Abstract
Background
Current technology has demonstrated that mutation and deregulation of non-coding RNAs (ncRNAs) are associated with diverse human diseases and important biological processes. Therefore, developing a novel computational method for predicting potential ncRNA-disease associations could benefit pathologists in understanding the correlation between ncRNAs and disease diagnosis, treatment, and prevention. However, only a few studies have investigated these associations in pathogenesis.
Results
This study utilizes a disease-target-ncRNA tripartite network, and computes prediction scores between each disease-ncRNA pair by integrating biological information derived from pairwise similarity based upon sequence expressions with weights obtained from a multi-layer resource allocation technique. Our proposed algorithm was evaluated based on a 5-fold-cross-validation with optimal kernel parameter tuning. In addition, we achieved an average AUC that varies from 0.75 without link cut to 0.57 with link cut methods, which outperforms a previous method using the same evaluation methodology. Furthermore, the algorithm predicted 23 ncRNA-disease associations supported by other independent biological experimental studies.
Conclusions
Taken together, these results demonstrate the capability and accuracy of predicting further biological significant associations between ncRNAs and diseases and highlight the importance of adding biological sequence information to enhance predictions.
Keywords: ncRNA-disease association predictions, Tripartite network, Resource allocation
Background
Recent studies have investigated the biological functions, transcriptome, and regulation of non-coding RNAs (ncRNAs) of all sizes in a wide range of organisms [1]. siRNA (short interfering RNA), miRNA (microRNA) and piRNA (piwi-interacting RNA) are the three main types of short ncRNAs (less than 30 nucleotides). They play an important role in histone modification, gene silencing, heterochromatin formation, and DNA methylation, targeting at the transcriptional and post-transcriptional levels. Long non-coding RNAs (lncRNAs), which are greater than 200 nucleotides, are relevant in fundamental processes of gene regulation, such as chromatin modification and transcriptional regulation [2]. Studies indicate that lncRNAs can be categorized in one or more of the four archetypes, which include signal archetype (molecular signal or transcriptional activity indicator), decoy archetype (measure and adjust the balance of RNA regulation), guide archetype (directs the localization of ribonucleoprotein complexes to their targets), and scaffold archetype (a structural role for relevant proteins or RNAs to resemble). However, the main functions, structures, and mechanisms of lncRNAs remain unknown [3].
Because of their categorization into the four archetypes, ncRNAs have been proposed to have strong connections with the development and pathophysiology of diseases. Computational and experimental studies have demonstrated that alteration and deregulation of both short ncRNAs and lncRNAs at the structural and expression levels, can cause various types of cancer, such as breast cancer, leukemia, hepatocellular, and colon cancer, as well as neurodegenerative disorders, cardiovascular diseases, and immune-mediated diseases [4].
X-inactive specific transcript (Xist) is a lncRNA located on the X chromosome of the placental mammals that plays an important role in the X inactivation process. Lee et al. [5] demonstrated that Xist is a potent suppressor of hematological cancer in mice. Xist is required for hematopoietic stem cell survival and function. Therefore, Xist deletion results in leukemia, marrow fibrosis, and histiocytic sarcoma.
Yang et al. [6] also demonstrated a strong correlation between lncRNAs in tumor tissues and hepatocellular carcinoma (HCC). According to their experimental results, Cox regression analysis showed that both lncRNAs H19 and UCA1 were serious risk factors for HCC. Furthermore, logistic regression also indicated that H19 was overexpressed in hepatitis B virus-infected individuals.
A study on lncRNA PRINS (Psoriasis susceptibility-related RNA Gene Induced by Stress) provides further evidence on an association between ncRNAs and diseases. PRINS is transcribed by RNA polymerase II and expressed at various levels of human tissues [7]. PRINS is believed to play a role in susceptibility to psoriasis and has been linked to autoimmune diseases [8].
These findings are representative of the small number of ncRNA-disease connections that have been functionally established. This poses a major obstacle for bio-physicians in their quest to formulate new hypotheses for molecular mechanisms underlying complexes diseases, and to enhance the efficacy and efficiency of disease diagnosis and treatment. For the purpose of determining an association, bio-physicians must partition patients into appropriate groups and accurately investigate them. This method indeed establishes the correlation and expands the acknowledgement of the association in various phenomena. Although it is necessary to predict and infer ncRNA-disease associations, it is an incredible cost and time burden for bio-physicians.
To address this problem, some computational models have been proposed for ncRNA-disease association inference. Chen et al. [9] introduced Laplacian Regularized Least Squares for LncRNA-Disease Association (LRLSLDA), a semi-supervised learning method based on the framework of Laplacian Regularized Least Square and the assumption that ncRNAs with similar functions tend to interact with similar diseases. LRLSLDA can predict ncRNA-disease associations reliably. Yet, there are obstacles with parameter selection and classifier combinations. Another method called RWRLncD has been proposed by Sun et al. [10] that infers ncRNA-disease associations by integrating ncRNAs functional similarity network, disease similarity network, and known ncRNA-disease associations. RWRLncD cannot be employed without any known ncRNA-disease associations. Li et al. [11] introduced a genomic location-based computational method for ncRNA-disease association prediction. However, the approach has not been evaluated with statistical tests, and not all ncRNAs were associated with their neighbor genes, a major limitation of the method. Yang et al. [12] introduced a method employing a bipartite network with resource-allocation technique to infer new ncRNA-disease connections. Their experiment to validate the performance of their method mainly focused on only one dataset collected from Chen et al. (2013), with roughly 1028 interactions between 322 ncRNAs and 221 diseases. They also included additional interactions via deep literature mining. Alaimo et al. then proposed a new ncRNA-disease association prediction method called ncPred, which was shown to outperform Yang et al.’s method [13]. ncPred is a resource-propagation-based method applied on an ncRNA-target-disease tripartite network. The tripartite network, formed up by ncRNA-target and target-disease interaction bipartite networks, guided the resource transferring process to infer new associations. Targets refer to a group of genes, microRNAs or proteins that are related to particular ncRNAs in terms of expression regulation and binding activities etc. With inclusion of the targets, ncPred was experimentally shown to infer more biological information with higher reliability than Yang et al.’s method. Yet, there is still some room for improvement, particularly with regards to establishing the biological significance of identified associations. Chen et al. [14] developed hypergeometric distribution for lncRNA-disease association inference (HGLDA) to predict lncRNA-disease associations by integrating miRNA-disease interactions and lncRNA-miRNA associations. HGLDA applied p-value matrices obtained from interaction networks, with false discovery rate (FDR) correction. lncRNA-disease pairs with FDR less than 0.05 were predicted to be potential lncRNA-disease associations. However, HGLDA provided the least biological information, which led to weaker biological significance reliability.
Here, we introduce a method that improves on Alaimo et al.’s ncPred. The proposed method integrates a resource-allocation technique in the ncRNA-target-disease tripartite network, with pairwise similarity information obtained from sequence expressions. The prior knowledge combined with the biological information is carried and transferred throughout the network, and finally utilized to infer new associations.
To validate the performance of our proposed method, we conducted a 5-fold-cross-validation procedure to demonstrate its reliability, accuracy and efficiency, on a dataset reconstructed from Chen et al. (2013) [15]. In addition, by using a database of experimentally confirmed interactions between ncRNAs and miRNAs shown in Helwak et al. [16], we performed another validation of our proposed method. Our results demonstrate that our proposed method outperforms ncPred.
Methods
Data sources
In order to evaluate the performance of the approach, we prepared two kinds of data, ncRNA-target interaction matrix (LncRNADisease database (2015)) and target-disease interaction matrix (DisGeNET database) which are the same database sources used in Chen et al. (2013) [15], to form the tripartite network, as well as the sequence expression of those targets and ncRNAs (extracted from Uniprot and LncRNADisease databases, respectively). Since there are ncRNA sequence expressions and targets that are still unknown, we could only collect 76 ncRNAs, 109 targets, and 514 diseases (see Table 1). In Fig. 1, the degree distribution of the resulting network is shown. The results indicate that the entire network may follow a power law distribution. Moreover, we collected experimentally confirmed interactions between ncRNAs and miRNAs from the database shown in [16] and from Alaimo [13] Supplementary Information files and reconstructed another dataset composed on 151 ncNRAs, 179 targets and 134 diseases (see Helwak dataset in Table 1).
Table 1.
Description of the datasets
| Type of Data | Chen et al | Helwak et al. |
|---|---|---|
| Diseases | 514 | 134 |
| Targets | 109 | 179 |
| ncRNAs | 76 | 151 |
| Disease-target interactions | 580 | 1572 |
| Target-ncRNA interactions | 111 | 610 |
| Average degree | 1.977 | 9.405 |
Fig. 1.
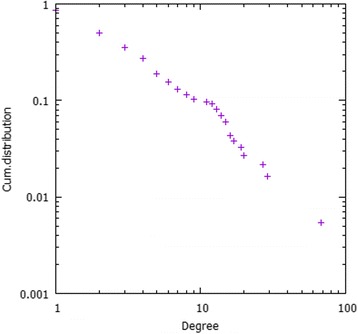
Cumulative degree distribution of the tripartite ncRNA-target disease network for Chen et al. dataset
Computational approach
In our approach, disease-target and target-ncRNA interaction matrices are required to construct the tripartite network (See Fig. 2). Sequence expressions of both targets and ncRNAs are also needed to generate string-kernel features for the prediction method. The overall algorithm of our method is shown in Fig. 3.
Fig. 2.
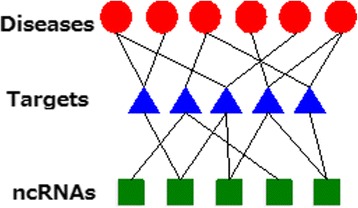
Example of a tripartite network used in this work. Targets integrate information from ncRNAs and diseases
Fig. 3.
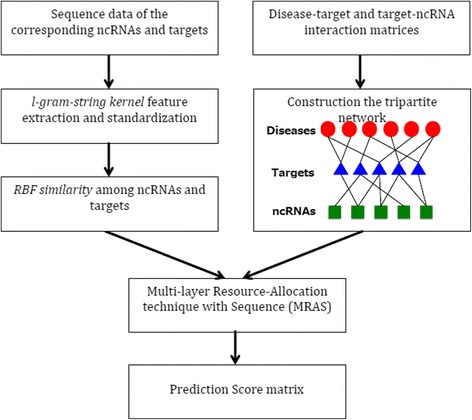
Illustration of the proposed computational method. Sequence information analysis is combined with a tripartite network structure. A multi-layer resource-allocation technique integrates the information and predicts associations between ncRNAs and human diseases
Let D = {d1, d2, …, dm} be a set of diseases, let T = {t1, t2, …, tn} be a set of targets which refer to genes or microRNA, and let R = {r1,r2, …, rp} be a set of non-coding RNAs. Let and let be sets of the sequence expressions of the targets and ncRNAs respectively.
L-gram-string kernel feature extraction and standardization
In our approach, the features of sequence expressions were extracted via the l-gram-string kernel method [17], in which each string is transformed into a vector consisting of the number of occurrences of each substring of length l. In bioinformatics, string kernel is considered a function that measures the similarity of a pair of sequence expressions (strings) with finite length, for the purpose of generating real-value feature vectors. In our case, the method was evaluated experimentally using length l = 1, 2, 3 and 4.
In order to improve the quality and reduce the redundancy of the computed features from the previous step, the standardization method below was applied [18].
where x is the original feature vector, is the mean and σ is the standard deviation.
Thus, let and be sets of the normalized real-value feature vectors computed from the sequence expressions of the targets and ncRNAs respectively.
RBF kernel similarity among ncRNAs and targets
To effectively compute the similarity among each pair of ncRNA-ncRNA and target-target, the RBF kernel similarity technique was employed. In statistical learning, RBF kernel can be viewed as a common kernel function to measure the similarity [19].
Let k_rna (i, j) and k_target (i, j) be the RBF kernel similarity function of pair ncRNA-ncRNA and target-target respectively.
Thus, the RBF similarity of ith ncRNA and jth ncRNA can be computed as
where
The RBF similarity of ith target and jth target can be computed as
where
Constructing the tripartite network disease-target-ncRNA
G (D, T, R, E), where E denotes a set of edges, is a tripartite network (Fig. 2) representing the disease-target interactions and target-ncRNA interactions. denotes the adjacency matrix of the disease-target network, and represents the adjacency matrix of target-ncRNA network.
Targets represent a group of biomolecules functionalized by non-coding RNAs. Targets can be genes, proteins, microRNAs, etc., whose activities include expression regulation, binding, and complex formation. In our method, targets function as a bridge allowing us to extract further biological information, for enhancing the accuracy of disease-ncRNA association inference.
Our method integrates the resource-allocation algorithm in the network with pairwise similarity information derived from sequence expressions to produce scores showing the level of certainty of the interaction. Essentially, the resource-allocation algorithm carries prior understanding of the bipartite network, which can be employed to predict the interactions.
Multi-layer resource-allocation technique with sequence (MRAS) information
Since the tripartite network consists of two bipartite networks, two-layer resource-allocation techniques were applied. For the first-layer (disease-target bipartite network), the resource will be transferred from the nodes in T (targets) to the nodes in D (diseases) integrating with pairwise similarity information between each pair of the diseases, then move back to the nodes in T. For the second-layer (target-ncRNA bipartite network), the resource integrated with pairwise similarity information between each pair of the ncRNAs, is allocated from the nodes in R (ncRNAs) to the nodes in T, and then combined with the resource from the previous layer. Finally, the weights computed within the two layers were merged into one, called combined weight (WC), indicating the likelihood that in case a disease associates with a target ti, it then possibly interacts with ncRNA rj. Prediction scores (P) can then be computed from the combined weight WC, and the higher the score, the greater the certainty that the ncRNA will associate with a particular disease.
Let deg(x) be the degree of node x in the disease-target network, and deg′(y) be the degree of node y in the target-ncRNA network.
-
Layer 1: disease-target bipartite network
Let be the probability that the ith target interacts with the jth target when both of them interact with the same disease:
-
Layer 2: target-ncRNA bipartite network
Let be the probability that the ith ncRNA interacts with the jth ncRNA when both of them interact with the same target:
Hence, the combined weight of the two bipartite networks together with the similar neighborhood of both targets and ncRNAs can be obtained as follows. The formula assigned more weight to the path with higher frequency.
Finally, the prediction score matrix can be computed based on the following formula: P = {pij}m × p = ADT ⋅ WC
Based on the above procedures, the prediction score matrix was produced via the integration of biological similarities of sequence expressions and the resource-allocation technique in the tripartite network. The score matrix thus predicted associations between ncRNAs and diseases. The higher the score, the higher the certainty of ncRNA-disease connectivity.
Results
Algorithm prediction performance and evaluation
As mentioned, the performance of our method was measured by applying a 5-fold-cross-validation procedure as well as other computational operations. The evaluation algorithm is described as follows:
First, we consider all pairs of ncRNA and disease (N pairs) exiting in the tripartite network. These pairs are expected to be predicted by the algorithm in the score matrix P.
Fix a pair of values for σ1 and σ2 .
Consider k-fold cross validation with k = 5. Therefore, all N pairs are divided into 5 groups.
Each group of N/5 elements becomes a test data one time. The rest four groups are training data
From each test data group, we scan those pairs that are connected through targets and disconnect them (see Fig. 4b)
Run the algorithm and make predictions only for the pairs in the test group Ptest.
Construct a ROC curve for the test group based on N/5 scores for Ptest.
Repeat 3~ 7 for different folds. We average all five ROC curves.
Repeat 3~ 8 for randomization of test and training groups five more times and obtained an averaged ROC curve.
Repeat 2~ 9 for each σ1 [0.1,0.2,...,0.9] x σ2 [0.1,0.2,..,0.9] and pick up the best AUROC among all results.
Fig. 4.
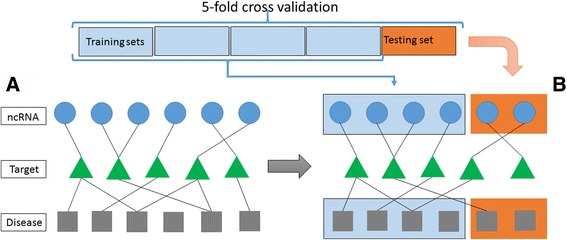
Algorithm performance evaluation methodology using 5-fold cross validation. From the constructed network as shown in (a), we divide all ncRNA-disease pairs into 5 groups. Each time one group performs as a testing dataset. To evaluate whether the algorithm is able to predict the interacting pairs without memory of existing interactions, we delete links that connect ncRNAs with disease if they belong to the testing set as shown in (b)
Evaluation results
We took into account the area under receiver operating characteristic (ROC) curve (AUC) values to assess the reliability, credibility, and accuracy of the prediction method. Table 2 demonstrates that our proposed method clearly outperforms ncPred in terms of the average values of AUC for the analysed datasets as shown in Table 1. These results emphasize the improved reliability and accuracy of predicting new ncRNA-disease associations or biological relevance using our approach, compared to that of Alaimo et al. which uses ncPred.
Table 2.
Comparison of the proposed method and ncPred through averaged area under ROC curve (ROC)
| AUC | ||
|---|---|---|
| Proposed method | ncPred | |
| Chen et al.(2013) | 0.57135 | 0.50242 |
| Helwak et al.(2013) | 0.77444 | 0.50295 |
Italic numbers indicate the best performance
In our approach, the RBF kernel technique was selected to conduct the similarity measure. In order to prove that RBF kernel can interpret the similarity of biological significance more appropriately, we conducted comparison experiments on the dataset listed in Table 1 with three representative kernel functions: linear kernel, polynomial (degree = 2) kernel and RBF kernel. Table 3 clearly illustrates the prediction performance of our approach underlying each kernel function. The results demonstrate that RBF kernel successfully outperforms the other two kernel techniques, showing the highest average AUC values (see Fig. 5b).
Table 3.
Comparison of performance using three representative kernel functions
| Dataset | AUC | ||
|---|---|---|---|
| Linear Kernel | Polynomial Kernel (d = 2) | RBF Kernel | |
| Chen et al. (2013) | 0.51561 | 0.54175 | 0.57135 |
Performance results for the following three kernel functions: linear kernel, polynomial (degree = 2) kernel and RBF kernel. The length of l-gram is set to l = 3 in this experiment
Fig. 5.
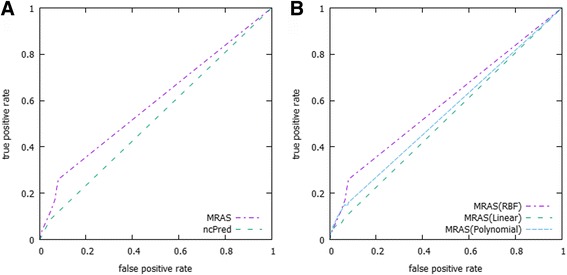
Comparison between the proposed method (MRAS) and ncPred. a shows an ROC curves comparison between the proposed method (MRAS) and ncPred with evaluation of the Chen et al. dataset. The ROC curves were drawn up based on the average results of the simulation, and repeated to ensure reliable estimates as described in the evaluation method procedure. The results also convey that the proposed method produced a higher true positive rate, which demonstrates its superior performance over its competitor. b shows the results of the proposed method using different kernel functions
The l-gram-string kernel technique was applied to extract biological features from the sequence data of ncRNAs and targets in the proposed method. In order to investigate which length of l can interpret and extract better information from the sequence data, we validated the performances underlying different lengths of l. We also compared the results of the proposed method under two assumptions. By using the cut link method described in algorithm performance evaluation section and without cut link. Table 4 shows the comparison for both methods and for l-gram-string kernel (when l = 1, 2, 3 and 4) in terms of AUC values using the dataset described in Table 1. The results show that link cut notably decreases the prediction of the algorithm as expected from 0.75 to 0.57 (see Fig. 6a and b). However, even though we perform this strict procedure, the algorithm is able to predict correctly interactions with an AUC above 0.5. The results also show that performance slightly improved at l = 3. Thus, l = 3 is the optimal parameter for the method (see Table 4 and Fig. 6b). Note that for Helwak dataset the optimal length is l = 1.
Table 4.
Comparison of l-gram-string kernel in terms of average Area Under the Curve (AUC) values
| l = 1 | l = 2 | l = 3 | l = 4 | ||
|---|---|---|---|---|---|
| Chen et al.(2013) | AUC | 0.7507 | 0.7506 | 0.7513 | 0.7433 |
| Cut link AUC | 0.5706 | 0.5644 | 0.5713 | 0.5476 | |
| Helwak et al.(2013) | AUC | 0.8543 | 0.7855 | 0.7338 | 0.7080 |
| Cut link AUC | 0.7744 | 0.7098 | 0.6430 | 0.6441 |
The performance of the l-gram-string kernel for l = 1, 2, 3 and 4 was examined using the AUC metric. The computation was done using the RBF kernel. Table also shows the prediction performance when the links between a disease and their associated ncRNAs are deleted (Cut link UAC). Italic numbers indicate the best performance
Fig. 6.
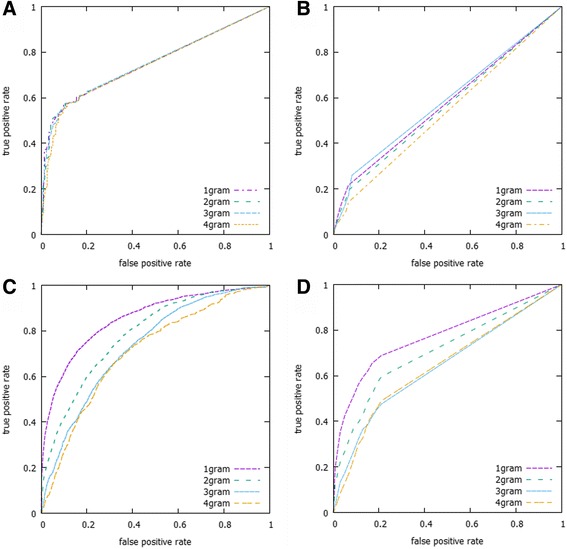
Illustration of the ROC curves for the proposed algorithm. ROC curves for the proposed algorithm MRAS using different values of l-gram string (l = 1, 2, 3 and 4) and using RBF kernel (a) without link cut (AUC = 0.75) and (b) with link cut (AUC = 0.57) for the Chen dataset. The results for the Helwak dataset using RBF kernel (c) without link cut (AUC = 0.85) and (d) with link cut (AUC = 0.77)
For parameter tuning of RBF kernel similarity function σ1 and σ2 we selected the optimal parameters as explained in the evaluation method procedure. Table 5 shows the optimal values identified in our computation.
Table 5.
Optimal values of σ1 and σ2 parameters
| σ 1 | σ 2 | |
|---|---|---|
| Chen et al.(2013) | 0.6 | 0.7 |
| Helwak et al.(2013) | 0.9 | 0.1 |
Case studies
In order to demonstrate the credibility and functionality of the proposed method, we compared predicted disease-ncRNA pairs using Chen et al. dataset with recent experimental studies, as shown in Table 6, on six diseases, namely lung cancer, liver cancer, breast cancer, urinary bladder neoplasms, prostatic neoplasms and stomach neoplasms.
Table 6.
The predicted ncRNAs related to the six diseases examined in this study
| Disease | ncRNA | Evidence (PMID) |
|---|---|---|
| Breast Neoplasms |
H19 | 15985428;27540977;9811352;24780616 |
| Breast Neoplasms |
PVT1 | 17908964;10485452;23907597 |
| Breast Neoplasms |
SRA1 | 17710122;26460974 |
| Breast Neoplasms |
WRAP53 | 27525856;15736456 |
| Carcinoma, Hepatocellular | H19 | 9570364 |
| Carcinoma, Hepatocellular | PCNA-AS1 | 24704293 |
| Carcinoma, Hepatocellular | WRAP53 | 23836507 |
| Lung Neoplasms |
HOTAIR | 25010625;22088988 |
| Lung Neoplasms |
MALAT1 | 23243023;26490983 |
| Lung Neoplasms |
PVT1 | 25400777 |
| Lung Neoplasms |
XIST | 26339353 |
| Prostatic Neoplasms | CDKN2B-AS1 | 27197191;24988946 |
| Prostatic Neoplasms | H19 | 19513555;25895025 |
| Prostatic Neoplasms | HOTAIR | 26411689 |
| Prostatic Neoplasms | MALAT1 | 23845456 |
| Prostatic Neoplasms | PCAT1 | 21804560 |
| Prostatic Neoplasms | SRA1 | 16607388 |
| Prostatic Neoplasms | XIST | 16261845 |
| Stomach Neoplasms | CDKN2B-AS1 | 24810364;26187665 |
| Stomach Neoplasms | HOTAIR | 25063030;23898077 |
| Stomach Neoplasms | MALAT1 | 24857172 |
| Urinary Bladder Neoplasms | H19 | 23354591 |
| Urinary Bladder Neoplasms | WRAP53 | 27912092 |
The predicted ncRNAs related to lung cancer, liver cancer, breast cancer, urinary bladder neoplasms, prostatic neoplasms and stomach neoplasms based upon our method. Each research article reporting on a specific ncRNA is shown with a unique ID (PMID) by PubMed
Lung cancer
Lung Cancer (LC) is one of the most deadly diseases affecting both men and women worldwide. There are three types of lung cancer: non-small cell lung cancer, small cell lung cancer, and lung carcinoid tumor. In 2015 lung and bronchus cancer accounted for 13.3% of all cancer cases in the US. The proposed method predicted that seven ncRNAs, H19, HOTAIR, MALAT1, PVT1, WRAP53, XIST, CDKN2B-AS1 are associated with LC. Kondo et al. reported that the alteration and deregulation of H19 impacts lung cancer cell growth [20]. HOX transcript antisense RNA (HOTAIR) prevents gene expression via collection of chromatin modifiers. Loewen et al. demonstrated that HOTAIR plays an important role in the intervention of lung cancer [21]. lncRNA metastasis associated lung adenocarcinoma transcript 1 (MALAT1) can impair in vitro cell motility of lung cancer cells and simultaneously influence a number of genes (Tano et al. & Tseng et al. [22, 23]). Yang et al. reported that an increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer [24]. Tantai et al. suggested a combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer [25]. Park et al., observed evidences for lung cancer with two variants located in cancer pleiotropic regions, namely TERT and risk of lung adenocarcinoma and CDKN2BAS1 with risk of lung squamous cell carcinoma [26].
Liver cancer
Liver cancer is the third most deadly cancer worldwide [27]. Very few patients receive curative treatments, while the majority do not recover since they are diagnosed at later stages. Our method predicted that ncRNAs H19, PCNA-AS1 and WRAP51 are correlated with liver cancer. H19 ncRNA expression was shown to result in high H19 protein expression in liver cancer whenever there is a loss of imprinting [28]. Iizuka et al. investigated further the epigenetic abnormalities in the insulin-like growth factor 2 (IGF2) and H19 genes observed in hepatocellular carcinoma (HCC) [29]. Yuan et al. reported that antisense long non-coding RNA PCNA-AS1 promotes tumor growth in hepatocellular carcinoma [30]. Several studies have also linked WRAP51 with HCC [31].
Breast cancer
Breast cancer is a ductal carcinoma beginning in the cells of lobules, ducts and other tissues of the breast. In the United States, breast cancer is the second most common cancer, just after skin cancer. Both women and men can suffer from this severe disease. Our method predicted that four ncRNAs, H19, PVT1, SRA1 and WRAP53 may be associated with this disease. Berteaux et al. found out that H19 transcript antisense RNA stabilize breast cancer cells and is overexpressed in breast tumors [32]. Others evidences were found across literature [33, 34]. Zhang et al. reported that the non-protein coding plasmacytoma variant translocation 1 (PVT1) has been implicated in human cancers [35, 36]. In addition, Hube et al. reported that alternative splicing of SRA1 could lead to the generation of coding and non-coding RNA isoforms in breast cancer cell lines [37]. Cao et al. recently reported about the association between the WRAP53 gene rs2287499 C > G polymorphism and cancer risk [38].
Urinary bladder Neoplasms
Urinary bladder neoplasms are the result of abnormal growth of bladder cells, and are considered as one of the common cancers. Men are at higher risk for this disease than women. The predictive results derived from our method inferred that ncRNAs H19 and WRAP53 might be associated with urinary bladder cancer. Luo et al. demonstrated that the abnormal expression of H19, particularly its up-regulation, contributed to cell proliferation in bladder neoplasms [39]. Wrap53 is a multi-functional gene which is capable of regulating p53 levels in both normal and cancer cell lines [40].
Prostatic Neoplasms
Prostatic neoplasm is caused by uncontrolled growth of cells located in the prostate causing tumors. Based upon our proposed method, prostate cancer was predicted to be associated with H19, MALAT1, CDKN2B-AS1, HOTAIR, PCAT1, SRA1, XIST. Zhu et al. demonstrated that H19 was significantly downregulated in the metastatic prostatic tumor cell line M12 [41]. Perez et al. showed that the antisense intronic transcript of MALAT1 is correlated with tumor differentiation in prostate cancer [42]. Fehringer reported the involvement of CDKBN2B-AS1in Cross-cancer genome-wide analysis of lung, ovary, breast, prostate and colorectal cancer using a cross-cancer genome-wide analysis [43]. Zhang et al. discovered that LncRNA HOTAIR enhances the Androgen-Receptor-Mediated Transcriptional Program and Drives Castration-Resistant Prostate Cancer [44]. Ren et al. suggested the Long noncoding RNA MALAT-1 as a potential therapeutic target for castration resistant prostate cancer [45]. Presner et al. results implied that rhe Long Non-Coding RNA PCAT-1 Promotes Prostate Cancer Cell Proliferation through cMyc [46]. The same research group used a transcriptome sequencing across a prostate cancer cohort to identify PCAT-1 as an unannotated lincRNA implicated in disease progression [47]. Several works also reported on the involvement of SRA1 [48] and XIST [49] in prostatic neoplasm.
Stomach Neoplams
Stomach neoplams that occurs as a consequence of abnormal grothw of stomach cells have also been associated to ncRNAs in several works. Our method predicted CDKN2B-AS1, HOTAIR and MALAT1 as main correlated molecules with stomach neoplasms. Zhang et al., reported that ANRIL (CDKN2B-AS1) which recruits and binds to PRC2 is usually observed upregulated in human gastric cancer (GC) tissues [50]. Lee et al. found that long non-coding RNA HOTAIR tends to promote not only carcinogenesis but also invasion of gastric adenocarcinoma [51]. Wang et al. also observed that MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF [52].
Discussion
Our work has improved the prediction quality and performance of Alaimo et al.’s method ncPred by integrating a biological feature information derived from sequence data with weight calculated by the multi-layer resource allocation technique, throughout the whole disease-target-ncRNA tripartite network.
The proposed method is unique because involves three types of biological information, namely ncRNAs sequences, target sequences and diseases. This rich biological information is also organized in a complex tripartite network in which targets integrate information from ncRNAs and diseases. Our study extended Alaimo’s approach [13] because we integrated biological sequence information. The uniqueness of the datasets and the complexity of the networks structure make it difficult to perform a straightforward comparison of our predictions with other approaches besides ncPred algorithm [13]. Indeed, most of the previous works have been done using a more simple approach involving a bipartite network. For example, Yang el al [12] constructed a bipartite network composed of only lncRNAs and diseases. On this network, a propagation algorithm navigated to infer lncRNAs implicated in diseases. Alaimo et al. [13] have already shown that their method outperformed the method by Yang et al. [12].
Our computational analyses indicate that our approach could result in more reliable and biologically efficient disease-ncRNA associations prediction than ncPred (See Tables 2 and 6). The prediction scores were obtained based on significant biological information, which provided useful suggestions of which ncRNA-disease associations have stronger interactions. This paves an easy path for pathologists to analyze and interpret ncRNA-disease associations. However, in order to accurately determine and confirm the associations, suitable patients and document cases are still needed.
The proposed method still has limitations that need to be considered. For instance, it lacks ncRNA-target interaction data and their particular sequence data. Those sequence data are required to extract the feature vector, which could extend the computational time. Furthermore, the lengths of ncRNAs and targets vary, ranging from less than 100 nucleotides to more than 10,000 nucleotides. This impacts the ranking of predicted ncRNA-disease associations. Accordingly, more related datasets are required to improve and expand the reliability and quality of ncRNA-disease association inference.
Acknowledgments
JCN was partially supported by JSPS KAKENHI Grant Number JP25330351. MH was partially supported by JSPS KAKENHI Grant Number 16 K00392. TA was partially supported by JSPS KAKENHI Grant Numbers 26240034 and 26540125. This research was also partially supported by the Collaborative Research Program of the Institute for Chemical Research, Kyoto University.
Funding
Publication of this article was funded by Toho University’s research budget.
Availability of data and materials
The datasets used in our study were downloaded from the publicly available databases mentioned in the text. The source code is available from the corresponding author on reasonable request.
About this supplement
This article has been published as part of BMC Systems Biology Volume 12 Supplement 1, 2018: Selected articles from the 16th Asia Pacific Bioinformatics Conference (APBC 2018): systems biology. The full contents of the supplement are available online at https://bmcsystbiol.biomedcentral.com/articles/supplements/volume-12-supplement-1 .
Authors’ contributions
TM and JCN made available computer programs and performed rigorous computational experiments by designing the algorithmic evaluation. HN designed the method and performed preliminary computational experiments. TM, JCN and MH contributed to the data acquisition, data analysis and interpretation of data. TA supervised HN’s work with giving various advices and comments. TM, HN and JCN wrote the draft. All of the authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 2.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Lee JT, Bartolomei MS. X-inactivation, imprinting and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z, Lu Y, Xu Q, Tang B, Park C-K, Chen X. HULC and H19 played different roles in overall and disease-free survival from hepatocellular carcinoma after curative hepatectomy: a preliminary analysis from gene expression omnibus. Di Markers. 2015;2015:191029. doi: 10.1155/2015/191029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scaria V, Pasha A. Long non-coding RNAs in infection biology. Front Genet. 2012;3:308.8. doi: 10.3389/fgene.2012.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Xuan Z, Liu C. Long non-coding RNAs and complex human diseases. Int J Mol Sci. 2013;14(9):18790–18808. doi: 10.3390/ijms140918790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Yan GY. Novel human lncRNA-disease association inference based onlncRNA expression profiles. Bioinformatics. 2013;29:2617–2624. doi: 10.1093/bioinformatics/btt426. [DOI] [PubMed] [Google Scholar]
- 10.Sun J, et al. Inferring novel lncRNA–disease associations based on a random walk model of a lncRNA functional similarity network. Mol BioSyst. 2014;10:2074–2081. doi: 10.1039/C3MB70608G. [DOI] [PubMed] [Google Scholar]
- 11.Li J, et al. A bioinformatics method for predicting long noncoding RNAs associated with vascular disease. Sci China Life Sci. 2014;57:852–857. doi: 10.1007/s11427-014-4692-4. [DOI] [PubMed] [Google Scholar]
- 12.Yang X, Gao L, Guo X, et al. A network based method for analysis of lncRNA-disease associations and prediction of lncRNAs implicated in diseases. PLoS One. 2014;9(1):e87797. doi: 10.1371/journal.pone.0087797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alaimo S, Giugno R, Pulvirenti A. ncPred: ncRNA-disease association prediction through tripartite network-based inference. Front Bioeng Biotechnol. 2014;2:71. doi: 10.3389/fbioe.2014.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X. Predicting lncRNA-disease associations and constructing lncRNA functional similarity network based on the information of miRNA. Sci Rep. 2015;5:13186. doi: 10.1038/srep13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153(3):654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sören S, Gunnar R, Konrad R, Bottou L, Chapelle O, DeCoste D, Weston J, editors. In: Large-Scale Kernel Machines, chap. 4. Cambridge: MIT Press; 2007. p. 73–104.
- 18.Ismail B, Dauda U. Standardization and its effects on k-means clustering algorithm. Res J Appl Sci Eng Technol. 2013;6(17):3299–3303. [Google Scholar]
- 19.Vert JP, Tsuda K, Scholkopf B. A primer on kernel methods. Kernel Methods in Comput Biol. 2004;35–70:7. [Google Scholar]
- 20.Kondo M, Suzuki H, Ueda R, Osada H, Takagi K, Takahashi T, Takahashi T. Frequent loss of imprinting of the H19 gene is often associated with its overexpression in human lung cancers. Oncogene. 1995;10(6):1193–1198. [PubMed] [Google Scholar]
- 21.Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3:219. doi: 10.3389/fgene.2012.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tseng JJ, Hsieh YT, Hsu SL, Chou MM. Metastasis associated lung adenocarcinoma transcript 1 is up-regulated in placenta previa increta/percreta and strongly associated with trophoblast-like cell invasion in vitro. Mol Hum Reprod. 2009;15(11):725–731. doi: 10.1093/molehr/gap071. [DOI] [PubMed] [Google Scholar]
- 24.Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS, Feng XJ. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7(10):6929–6935. [PMC free article] [PubMed] [Google Scholar]
- 25.Tantai J, Hu D, Yang Y, Geng J. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8(7):7887–7895. [PMC free article] [PubMed] [Google Scholar]
- 26.Park SL, et al. Pleiotropic associations of risk variants identified for other cancers with lung cancer risk: the PAGE and TRICL consortia. J Natl Cancer Inst. 2014;106(4):dju061. doi: 10.1093/jnci/dju061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Llovet JM, Lok A. Hepatitis B virus genotype and mutants: risk factors for hepatocellular carcinoma. JNCI J Natl Cancer Ins. 2008;100(16):1121–1123. doi: 10.1093/jnci/djn261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26:155–165. doi: 10.1038/modpathol.2012.160. [DOI] [PubMed] [Google Scholar]
- 29.Iizuka N, Oka M, Tamesa T, Hamamoto Y, Yamada-Okabe H. Imbalance in expression levels of insulin-like growth factor 2 and H19 transcripts linked to progression of hepatocellular carcinoma. Anticancer Res. 2004;24(6):4085–4089. [PubMed] [Google Scholar]
- 30.Yuan SX, Tao QF, Wang J, Yang F, Liu L, Wang LL, Zhang J, Yang Y, Liu H, Wang F, Sun SH, Zhou WP. Antisense long non-coding RNA PCNA-AS1 promotes tumor growth by regulating proliferating cell nuclear antigen in hepatocellular carcinoma. Cancer Lett. 2014;349(1):87–94. doi: 10.1016/j.canlet.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 31.Ortiz-Cuaran S, Cox D, Villar S, Friesen MD, Durand G, Chabrier A, Khuhaprema T, Sangrajrang S, Ognjanovic S, Groopman JD, Hainaut P, Le Calvez-Kelm F. Association between TP53 R249S mutation and polymorphisms in TP53 intron 1 in hepatocellular carcinoma. Genes Chromosomes Cancer. 2013;52(10):912–919. doi: 10.1002/gcc.22086. [DOI] [PubMed] [Google Scholar]
- 32.Berteaux N, Lottin S, Monté D, Pinte S, Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T, Adriaenssens E. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem. 2005;280(33):29625–29636. doi: 10.1074/jbc.M504033200. [DOI] [PubMed] [Google Scholar]
- 33.Zhang K, Luo Z, Zhang Y, Zhang L, Wu L, Liu L, Yang J, Song X, Liu J. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomark. 2016;17(2):187–194. doi: 10.3233/CBM-160630. [DOI] [PubMed] [Google Scholar]
- 34.Adriaenssens E, Dumont L, Lottin S, Bolle D, Leprêtre A, Delobelle A, Bouali F, Dugimont T, Coll J, Curgy JJ. H19 overexpression in breast adenocarcinoma stromal cells is associated with tumor values and steroid receptor status but independent of p53 and Ki-67 expression. Am J Pathol. 1998;153(5):1597–1607. doi: 10.1016/S0002-9440(10)65748-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z, Zhu Z, Zhang B, Li W, Li X, Wu X, Wang L, Fu L, Fu L, Dong JT. Frequent mutation of rs13281615 and its association with PVT1 expression and cell proliferation in breast cancer. J Genet Genomics. 2014;41(4):187–195. doi: 10.1016/j.jgg.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, Kalloger SE, Carlson JW, Ginzinger DG, Celniker SE, Mills GB, Huntsman DG, Gray JW. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13(19):5745–5755. doi: 10.1158/1078-0432.CCR-06-2882. [DOI] [PubMed] [Google Scholar]
- 37.Hube F, Guo J, Chooniedass-Kothari S, Cooper C, Hamedani MK, Dibrov AA. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 2006;25:418–428. doi: 10.1089/dna.2006.25.418. [DOI] [PubMed] [Google Scholar]
- 38.Cao HY, Wang S, Zhang ZY, Lou JY. Association between the WRAP53 gene rs2287499 C>G polymorphism and cancer risk:a meta-analysis. Genet Mol Res. 2016;15(3):25. doi: 10.4238/gmr.15037976. [DOI] [PubMed] [Google Scholar]
- 39.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Upregulated H19 contributes to bladder cancer cell proliferation by regulating ID2 expression. FEBS J. 2013;280(7):1709–1716. doi: 10.1111/febs.12185. [DOI] [PubMed] [Google Scholar]
- 40.Mahmoudi S, Henriksson S, Corcoran M, Méndez-Vidal C, Wiman KG, Farnebo M. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol Cell. 2009;33(4):462–471. doi: 10.1016/j.molcel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 41.Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng R, Wang Y, Huang J, Xu M, Yan J, et al. LncRNA h19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281:3766–3775. doi: 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
- 42.Perez DS, Hoage TR, Pritchett JR, Ducharme-Smith AL, Halling ML, Ganapathiraju SC, Streng PS, Smith DI. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum Mol Genet. 2008;17:642–655. doi: 10.1093/hmg/ddm336. [DOI] [PubMed] [Google Scholar]
- 43.Fehringer G, et al. Cross-cancer genome-wide analysis of lung, ovary, breast, prostate and colorectal cancer reveals novel pleiotropic associations. Cancer Res. 2016;76(17):5103–5114. doi: 10.1158/0008-5472.CAN-15-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang A, Zhao JC, Kim J, Fong KW, Yang YA, Chakravarti D, Mo YY, Yu J. LncRNA HOTAIR enhances the androgen-receptor-mediated transcriptional program and drives castration-resistant prostate cancer. Cell Rep. 2015;13(1):209–221. doi: 10.1016/j.celrep.2015.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren S, Liu Y, Xu W, Sun Y, Lu J, Wang F, Wei M, Shen J, Hou J, Gao X, Xu C, Huang J, Zhao Y, Sun Y. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol. 2013;190(6):2278–2287. doi: 10.1016/j.juro.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 46.Prensner JR, Chen W, Han S, Iyer MK, Cao Q, Kothari V, Evans JR, Knudsen KE, Paulsen MT, Ljungman M, Lawrence TS, Chinnaiyan AM, Feng FY. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 2014;16(11):900–908. doi: 10.1016/j.neo.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, Cao X, Jing X, Wang X, Siddiqui J, Wei JT, Robinson D, Iyer HK, Palanisamy N, Maher CA, Chinnaiyan AM. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29(8):742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurisu T, Tanaka T, Ishii J, Matsumura K, Sugimura K, Nakatani T, Kawashima H. Expression and function of human steroid receptor RNA activator in prostate cancer cells: role of endogenous hSRA protein in androgen receptor-mediated transcription. Prostate Cancer Prostatic Dis. 2006;9(2):173–178. doi: 10.1038/sj.pcan.4500867. [DOI] [PubMed] [Google Scholar]
- 49.Laner T, Schulz WA, Engers R, Müller M, Florl AR. Hypomethylation of the XIST gene promoter in prostate cancer. Oncol Res. 2005;15(5):257–264. doi: 10.3727/096504005776404607. [DOI] [PubMed] [Google Scholar]
- 50.Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W, Jf C. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5(8):2276–2292. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee NK, Lee JH, Park CH, Yu D, Lee YC, Cheong JH, Noh SH, Lee SK. Long non-coding RNA HOTAIR promotes carcinogenesis and invasion of gastric adenocarcinoma. Biochem Biophys Res Commun. 2014;451(2):171–178. doi: 10.1016/j.bbrc.2014.07.067. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, Su L, Chen X, Li P, Cai Q, Yu B, Liu B, Wu W, Zhu Z. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed Pharmacother. 2014;68(5):557–564. doi: 10.1016/j.biopha.2014.04.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used in our study were downloaded from the publicly available databases mentioned in the text. The source code is available from the corresponding author on reasonable request.


