Abstract
Memory encoding is an essential step for all learning. However, the genetic and molecular mechanisms underlying human memory encoding remain poorly understood, and how this molecular framework permits the emergence of specific patterns of brain oscillations observed during mnemonic processing is unknown. Here, we directly compare intracranial electroencephalography recordings from the neocortex in individuals performing an episodic memory task with human gene expression from the same areas. We identify genes correlated with oscillatory memory effects across 6 frequency bands. These genes are enriched for autism-related genes and have preferential expression in neurons, in particular genes encoding synaptic proteins and ion channels, supporting the idea that the genes regulating voltage gradients are involved in the modulation of oscillatory patterns during successful memory encoding across brain areas. Memory-related genes are distinct from those correlated with other forms of cognitive processing and resting state fMRI. These data are the first to identify correlations between gene expression and active human brain states as well as provide a molecular window into memory encoding oscillations in the human brain.
Keywords: gene expression, intracranial EEG, memory, neocortex
Introduction
Comparing brain activity during successful versus unsuccessful memory encoding permits exploration of the neurophysiological mechanisms that underlie episodic memory formation. Observed differences in activity based on encoding success are termed subsequent memory effects (SMEs), and the spatial and temporal resolution afforded through the use of intracranial electroencephalography (iEEG) allows for the identification of SMEs across the brain (Sederberg et al. 2007; Lega et al. 2016). Successful encoding is often associated with a low frequency power decrease (local desynchronization) and gamma band power increase, however, important differences in the magnitude and preferred frequency range for this effect have been observed both within a core network for item encoding and in adjacent brain regions (Sederberg et al. 2007; Hanslmayr et al. 2012).
We sought to link for the first time the expanding literature for intracranial SME with gene expression datasets. We modeled our analytic methods on recent work by our group and others suggesting there are gene expression patterns that match resting state brain activity across multiple cortical locations (Hawrylycz et al. 2015; Richiardi et al. 2015; Wang et al. 2015; Krienen et al. 2016). We used similar methodology for this analysis, using a large existing database of oscillatory SMEs across the brain. We used RNA-seq data and iEEG to identify genes correlated with oscillatory activity across 14 neocortical regions, suggesting several gene targets that may influence memory performance. We also used co-expression network analysis to ascertain the co-expression of oscillatory SME-correlated genes together with genes involved in several brain disorders, highlighting potential therapeutic inroads into diseases for which memory disorders are often comorbid. Finally, we performed an additional control analysis by correlating gene expression separately with brain oscillatory activity recorded as the same group of subjects performed basic mathematical calculations (data collected as part of the distractor portion of the free recall task). This revealed a distinct set of nonoverlapping genes as compared with the memory analysis, giving confidence to the assessment that our methods were able to identify a set of genes unique to mnemonic processing rather than genes that were correlated with cognitive activity more generally.
Such an analysis fills an important gap in the literature: the molecular mechanisms and regional differences in gene expression that may support these observed differences in oscillatory patterns are currently unknown (Wang 2010; Buzsaki and Wang 2012). Previous work has suggested that there is a genetic component to memory performance (McClearn et al. 1997; Volk et al. 2006; Panizzon et al. 2011). Most previous genetic studies of memory have focused on uncovering variants associated with disorders that have associated memory deficits such as schizophrenia or Alzheimer's disease (AD). At least one gene, WWC1 (also known as KIBRA), has been associated not only with these disorders, but also with memory performance in control cohorts (Papassotiropoulos et al. 2006; Schwab et al. 2014; Vyas et al. 2014; Kos et al. 2016). However, outside of the identification and study of WWC1, there have been few studies linking specific genes to normal memory function, and no studies directly linking genes in humans, either at the level of variants in the DNA or in their mRNA expression in the brain, to memory-related oscillations.
Fundamentally, the type of investigation we report here (correlating SME patterns and gene expression across the brain using large, existing datasets) is hypothesis-generating, designed to identify gene targets for further experimentation in animal models of memory function. However, our efforts were guided by several testable hypotheses. Foremost we hypothesized that the correlated genes we observed would be distinct from those correlated with resting state brain activity and different from those linked to mathematical processing based on our expectation of differences in brain activity for task based memory activation. Previous studies seeking to correlate EEG evoked potentials (outside of a memory task) with genetic variations posited an association with genes encoding ion channels and other receptor proteins such as GABRA2, CHRM2, and GRM8 (Rangaswamy and Porjesz 2008). We therefore hypothesized that the brain oscillation pattern across cortical regions would be correlated with genes known to affect ion channel function, along with genes previously linked to memory disorders. Finally, we hypothesized a priori that the most gene correlations would be related to gamma band effects given their hypothesized association with local cortical activation in cognition ((Lachaux et al. 2007) but see also (Hanslmayr et al. 2016)). In the work described here, we present unique data derived from human brain datasets. We believe our results will provide researchers with a set of gene targets for memory-related experimentation, some of which are nonintuitive and might otherwise be overlooked as gene candidates. Animal studies investigating these genes will further explicate how those we identify are related to memory function and the genesis of SMEs across the brain.
Materials and Methods
iEEG Data Processing and Analysis
iEEG data was collected over a period of 10 years across 2 institutions (University of Pennsylvania, IRB# 820 461 and Thomas Jefferson University, IRB# 08 F.464 R). Overall, 45 of the participants have contributed data to previous publications (Burke et al. 2014; Long and Kahana 2015; Lega et al. 2016). While the total study contains data from 29 cortical regions in 183 patients, the analysis reported here only uses data from 14 neocortical regions in the left hemisphere of 66 male patients. This is because this subset of data matches the left neocortical gene expression dataset from 2 male postmortem brain expression datasets (Kang et al. 2011; Wang et al. 2015). Participants performed the free recall task, a standard test of episodic memory. The experimental design has been described in several previous publications (Lega et al. 2012). In brief, participants were presented with 25 lists of memory items (high frequency nouns, 15 items per list) and instructed to remember as many as possible. Following a distractor task consisting of simple addition problems, they were given 30 s to retrieve as many memory items as they could in any order. Words vocalized at the retrieval period were used to classify encoding events as successful or unsuccessful; the magnitude of the SME for each frequency band was calculated using this classification (difference in oscillatory power between recalled and nonrecalled events). None of the patients had a known diagnosis of a heritable epilepsy disorder. Participants contributed Z values for a given Brodmann area only if they had electrodes placed in this area for phase II monitoring.
Cortical oscillatory signals were obtained from brain surface contacts in subdural electrode grid and strip arrays (only surface electrodes were used for this study due to the availability of matching gene data). Contacts were standard platinum-iridium contacts 2 mm in diameter placed via open craniotomy. Electrodes were localized to Brodmann areas using standard methods previously published (Sederberg et al. 2007; Long and Kahana 2015). In brief, Talairach coordinates were determined by expert identification of electrode contacts after fusion of patient brains with standard atlas brains using FSL software. These coordinates were used to assign electrodes to Brodmann areas using the standard atlas (Jenkinson et al. 2012). All electrodes within the Brodmann areas of interest for each of the 66 male participants in the analysis were included, except for electrodes located at or adjacent to the ictal onset location or for which manual review of EEG signal at the time of data collection revealed significant artifact. In total, 5 153 electrodes were available for analysis with each patient contributing an average of 5.6 electrodes in each Brodmann area. We did not restrict our analysis to a minimum number of electrodes or a minimum number of patients for each area for the analysis. Table S1 contains the number of electrodes from each Brodmann area used per patient. In total, 14 Brodmann regions were used from the participants (BA8, BA9, BA10, BA11, BA20, BA21, BA22, BA39, BA40, BA42, BA44, BA45, BA46, and BA47) to carry out correlation analyses. We made the a priori decision to exclude primary motor, sensory, and visual cortex from the correlation analysis based on these regions exhibiting quite different expression profiles compared with relatively homogenous gene expression across other regions of the human cortex (Hawrylycz et al. 2015), along with relatively less sampling from these regions in the iEEG memory dataset.
Intracranial EEG activity was initially recorded between 256 and 1000 Hz and rereferenced using a global average re-referencing scheme. Electrodes located immediately within seizure onset locations were excluded from the analysis. Inter-ictal spiking activity and other events with signal artifact were further excluded by applying a kurtosis rejection algorithm that has been previously published (Sederberg et al. 2003, 2007) with a (conservative) threshold of 4. Morlet wavelets (wave number 6) were used to extract oscillatory power at each frequency with a 500 ms buffer to eliminate edge artifacts. Power values were averaged across the entire encoding epoch (1600 ms) that followed the onset of the presentation of memory items. We compared the extracted oscillatory power after log transforming the raw values between recalled and nonrecalled distributions using a two-tailed t-test. We used non-normalized data for the power analysis to avoid the influence of different normalization schemes on the results. This generated a P-value of the difference between the distribution of oscillatory power for remembered and forgotten words for each electrode and at each frequency. The P-values were then Z-transformed using the cumulative distribution function to permit combining data across regions, electrodes, and bands.
The Z values for each electrode were averaged across 6 frequency bands defined based on standard human EEG categorization (Hanslmayr and Staudigl 2014): delta (2–4 Hz), theta (4–8 Hz), alpha (8–12 Hz), beta (16–20 Hz), low gamma (35–70 Hz), and high gamma (70–150 Hz). Frequencies from 20 to 35 Hz were not initially included as these fall between the standard beta and low gamma frequency ranges. However, we did subsequently include data from these frequencies and obtained additional gene correlations (Supplemental Methods). While we acknowledge that the demarcated limits of specific frequency bands are somewhat arbitrary, we used standard definitions for these bands in human EEG (Hanslmayr and Staudigl 2014) to facilitate comparisons among different genes and limit the number of correlations to evaluate. The SME for each frequency band was averaged across all electrodes in a given Brodmann area to obtain a single SME value for each region to be used in the correlation with the gene expression dataset. SME values from 9 Brodmann regions (BA9, BA21, BA22, BA39, BA40, BA44, BA45, BA46, and BA47) were compared with the dataset from Wang et al. and SME values from Brodmann regions (BA8, BA9, BA10, BA11, BA20, BA21, BA22, BA39, BA40, BA42, BA44, BA45, BA46, and BA47) were compared with the 7 brain regions denoted by gyral landmarks in the dataset from Kang et al. (Table S2).
The gene datasets available for comparison with the iEEG data presented certain challenges. The first is the spatial scale of the tissue data that was used to generate the gene expression compilations, which combines data within one or several Brodmann areas to define a cortical region. For high frequency oscillations, Brodmann areas are a relatively blunt scale of spatial segregation for analysis, and the regions typically employed in aggregating data for iEEG studies are smaller. We needed a method to rationally combine data across the cortical areas that contribute to the regions from the gene expression dataset. We elected to employ a two-sided statistical test to capture memory-relevant information from electrodes that exhibited a significant difference in oscillatory power in both directions (recalled > nonrecalled power and vice versa). This also allowed us to use an unbiased method of including all information from a given region without filtering for electrodes exhibiting an effect in only one direction and to capture the magnitude of memory processing in a region with heterogeneous properties for the electrodes. An example of the logic underlying this a priori decision is shown in FigureS2.
This means that the correlations we identify cannot disambiguate genes linked to positive versus negative SMEs, but rather quantify how gene expression differences across the brain match cortical differences in the strength of oscillatory correlation of successful encoding.
Brain Tissue and RNA Sequencing
All gene expression data are derived from different individuals than the ones that participated in the iEEG study. Gene expression data are limited by tissue availability, thereby focusing the comparisons to specific brain regions and only male samples. Postmortem brain tissue processing and RNA-seq analysis has been previously reported (Wang et al. 2015). Samples are from the left hemisphere of 5 male donors. For this dataset, we used RPKM (reads per kilobase of transcript per million reads mapped) values for gene expression measurements. Analysis of the replication gene expression dataset was previously reported (Kang et al. 2011). For our analysis, we used data from the left hemisphere of 22 male donors. The average Z values of each brain region from both studies are included in Table S2. The number of expressed genes in both studies that were used for comparisons was 12 018.
Identification of Individual SME-Correlated Genes
The Z values were estimated across participants for each of the 6 frequency bands and placed in a correlation matrix with gene expression values for the neocortical areas included in the analysis of each dataset. Only genes longer than 500 bp and annotated as “protein_coding” were included. RNA-seq expression values were log2 (RPKM+1) scaled to make them comparable with RMA values from the Kang et al. microarray study. A Spearman's rank correlation test was used. We assumed the relationship between oscillations and gene expression as monotonic. P < 0.05 from a Spearman's rank correlation was used as the cutoff to identify the significantly correlated genes in each oscillation. Genes that met these criteria for both gene expression datasets and were correlated in the same direction in both datasets (i.e., positive or negative) were kept for further analyses. By correlating gene expression across brain regions, this means that any genes identified are associated with how SME information varies across the brain not within a given brain region
To confirm the predictions, we applied a general linear model estimating the linear relationship between oscillations and gene expression using lm(log2(Oscillations+1) ~ log2(Gene expression level+1)). To note, one was added to maintain 0 in the downstream analysis. We then assessed the adjusted R squared from the linear model and compared with the rho squared from the Spearman's rank correlation (Pearson's R = 0.78, P = 0.002; permutational Pearson's correlation test; 1000 permutations). To test the robustness of the results, permutations were applied for each dataset. We shuffled the expression of both datasets 1000 times and recalculated the Spearman's rank correlation between oscillations and gene expression. We then assessed the difference between predicted nominal Spearman's rank correlation estimated and the permuted one. Permuted P-values are in Table S3.
Along with the permutation testing and use of confirmatory gene expression dataset described above, we conducted a control analysis in which we examined the genes correlated with a different (nonmnemonic) cognitive task. We quantified oscillatory activity across the same cortical regions recorded as subjects solved simple arithmetic problems. These are performed as the distractor portion of the free recall task. We generated a Z value comparing oscillatory power across the frequency spectrum for a 900 ms period after math item presentation to a baseline period from the beginning of each word list (cross fixation). We performed a correlation between this measure of oscillatory activity (during mathematical processing) following the same methods as for the SME analysis. We quantified the fraction of overlapping genes between these 2 analyses in an effort to ensure the genes we report as memory-related are not actually linked to nonmnemonic oscillatory activity such as more general visual processing or nonspecific cognitive engagement.
Results
Identification of SMEs in Fronto-temporal-Parietal Cortex
We analyzed human data obtained via direct intracranial electrophysiological recordings from 29 neocortical areas in 183 participants as they performed a delayed free recall task while undergoing phase II monitoring for identification of an epileptic focus (Fig. 1 and Fig. S1) (Lega et al. 2012; Burke et al. 2014). Subjects had an average recall probability of 22.5% of items from the 15 word lists, with a range from 4.2% to 51.7%. The results of a binomial test on the proportion of electrodes in each region exhibiting a significant SME for each frequency band are included in Table S4. The significance of these P-values (the majority exceed an FDR < 0.01) indicates that the identified SMEs are robust and valid for comparing to gene expression. FigureS2 shows the average effect size of the difference between oscillatory power for recalled and nonrecalled events (normalized across regions), with expected stronger effects for certain bands (such as gamma) in the temporal cortex for example. However, there were significant SMEs in many cortical regions for all frequency bands, consistent with existing data (Sederberg et al. 2007; Burke et al. 2014). These values demonstrate the robustness of the memory effects observed in our data, but for the purposes of the oscillation/gene expression correlations, the important property is how these memory effects varied across the cortex, including regions with overall weaker SMEs (Fig. S2).
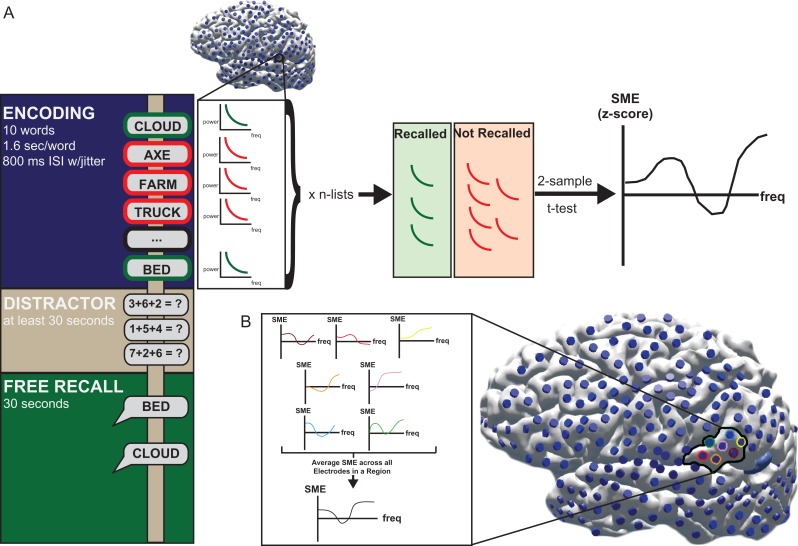
Figure 1.
Schematic of SME task and analysis. (A) Patients were presented with lists of 15 words to remember during the encoding period. After a distraction period comprised of basic math problems in the format X + Y + Z = ?, patients were asked to verbally recall the previously presented words. Throughout the encoding, distraction, and recall period, intracranial electrodes were recording activity throughout the neocortex. A Z-score characterizing the differences in activity during encoding of items that were subsequently recalled and during encoding of items that were forgotten (SME) was calculated for each electrode. (B) The SME for each region of interest was calculated by averaging the SME from all of the electrodes within that region. The SME was binned into distinct frequency ranges corresponding to known frequency oscillations.
Identification of SME-Correlated Genes
For comparison with the gene expression dataset, we examined the correlation between the pattern of variation in gene expression across cortical regions and the pattern of variation in SME values across these same regions using a nonparametric statistical test (Spearman's rank correlation). This variation was different for each frequency band (Table S2) and is the reason the gene correlations are different for each band. This method was modeled on a comparison of gene expression and fMRI BOLD signal variation across brain regions (Wang et al. 2015), and we discuss our results versus this previous analysis below.
The postmortem gene expression data we used for comparison with the SME data are derived from a separate cohort of 5 individuals, each with mRNA samples from 9 regions for a total of 45 samples. We also compared the SME values to an independent postmortem gene expression neocortical dataset derived from 22 individuals, each with mRNA samples from 7 regions (aggregated from 14 Brodmann areas) for a total of 154 samples (Kang et al. 2011) for replication (Fig. 2A). SME values calculated from either cohort show high similarity (Fig. S3A; P = 1.3e–03, Mantel Test for similarity with 10 000 Monte Carlo simulations). Thus, in total, the SME values from 66 individuals were compared with gene expression from the union of 14 regions in 27 different individuals. For the correlation with SME values, only genes that met our criteria for significance (P < 0.05, Spearman's rank correlation) in the same direction in both datasets were considered validated and used in further analyses (Fig. S4). We confirmed this empirical correlation using a linear regression model between SMEs and gene expression (Fig. 2B). The linear regression recapitulates the correlation, supporting a strong linear relationship between SMEs and the identified genes (Table S3). In addition, we also compared the rho2 values between the 2 datasets, and observed similar correlations between oscillations and gene expression (Fig. S5). Finally, we carried out permutation testing to compare the observed correlations to random distributions of each dataset. We found that the predicted nominal Spearman's rho values were highly discordant from the permuted distributions in both datasets (Fig. 2C and Table S3). Thus, we controlled for the false positives among the nominal correlations of the 12 018 expressed genes using a 3-step approach: (1) independent running of the correlation analysis (with shuffle) on a second gene expression dataset, (2) using an independent verification of the identified genes with a linear modeling approach, and (3) using a permutation procedure to identify genes whose rho values exceeded those of the type one error rate (5%) and control for the false discovery rate. We believe these methods provide robust statistical confirmation of the genes correlated with SMEs across cortical brain areas.
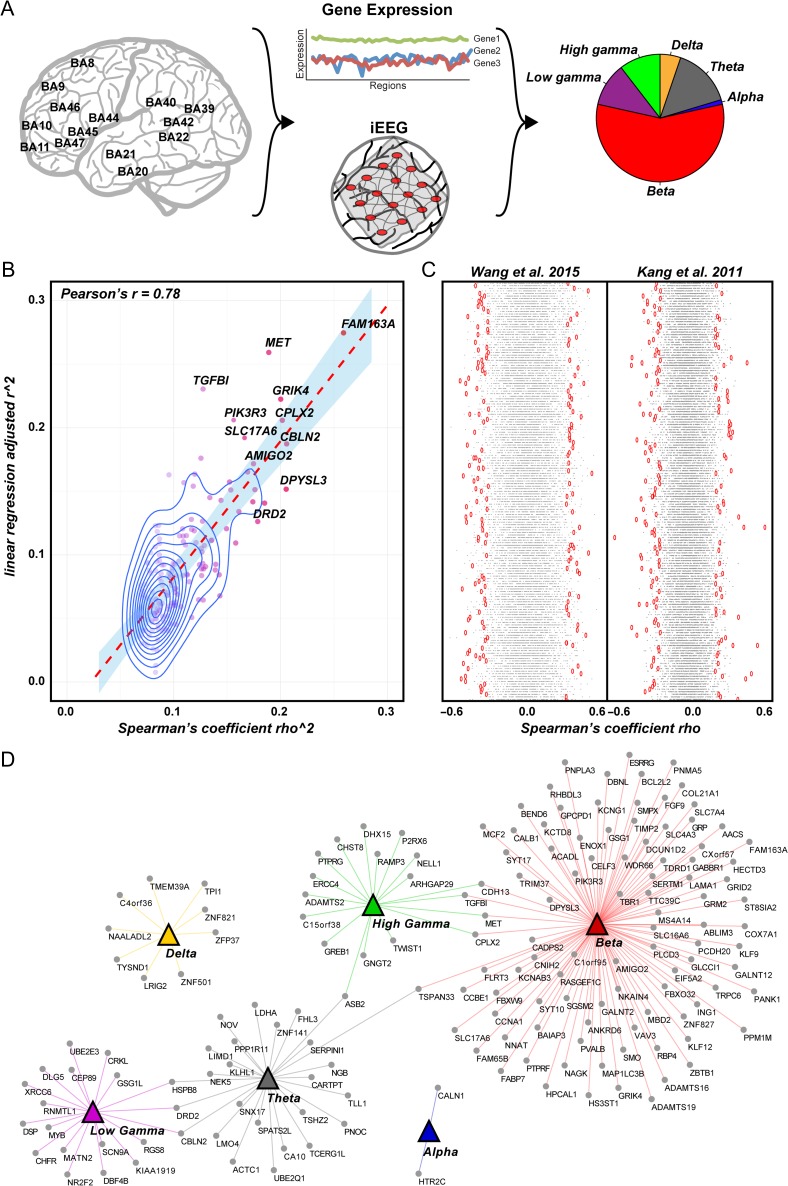
Figure 2.
Identification of SME-correlated genes. (A) Gene expression (e.g., RPKM, reads per kilobase of transcript per million reads mapped) from neocortical areas was correlated with iEEG signals that corresponded to an SME (Z-score) in the same neocortical areas. Expression of 163 genes correlated with SME frequency oscillations. The majority of these genes correlated with beta oscillations. (B) Plot of the adjusted R-squared from a linear model compared with the rho-squared of the Spearman's co-efficient. Highlighted are the genes with the greatest value for both calculations. (C) Visualization of the permutation testing (1 000) for both Wang et al. and Kang et al. gene expression datasets. In red, displayed are the real nominal values from the Spearman's rank correlation test, while in grey displayed are the permuted values. (D) Visualization of the genes correlated with each frequency oscillation.
With this approach, we found that the expression of 163 out of a total of 12 018 expressed genes is correlated with iEEG SMEs (Fig. 2D, Table S3). There was an equivalent distribution of genes positively and negatively correlated with SMEs using both expression datasets (Fig. S6A,B). This indicates that we identified genes for which expression increases are correlated with larger magnitude SMEs as well as those genes for which expression decreases in a corresponding manner, suggesting that the expression of these genes might underlie the ability to generate SMEs. In addition, we observed similar co-expression (Spearman's rank correlation) among these 163 genes in the human neocortex using both gene expression datasets (P = 4.5e–02, Mantel Test for similarity with 10 000 Monte Carlo simulations; Fig. S6C).
Gene Correlations Differ Among Frequency Bands
SMEs in the gamma band, especially in the hippocampus, have been linked to long term potentiation and learning in animal studies (Lisman and Buzsaki 2008). These oscillations are also found in human neocortex, but their underlying mechanism in humans is not as well described (Fries 2009). Encoding-related desynchronization that is a hallmark of successful item encoding throughout the brain often involves multiple frequency bands (Hanslmayr et al. 2016), and in our data we identified 9 genes correlated with more than 2 oscillations (Fig. 2D and Table S5). Interestingly, CBLN2 and HSPB8 are positively correlated with low gamma SMEs but negatively correlated with lower frequency oscillations, suggesting a gene-level preservation of a desynchronization/gamma synchronization pattern often observed during successful encoding (Hanslmayr et al. 2012).
Beta oscillations are thought to contribute to information processing as part of the prevalent low frequency desynchronization although they have received less focus than gamma or theta frequency oscillations in memory experiments (Fries 2009). We observed beta-band SMEs throughout the cortex, and 61% (98 genes) of the SME-correlated genes we identified were correlated with oscillatory effects in this frequency range with 37 genes positively correlated and 61 genes negatively correlated (Tables S3 and S5).
SME-Correlated Genes are Enriched for Neuronal Markers
Previous investigations have postulated that interneurons predominately modulate sub-gamma frequency oscillations, while gamma oscillations may emerge from interactions between inhibitory interneurons and excitatory pyramidal neurons (Buzsaki and Wang 2012; Lee et al. 2013). We therefore asked whether SME-correlated genes are enriched for specific cell types. We used 3 cell marker transcriptome resources with detailed annotations of genes linked to cell types (Zhang et al. 2014; Zeisel et al. 2015; Lake et al. 2016) to perform enrichment analyses of genes with preferential cell types expression patterns among the SME-correlated genes (Fig. 3A and Table S5; Supplemental Methods). The Zhang et al. study used a combination of immunopanning and FACS from cortical tissue of neonatal mice, the Zeisel et al. study used single cell microfluidics from somatosensory cortex and hippocampal CA1 tissue from adult mice, and the Lake et al. study used FACS combined with microfluidics to obtain neuronal nuclei from 6 cortical regions of an adult human brain. Therefore these cell-type expression datasets are independent from the human neocortical expression datasets we used to generate the SME correlations. We found that SME-correlated genes show the highest expression level in neurons compared with 6 other cell types in the cerebral cortex (Zhang et al. 2014), which is significantly higher than expected by chance (P = 8.23e–08; hypergeometric test, Benjamini–Hochberg corrected). In the second cell marker study, 9 cell type markers are available from the single-cell expression analysis of hippocampus and somatosensory cortex (Zeisel et al. 2015). We found that SME-correlated genes are significantly overrepresented among interneurons (P = 1.2e–02; hypergeometric test, Benjamini–Hochberg corrected). Analyzing the human single nuclei expression transcriptome dataset derived exclusively from cortical neurons (Lake et al. 2016), we find that SME-correlated genes are significantly overrepresented in excitatory neuron genes (P = 9.0e–03; hypergeometric test, Benjamini–Hochberg corrected) and that these are mostly correlated with theta and beta oscillations. Since this cell-type marker dataset is the only one we analyzed derived from human brain, it is possibly the most relevant to our analyses. The genes we identified in both of these cell types may provide clues to the mechanism of beta frequency modulation during memory encoding. It is also worth noting that we did not observe non-neuronal gene markers correlating with SMEs. However, as only the nonhuman cell-type marker datasets (Zhang et al. 2014; Zeisel et al. 2015) contained information from non-neuronal genes, our calculations might have been underpowered, especially as several studies have uncovered functional and expression differences between human and mouse astrocytes (Oberheim et al. 2009; Han et al. 2013; Zhang et al. 2016).
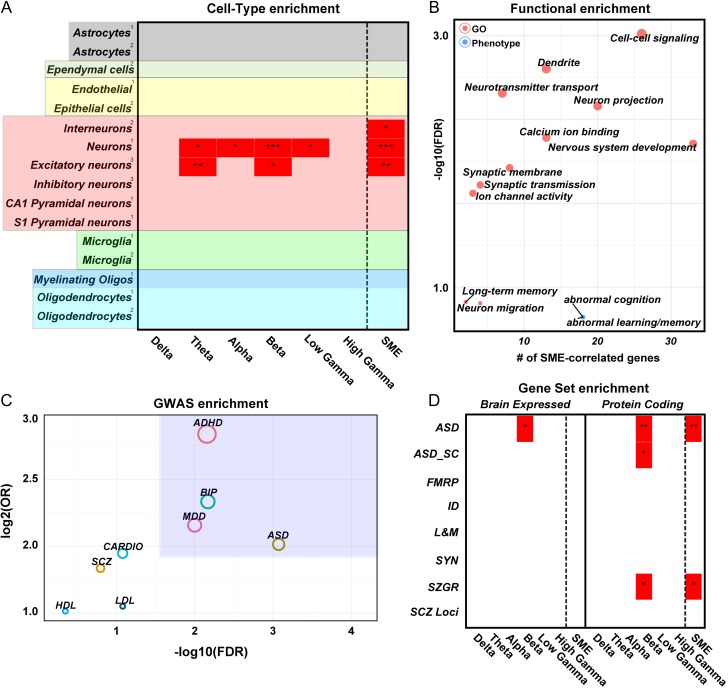
Figure 3.
Functional characterization of SME-correlated genes. (A) Cell-type enrichment for SME-correlated genes (SME) and oscillation-specific correlated genes using 3 independent datasets: 1(Zhang et al. 2014), 2(Zeisel et al. 2015), and 3(Lake et al. 2016). Enrichment is based on brain-expressed genes as background. (B) Gene ontology and phenotype enrichment for SME-correlated genes. On the x-axis is the number of SME-correlated genes per category; on the y-axis are the –log10(FDR) values. (C) Genome-wide association study enrichment for SME-correlated genes. On the x-axis are the –log10(FDR) values; on the y-axis are the log2 values of the odds ratio from the Fisher's exact test. (D) Gene set enrichment for SME-correlated genes. Left panel: enrichment based on brain-expressed genes. Right panel: enrichment based on protein-coding genes. ASD = autism genes in the SFARI database, ASD_SC = autism genes from the SFARI database with scores 1–4, FMRP = targets of FMRP from the Darnell et al. 2011 study, ID = genes associated with intellectual disability, L&M = genes in the ontology category “learning and memory,” SYN = genes encoding for synaptic genes, SZGR = genes associated with schizophrenia from the SZGR database, and SCZ_loci = genes near the 108 loci identified by the Schizophrenia Working Group of the Psychiatric Genomics 2014 study.
SME-Correlated Genes are Linked to Cognitive Disorders
We next carried out a functional ontological analysis of the SME-correlated genes. We found that those genes are significantly enriched for synaptic transmission, ion channel activity, and neurotransmitter transport (Fig. 3B and Table S6), further refining the SME-correlated genes to neuronal functions related to activity. Moreover, we identified enrichment in specific human and mouse phenotypes, such as memory disorders, learning disorders, and seizures.
To further confirm our prediction about the role of SME-correlated genes in cognition, we next asked whether SME-correlated genes are enriched for genetic variants implicated in multiple cognitive and neurodevelopmental disorders (Supplemental Methods). Using a Fisher's exact test model containing the brain-expressed genes as background, genome-wide association studies were filtered using a minor allele frequency (MAF) of 0.05 and P-value of 0.05. We found that SME-correlated genes are enriched for single-nucleotide polymorphisms (SNP) in genes associated with attention-deficit/hyperactivity disorder (ADHD), autism spectrum disorder (ASD), bipolar disorder (BIP), and major depressive disorder (MDD), while variants in genes associated with schizophrenia (SCZ) and nonbrain disorders such as cholesterol levels (HDL and LDL) and cardiac function (CARDIO) were not enriched (Fig. 3C).
We next asked whether any SME-correlated genes have previously been linked to memory and identified 3 SME-correlated genes associated with the ontology term “learning and memory” (CALB1, DRD2, and TBR1) (Table S5). These genes are noteworthy because all 3 have been linked to specific brain disorders that can include memory or cognitive impairments: CALB1 with Huntington's disease (Seto-Ohshima et al. 1988), DRD2 with schizophrenia (Schizophrenia Working Group of the Psychiatric Genomics 2014), and TBR1 with ASD (Sanders et al. 2015). We therefore investigated whether SME-genes are enriched for cognitive disorder genes, in general. Only beta-frequency SME-correlated genes are significantly enriched for ASD genes (Fig. 3D; P = 3.8e–02; hypergeometric test, Benjamini–Hochberg corrected). These genes include examples such as CADPS2, GRID2, GRIK4, and MET, all genes important for propagating information at synaptic membranes.
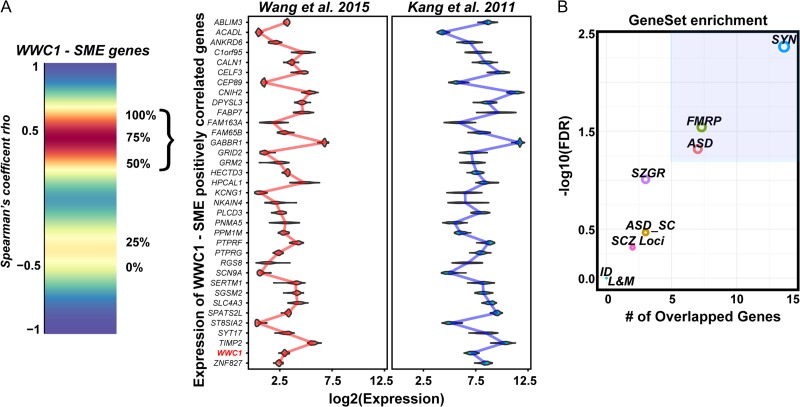
We next investigated whether one of the few genes to be previously linked specifically to human memory capabilities, WWC1, had association with the observed SME correlations. Variants in WWC1 have been linked to episodic memory and synaptic plasticity as well as cognitive disorders such as schizophrenia and AD (Papassotiropoulos et al. 2006; Schwab et al. 2014; Vyas et al. 2014; Kos et al. 2016). As we had observed an enrichment of synaptic genes among SME-correlated genes, this raised the possibility that WWC1 related pathways might be observed within the SME data. We therefore investigated whether SME-correlated genes are linked to WWC1 co-expression. We assessed the correlation of SME genes with the genes most highly connected to WWC1 expression in our dataset. We found a significant correlation of SME genes with WWC1 related genes (odds ratio = 4.85, P = 1.81e–16, Fisher's exact test with 10 000 Monte Carlo simulations), and most of these genes are positively correlated with WWC1 expression (odds ratio = 9.51, P = 9.10e–15, Fisher's exact test with 10 000 Monte Carlo simulations; Fig. 4A). The relative expression level of these positively correlated genes follows a similar expression pattern in both the Wang et al. and Kang et al. datasets. Moreover, only the positively correlated genes are enriched for ASD and synaptic genes (Fig. 4B). These results further strengthen the relevance of the identified SME-correlated genes with cognitive functions.
Figure 4.
Identification of SME-correlated genes co-expressed with WWC1. (A) Left panel: Density plot showing the percentage of SME-correlated genes. The majority of SME-correlated genes are positively correlated with WWC1 in both gene expression datasets analyzed. Right panel: Expression level of the positively correlated SME genes in both datasets analyzed. (B) Enrichment of synaptic and ASD genes among the WWC1-SME-correlated genes. ASD = autism genes in the SFARI database, ASD_SC = autism genes from the SFARI database with scores 1–4, FMRP = targets of FMRP from the Darnell et al. 2011 study, ID = genes associated with intellectual disability, L&M = genes in the ontology category “learning and memory,” SYN = genes encoding for synaptic genes, SZGR = genes associated with schizophrenia from the SZGR database, and SCZ_loci = genes near the 108 loci identified by the Schizophrenia Working Group of the Psychiatric Genomics 2014 study.
Co-expression Network Analysis Identifies Additional Genes Linked to SMEs
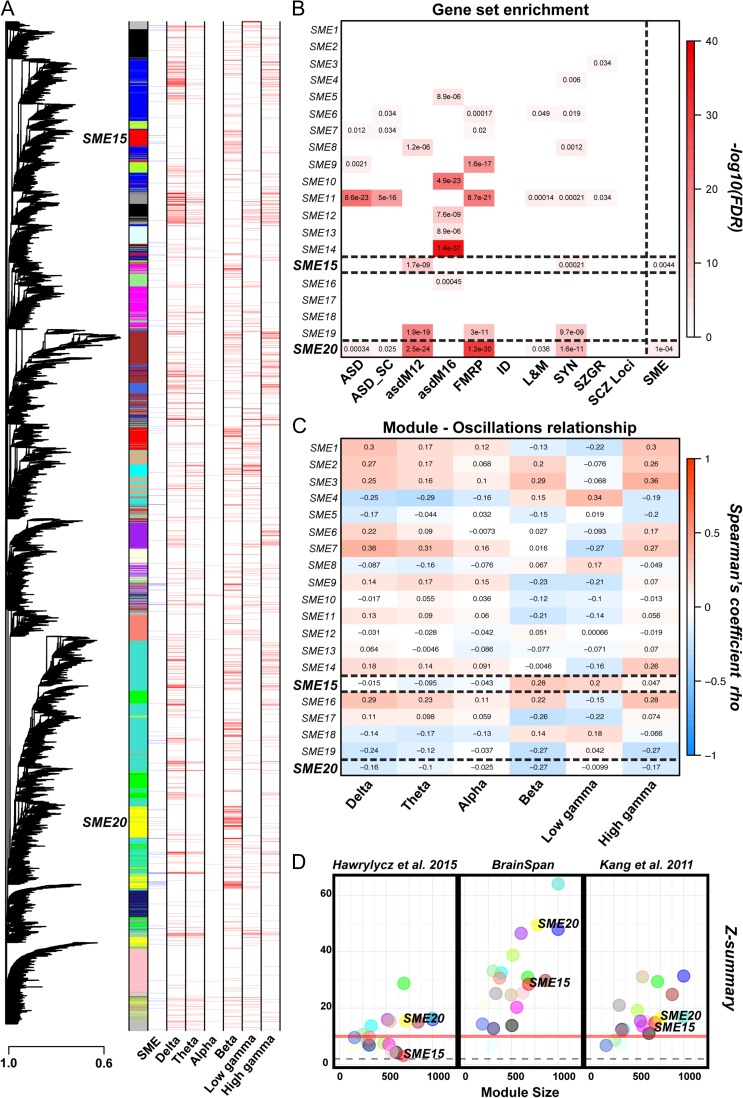
To further expand our understanding of gene expression patterns correlated with an SME, we carried out weighted gene co-expression network analysis (WGCNA). We identified 20 modules containing highly co-expressed genes (Fig. 5A, Table S5). Two modules were significantly enriched for SME-correlated genes, SME15 (P = 4.3e–03; hypergeometric test, Benjamini–Hochberg corrected) and SME20 (P = 1.0e–04; hypergeometric test, Benjamini–Hochberg corrected) (Fig. 5B and Table S7). We also found that genes in these 2 modules are enriched for genes associated with ASD, genes that are targets of the Fragile X Mental Retardation Protein (FMRP), genes encoding synaptic proteins, or genes associated with learning and memory (Fig. 5B and Table S7). Upon examining the correlations at the level of oscillations, we observed that SME15 has strong positive associations with beta and low gamma oscillations, while SME20 has negative associations (Fig. 5C). Visualization of the genes connected to SME-correlated genes in SME15, and SME20 reveals a number of connections to synaptic-protein encoding genes and ASD genes (Fig 6A,B). In contrast, the gene ontology of these 2 modules reveals quite different enrichments as SME20 is enriched for neuron-specific categories such as synaptic transmission and ion channel function, while SME15 is enriched for protein modification, protein binding, and RNA binding (Fig 6A,B and Table S8). To determine the applicability of our findings to the cortex in general, we carried out a module preservation analysis of our data with another human brain transcriptome study containing 44 cortical regions in the left hemisphere of 5 individuals (Hawrylycz et al. 2015), an additional dataset comprising 5 cortical regions of 5 individuals (BrainSpan), and the Kang et al. dataset. All modules are preserved; however, SME20 has a much greater relative preservation than SME15 (Fig. 5D). This suggests that SME15 may capture the genes relevant to memory encoding in the specific cortical regions we assessed, whereas SME20 may identify general neuronal processes for memory encoding, in line with the ontology categories identified. In addition, we assessed cell type enrichment within the SME-enriched modules. Only SME15 is enriched for neuronal genes representative of both excitatory and inhibitory neurons (Fig. 6C), again perhaps suggesting some specificity of neuronal networks underlying SMEs in the cortical regions we studied. Upon carrying out enrichment analyses using genes categorized by oscillation correlations, we only observed a significant enrichment of beta-correlated SME genes among the “Inhibitory group 6” list of genes (P = 4.5e–02; hypergeometric test, Benjamini–Hochberg corrected).
Figure 5.
Co-expression network analysis of SME-correlated genes. (A) Network dendrogram of all identified modules. (B) Functional enrichments in the 2 SME-enriched modules. ASD = autism genes in the SFARI database, ASD_SC = autism genes from the SFARI database with scores 1–4, asdM12/16 = autism-associated co-expression modules from the Voineagu et al. 2011 study, FMRP = targets of FMRP from the Darnell et al. 2011 study, ID = genes associated with intellectual disability, L&M = genes in the ontology category “learning and memory,” SYN = genes encoding for synaptic genes, SZGR = genes associated with schizophrenia from the SZGR database, SCZ_loci = genes near the 108 loci identified by the Schizophrenia Working Group of the Psychiatric Genomics 2014 study, and SME = SME-correlated genes. FDR are based on a hypergeometric test, corrected using the Benjamini–Hochberg method. (C) The relationship of each module across oscillations. In blue, are displayed negative correlations, whereas in red are displayed positive correlations. The 2 SME-enriched modules are outlined with dashed lines. (D) Module preservation of the network in 3 independent datasets ((Hawrylycz et al. 2015), (Kang et al. 2011) and BrainSpan) demonstrates the robustness of the SME-correlated modules.
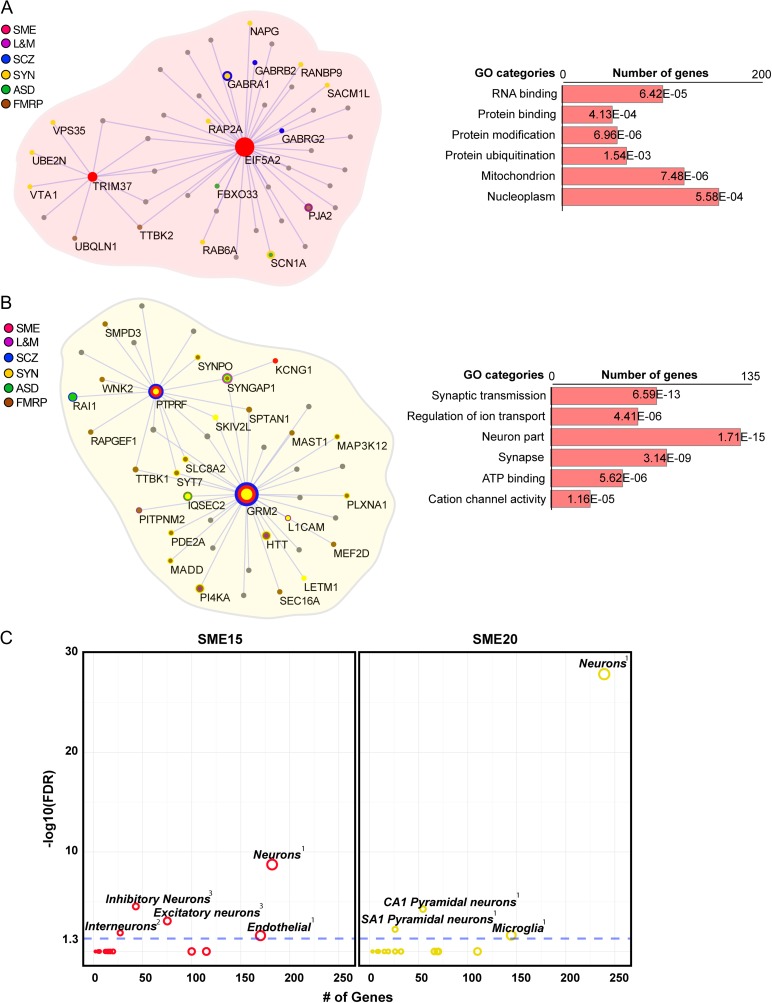
Figure 6.
Characterization of SME15 and SME20. (A) Visualization of SME15 module showing EIF5A2 and TRIM37 SME-correlated genes as hubs. The gene ontology functional enrichments are displayed on the right. (B) Visualization of SME20 module showing GRM2 and PTPRF SME-correlated genes as hubs. The gene ontology functional enrichments are displayed on the right. (C) Cell type enrichments among genes in SME15 and SME20 using excitatory and inhibitory single-nuclei transcriptome data (Lake et al. 2016).
SME-Correlated Genes are Distinct From Those Corresponding to Resting-State fMRI
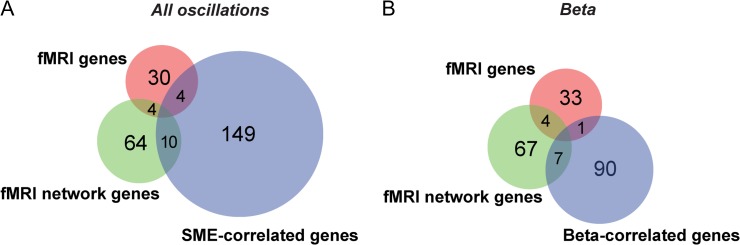
Previous work has correlated postmortem human brain gene expression with fMRI resting state activity identified through measurements of both functional connectivity and fractional amplitude of low frequency fluctuations (fALFF), highlighting molecular mechanisms of fMRI signals (Hawrylycz et al. 2015; Richiardi et al. 2015; Wang et al. 2015; Krienen et al. 2016). We found that 4 (11%) of the 38 reported fALFF-correlated genes significantly overlapped with the SME-correlated genes (DRD2, HTR2C, NR2F2, and PVALB) (P = 3.0e–02; hypergeometric test, Benjamini–Hochberg corrected) (Fig. 7A and Table S5). Among the 78 genes most stably correlated with fMRI connectivity (Richiardi et al. 2015), 10 (12%) of them also significantly overlapped with the SME-correlated genes (ANKRD6, BAIAP3, CARTPT, GNGT2, GRP, NOV, RBP4, SLC16A6, SYT10, and TGFBI) (P = 5.68e–08, Fisher's exact test) (Fig. 7A and Table S5). Interestingly, among those overlapped genes, there are differences among the oscillation frequencies with which the genes are correlated. The fALFF-correlated genes are correlated with alpha, beta, low gamma, and theta oscillations and the fMRI connectivity correlated genes are correlated with beta, high gamma, and theta oscillations (Table S5). However, none of the shared fMRI-correlated genes are correlated with beta oscillations (Fig. 7B). The comparison to the Wang et al. 2015 study is potentially the most relevant to the genes identified correlated to SMEs. This is because the Wang et al. study focused on region-specific functional activity as measured by fALFF in only the default mode network, while the other studies used measurements of functional connectivity across multiple brain networks. These findings underscore that these 2 different methods of quantifying resting state activity (fALFF and functional connectivity) may provide complementary information about memory encoding networks.
Figure 7.
Comparison of SME-correlated genes with fMRI-correlated genes. (A) Venn diagram indicating the overlaps among the SME-correlated genes and those correlated with either fALFF (“fMRI genes”; (Wang et al. 2015)) or functional connectivity (“fMRI network genes”; (Richiardi et al. 2015)) using resting state fMRI. The majority of SME-correlated genes do not overlap with either fMRI study. (B) Venn diagram indicating the overlap of beta-correlated genes specifically with each fMRI study.
Genes Correlated with SME Oscillations are Distinct From Those Correlated with a Mathematical Cognitive Task
We conducted a control analysis following the same methods as above using oscillatory activity recorded as the same intracranial EEG subjects answered simple arithmetic problems. We sought to address the possibility that the correlations we observed were due to nonmnemonic effects such oscillatory activity linked to visual processing or more general engagement in a cognitive task not specific for memory. We hypothesized that the genes identified in this analysis would be mostly distinct from those observed in the SME analysis. This control analysis revealed 210 genes across all frequency bands that were correlated with brain activity during mathematical processing. The correlation of these 210 genes was similar in each of the gene expression datasets (Fig. S7A; P = 1.3e–02, Mantel Test for similarity with 10 000 Monte Carlo simulations). Unlike the SME-correlated genes, these genes correlated with mathematical processing were primarily associated with low gamma oscillations (Fig. S7B). Only 27 overlapped with SME-related genes (there are 36 overlapping values as some genes are correlated with multiple oscillations), none of which were associated with neuronal function, cognitive disorders or ion channels (Fig. S7C). Moreover, if we filtered these 27 genes for oscillation association and direction of correlation (positive or negative correlation in both memory and math tasks), only 4 genes overlapped (Fig. S7C). Interestingly, there was greater overlap between the math correlated genes and resting state fMRI correlations (Fig. S7D,E), perhaps reflecting the contributions of the parietal regions to both math function and resting state activation (Fransson 2005; Wintermute et al. 2012).
Discussion
Our study is the first attempt to link memory-related brain oscillations to underlying gene expression in humans. iEEG has revealed a number of oscillatory patterns across the frequency spectrum that are reliably associated with successful memory encoding and retrieval. These include localized changes in gamma band activity and low-frequency desynchronization (Hanslmayr et al. 2012; Burke et al. 2013, 2014). To date, knowledge of these patterns, no matter how robust, has not translated into strategies for using oscillatory information to improve memory performance in patients suffering memory deficits (Laxton et al. 2010). Our data have implications for both of these issues. By identifying genes correlated with oscillatory SMEs, our data may suggest mechanisms common to effects observed in different frequency bands. With a set of target genes implicated for such patterns, our data should stimulate controlled investigations of how manipulations of these genes influence neurophysiology and behavior in animal models of human memory and thereby help understand the mnemonic roles of changes in specific frequency ranges. These experiments in turn could inform basic questions of how oscillatory patterns arise and how differences in specific gene function can impact memory performance. The genes we identify may also stimulate investigations of new therapeutic targets to alter memory dysfunction in diseased states. For example, GRIK4, a gene encoding an ionotropic glutamate receptor, that we show is correlated with beta oscillations, has association with both ASD and schizophrenia (De Rubeis et al. 2014; Greenwood et al. 2016). Therefore, pharmacological agents with preferential activity for this receptor may be helpful for treating cognitive problems in these patient populations. This same approach may also suggest new ways to understand side effect profiles of medications that impact the gene targets we have identified, notably ion channels.
The rationale for our study was to determine whether human brain gene expression profiles could correlate with a robust and quantifiable measurement of active brain activity such as SMEs. We expected to identify genes that were associated with ion channels and cognitive disorders, and that were mostly nonoverlapping with those whole expression correlates with fMRI signals and other cognitive processes, namely arithmetic calculation. We were successful in identifying over 100 correlated genes and the genes identified here are among the first genes to be linked to memory encoding in humans. We believe further experimentation in which expression of these genes is manipulated in a controlled setting will help explicate how they influence oscillatory patterns across the brain. The enrichment of these genes for neuronal and synaptic function makes their participation in oscillatory activity quite plausible and suggests that manipulation will be associated with observable differences in electrophysiological activity.
Our results also highlight both the utility and limitations of using resting-state fMRI to understand memory-encoding networks. Several of the genes that are correlated with both iEEG and resting fMRI notably show a strong correlation (e.g., BAIAP3 and DRD2; Table S3), implying they may serve a central role in both networks and further that there may be fundamental brain processes important in memory encoding that are also identified via resting state fMRI. This would be consistent with the observation that fMRI resting state networks identify a number of nodes that overlap with sites identified during fMRI studies of memory retrieval (Kim 2010). However, the overall fraction of genes that exhibit significant overlap between iEEG SMEs and resting state fMRI was low, suggesting that there are different electrophysiological processes involved in each. The brain regions that are principally involved in default mode network activity (especially prominent during resting state fMRI) (Greicius et al. 2009) were not well-sampled in the gene expression data we have available. Sampling from regions such as precuneus and midline prefrontal areas would perhaps reveal more consistent overlap between fMRI resting state data and iEEG SME data.
Given the strong relationship between SME-correlated genes and WWC1 gene co-expression (Fig. 4), it would have been interesting to identify AD-relevant SNPs within the SME genes as variants within WWC1 itself have been associated with AD (Schwab et al. 2014). However, the dataset available for comparison (International Genomics of Alzheimer's Disease 2015) did not contain the same statistical criteria (MAF cutoff, etc.) that was readily available for the other datasets. Discovery of variants relevant to AD may still be underpowered, as recent large-scale GWAS have demonstrated the need for tens of thousands of subjects to uncover significant variants (Schizophrenia Working Group of the Psychiatric Genomics 2014; CONVERGE Consortium 2015). Our focus on fronto-temporal activity and gene expression outside of the medial temporal lobe and the hippocampus may also limit our ability to observe AD-relevant correlations. Moreover, gene networks relevant to SMEs may be different from those disrupted in late-onset neurodegenerative disorders such as AD. In contrast, the enrichment of variants and genes associated with neurodevelopmental disorders such as ASD among the SME-correlated genes suggest that we have uncovered components of memory-relevant pathways important for early brain development. Intriguingly, several studies have shown deficits in working memory in patients with ASD (Williams et al. 2006), MDD (Schwarz et al. 2016), BIP (Buoli et al. 2016), and ADHD (Rapport et al. 2008; Kofler et al. 2011; Roman-Urrestarazu et al. 2016), suggesting that SME-correlated genes might be connected with this particular endophenotype in these neuropsychiatric disorders. Thus our findings are even more remarkable considering that the data from patients undergoing iEEG as well as the source of the postmortem tissues are all from adult individuals. As memory encoding is a fundamental property of most, if not all, cognitive functions, a convergence on genes relevant to early brain development among the SME-correlated genes are not unexpected. Moreover, the specific enrichment of beta-correlated genes among a human cell-type specific cluster of inhibitory genes (Fig. 6C) provides an entry point for testing the roles of these genes in specific cortical functions. These findings as well as the enrichment of ASD genes in those modules may underscore a gene network important for the disrupted functional neocortical circuits and working memory observed in patients with ASD (Williams et al. 2006).
Our gene expression data does not permit gene-SME comparisons with memory-relevant structures such as the hippocampus, parahippocampus, entorhinal cortex, and posterior cingulate region. However, numerous studies have observed robust SME patterns outside the mesial temporal lobe, especially a simultaneous gamma band power increase and low-frequency power decrease (most prominent in alpha and beta frequency ranges) observed in different experiments throughout the cortex using iEEG (Sederberg et al. 2007; Burke et al. 2014; Long et al. 2014). In addition, differences in gene expression have been linked indirectly to oscillatory differences in theta oscillations in the rodent hippocampus along the dorsal-ventral axis (Dong et al. 2009). Interestingly, when we include gene expression data from “hippocampus” that is available in the Kang et al. dataset with hippocampal oscillatory data, we find that the number of genes correlated with delta, low gamma, and high gamma increases (data not shown). Since the gene expression data are not distinguished at the level of hippocampal subfields, these findings await further exploration with such refined human expression datasets.
An important theory of information processing stimulated by such findings from intracranial EEG is the “information via desynchronization” hypothesis (Hanslmayr and Staudigl 2014). We identified a subset of genes that are correlated with oscillations in multiple frequency bands, lending support to the view that common neurophysiological mechanisms underlie changes across the spectrum. Our results immediately suggest several follow up analyses such as the identification of gene correlates of attentional processes that support memory encoding (especially 3 Hz synchrony, see Haque et al. (2015)), genes associated more specifically with the primacy effect for episodic memory (Sederberg et al. 2006), and working (and recognition) memory (Axmacher et al. 2008).
The large fraction of genes correlated with beta frequency oscillations may be an indication of the relative importance of desynchronization at these frequencies to information processing in the cortex (Engel and Fries 2010; Hanslmayr et al. 2016). The contributions of beta oscillations to mnemonic processing have been studied in nonhuman primates (Siegel et al. 2009) and human intracranial oscillations as well as surface EEG (Haenschel et al. 2000; Hanslmayr et al. 2014). Beta oscillations have been characterized most comprehensively via their contribution to movement, where pathological beta frequency activity has been linked to abnormalities in Parkinson's Disease. This observation and others led to the hypothesis from Engel and Fries that beta frequency activity signals baseline “status quo” activity in the cortex (Engel and Fries 2010). However, primate studies examining frontal cortex spike-field coherence during working memory and work by Hanslmayr (Siegel et al. 2009; Hanslmayr et al. 2016) has suggested there may be unique contributions to mnemonic processing from oscillations within the beta range. Our results support the latter conclusion. Our data may also reflect a role for beta oscillations in both desynchronization and local activation, as beta-to-gamma oscillatory changes are hypothesized to support memory encoding (Engel and Fries 2010), or an attention-related contribution to encoding success in tasks such as free recall (Donner and Siegel 2011; Kopell et al. 2014).
When formulating methods for this experiment, we considered the issue of combining data across frequency bands. The exact frequency range that defines a band is often subject for debate, and previous work has suggested that different individuals and brain areas exhibit oscillations with similar functional properties as slightly different frequencies (Zhang and Jacobs 2015). This heterogeneity in the edges of frequency bands is another reason we favored preserving SME information for as long as possible within our analysis before calculating a single test statistic for each brain area (see Methods), but averaging effects across the bands allowed us to make more straightforward comparisons with gene expression. For example, the genes TBR1 and TWIST1 are correlated with beta and gamma separately. Both genes encode transcription factors and while much work has been done to study the role of TBR1 in brain development and cognitive disorders such as ASD, there is little known about the functional relevance of TWIST1 to human brain development and function. Moreover, TBR1 is also enriched in synapses in the adult brain (Hong and Hsueh 2007), and thus its correlation with SMEs in adult human brain might be linked to the synaptic function of TBR1. Averaging across bands allows us to prioritize genes that exhibit preferentially high or low frequency effects for future functional analyses.
Given the differences in memory and math-related tasks that we contrasted using gene expression correlations, we would have been surprised if the correlated genes overlapped as this would have diminished the reliability of our claim to have identified genes related to memory for future experimental exploration. However, investigation of the few overlapping genes between SMEs and math suggest several interesting avenues of research such as whether these specific genes are relevant to attentional networks in the brain. Again, future studies in animal models on these genes are warranted.
Finally, these data are the first to demonstrate that gene expression patterns linked to specific cognitive tasks such as memory encoding can be identified in humans. Such correlations are essentially limited only by the quantitative measures that can be obtained during a given behavior. The excellent spatial and temporal resolution and favorable signal to noise characteristics of intracranial EEG likely facilitated the identification of robust correlations in our data. However, analyses performed using noninvasive modalities such as MEG may also prove fruitful and deserve investigation. In either case though, comparisons to gene expression will primarily be limited to postmortem tissue where temporal changes in gene expression cannot be assessed. Such a caveat might someday be overcome by new technology such as molecular fMRI (Bartelle et al. 2016), however this technology is far from being used in humans. There are many other possible sources of variation in gene expression that could serve as the basis of this type of analysis; investigators are limited only by the nature the available data. The only human gene expression datasets currently available utilize brain samples with relatively broad spatial specificity, and as such we developed methods for quantifying and aggregating SME data that matched the spatial scale of these gene data. Other types of variation in gene expression, such as variation within a region across subjects, or variation across time within a region during cognition, will be exceptionally interesting avenues for future investigation as these data become available. While finer-scale gene expression data is desirable, the brain regions we included have been shown to participate in memory networks in previous studies. Finally, our results are necessarily limited by the fact that only patients suffering from intractable epilepsy undergo invasive electrode implantation. It is possible that the gene/memory effect correlations we observed are unique to epilepsy patients. However, our methods of excluding electrodes in epileptogenic cortex, our imposition of a conservative artifact rejection algorithm, and the properties of the genes we identified (genes implicated in learning and memory in humans) provide some confidence of the wider generalizability of our findings.
Authors’ Contributions
B.L. and G.K. designed the study. S.B., G.-Z.W., J.G., and B.L. analyzed data. S.B., G.-Z.W., B.L., and G.K. wrote the article. All authors discussed and commented on the article. The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to G.K. (genevieve.konopka@utsouthwestern.edu) or B.L. (bradley.lega@utsouthwestern.edu).
Supplementary Material
Notes
We are grateful to our colleagues Brad Pfeiffer and Joseph Takahashi for suggestions and comments on this study. Conflict of Interest: None declared.
Supplementary Material
Supplementary material are available at Cerebral Cortex online.
Funding
UT Southwestern Jon Heighten Scholar in Autism Research, grants from the National Institutes of Health (R01DC014702, R01MH102603, R01MH103517, and R21MH107672), the James S. McDonnell Foundation 21st Century Science Initiative in Understanding Human Cognition Scholar Award (220 020 467), and Friends of the Alzheimer's Disease Center at UT Southwestern to G.K., the David M. Crowley Foundation to B.L., and a UT BRAIN Initiative Seed Grant (366 582) to G.K. and B.L.
References
- Axmacher N, Elger CE, Fell J. 2008. Ripples in the medial temporal lobe are relevant for human memory consolidation. Brain. 131:1806–1817. [DOI] [PubMed] [Google Scholar]
- Bartelle BB, Barandov A, Jasanoff A. 2016. Molecular fMRI. J Neurosci. 36:4139–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buoli M, Caldiroli A, Cumerlato Melter C, Serati M, de Nijs J, Altamura AC. 2016. Biological aspects and candidate biomarkers for psychotic bipolar disorder: a systematic review. Psychiatry Clin Neurosci. 70:227–244. [DOI] [PubMed] [Google Scholar]
- Burke JF, Long NM, Zaghloul KA, Sharan AD, Sperling MR, Kahana MJ. 2014. Human intracranial high-frequency activity maps episodic memory formation in space and time. Neuroimage. 85(Pt 2):834–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JF, Zaghloul KA, Jacobs J, Williams RB, Sperling MR, Sharan AD, Kahana MJ. 2013. Synchronous and asynchronous theta and gamma activity during episodic memory formation. J Neurosci. 33:292–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsaki G, Wang XJ. 2012. Mechanisms of gamma oscillations. Annu Rev Neurosci. 35:203–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONVERGE Consortium 2015. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 523:588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. 2011. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 146:247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Ercument Cicek A, Kou Y, Liu L, Fromer M, Walker S, et al. 2014. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 515:209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong HW, Swanson LW, Chen L, Fanselow MS, Toga AW. 2009. Genomic-anatomic evidence for distinct functional domains in hippocampal field CA1. Proc Natl Acad Sci U S A. 106:11794–11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner TH, Siegel M. 2011. A framework for local cortical oscillation patterns. Trends Cogn Sci. 15:191–199. [DOI] [PubMed] [Google Scholar]
- Engel AK, Fries P. 2010. Beta-band oscillations—signalling the status quo? Curr Opin Neurobiol. 20:156–165. [DOI] [PubMed] [Google Scholar]
- Fransson P. 2005. Spontaneous low-frequency BOLD signal fluctuations: an fMRI investigation of the resting-state default mode of brain function hypothesis. Hum Brain Mapp. 26:15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries P. 2009. Neuronal gamma-band synchronization as a fundamental process in cortical computation. Annu Rev Neurosci. 32:209–224. [DOI] [PubMed] [Google Scholar]
- Greenwood TA, Lazzeroni LC, Calkins ME, Freedman R, Green MF, Gur RE, Gur RC, Light GA, Nuechterlein KH, Olincy A, et al. 2016. Genetic assessment of additional endophenotypes from the Consortium on the Genetics of Schizophrenia Family Study. Schizophr Res. 170:30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Supekar K, Menon V, Dougherty RF. 2009. Resting-state functional connectivity reflects structural connectivity in the default mode network. Cereb Cortex. 19:72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenschel C, Baldeweg T, Croft RJ, Whittington M, Gruzelier J. 2000. Gamma and beta frequency oscillations in response to novel auditory stimuli: a comparison of human electroencephalogram (EEG) data with in vitro models. Proc Natl Acad Sci U S A. 97:7645–7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Chen M, Wang F, Windrem M, Wang S, Shanz S, Xu Q, Oberheim NA, Bekar L, Betstadt S, et al. 2013. Forebrain engraftment by human glial progenitor cells enhances synaptic plasticity and learning in adult mice. Cell Stem Cell. 12:342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanslmayr S, Matuschek J, Fellner MC. 2014. Entrainment of prefrontal beta oscillations induces an endogenous echo and impairs memory formation. Curr Biol. 24:904–909. [DOI] [PubMed] [Google Scholar]
- Hanslmayr S, Staresina BP, Bowman H. 2016. Oscillations and episodic memory: addressing the synchronization/desynchronization conundrum. Trends Neurosci. 39:16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanslmayr S, Staudigl T. 2014. How brain oscillations form memories—a processing based perspective on oscillatory subsequent memory effects. Neuroimage. 85(Pt 2):648–655. [DOI] [PubMed] [Google Scholar]
- Hanslmayr S, Staudigl T, Fellner MC. 2012. Oscillatory power decreases and long-term memory: the information via desynchronization hypothesis. Front Hum Neurosci. 6:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque RU, Wittig JH Jr., Damera SR, Inati SK, Zaghloul KA. 2015. Cortical low-frequency power and progressive phase synchrony precede successful memory encoding. J Neurosci. 35:13577–13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz M, Miller JA, Menon V, Feng D, Dolbeare T, Guillozet-Bongaarts AL, Jegga AG, Aronow BJ, Lee CK, Bernard A, et al. 2015. Canonical genetic signatures of the adult human brain. Nat Neurosci. 18:1832–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CJ, Hsueh YP. 2007. Cytoplasmic distribution of T-box transcription factor Tbr-1 in adult rodent brain. J Chem Neuroanat. 33:124–130. [DOI] [PubMed] [Google Scholar]
- International Genomics of Alzheimer's Disease C 2015. Convergent genetic and expression data implicate immunity in Alzheimer's disease. Alzheimers Dement. 11:658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. 2012. Fsl. Neuroimage. 62:782–790. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. 2011. Spatio-temporal transcriptome of the human brain. Nature. 478:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. 2010. Dissociating the roles of the default-mode, dorsal, and ventral networks in episodic memory retrieval. Neuroimage. 50:1648–1657. [DOI] [PubMed] [Google Scholar]
- Kofler MJ, Rapport MD, Bolden J, Sarver DE, Raiker JS, Alderson RM. 2011. Working memory deficits and social problems in children with ADHD. J Abnorm Child Psychol. 39:805–817. [DOI] [PubMed] [Google Scholar]
- Kopell NJ, Gritton HJ, Whittington MA, Kramer MA. 2014. Beyond the connectome: the dynome. Neuron. 83:1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos MZ, Carless MA, Peralta J, Blackburn A, Almeida M, Roalf D, Pogue-Geile MF, Prasad K, Gur RC, Nimgaonkar V, et al. 2016. Exome sequence data from multigenerational families implicate AMPA receptor trafficking in neurocognitive impairment and schizophrenia risk. Schizophr Bull. 42:288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krienen FM, Yeo BT, Ge T, Buckner RL, Sherwood CC. 2016. Transcriptional profiles of supragranular-enriched genes associate with corticocortical network architecture in the human brain. Proc Natl Acad Sci U S A. 113:E469–E478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaux JP, Fonlupt P, Kahane P, Minotti L, Hoffmann D, Bertrand O, Baciu M. 2007. Relationship between task-related gamma oscillations and BOLD signal: new insights from combined fMRI and intracranial EEG. Hum Brain Mapp. 28:1368–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung HL, Chen S, et al. 2016. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 352:1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxton AW, Tang-Wai DF, McAndrews MP, Zumsteg D, Wennberg R, Keren R, Wherrett J, Naglie G, Hamani C, Smith GS, et al. 2010. A phase I trial of deep brain stimulation of memory circuits in Alzheimer's disease. Ann Neurol. 68:521–534. [DOI] [PubMed] [Google Scholar]
- Lee JH, Whittington MA, Kopell NJ. 2013. Top-down beta rhythms support selective attention via interlaminar interaction: a model. PLoS Comput Biol. 9:e1003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lega B, Burke J, Jacobs J, Kahana MJ. 2016. Slow-theta-to-gamma phase-amplitude coupling in human hippocampus supports the formation of new episodic memories. Cereb Cortex. 26:268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lega BC, Jacobs J, Kahana M. 2012. Human hippocampal theta oscillations and the formation of episodic memories. Hippocampus. 22:748–761. [DOI] [PubMed] [Google Scholar]
- Lisman J, Buzsaki G. 2008. A neural coding scheme formed by the combined function of gamma and theta oscillations. Schizophr Bull. 34:974–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long NM, Burke JF, Kahana MJ. 2014. Subsequent memory effect in intracranial and scalp EEG. Neuroimage. 84:488–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long NM, Kahana MJ. 2015. Successful memory formation is driven by contextual encoding in the core memory network. Neuroimage. 119:332–337. [DOI] [PubMed] [Google Scholar]
- McClearn GE, Johansson B, Berg S, Pedersen NL, Ahern F, Petrill SA, Plomin R. 1997. Substantial genetic influence on cognitive abilities in twins 80 or more years old. Science. 276:1560–1563. [DOI] [PubMed] [Google Scholar]
- Oberheim NA, Takano T, Han X, He W, Lin JH, Wang F, Xu Q, Wyatt JD, Pilcher W, Ojemann JG, et al. 2009. Uniquely hominid features of adult human astrocytes. J Neurosci. 29:3276–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizzon MS, Lyons MJ, Jacobson KC, Franz CE, Grant MD, Eisen SA, Xian H, Kremen WS. 2011. Genetic architecture of learning and delayed recall: a twin study of episodic memory. Neuropsychology. 25:488–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, Huynh KD, Brunner F, Corneveaux J, Osborne D, et al. 2006. Common Kibra alleles are associated with human memory performance. Science. 314:475–478. [DOI] [PubMed] [Google Scholar]
- Rangaswamy M, Porjesz B. 2008. Uncovering genes for cognitive (dys)function and predisposition for alcoholism spectrum disorders: a review of human brain oscillations as effective endophenotypes. Brain Res. 1235:153–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapport MD, Alderson RM, Kofler MJ, Sarver DE, Bolden J, Sims V. 2008. Working memory deficits in boys with attention-deficit/hyperactivity disorder (ADHD): the contribution of central executive and subsystem processes. J Abnorm Child Psychol. 36:825–837. [DOI] [PubMed] [Google Scholar]
- Richiardi J, Altmann A, Milazzo AC, Chang C, Chakravarty MM, Banaschewski T, Barker GJ, Bokde AL, Bromberg U, Buchel C, et al. 2015. BRAIN NETWORKS. Correlated gene expression supports synchronous activity in brain networks. Science. 348:1241–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman-Urrestarazu A, Lindholm P, Moilanen I, Kiviniemi V, Miettunen J, Jaaskelainen E, Maki P, Hurtig T, Ebeling H, Barnett JH, et al. 2016. Brain structural deficits and working memory fMRI dysfunction in young adults who were diagnosed with ADHD in adolescence. Eur Child Adolesc Psychiatry. 25:529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, Murtha MT, Bal VH, Bishop SL, Dong S, et al. 2015. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 87:1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics C 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab LC, Luo V, Clarke CL, Nathan PJ. 2014. Effects of the KIBRA single nucleotide polymorphism on synaptic plasticity and memory: a review of the literature. Curr Neuropharmacol. 12:281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E, Tost H, Meyer-Lindenberg A. 2016. Working memory genetics in schizophrenia and related disorders: an RDoC perspective. Am J Med Genet B Neuropsychiatr Genet. 171B:121–131. [DOI] [PubMed] [Google Scholar]
- Sederberg PB, Gauthier LV, Terushkin V, Miller JF, Barnathan JA, Kahana MJ. 2006. Oscillatory correlates of the primacy effect in episodic memory. Neuroimage. 32:1422–1431. [DOI] [PubMed] [Google Scholar]
- Sederberg PB, Kahana MJ, Howard MW, Donner EJ, Madsen JR. 2003. Theta and gamma oscillations during encoding predict subsequent recall. J Neurosci. 23:10809–10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sederberg PB, Schulze-Bonhage A, Madsen JR, Bromfield EB, McCarthy DC, Brandt A, Tully MS, Kahana MJ. 2007. Hippocampal and neocortical gamma oscillations predict memory formation in humans. Cereb Cortex. 17:1190–1196. [DOI] [PubMed] [Google Scholar]
- Seto-Ohshima A, Emson PC, Lawson E, Mountjoy CQ, Carrasco LH. 1988. Loss of matrix calcium-binding protein-containing neurons in Huntington's disease. Lancet. 1:1252–1255. [DOI] [PubMed] [Google Scholar]
- Siegel M, Warden MR, Miller EK. 2009. Phase-dependent neuronal coding of objects in short-term memory. Proc Natl Acad Sci U S A. 106:21341–21346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. 2011. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 474:380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk HE, McDermott KB, Roediger HL 3rd, Todd RD. 2006. Genetic influences on free and cued recall in long-term memory tasks. Twin Res Hum Genet. 9:623–631. [DOI] [PubMed] [Google Scholar]
- Vyas NS, Ahn K, Stahl DR, Caviston P, Simic M, Netherwood S, Puri BK, Lee Y, Aitchison KJ. 2014. Association of KIBRA rs17070145 polymorphism with episodic memory in the early stages of a human neurodevelopmental disorder. Psychiatry Res. 220:37–43. [DOI] [PubMed] [Google Scholar]
- Wang GZ, Belgard TG, Mao D, Chen L, Berto S, Preuss TM, Lu H, Geschwind DH, Konopka G. 2015. Correspondence between resting-state activity and brain gene expression. Neuron. 88:659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ. 2010. Neurophysiological and computational principles of cortical rhythms in cognition. Physiol Rev. 90:1195–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DL, Goldstein G, Minshew NJ. 2006. The profile of memory function in children with autism. Neuropsychology. 20:21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermute S, Betts S, Ferris JL, Fincham JM, Anderson JR. 2012. Brain networks supporting execution of mathematical skills versus acquisition of new mathematical competence. PLoS One. 7:e50154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, et al. 2015. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 347:1138–1142. [DOI] [PubMed] [Google Scholar]
- Zhang H, Jacobs J. 2015. Traveling theta waves in the human hippocampus. J Neurosci. 35:12477–12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. 2014. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 34:11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G, et al. 2016. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 89:37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.