.
Four proteins, 75‐, 70‐, 60‐ and 50‐kDa protein, were phosphorylated by A‐kinase (Fig. 7). Several studies have shown that sperm capacitation and hyperactivation were regulated with A‐kinase. 1 , 5 , 10 Moreover, sperm motility is also regulated with A‐kinase. 20 Because A‐kinase is one of the serine/threonine kinases, it seemed that 75‐, 70‐, 60‐ and 50‐kDa protein were phosphorylated at serine or threonine residues. Phosphoproteins of similar size detected in the present study were pS75u, pT70u, pT60u, pS50u and pT50u. Furthermore, similar phosphorylation patterns were pS75u, pT70u, pT60u and pS50u. Therefore, it was likely that pS75u, pT70u, pT60u and pS50u were phosphorylated by A‐kinase and associated with motility and/or hyperactivation.
In the present study, we comprehensively detected proteins phosphorylated or dephosphorylated during hyperactivation via activation. Many proteins were phospho‐ or dephosphorylated in a time‐dependent manner. Proteins detected in the present study consisted of sperm head proteins and flagellar proteins because proteins were extracted from whole spermatozoa. Although proteins were phosphorylated or dephosphorylated during hyperactivation, it was likely that they were associated with sperm functions such as ATP synthase, acrosomal reaction, motility activation and motility hyperactivation. In future studies, we should classify protein phosphorylation and dephosphorylation associated with the above sperm functions. Moreover, we must to characterize and identify proteins detected in the present study. From the results of analysis of proteins detected in the present study, we can understand the regulatory mechanisms that regulate sperm hyperactivation via activation.
ACKNOWLEDGMENTS
WE THANK STAFF of the Laboratory Animal Research Center of Dokkyo University School of Medicine for their support. This work was supported by a Grant‐in‐Aid for Scientific Research (No. 15790860) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to M. F.
REFERENCES
- 1. Yanagimachi R. Mammalian fertilization. In: Neill K, Pfaff GM, eds. The Physiology of Reproduction , 2nd edn, Vol. 1 New York: Raven Press, 1994; 189–317. [Google Scholar]
- 2. Langlais J, Roberts KD. A molecular membrane model of sperm capacitation and the acrosome reaction of mammalian spermatozoa. Gamete Res 1985; 13: 183–224. [Google Scholar]
- 3. Okamura N, Tajima Y, Soejima A, Masuda H, Sugita Y. Sodium bicarbonate in semal plasma stimulates the motility of mammalian spermatozoa through the direct activation of adenylate cyclase. J Biol Chem 1985; 260: 9699–9705. [PubMed] [Google Scholar]
- 4. Visconti PE, Bailey JL, Moore GD, Pan D, Old‐Clarke P, Kopf GS. Capacitation of mouse spermatozoa I. Correlation between the capacitation state and protein tyrosine phosphorylation. Development 1995; 121: 1129–1137. [DOI] [PubMed] [Google Scholar]
- 5. Visconti PE, Stewart‐Sawage J, Blasco A et al. Roles of bicarbonate, cAMP and protein tyrosine phosphorylation on capacitation and the spontaneous acrosome reaction of hamster sperm. Biol Reprod 1999; 61: 76–84. [DOI] [PubMed] [Google Scholar]
- 6. Visconti PE, Kopf GS. Regulation of protein phosphorylation during sperm capacitation. Biol Reprod 1998; 59: 1–6. [DOI] [PubMed] [Google Scholar]
- 7. Visconti PE, Galantino‐Homer H, Ning X et al. The molecular basis of capacitation. J Androl 1998; 19: 242–248. [PubMed] [Google Scholar]
- 8. Si Y, Okuno M. Regulation of microtubule sliding by a 36‐kDa phosphoprotein in hamster sperm flagella. Mol Reprod Dev 1999; 52: 328–334. [DOI] [PubMed] [Google Scholar]
- 9. Marin‐Briggiler CI, Jha KN, Chertihin O et al. Evidence of the presence of calmodulin/calmodulin‐dependent protein kinase IV in human sperm and its involvement in motility regulation. J Cell Sci 2005; 118: 2013–2022. [DOI] [PubMed] [Google Scholar]
- 10. Kaláb P, Pêknicová J, Geussová G, Moos J. Regulation of protein tyrosine phosphorylation in boar sperm through a cAMP‐dependent pathway. Mol Reprod Dev 1998; 51: 304–314. [DOI] [PubMed] [Google Scholar]
- 11. Si Y, Okuno M. Role of tyrosine phosphorylation of flagellar proteins in hamster sperm hyperactivation. Biol Reprod 1999; 61: 240–246. [DOI] [PubMed] [Google Scholar]
- 12. Fujinoki M, Ohtake H, Okuno M. Tyrosine phosphorylation and dephosphorylation associated with motility of hamster spermatozoa. Biomed Res 2001; 22: 147–155. [Google Scholar]
- 13. Jha KN, Shivaji S. Capacitation‐associated changes in protein tyrosine phosphorylation, hyperactivation and acrosome reaction in hamster spermatozoa. Andorologia 2001; 33: 95–104. [DOI] [PubMed] [Google Scholar]
- 14. Jha KN, Shivaji S. Protein serine and threonine phosphorylation, hyperactivation and acrosome reaction in in vitro capacitated hamster spermatozoa. Mol Reprod Dev 2002; 63: 119–130. [DOI] [PubMed] [Google Scholar]
- 15. Fujinoki M, Ishimoda‐Takagi T, Ohtake H. Serine/ threonine phosphorylation associated with hamster sperm hyperactivation. Reprod Med Biol 2004; 3: 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Naz RK. Involvement of protein serine and threonine phosphorylation in human sperm capacitation. Biol Reprod 1999; 60: 1402–1409. [DOI] [PubMed] [Google Scholar]
- 17. Carrera A, Moos J, Ning XP et al. Regulation of protein tyrosine phosphorylation in human sperm by a calcium/calmodulin‐dependent mechanism: Identification of A kinase anchoring proteins as major substrates for tyrosine phosphorylation. Dev Biol 1996; 180: 284–296. [DOI] [PubMed] [Google Scholar]
- 18. Visconti PE, Johnson LR, Oyaski M et al. Regulation, localization, and anchoring of protein kinase A subunits during mouse sperm capacitation. Dev Biol 1997; 192: 351–363. [DOI] [PubMed] [Google Scholar]
- 19. Galantino‐Homer HL, Visconti PE, Kopf GS. Regulation of protein tyrosine phosphorylation during bovine sperm capacitation by a cyclic adenosine 3′,5′‐monophosphate‐dependent pathway. Biol Reprod 1997; 56: 707–719. [DOI] [PubMed] [Google Scholar]
- 20. Inaba K. Molecular architecture of the sperm flagella: molecules for motility and signaling. Zool Sci 2003; 20: 1043–1056. [DOI] [PubMed] [Google Scholar]
- 21. Fujinoki M, Ohtake H, Okuno M. Serine phosphorylation of flagellar proteins associated with the motility activation of hamster spermatozoa. Biomed Res 2001; 22: 45–58. [Google Scholar]
- 22. Fujinoki M, Kawamura T, Toda T et al. Identification of 36‐kDa flagellar phosphoproteins associated with hamster sperm motility. J Biochem 2003; 133: 361–369. [DOI] [PubMed] [Google Scholar]
- 23. Fujinoki M, Kawamura T, Toda T et al. Identification of 66 kDa phosphoprotein associated with motility initiation of hamster spermatozoa. Reprod Med Biol 2004; 3: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fujinoki M, Kawamura T, Toda T et al. Identification of the 58‐kDa flagellar phosphoprotein associated with motility initiation of hamster spermatozoa. J Biochem 2003; 134: 559–565. [DOI] [PubMed] [Google Scholar]
- 25. Fujinoki M, Kawamura T, Toda T et al. Identification of 36 kDa phosphoprotein in fibrous sheath of hamster spermatozoa. Comp Biochem Physiol 2004; 137B: 509–520. [DOI] [PubMed] [Google Scholar]
- 26. Jha KN, Shivaji S. Identification of nthe major tyrosine phosphorylated protein of capacitated hamster spermatozoa as a homologue of mammalian sperm A kinase anchoring protein. Mol Reprod Dev 2002; 61: 258–270. [DOI] [PubMed] [Google Scholar]
- 27. Itoh A, Inaba K, Fujinoki M, Morisawa M. Motility‐associated and cyclic AMP‐dependent protein phosphorylation in the sperm of the chum salmon, Oncorhynchus keta . Biomed Res 2001; 22: 241–248. [Google Scholar]
- 28. Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 1970; 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 29. Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 1979; 76: 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Posewitz MC, Tempst P. Immobilized gallium (III) affinity chromatography of phosphopeptides. Anal Chem 1999; 71: 2883–2892. [DOI] [PubMed] [Google Scholar]
- 31. Brito M, Burzio LO. Isolation of the fibrous sheath of mammalian sperm flagella In: Dentler W, Witman G, eds. Methods in Cell Biology, Vol. 47 New York: Academic Press, 1995; 391–395. [DOI] [PubMed] [Google Scholar]
- 32. http://www.ciphergen.com/.


