Abstract
We report on the isolation and characterization of full-length cDNA sequences coding for N-acetylglucosaminyltransferase I (GnTI) from potato (Solanum tuberosum L.), tobacco (Nicotiana tabacum L.), and Arabidopsis. The deduced polypeptide sequences show highest homology among the solanaceous species (93% identity between potato and tobacco compared with about 75% with Arabidopsis) but share only weak homology with human GnTI (35% identity). In contrast to the corresponding enzymes from animals, all plant GnTI sequences identified are characterized by a much shorter hydrophobic membrane anchor and contain one putative N-glycosylation site that is conserved in potato and tobacco, but differs in Arabidopsis. Southern-blot analyses revealed that GntI behaves as a single-copy gene. Northern-blot analyses showed that GntI-mRNA expression is largely constitutive. Arabidopsis cgl mutants deficient in GnTI activity also possess GntI mRNA, indicating that they result from point mutations. GntI-expression constructs were tested for the ability to relieve the GnTI block in protoplasts of the Arabidopsis cgl mutant and used to obtain transgenic potato and tobacco plants that display a substantial reduction of complex glycan patterns. The latter observation indicates that production of heterologous glycoproteins with little or no antigenic glycans can be achieved in whole plants, and not in just Arabidopsis, using antisense technology.
In the Golgi apparatus of both plant and animal cells, N-acetylglucosaminyltransferase I (GnTI) initiates the formation of complex N-linked glycans on secretory glycoproteins that are derived from high-Man glycan precursors synthesized in the endoplasmic reticulum (Kornfeld and Kornfeld, 1985; Kaushal et al., 1988). Early blocks in glycan synthesis in the endoplasmic reticulum are fatal as shown by mutants selected for tunicamycin resistance in the yeast Saccharomyces cerevisiae (Barnes et al., 1984), which demonstrates that N-glycosylation of proteins is an essential step in eukaryotic cells. At the single cell level, several pleiotropic mutants with defects in Golgi-resident glycan trimming and modification enzymes were isolated by screening mutagenized mammalian cell lines for surviving exposure to specific plant lectins, of which the lec1 mutation represents a defect in GnTI (for review, see Stanley, 1984). At the whole-organism level, most of these mutations were lethal (e.g. GntI-knockout mice die at mid-gestation; Ioffe et al., 1996). This is probably because in mammals, glycoprotein glycans exhibit essential functions such as terminal sialic acid residues are involved in recognition events at the cell surface (Varki, 1997), terminal Man-6-P modification mediates in targeting of acid hydrolases to lysosomes (Dahms et al., 1989), and carbohydrate moieties also play a role in protein sorting to the apical membrane in epithelial cells (Scheiffele et al., 1995).
The isolation and characterization of GnTI-deficient Arabidopsis cgl mutants provided the first evidence that the viability of whole plants can be maintained if small N-linked glycoprotein glycans of uniform N-acetylglucosamine (GlcNAc)2Man5 structure are present (von Schaewen et al., 1993). Recently, however, Fitchette-Lainé et al. (1997) detected complex glycans with mammalian-type Lewis-a (Lea, terminal GalFuc-GlcNAc) modification in higher plants, except in some members of the Cruciferae family like Arabidopsis, and postulated a special role for this epitope in the cell-to-cell communications of most gymnosperms, monocots, and dicots. Hence, it was unclear whether plant species other than Arabidopsis would tolerate the loss of complex glycan modification imposed by, for instance, antisense suppression of the gene encoding GnTI.
To assess GntI expression and to devise gene-silencing strategies in higher plants, we isolated several plant-specific GntI-cDNA sequences: one from Arabidopsis to allow for further characterization of Arabidopsis cgl mutants that are deficient in GnTI-enzyme activity, and those from potato (Solanum tuberosum L.) and tobacco (Nicotiana tabacum L.) with the goal of generating transgenic solanaceous plants with reduced GnTI levels. The observations indicate that although GntI is present as a single-copy gene in all three plant species investigated, regulation of its expression seems to be complex. GnTI-deficient Arabidopsis cgl mutant plants also proved useful in expression and complementation analyses. We demonstrate that a drastic reduction of complex-glycan modification by stable antisense suppression of GntI did not interfere with the viability of either potato or tobacco transformants, suggesting that the production of therapeutic glycoproteins in agronomically important plant species is feasible.
RESULTS
Analysis of the Different Plant GntI-cDNA Sequences
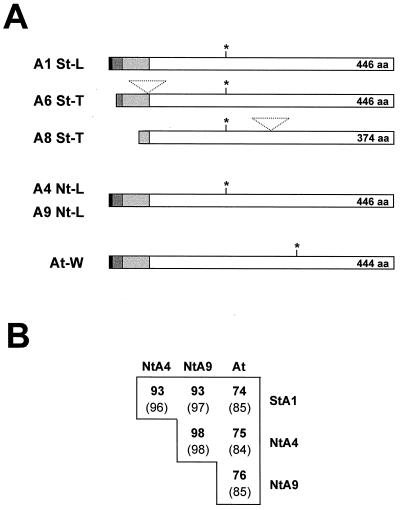
Using specific reverse transcriptase (RT)-PCR probes we isolated GntI-cDNA sequences from three higher plant species. The characteristics of the deduced GnTI primary structures are summarized schematically in Figure 1. From a potato leaf cDNA library, three clones were isolated. Sequence analyses showed that two of the clones were shorter versions of clone A1 (Fig. 1A, A1 St-L). The deduced amino acid sequence of A1 was complete (including 38 bp of 5′-untranslated leader sequence) and matched the general domain structure of a type II membrane protein (Nin/Cout; for review, see Singer, 1990) comprising a short NH2-terminal cytoplasmic tail, a single hydrophobic domain (transmembrane anchor), a lumenal hinge region, and a large C-terminal catalytic domain. Clone A1 was, therefore, used for further analyses.
Figure 1.
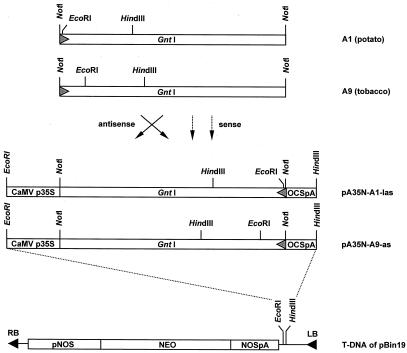
Characteristics of the isolated plant GntI-cDNA clones. A, Schematic representation of the deduced polypeptide sequences of the isolated plant GntI-cDNA clones. Potato leaf (A1 St-L) and tuber clones (A6 and A8 St-T), tobacco leaf (A4 and A9 Nt-L), and the assembled cDNA clone from Arabidopsis whole-plant tissue (At-W). Color code: black, cytosolic region; dark gray, membrane anchor; light gray, stalk region; white box, catalytic domain. The N-glycosylation sites are indicated by asterisks. Intron sequences in A6 and A8 are depicted as triangles. B, Homology matrix of the deduced amino acid sequences of the complete plant GntI-cDNA clones. Paired alignments were conducted with the GAP option of the Genetics Computer Group software package (Devereux et al., 1984). Percentage values for identical positions are shown in bold and those for similar ones in brackets. St, S. tuberosum (potato); Nt, N. tabacum (tobacco); At, Arabidopsis.
In addition to the above, two 5′ incomplete clones were isolated from a potato tuber cDNA library (Fig. 1A, A6/A8 St-T). Compared with the A1 leaf sequence, tuber cDNA A6 lacked 15 NH2-terminal amino acids and contained an insertion of 103 bp (between DV104Q and 105M, position numbers refer to A1, compare Fig. 2) that matched class II-intron characteristics (data not shown). Tuber cDNA A8 also contained an intron of 106 bp (between DV338Q and 339V, Fig. 2) with an imperfect 5′-splice junction (GC instead of GT, data not shown). Compared with the leaf A1 and tuber A6 cDNA sequences, A8 displayed a completely unrelated 5′ region, starting 6 bp upstream of Met codon 74M in A1 (5′-[EcoRI/NotI-cDNA linker]-CTG CTC TTG AAG GTG CAC TGT GTT CTC CGG TAA CAA ATG AAG … ; the deviating 5′ sequence is shown in italics, the in-frame stop codon is highlighted in bold, and the Met codon is underlined). The encoded isoform thus lacked the NH2-terminal transmembrane anchor, plus most of the hinge region (Fig. 1A), which should indicate a cytosolic location of the encoded protein. The two tuber sequences, A6 and A8, differed in four amino acid positions from each other (position numbers refer to A1: 123K↔R, 256Y↔D, 305R↔H, 396D↔A), and in five residues from the A1 leaf sequence (76R↔H, 118A↔T, 160D↔G, 170F↔Y, 434A↔S; see Fig. 2). The 3′-untranslated regions (UTRs) of the three potato cDNA sequences differed more so. The coding regions shared 98.6% to 99.3% identity at the nucleotide level (starting from common Met codon 74M in A1). Comparison between the 3′-UTRs of the two tuber cDNAs A6/A8, and tuber cDNA A8/leaf cDNA A1 gives similar values (93.3% and 93.6% identity, respectively), whereas tuber cDNA A6 and leaf cDNA A1 differed most (89.4% identity; data not shown).
Figure 2.
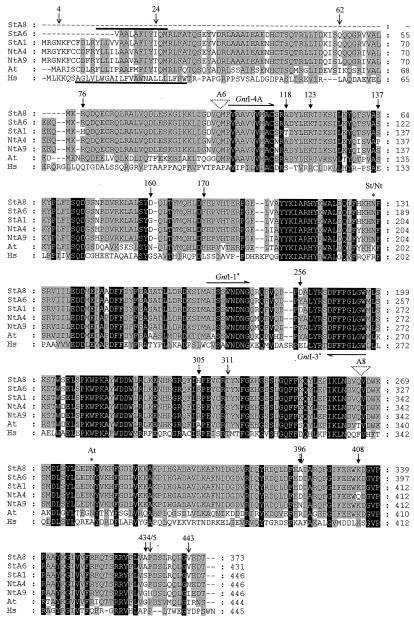
Alignment of the deduced plant GntI-polypeptide sequences with human GnTI. Amino acid residues identical in all sequences are highlighted by a black background, those conserved in most of the sequences are marked by a gray background. Hydrophobic regions in the membrane-anchor domains are indicated by a bold line above (plant GnTI) and underneath (human GnTI) the NH2-terminal sequence block. Primer binding sites are shown as arrows above (sense primers) and underneath (antisense primer) the sequence blocks. Sites of introns in tuber clones A6 and A8 are indicated by triangles. The N-glycosylation consensus sites in the plant sequences are marked by asterisks. St, S. tuberosum (potato); Nt, N. tabacum (tobacco); At, Arabidopsis (Arabidopsis); Hs, Homo sapiens (man). Arrows indicate positions of amino acid differences described in the text. [→] A6 and A8 compared with A1; [--▸] A6 compared with A8; [→] A4 compared with A9.
Two complete but different GntI-cDNA sequences were isolated from a tobacco leaf library (Fig. 1A, A4/A9 Nt-L). A4 displayed a short 5′-untranslated leader (only 26 bp with one mismatch compared with 112 bp in A9) but a much longer 3′-UTR (469 bp compared with 256 bp in A9). Except for 15 bp downstream of the stop codon, the two 3′-UTRs of A4 and A9 differ (91.4% identity compared with 98.75% identity in the coding regions). In a manner similar to the potato leaf and tuber sequences, there were nine amino acid differences between the polypeptides of the A4 and A9 tobacco leaf clones (4Y↔N, 24I↔T, 62Q↔L, 137P↔S, 311S↔T, 396D↔N, 408Q↔K, 435P↔H, 443N↔I; see Fig. 2), compared with over 30 differences detected between the leaf sequences of the two solanaceous species (potato A1 compared with tobacco A4 and A9; EMBL accession nos. AJ249878 and AJ249882; Fig. 2). However, in the amphidiploid tobacco genome these differences are thought to be a relic of two separate genomes after hybridization of the two diploid species Nicotiana sylvestris and Nicotiana tomentosiformis (Narayan, 1987).
Several GntI-cDNA clones with incomplete 5′ ends were isolated from a whole-plant Arabidopsis library. The clone with the longest insert was completed by assembly with the missing 5′ region (including 116 bp of 5′-untranslated leader sequence) obtained through vector-insert PCR (see “Materials and Methods”; Fig. 1A, At-W).
Figure 1B shows that the overall homologies shared between Arabidopsis GnTI and the two solanaceous species (about 75% identity and 85% similarity), as well as between potato and tobacco (93% identity and 96% similarity), and within tobacco (98% identity and 98% similarity), were within the range of values previously determined for other enzymes of primary and secondary plant metabolism (for review, see Wendt et al., 1999, and refs. therein). In contrast, homologies were extremely low, in the range of only 33% to 36% identity and 57% to 59% similarity (data not shown), compared with known animal GnTI sequences from man (Kumar et al., 1990), mouse (Pownall et al., 1992), rat (Fukada et al., 1994), and the worm Caenorhabditis elegans (Wilson et al., 1994). This is obvious in the alignment of the plant sequences with the human protein, although conserved amino acid blocks are also present (Fig. 2). Compared with human GnTI, the NH2-terminal transmembrane anchor of the plant sequences is characterized by a much shorter hydrophobic region (15 amino acids compared with 23 amino acids in human GnTI; Kumar et al., 1990), which is flanked by conserved positively charged Arg residues on both sides. In addition, all plant GnTI sequences identified to date contain one putative N-glycosylation site, which is the same in potato and tobacco (203NFS), but differs in context and position from the one in Arabidopsis (351NYT).
Estimation of GntI Gene Copy Number in Potato and Arabidopsis
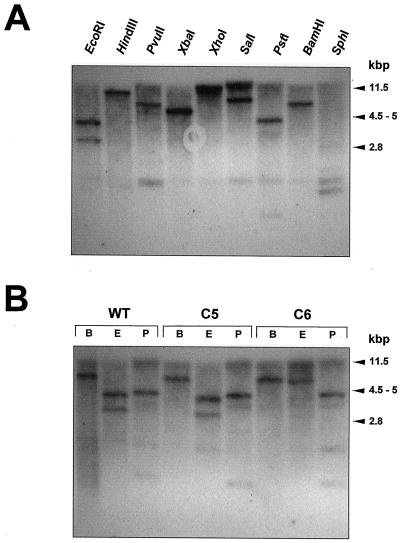
In light of the differences registered between the potato leaf- and tuber-cDNA sequences, we assessed GntI gene copy numbers by Southern-blot analyses using the respective plant-specific hybridization probes. With most restriction enzymes employed on genomic DNA, only one band lit up with potato and two with tobacco, which indicated that in the allotetraploid potato genome, GntI is present as single-copy gene (data not shown). Figure 3A shows the result obtained with genomic DNA from Arabidopsis: Five of the nine hexamer cutters give rise to a single hybridization signal. The two allelic Arabidopsis cgl mutants, C5 and C6, displayed differences in their EcoRI restriction patterns (Fig. 3B), which is in agreement with the fact that they result from independent mutation events (von Schaewen et al., 1993).
Figure 3.
Assessment of GntI-copy number in Arabidopsis. Total DNA was isolated from Arabidopsis leaves and 10-μg aliquots were digested overnight with the restriction enzymes (100 units of each) indicated above the blots. Samples were concentrated by sodium acetate-ethanol precipitation prior to separation in a 0.7% (w/v) agarose gel and subjected to Southern-blot analyses. A fragment comprising the entire Arabidopsis GntI cDNA was radioactively labeled and used as hybridization probe. Sizes of the molecular mass standard (λPstI) are indicated. A, Southern-blot analysis of Arabidopsis wild type; B, comparison of the restriction patterns in Arabidopsis wild type and in cgl mutant lines, C5 and C6. B, BamHI; E, EcoRI; P, PstI.
Analysis of GntI-mRNA Expression
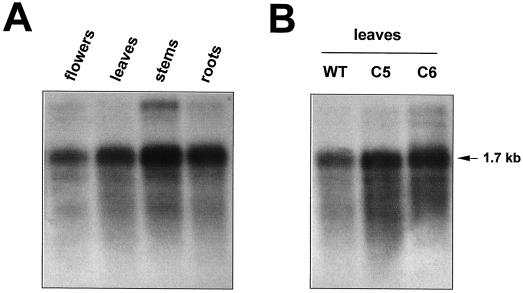
Northern-blot analyses conducted with total RNA indicated that in most plant tissues GntI expression is low and largely constitutive (data not shown). Detection improved upon using poly(A+) mRNA, which is shown here for Arabidopsis. The highest GntI-mRNA levels were detected in stems and the lowest was in flowers, whereas comparable expression levels are present in leaf and root tissue (Fig. 4A). In leaves of the two Arabidopsis cgl mutant lines, C5 and C6, GntI mRNA was also present. The elevated steady-state mRNA levels in the mutants compared with those in wild-type leaves (Fig. 4B) suggest that in the Arabidopsis cgl mutants, GntI-mRNA expression is up-regulated to compensate for missing GnTI activity.
Figure 4.
Analysis of GntI-mRNA expression in Arabidopsis wild type and the cgl mutant. Poly(A+) mRNA was isolated from 250 μg of total RNA and subjected to northern-blot analysis. The radioactively labeled hybridization probe was the same as described for Southern-blot analyses (Fig. 3). A, Different tissues of Arabidopsis wild-type plants; B, leaf samples of Arabidopsis wild type (WT) and Arabidopsis cgl mutants C5 and C6. The approximate size of the Arabidopsis GntI mRNA is indicated.
Complementation Test of GntI Expression Constructs
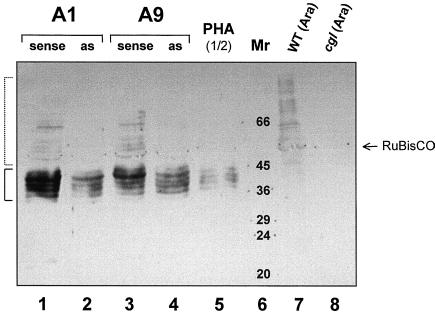
Complementation of the complex glycan defect in the Arabidopsis cgl mutant by a cDNA coding for human GnTI has been reported previously (Gomez and Chrispeels, 1994). To check for functionality of the isolated cDNA sequences prior to generating transgenic plants, cauliflower mosaic virus (CaMV)-35S promoter-driven potato and tobacco GntI-expression cassettes were constructed (Fig. 5). Both potato and tobacco sense and antisense constructs were used for transient expression analyses in protoplasts isolated from leaf tissue of back-crossed “non-stainer” progeny of Arabidopsis cgl mutant C5 (von Schaewen et al., 1993). The immunoblot in Figure 6 shows that, compared with a control mock-transformed with phytohemagglutinin (PHA), restoration of complex-glycan staining on high Mr secretory proteins (compare dotted bracket on the left side of the panel) occurred exclusively with constructs carrying the GntI-cDNA sequence in the sense, but not in the antisense orientation. Strong labeling of bands in the range of 36 to 45 kD was also observed, but only in protoplast samples cultivated for more than 48 h (Fig. 6, lanes 1–5) and not in whole-leaf extracts of Arabidopsis wild-type (Fig. 6, lane 7) or cgl mutant plants (Fig. 6, lane 8). The labeling of the cultivated protoplasts was most likely due to hemicellulose cell wall precursors of the xyloglucan type (for review, see Hayashi, 1989) synthesized in the Golgi and bound to proteins cross-reacting with the complex glycan antiserum. Staining increased with incubation time (data not shown) and signal strengths were always higher in the sense samples compared with antisense or mock-transformed control samples (Fig. 6; compare sense constructs in lanes 1 and 3 with antisense constructs in lanes 2 and 4, and with the mock-transformed PHA control in lane 5). Relief of the GnTI block in protoplasts transformed with GntI-sense constructs (Fig. 6, lanes 1 and 3) resulted in stronger labeling of a subset of the low Mr bands, indicating that some of the hemicellulose cell wall precursors are bound to glycoproteins containing glycans subject to complex glycan modification.
Figure 5.
Cloning scheme of potato and tobacco GntI-sense and -antisense constructs. The complete potato and tobacco NotI-cDNA fragments (A1, 1,660 bp; A9, 1,720 bp) were inserted between CaMV-35S promoter (CaMV p35S, approximately 550 bp) and octopine synthase termination sequences (OCSpA, approximately 200 bp) in plant expression vector pA35N (see “Materials and Methods”). Clones with GntI-cDNA inserts in sense and antisense orientation, respectively (indicated by arrows), were used for complementation tests (Fig. 6). The EcoRI-HindIII fragments comprising the entire plant expression cassettes of constructs pA35N-A1-las and pA35N-A9as, respectively, were inserted into binary vector pBin19 (Bevan, 1984) and used for the generation of transgenic potato and tobacco plants. LB/RB, Left and right T-DNA borders, respectively; pNOS, nopaline synthase promoter driving the neomycin phosphotransferase gene (NEO); NOSpA, nopaline synthase termination sequences.
Figure 6.
Complementation test of potato and tobacco GntI-expression constructs in the Arabidopsis cgl mutant. Protoplasts were isolated from 3- to 4-week-old C5 plants and transformed with plasmid DNA using the polyethylene glycol method. Subsequent cultivation was for 115 h at 25°C in the dark. Afterward, cells were chilled on ice, pelleted, and prepared for SDS-PAGE along with leaf samples harvested from Arabidopsis wild-type and cgl mutant plants. The western blot was stained with Ponceau S to check for comparable transfer and protein amounts (not shown) and developed with a complex glycan antiserum (Laurière et al., 1989). Lanes 1 through 5, Protoplast samples transformed with plasmid DNA of the following plant expression constructs: 1, potato GntI (A1) sense; 2, potato GntI (A1) antisense; 3, tobacco GntI (A9) sense; 4, tobacco GntI (A9) antisense; and 5, mock-transformed control (modified PHA-L; von Schaewen et al., 1993). Note that only one-half of the amount used in lanes 1 through 4 was loaded in lane 5 (PHA ½). Lane 6, Molecular-mass standard (Sigma SDS VII-L). Lanes 7 and 8, Whole-leaf control samples (50 μg of total protein each): 7, Arabidopsis wild type (WT); and 8, Arabidopsis mutant C5 (cgl). Molecular masses are given in kD. The position of Rubisco (large subunit) is indicated by an arrow. High molecular mass signals (≥45 kD bands) resulting from relief of the GnTI block in cgl protoplasts are marked by a dotted bracket on the left side of the panel. For origin of low molecular mass signals (36–45 kD bands) marked by a solid bracket, see explanation in the text.
Analysis of GntI Suppression in Transgenic Plants
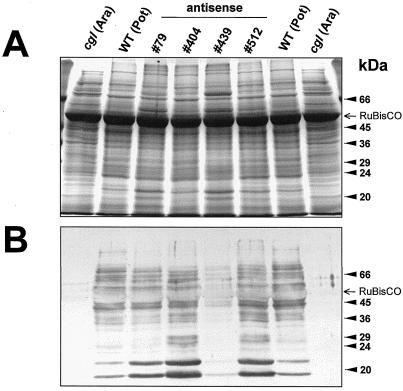
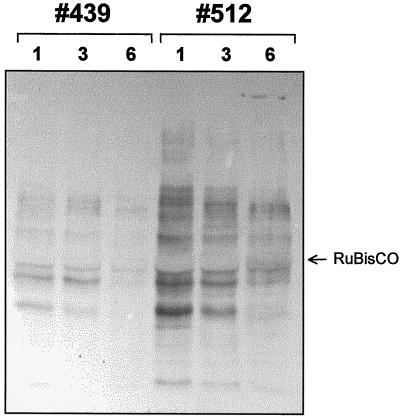
Upon verification of the potato and tobacco GntI-expression cassettes in the complementation test described above, binary vector constructs were generated (Fig. 5) and used for Agrobacterium tumefaciens-mediated delivery to leaf discs of the corresponding homologous plant species (i.e. tobacco-antisense constructs for tobacco and potato-antisense constructs for potato). After regeneration of transgenic plants, the extent of GntI suppression was assessed indirectly by immunoblot analyses using the complex glycan antiserum on leaf extracts of the primary transformants (T0 potato, n = 512; T0 tobacco, n = 120). In tissue culture, initially four (nos. 79, 404, 439, and 512) of the potato and three (nos. 1, 19, and 20) of the tobacco transformants showed substantial reduction in complex glycan staining compared with untransformed controls. After transfer of these candidates to soil and further propagation, immunoblot analysis was repeated with 3- to 4-week-old plants. The observations are shown for potato. Transformant number 439 repeatedly scored best for reduction of complex glycan formation (Fig. 7), indicating stability of transgene expression in this line. It is surprising that within the selected transformants, the extent of complex glycan reduction varied in leaves of different ages and developmental states. Figure 8 shows that antisense suppression of complex glycan formation progresses from primary to fully developed leaves in our strongest (no. 439) and weakest (no. 512) transformants. Thus, the GntI-antisense effect is much more pronounced in mature (source) compared with young (sink) leaves.
Figure 7.
Analysis of complex glycan patterns in leaf protein extracts of potato wild-type and selected antisense plants growing in tissue culture. A, Coomassie-stained SDS-gel reference (12% [w/v] polyacrylamide, 75 μg of total protein per lane); B, corresponding western blot developed with a complex glycan antiserum. WT (Pot), Untransformed potato wild type; cgl (Ara), Arabidopsis cgl mutant C5; numbers 79 through 512 (antisense), selected potato transformants. The positions of the molecular mass standard (Sigma SDS VII-L) and of Rubisco (large subunit) are indicated.
Figure 8.
Progression of complex glycan reduction in terminal leaves of potato GntI-antisense plants after growing for 4 weeks in soil. Western-blot analysis was analogous to Figure 7 (only that a 10% [w/v] polyacrylamide gel was used). Numbers 1, 3, and 6 refer to leaves counted from the top of the respective potato plants. Number 439, strongest; number 512, weakest potato GntI-antisense plant (compare Fig. 7). The position of Rubisco (large subunit) is indicated by an arrow.
DISCUSSION
Characteristics of the Different Plant GntI-cDNA Sequences
We report the isolation and characterization of several cDNA clones from three plant species encoding GnTI, the enzyme that initiates complex-type Asn-linked glycoprotein modification in the Golgi compartment of higher eukaryotes. A greater number of GntI-cDNA sequences with significant differences were obtained from potato leaf and tuber libraries of the same cultivar than were obtained for the two leaf cDNAs isolated from tobacco. The intron sequences identified in the two tuber GntI sequences indicate that they are derived from precursor mRNA molecules. The different 5′ region in tuber cDNA A8 could result from an imperfectly spliced and 5′ incomplete pre-mRNA, or it could indicate that GnTI is also present in the plant cytosol. Although the putative start codon in the A8 sequence (74M in A1, Fig. 2) does not fully comply with the Kozak rules (.ACCATGG..; Kozak, 1989), i.e. positions −3 and +4 should be purines (shown in bold, ACAAATGAAG), position −3 matches the consensus found in a compilation of 79 plant sequences (ACCAATGGCT; Joshi, 1987). In this context, it is noteworthy that an O-linked glycan structure has been identified on a plant nuclear-pore complex protein containing terminal GlcNAc residues preceded by a spacer of several hexoses (Heese-Peck et al., 1995). In contrast, on cytosolic glycoproteins of animals, GlcNAc residues are directly O-linked to the polypeptide backbone at alternatively phosphorylated sites (Hart et al., 1996). Thus, in plants a cytosolic GlcNAc transferase seems to exist. However, whether or not this activity is encoded by the A8 cDNA remains to be shown.
Compared with human GnTI, the significantly shorter hydrophobic membrane anchor that is flanked by conserved Arg residues on both sides in all plant GnTI sequences most likely reflects differences in Golgi-membrane structure and GnTI-retention mechanisms between animals and plants. The fact that the position of the single N-glycosylation site in the plant GnTI sequences is only conserved among the two solanaceous species, but not in Arabidopsis, argues against their actual use in planta, or at least indicates that they do not play a role other than modulation of the physicochemical properties of the polypeptide (e.g. stabilization of protein conformation; Olden et al., 1985).
Evidence for Complex Regulation of GntI-Gene Expression
The presence of two GntI genes in tobacco is consistent with the amphidiploid nature of the genome. The recently published GnTI sequence from tobacco (Strasser et al., 1999) is extremely similar to the A4 cDNA (one amino acid substitution at 408K↔Q, plus some base changes in the 3′-UTR) and differs in eight amino acid positions from cDNA A9. It is interesting that the differences between the two tobacco GntI-cDNA sequences lie in between those values determined for the potato leaf and tuber cDNAs (coding regions without introns starting from common Met codon 74M in the complete sequences: 98.6% ← 98.75% → 99.3%, and 3′-UTRs: 89.4% ← 91.4% → 93.6%). Despite the dissimilarities in GntI-cDNA sequences isolated from different potato tissues, Southern-blot analyses indicate that there is only one GntI locus present in all higher plant species investigated to date. For Arabidopsis, this was already suggested by the allelic character of the two independent cgl-mutant lines C5 and C6 (von Schaewen et al., 1993).
Due to the amphidiploid nature of the genome, two slightly different GntI copies are present in tobacco that are represented by the two cDNA clones A4 and A9 isolated from a tobacco leaf library of the same variety (for assessment of GntI-copy numbers in tobacco, see Strasser et al., 1999). However, differences in tissue-specific expression or additional GnTI variants could also be created by means of differential splicing. For example, a murine sialyltransferase gene is transcribed from at least three distinct promoter regions that are active in different tissues. Thus, a family of mRNAs with differing 5′-UTRs, but with identical protein-coding domains, is generated from the same gene (Lo and Lau, 1999).
Tissue-specific expression resulting from alternative splicing of the primary transcript has recently also been reported for a single-copy housekeeping gene in Arabidopsis (encoding enoyl-acyl carrier protein reductase; Boer et al., 1999). Another explanation for the differences observed between the potato leaf and tuber GntI cDNAs would be nuclear precursor mRNA editing (for review, see Keller et al., 1999). De-amination of adenosine to inosine bases, similar to that which occurs in primary transcripts of Glu receptor channels in mammals (for review, see Seeburg et al., 1998), could create alternative splice sites (as suggested by the imperfect 5′-splice junction of the intron in A8), or give rise to GnTI enzymes with slightly different amino acid composition, and thus enlarge the functional repertoire of the genome. Although a full understanding of the underlying mechanism(s) requires comparison with the corresponding genomic sequence, the differences between potato leaf- and tuber-GntI cDNAs provide circumstantial evidence that regulation of GntI-gene expression in plants is complex. Also, the fact that one of our tobacco GntI-cDNA sequences almost perfectly matches one described in a recent report (Strasser et al., 1999) argues strongly against them being merely the result of copy errors acquired during cDNA synthesis.
Molecular Analyses Employing Arabidopsis cgl Mutants
According to our northern-blot analyses, GntI-mRNA expression in plant tissues is largely constitutive, as has been reported for mammal (Kumar et al., 1990; Pownall et al., 1992; Fukada et al., 1994). This is not surprising for a Golgi-resident glycosyltransferase that belongs to the cellular housekeeping enzymes. Our observation that GntI mRNA is produced despite undetectable GnTI activity in the two Arabidopsis cgl mutants, C5 and C6, is consistent with the mutagen employed (ethyl methanesulfonate is known to predominantly create point mutations; von Schaewen et al., 1993). Moreover, the elevated steady-state mRNA levels detected in the two GnTI-deficient Arabidopsis cgl mutants suggest that GntI expression can be up-regulated on demand. A similar phenomenon has been reported for mutant mammalian cell lines with glycosylation defects that show up-regulation or induction of glycosyltransferase genes to alleviate the effects caused by the mutations (Stanley et al., 1996).
The complementation test of the GntI plant expression constructs in protoplasts of the Arabidopsis cgl mutant proved that the complete potato and tobacco GntI-cDNA sequences encode active enzymes. Besides the expected labeling of bands in the high molecular mass range (≥45 kD), labeling was enhanced in the range of 36 to 45 kD, but only upon long-term cultivation of protoplasts. This can be explained by a bona fide cross-reaction of protein-bound cell wall precursors massively synthesized in the Golgi of protoplasts that attempt to regenerate functional cell walls. It is known that hemicellulose of the xyloglucan-type constitutes about 20% in the primary cell wall of young growing dicotyledonous plants (Hayashi, 1989). Thus, it is feasible that the xyloglucan-carbohydrate moieties also bind antibodies of the complex glycan antiserum, which is largely an anti-Xyl/Fuc serum (Laurière et al., 1989). Signal strengths were always higher in sense compared with antisense or mock-incubated control samples. This shows that in protoplasts of the Arabidopsis cgl mutant, transiently expressed potato and tobacco GnTI enzymes act on components bound to these cell wall precursors, indicating that some of them are glycoproteins that acquire complex glycans due to relaxation of the GnTI block.
Developmental Progression of the GntI-Antisense Effect in Transgenic Plants
Subsequent use of the antisense constructs to generate transgenic plants resulted in the identification of potato and tobacco transformants with substantial reduction of complex glycan modification on their secretory glycoproteins. The phenomenon was more pronounced in fully developed source leaves compared with immature sink leaves. A comparable progression or gradient of the antisense effect from young to mature leaves was observed in tobacco following a similar antisense approach for suppressing genes encoding cytosolic Glc-6-P dehydrogenase (C. Lange, R. Hauschild, and A. von Schaewen, unpublished data) and cytosolic isocitrate dehydrogenase (S. Galvez and M. Hodges, personal communication). This was initially interpreted to result from some tissue specificity or sink-source response of the CaMV-35S promoter. However, as we now report similar observations for antisense suppression of GntI, it seems that the phenomenon may be associated with genes involved in primary metabolism (i.e. housekeeping genes), which are highly expressed in growing tissues (see Fig. 3 for GntI expression in Arabidopsis stem tissue and von Schaewen et al., 1995 for mRNA levels of cytosolic Glc-6-P dehydrogenase).
It is probable that a constant level of antisense message produced from the CaMV-35S promoter in the transgenic plants does not suffice for complete suppression of the corresponding endogenous genes in fast growing organs or metabolic sink situations. This is a potentially important observation that might explain the normal behavior of the potato and tobacco GntI-antisense plants under controlled growth conditions since it has been suggested that the Lewis-a epitope detected on complex-type glycans of most higher plant species—except in Arabidopsis and members of the Cruciferae family—might not only play a role in cell-to-cell recognition events in animals, but also in plants (Fitchette-Lainé et al., 1997). For a final judgement on this, specific plant-pathogen interaction experiments are necessary. Palacpac et al. (1999) recently demonstrated by a gain-of-function approach that expression of human β-1,4-galactosyltransferase also results in deficiency of Xyl and Fuc residues on plant complex glycans carrying novel terminal Gal residues. However, since the entire study was performed with undifferentiated tobacco BY-2 callus cells, viability of transgenic plants expressing human β-1,4-galacto-syltransferase remains to be shown.
Our observations demonstrate that effective reduction of GnTI activity can be achieved in mature tissues of solanaceous plants by means of GntI-mediated gene silencing. Thus, the resulting deficiency in highly immunogenic complex-type glycoprotein glycans is tolerated in plant species other than Arabidopsis. This now paves the way for the production of therapeutic glycoproteins in transgenic, agronomically important plant species carrying minimal compatible N-glycans of uniform GlcNAc2Man5 structure, a methodology previously reported for mammalian cell lines (Stanley, 1989).
MATERIALS AND METHODS
Plant Material
The following soil-grown plants were used in the experiments described: potato (Solanum tuberosum L. var Désirée), tobacco (Nicotiana tabacum L. var Wisconsin 38), Arabidopsis var Columbia, and Arabidopsis var Columbia cgl-mutant lines, C5 and C6 (von Schaewen et al., 1993).
Oligonucleotide Primers
Commercial sequencing primers KS and SK (Stratagene, Heidelberg) were used for cDNA-clone analyses. The following designed oligonucleotide primers were ordered from MWG BioTech (Ebersberg, Germany).
GntI-1*
Degenerate GntI-specific sense primer deduced from animal GntI sequences (human, rat, mouse, Caenorhabditis elegans, and Xenopus laevis), corresponding to region 239AISSWNDNG in potato GnTI; 26-mer, 5′-TG(CT) G(CT) I (AT)(GC) I GCI TGG (AC) A(CT) GA(CT) AA(CT) GG-3′.
GntI-3*
Degenerate GntI-specific antisense primer deduced from animal GntI sequences (human, rat, mouse, C. elegans, and X. laevis), corresponding to region 266DFFPGLGW in potato GnTI; 24-mer, 5′-CCA ICC IT(AG) ICC (AGCT) G(GC) (AG) AA (AG) AA (AG) TC-3′.
GntI-4A
Degenerate GntI-specific sense primer, including two 5′-restriction sites (HindIII, underlined; BamHI, in bold), corresponding to region 106PVAAVVVMAC in potato GnTI; 48-mer, 5′-ATC GGA AAG CTT GGA TCC CCA GTG GCC(A/G) GCT GTA GTT GTT ATG GCT TGC-3′.
pACT-Sense
Vector-specific primer based on the 5′ region in the multiple cloning site of two-hybrid vector pACT (Durfee et al., 1993), introducing two restriction sites (BamHI, underlined; XhoI, in bold); 27-mer, 5′-ATC TGG AAT TCG GAT CCT CGA GGC CAC-3′.
GntI-Bio
GntI-specific antisense primer with PstI restriction site (underlined) and 5′-biotin modification (designed for 5′ amplification of Arabidopsis GntI cDNA sequences), based on region 111MACSRADYL in Arabidopsis GnTI; 27-mer, bio5′-TTT CAA GAT AGT CTG CAC GAC TGC AGG-3′.
cDNA Synthesis and PCR Amplification (RT-PCR)
RT of poly(A+) mRNA from total plant RNA was performed as by Graeve et al. (1994), modified by von Schaewen et al. (1995).
Standard PCR reactions were run essentially as described by Graeve et al. (1994) using 30 to 50 pmol of each primer, 200 μm dNTPs, and 2 units of Taq-DNA polymerase per 50 μL reaction in the buffer supplied (Angewandte Geutechnologie Systeme, Heidelberg).
Cloning of Species-Specific GntI-PCR Products
To obtain optimal hybridization probes for the subsequent plant cDNA-library screenings, degenerate primers corresponding to highly conserved regions in animal GnTI sequences were used to amplify plant-specific GntI-cDNA fragments from potato and tobacco leaf RNA. The resulting approximately 90-bp RT-PCR products were gel-purified using the Qiaex II Gel Extraction Kit (Qiagen, Hilden, Germany), subcloned in EcoRV digested pBSK (Stratagene) by standard procedures (Sambrook et al., 1989), introduced into RbCl-competent (Hanahan, 1983) Escherichia coli XL1-Blue cells (Stratagene), and sequenced. The deduced primary structures showed similarity to known GnTI sequences from animals (→ Q(R/M)QFVQDP(D/Y)ALYRS ← primers depicted as arrows, homologous amino acid residues underlined). The potato fragment was used as a radiolabeled probe for screening the corresponding leaf- and tuber-cDNA libraries. After the complete amino acid sequence was deduced from potato, degenerate primer GntI-4A was designed and used in combination with GntI-3* to amplify longer specific cDNA fragments (492 bp) from Arabidopsis and tobacco RNA. RT-PCR products of the appropriate sizes were gel-purified, subcloned in pGEM-T (Promega, Mannheim, Germany), sequenced, and used as described above.
Labeling of cDNA Probes
For Southern and northern analyses and for screening of cDNA libraries, GntI-cDNA fragments were liberated by restriction digests, gel-purified, and radioactively labeled essentially as described in von Schaewen et al. (1995), using 32P(dCTP) and the HexaLabel DNA Labeling Kit (MBI Fermentas, St. Leon-Rot, Germany).
Isolation of GntI cDNA Clones
Screening of potato leaf- and tuber-cDNA, and tobacco leaf-cDNA libraries constructed in λ ZAP II (Stratagene) with EcoRI/NotI adapters (Pharmacia, Freiburg, Germany), and of a whole plant Arabidopsis cDNA library constructed in λ Uni-ZAP (Stratagene) with 5′-EcoRI and 3′-XhoI cDNA ends, was by standard procedures (Sambrook et al., 1989) using radiolabeled specific GntI-cDNA probes obtained from the appropriate species (described above). In vivo excision of the phagemid clones identified followed the instructions in the Stratagene manual.
Vector-Insert PCR
Amplification of the 5′ end of the Arabidopsis GntI sequence was conducted by vector-insert PCR according to Tillmann and Eschrich (1998) using a cDNA library constructed in λACT XhoI (Durfee et al., 1993) after quantitative conversion to pACT phagemid DNA. In a final volume of 40 μL, the initial reactions contained different amounts of plasmid DNA, 25 nmol primer GntI-bio, dNTPs (200 μm each), and 2 units of Pfu-DNA polymerase (Stratagene) in the buffer supplied. Incubation was for 2 min at 95°C followed by 30 cycles for 30 s at 95°C and 3 min at 70°C. Isolation of biotinylated DNA was with 25 μL of Dynabeads M-280 Streptavidin (Dynal, Hamburg, Germany) following the protocol of the supplier. Subsequent PCR conditions were as described above using aliquots of biotinylated DNA as template in combination with primers pACT-sense and GntI-bio. Amplified fragments of ≥300 bp were gel-purified, digested with XhoI and PstI, ligated to the respective ends in pBluescript SK (Stratagene), and cloned in E. coli XL1-Blue cells (Stratagene). The open reading frame of the clone with the longest insert (440 bp) contained the complete 5′ end of Arabidopsis-GntI, and served as the template for PCR amplification with vector primer SK (25-mer) and GntI-bio. To assemble the full-length cDNA sequence, the obtained fragments were digested with XbaI and SpeI, ligated to the respective sites in 5′ incomplete Arabidopsis GntI cDNA clone 10.2-1, and transformed into E. coli XL1-Blue cells. The resulting complete Arabidopsis GntI cDNA clone, termed pBSK-AraGntI-full, was used for further analyses.
DNA-Sequence Analysis
Sequence reactions were conducted as described in Wenderoth et al. (1997). Double stranded sequence analyses of the GntI cDNAs were conducted by a commercial sequencing service (MWG BioTech, Ebersberg, Germany). The data were analyzed with appropriate programs of the Genetics Computer Group (Madison, WI) software package (Devereux et al., 1984). Sequence alignments were performed using CLUSTAL W version 1.7 (Thompson et al., 1994) and GeneDoc (Nicholas and Nicholas, 1997) programs.
Nucleic Acid Analyses
For preparation of nucleic acids, tissue of 3- to 4-week-old plants was frozen in liquid nitrogen. Genomic DNA and total RNA were isolated using a RNA/DNA Midi Kit (Qiagen). Poly(A+) mRNA was purified from 250 μg of total RNA using the Oligotex mRNA Mini Kit (Qiagen) and separated in a denaturing agarose gel following standard techniques (Sambrook at al., 1989).
Southern analyses of genomic DNA and preparation of RNA samples for northern analyses were performed as described previously (von Schaewen et al., 1995). DNA fragments were transferred to Qiabrane Plus Nylon membranes (Qiagen) and denatured RNA to Hybond N Nylon membranes (Amersham/USB, Braunschweig, Germany). Nucleic acids were immobilized on the blots by UV-cross linking and probed with radioactively labeled full-length cDNA fragments of the corresponding GntI-cDNA clones. Filters were washed three times for 30 min at 68°C with 1× SSC (Sambrook et al., 1989) containing 0.1% (w/v) SDS.
Generation of Binary Vector Constructs for Plant Transformation
To allow for direct insertion of NotI digested cDNA sequences, the expression cassette of pA35S (Höfte et al., 1991) was modified by inserting a NotI linker (8-mer, NE-Biolabs, Bad Schwalbach, Germany) into the filled SalI site of the multiple cloning region, yielding plant expression vector pA35N. Subsequently, the full-length NotI cDNA fragment of potato clone A1 (1,660 bp) was introduced into pA35N. The resulting clones were analyzed for insert orientation with respect to the flanking CaMV-35S promoter and octopine synthase termination sequences, and termed pA35N-A1-s (for sense) and pA35N-A1-las (for long antisense), respectively. The cloning strategy for the tobacco-cDNA fragments was analogous. Insertion of the NotI-cDNA fragment of tobacco clone A9 (1,717 bp) into pA35N resulted in constructs pA35N-A9-s and pA35N-A9-as. The constructs were tested for complementation of the complex glycan defect in protoplasts of the Arabidopsis cgl mutant. Upon verification of their functionality, the complete expression cassettes were obtained by partial digests and introduced as EcoRI-HindIII fragments (2,415 bp for A1 and 2,472 bp for A9, respectively) into the T-DNA region of binary vector pBin19 (Bevan, 1984; compare Fig. 5). The resulting binary constructs were transformed into Agrobacterium tumefaciens strain GV2260 (Deblaere et al., 1985) using the freeze-thaw method described by Höfgen and Willmitzer (1988). Purified plasmid DNA of single transformants was checked by restriction digests prior to using selected clones for cocultivation with sterile potato and tobacco leaf discs.
Transient Expression in Protoplasts of the Arabidopsis cgl Mutant
Transient expression of pA35N-GntI-sense and -antisense constructs in protoplasts of Arabidopsis cgl mutant C5 was performed as described previously (von Schaewen et al., 1990) with the modification that 30 μg of plasmid DNA were used to transform 106 protoplasts.
Leaf-Disc Transformation and Regeneration of Transgenic Plants
Transgenic plants were regenerated in tissue culture under continuous kanamycin selection (100 μg mL−1 for potato and 200 μg mL−1 for tobacco). A. tumefaciens-mediated transformation of leaf explants was essentially as described by Rocha-Sosa et al. (1989) for potato and as given by Faske et al. (1997) for tobacco.
Protein Determination, SDS-PAGE, and Immunoblot Analyses
Estimation of protein concentrations, SDS-PAGE, and subsequent immunoblot analyses were conducted as described by Graeve et al. (1994) with the modification that protein extraction buffer without SDS was used (von Schaewen et al., 1993). Blots were developed using a complex glycan antiserum as described by von Schaewen et al. (1993). A fresh batch of antiserum with essentially the same characteristics as described previously (Laurière et al., 1989) was prepared by a commercial service lab (BioScience, Göttingen, Germany) using PHA-L (Sigma, Deisenhofen, Germany) for injection into rabbits.
ACKNOWLEDGMENTS
The potato and tobacco cDNA libraries were kind gifts from the group of Uwe Sonnewald (Institute für Pflanzengenetic und Kulturpflanzen-forschung, Gatersleben, Germany). The different Arabidopsis cDNA libraries (“Leiden” phage and “Walker” plasmid library) were provided by Csaba Koncz (Max-Planck Institut für Züchtungsforschung, Köln, Germany). The excellent technical assistance of Monika Nietschke and the help of Urte Wendt and Joachim Tjaden in computer-based sequence analyses are gratefully acknowledged. The authors thank Jan Mucha (Universität für Bodenkultur, Vienna) for sharing information on X. leavis GntI- and GntII-cDNA sequences prior to publication, and Susanna Galvez and Michael Hodges, Universite de Paris-Sud, France, for sharing information on CaMV 35S promoter-driven expression patterns in tobacco.
Footnotes
This work was financially supported by the Deutsche Forschungsgemeinschaft (Scha 541/4).
LITERATURE CITED
- Barnes G, Hansen WJ, Holcomb CL, Rine J. Asparagine-linked glycosylation in Saccharomyces cerevisiae: genetic analysis of an early step. Mol Cell Biol. 1984;4:2381–2388. doi: 10.1128/mcb.4.11.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan MW. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer GJ, Testerink C, Pielage G, Nijkamp HJ, Stuitje AR. Sequences surrounding the transcription initiation site in the Arabidopsis enoyl-acyl carrier protein reductase gene control seed expression in transgenic tobacco. Plant Mol Biol. 1999;39:1197–1207. doi: 10.1023/a:1006129924683. [DOI] [PubMed] [Google Scholar]
- Dahms NM, Lobel P, Kornfeld S. Mannose-6-phosphate receptors and lysosomal enzyme targeting. J Biol Chem. 1989;264:12115–12118. [PubMed] [Google Scholar]
- Deblaere R, Bytebier B, De Greve H, Debroeck F, Schell J, van Montagu M, Leemans J. Efficient octopine Ti plasmid-derived vectors of Agrobacterium mediated gene transfer to plants. Nucleic Acids Res. 1985;13:4777–4788. doi: 10.1093/nar/13.13.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Faske M, Backhausen JE, Sendker M, Singer-Bayerle M, Scheibe R, von Schaewen A. Transgenic tobacco plants expressing pea chloroplast Nmdh cDNA in sense and antisense orientation. Plant Physiol. 1997;115:705–715. doi: 10.1104/pp.115.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitchette-Lainé A-C, Gomord V, Cabanes M, Michalski J-C, Saint Macary M, Foucher B, Cavelier B, Hawes C, Lerouge P, Faye L. N-Glycans harboring the Lewis a epitope are expressed at the surface of plant cells. Plant J. 1997;12:1411–1417. doi: 10.1046/j.1365-313x.1997.12061411.x. [DOI] [PubMed] [Google Scholar]
- Fukada T, Iida K, Kioka N, Sakai H, Komano T. Cloning of a cDNA encoding N-acetylglucosaminyltransferase I from rat liver and analysis of its expression in rat tissues. Biosci Biotech Biochem. 1994;58:200–201. doi: 10.1271/bbb.58.200. [DOI] [PubMed] [Google Scholar]
- Gomez L, Chrispeels MJ. Complementation of an Arabidopsis thaliana mutant that lacks complex asparagine- linked glycans with the human cDNA encoding N-acetyl-glucosaminyltransferase I. Proc Natl Acad Sci USA. 1994;91:1829–1833. doi: 10.1073/pnas.91.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeve K, von Schaewen A, Scheibe R. Purification, characterization, and cDNA sequence of glucose-6-phosphate dehydrogenase from potato (Solanum tuberosum L.) Plant J. 1994;5:353–361. doi: 10.1111/j.1365-313x.1994.00353.x. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hart GW, Kreppel LK, Comer FI, Arnold CS, Snow DM, Ye Z, Cheng X, Della Manna D, Caine DS, Earles BJ, Akimoto Y, Cole RN, Hayes BK. O-GlcNAcylation of key nuclear and cytoskeletal proteins: reciprocity with O-phosphorylation and putative roles in protein multimerization. Glycobiology. 1996;6:711–716. doi: 10.1093/glycob/6.7.711. [DOI] [PubMed] [Google Scholar]
- Hayashi T. Xyloglucans in the primary cell wall. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:139–168. [Google Scholar]
- Heese-Peck A, Cole RN, Borkhsenious ON, Hart GW, Raikhel NV. Plant nuclear pore complex proteins are modified by novel oligosaccharides with terminal N-acetylglucosamine. Plant Cell. 1995;7:1459–1471. doi: 10.1105/tpc.7.9.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfgen R, Willmitzer L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988;16:9877. doi: 10.1093/nar/16.20.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfte H, Faye L, Dickinson C, Herman EM, Chrispeels MJ. The protein-body proteins phytohemagglutinin and tonoplast intrinsic protein are targeted to vacuoles in leaves of transgenic tobacco. Planta. 1991;184:431–437. doi: 10.1007/BF00197889. [DOI] [PubMed] [Google Scholar]
- Ioffe E, Liu Y, Stanley P. Essential role for complex N-glycans in forming an organized layer of bronchial epithelium. Proc Natl Acad Sci USA. 1996;93:11041–11046. doi: 10.1073/pnas.93.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi CP. An inspection of the domain between the putative TATA box and translation start in 79 plant genes. Nucleic Acids Res. 1987;15:6643–6653. doi: 10.1093/nar/15.16.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal GP, Szumilo T, Elbein AD. Structure and biosynthesis of plant N-linked glycans. In: Preiss J, editor. The Biochemistry of Plants. 14: Carbohydrates. San Diego: Academic Press; 1988. pp. 421–463. [Google Scholar]
- Keller W, Wolf J, Gerber A. Editing of messenger RNA precursors and of tRNAs by adenosine to inosine conversion. FEBS Lett. 1999;452:71–76. doi: 10.1016/s0014-5793(99)00590-6. [DOI] [PubMed] [Google Scholar]
- Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Yang J, Larsen RD, Stanley P. Cloning and expression of N-acetylglucosaminyltransferase I, the medial Golgi transferase that initiates complex amino-linked carbohydrate formation. Proc Natl Acad Sci USA. 1990;87:9948–9952. doi: 10.1073/pnas.87.24.9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurière M, Laurière C, Chrispeels MJ, Johnson KD, Sturm A. Characterization of a xylose-specific antiserum that reacts with the complex asparagine-linked glycans of extracellular and vacuolar glycoproteins. Plant Physiol. 1989;90:1182–1188. doi: 10.1104/pp.90.3.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo NW, Lau JT. Transcription of the β-galactoside α-2,6-sialyltransferase gene (SIAT1) in B-lymphocytes: cell type-specific expression correlates with presence of the divergent 5′-untranslated sequence. Glycobiology. 1999;9:907–914. doi: 10.1093/glycob/9.9.907. [DOI] [PubMed] [Google Scholar]
- Narayan RKJ. Nuclear DNA changes, genome differentiation and evolution in Nicotiana (Solanaceae) Plant Syst Evol. 1987;157:161–180. [Google Scholar]
- Nicholas KB, Nicholas HB. GeneDoc: a tool for editing and annotating multiple sequence alignments. 1997. Distributed by the authors. [Google Scholar]
- Olden K, Bernard BA, Humphries MJ, Yeo T-K, Yeo K-T, White SL, Newton SA, Bauer HC, Parent JB. Function of glycoprotein glycans. Trends Biochem Sci. 1985;10:78–82. [Google Scholar]
- Palacpac NQ, Yoshida S, Sakai H, Kimura Y, Fujiyama K, Yoshida T, Seki T. Stable expression of human β-1,4-galactosyltransferase in plant cells modifies N-linked glycosylation patterns. Proc Natl Acad Sci USA. 1999;96:4692–4697. doi: 10.1073/pnas.96.8.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pownall S, Kozak CA, Schappert K, Sakar M, Hull E, Schachter H, Marth JD. Molecular cloning and characterization of the mouse UDP-N-acetylglucosamine α-3-d-mannoside β-1,2-N-acetylglucosaminyltransferase I gene. Genomics. 1992;12:699–704. doi: 10.1016/0888-7543(92)90297-6. [DOI] [PubMed] [Google Scholar]
- Rocha-Sosa M, Sonnewald U, Frommer W, Stratmann M, Schell J, Willmitzer L. Both developmental and metabolic signals activate the promoter of a class I patatin gene. EMBO J. 1989;8:23–29. doi: 10.1002/j.1460-2075.1989.tb03344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scheiffele P, Peränen J, Simons K. N-Glycans as apical sorting signals in epithelial cells. Nature. 1995;378:96–98. doi: 10.1038/378096a0. [DOI] [PubMed] [Google Scholar]
- Seeburg PH, Higuchi M, Sprengel R. RNA editing of brain glutamate receptor channels: mechanism and physiology. Brain Res Brain Res Rev. 1998;26:217–229. doi: 10.1016/s0165-0173(97)00062-3. [DOI] [PubMed] [Google Scholar]
- Singer SJ. The structure and insertion of integral proteins in membranes. Annu Rev Cell Biol. 1990;6:247–296. doi: 10.1146/annurev.cb.06.110190.001335. [DOI] [PubMed] [Google Scholar]
- Stanley P. Glycosylation mutants of animal cells. Annu Rev Genet. 1984;18:525–552. doi: 10.1146/annurev.ge.18.120184.002521. [DOI] [PubMed] [Google Scholar]
- Stanley P. Chinese hamster ovary cell mutants with multiple glycosylation defects for production of glycoproteins with minimal carbohydrate heterogeneity. Mol Cell Biol. 1989;9:377–383. doi: 10.1128/mcb.9.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P, Raju TS, Bhaumik M. CHO cells provide access to novel N-glycans and developmentally regulated glycosyltransferases. Glycobiology. 1996;6:695–699. doi: 10.1093/glycob/6.7.695. [DOI] [PubMed] [Google Scholar]
- Strasser R, Mucha J, Schwihla H, Altmann F, Glössl J, Steinkellner H. Molecular cloning and characterization of cDNA coding for β-1,2-N-acetylgluco-saminyltransferase I (GlcNAc-TI) from Nicotiana tabacum. Glycobiology. 1999;9:779–785. doi: 10.1093/glycob/9.8.779. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmann H, Eschrich K. Isolation and characterization of an allelic cDNA for human muscle fructose-1,6-bisphosphatase. Gene. 1998;212:295–304. doi: 10.1016/s0378-1119(98)00181-4. [DOI] [PubMed] [Google Scholar]
- Varki A. Sialic acids as ligands in recognition phenomena. FASEB J. 1997;11:248–255. doi: 10.1096/fasebj.11.4.9068613. [DOI] [PubMed] [Google Scholar]
- von Schaewen A, Langenkämper G, Graeve K, Wenderoth I, Scheibe R. Molecular characterization of the plastidic glucose-6-phosphate dehydrogenase from potato in comparison to its cytosolic counterpart. Plant Physiol. 1995;109:1327–1335. doi: 10.1104/pp.109.4.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schaewen A, Stitt M, Schmidt R, Sonnewald U, Willmitzer L. Expression of a yeast-derived invertase in the cell wall of tobacco and Arabidopsis plants leads to accumulation of carbohydrate and inhibition of photosynthesis and strongly influences growth and phenotype of transgenic tobacco plants. EMBO J. 1990;9:3033–3044. doi: 10.1002/j.1460-2075.1990.tb07499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schaewen A, Sturm A, O'Neill J, Chrispeels MJ. Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiol. 1993;109:1327–1335. doi: 10.1104/pp.102.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenderoth I, Scheibe R, von Schaewen A. Identification of the cysteine residues involved in redox modification of plant plastidic glucose-6-phosphate dehydrogenase. J Biol Chem. 1997;272:26985–26990. doi: 10.1074/jbc.272.43.26985. [DOI] [PubMed] [Google Scholar]
- Wendt UK, Hauschild R, Lange C, Pietersma M, Wenderoth I, von Schaewen A. Evidence for functional convergence of redox regulation in G6PDH isoforms of cyanobacteria and higher plants. Plant Mol Biol. 1999;40:487–494. doi: 10.1023/a:1006257230779. [DOI] [PubMed] [Google Scholar]
- Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J, Coulson A, Craxton M, Dear S, Du Z, Durbin R, Favello A, Fraser A, Fulton L, Gardner A, Green P, Hawkins T, Hillier L, Jier M, Johnston L, Jones M, Kershaw J, Kirsten J, Laisster N, Latreille P, Lightning J, Lloyd C, Mortimore B, O'Callaghan M, Parsons J, Percy C, Rifken L, Roopra A, Saunders D, Shownkeen R, Sims M, Smaldon N, Smith A, Vaughan K, Waterston R, Watson A, Weinstock L, Wilkinson-Sproat J, Wohldman P. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]