Abstract
Auxin biosynthesis was analyzed in a maize (Zea mays) kernel culture system in which the seeds develop under physiological conditions similar to the in vivo situation. This system was modified for precursor feeding experiments. Tryptophan (Trp) is efficiently incorporated into indole-3-acetic acid (IAA) with retention of the 3,3′ bond. Conversion of Trp to IAA is not competed by indole. Labeling with the general precursors [U-13C6]glucose and [1,2-13C2]acetate followed by retrobiosynthetic analysis strongly suggest that Trp-dependent IAA synthesis is the predominant route for auxin biosynthesis in the maize kernel. The synthesis of IAA from indole glycerol phosphate and IAA formation via condensation of indole with an acetyl-coenzyme A or phosphoenolpyruvate derived metabolite can be excluded.
The phytohormone auxin is a key regulator of numerous processes during plant growth and development. Although the structure of indole-3-acetic acid (IAA), the most abundant plant auxin, was elucidated in the 1930s (Thimann, 1977), IAA biosynthesis is not completely understood. The existence of multiple pathways that can differ according to the plant's developmental stage and response to environmental stimuli has complicated the elucidation of IAA biosynthesis (for review, see Bartel, 1997; Normanly and Bartel, 1999).
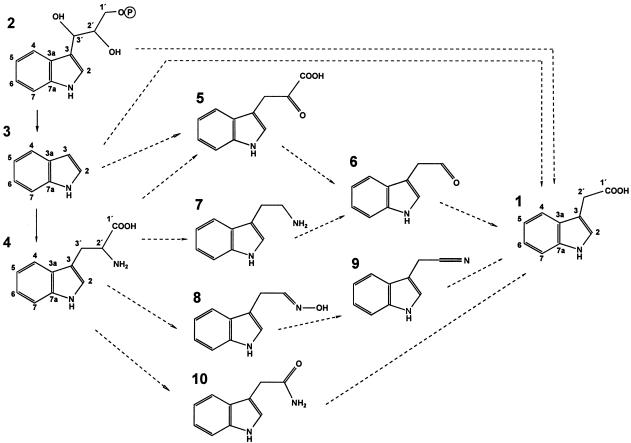
Originally, the amino acid Trp was identified as precursor of IAA, and IAA biosynthesis was proposed to occur via deamination and decarboxylation of Trp (Fig. 1; for review, see Bandurski et al., 1995). At least two other Trp-dependent pathways have been proposed based on the detection of specific enzymes or metabolites (Fig. 1). The indole-3-acetonitrile pathway was suggested on the basis of isolation and characterization of a nitrilase gene family (Bartel and Fink, 1994; Bartling et al., 1994; Hillebrand et al., 1998) in Arabidopsis. Indole-3-pyruvate and indole-3-acetaldehyde oxidase (Tam and Normanly, 1998; Sekimoto et al., 1998) have been found in several plants and are perhaps involved in yet another route for IAA biosynthesis.
Figure 1.
Hypothetical pathways of the IAA biosynthesis: 1, IAA; 2, IGP; 3, indole; 4, Trp; 5, indole-3-pyruvate; 6, indole-3-acetaldehyde; 7, tryptamine, 8: indole-3-acetaldoxime; 9, indole-3-acetonitrtile; and 10, indole-3-acetamide. The carbon positions of IAA, IGP, indole, and Trp are designated.
The characterization of Trp auxotrophs and stable isotope labeling of intact plants in maize (Zea mays; Wright et al., 1991) and Arabidopsis (Normanly et al., 1993) led to the postulation of Trp-independent IAA biosynthesis starting from indole or indole-3-glycerol phosphate (IGP; for review, see Bartel, 1997). It was shown in carrot (Michalczuk et al., 1992), Arabidopsis (Normanly, 1997), and maize (Östin et al., 1999) that both Trp-dependent and Trp-independent pathways exist in a single plant. The picture is further complicated by the finding that multiple Trp pools may exist and that exogenous IAA precursors are preferentially metabolized over their endogenous counterparts (Rapparini et al., 1999). Therefore, the molecular elucidation of IAA biosynthesis in a given plant tissue first requires the exact determination of the relevant in vivo situation.
The endosperm of the maize kernel has been used extensively for the biochemical analysis of IAA metabolism (Bandurski et al., 1998), since relatively high concentrations of IAA accumulate in this tissue. In vitro, conversion of both indole (Rekoslavskaya and Bandurski, 1994) and Trp (Östin et al., 1999) to IAA was reported to occur in the maize endosperm. Developing maize kernels can be matured in tissue culture (Gengenbach and Jones, 1994) under physiological conditions similar to the in vivo situation (Cobb and Hannah, 1983; Cully et al., 1984). Here we demonstrate that this system is suitable for the investigation of auxin biosynthesis. Precursor feeding experiments showed that the label from Trp is efficiently incorporated into IAA. To quantify the metabolite flux into IAA we used the general metabolites [U-13C6]Glc and [1,2-13C2]acetate for labeling. The IAA biosynthetic route was determined by retrobiosynthetic analysis (for review, see Bacher et al., 1999; Eisenreich and Bacher, 2000). These experiments clearly indicate a predominant Trp-dependent biosynthetic pathway in the maize endosperm. No indication of a Trp-independent IAA biosynthesis was observed. The existence of two separate Trp pools for protein and auxin biosynthesis is suggested.
RESULTS
Incorporation of l-[Ring-2H5]Trp into IAA and Proteinogenic Trp
The standard protocol for maize kernel culture was modified to make a more efficient use of labeled precursor. These changes allowed the use of only 1 to 2 mL of medium per harvested kernel. No change in kernel growth rate was observed compared to the standard protocol. To test whether developing maize kernels incorporated label from Trp into IAA, kernels were grown for 19 d on standard medium containing 20 mm l-[ring-2H5]Trp (experiment A) or 2 mm l-[ring-2H5]Trp (experiment B). In a third experiment (experiment C), kernels were incubated with a medium containing 2 mm l-[ring-2H5]Trp and 2 mm unlabeled indole, to test whether indole is an efficient competitor for Trp as IAA precursor.
IAA was isolated from each sample and the incorporation of 2H from l-[2H5]Trp was monitored by gas chromatography-mass spectrometry (GC-MS). In addition, Trp was isolated from total protein and analyzed by GC-MS to monitor the uptake of the amino acid Trp. The mass distribution of the quinoline ion region of the different IAA and Trp samples is given in Table I. In all samples derived from kernels labeled with l-[2H5]Trp a high abundance of the m + 5 peak was observed. To confirm that the mass of unlabeled ion (m) + 5 ion derived from the incorporation of five 2H atoms, aliquots of the IAA (A) sample were subjected to high resolution electron impact mass spectroscopy (HR EI-MS). For the fully protonated quinoline ion the mass of 130.0654 was observed (calculation 130.0657). For the [2H5]quinoline ion the mass of 135.0971 was observed (calculation: 135.0971). In addition, the abundant [2H4]quinoline (m + 4) ions, probably derived from the partial exchange of 2H during the purification of [2H5]IAA, were observed in labeled IAA samples (calculated: m = 134.0908, observed: m = 134.0903). The same isotope shift was observed for the m/z 189 IAA molecular ion. From the quinolinium isotope distribution the relative enrichment was determined: IAA (A), 89%; IAA (B), 25%; IAA (C), 39%; Trp (A), 64%; Trp (B), 47%; Trp (C), 47%. The incorporation of 2H from l-[2H5]Trp into IAA indicated that the indole ring of Trp was efficiently incorporated into IAA. A reduction of the incorporation rate by indole was not observed. It was additionally shown that this lack of competition cannot be due to an extremely low uptake rate of indole. For Trp isolated from kernels that were cultured on standard medium, containing 2 mm [2-13C]indole a relative enrichment of 20% [2-13C]Trp was determined (data not shown).
Table I.
Incorporation of deuterium from l-[2H5]Trp into IAA and Trp
| Compound | Ion | A | B | C |
|---|---|---|---|---|
| % | ||||
| Trp | 202 | 36 | 53 | 53 |
| 207 | 64 | 47 | 47 | |
| IAA | 130 | 11 | 76 | 61 |
| 134 | 42 | 21 | 29 | |
| 135 | 47 | 3 | 10 | |
| 134 + 135 | 89 | 24 | 39 | |
| 189 | 11 | 75 | 62 | |
| 193 | 41 | 20 | 23 | |
| 194 | 48 | 5 | 15 | |
| 193 + 194 | 89 | 25 | 38 | |
IAA and tryptophan isolated from kernels labeled with 20 mm l-[2H5]Trp (A), 2 mm l-[2H5]Trp (B), 2 mm l-[2H5]Trp, and 2 mm unlabeled indole as competitor (C) were analyzed by GC-MS. For Trp the relative mass distribution of the N-sylilated quinolinium ion is shown (m202 + m207 = 100%). For IAA the relative mass distribution of the quinolinium ion (m189 + m193 + m194 = 100%) and the molecular ion (m189 + m193 + m194 = 100%) is shown. The identity of [ring-1H5]IAA (m = 130), [ring-2H4]IAA (m = 134), and [ring-2H5]IAA (m = 135) was confirmed by HR EI-MS.
Incorporation of [3,3′-13C2]Trp into IAA and Proteinogenic Trp
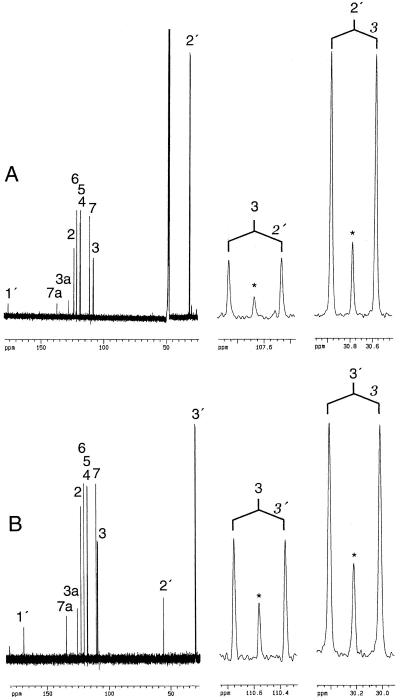
The next question was whether the side chain of IAA is also derived from Trp or if IAA is synthesized via a hypothetical pathway that involves the breakage of the Trp 3,3′ bond. Kernels were labeled with 1 mm d,l-[3,3′-13C2]Trp. IAA and Trp were isolated from this material and analyzed by 13C-NMR spectroscopy (Fig. 2). Satellite signals caused by 13C13C couplings were observed for IAA (C-3 and C-2′; JCC [one-bond carbon-carbon coupling constant] = 50.2 Hz) and Trp (C-3 and C-3′; JCC = 49.3 Hz). The relative signal intensities were compared to a natural abundance sample and a concentration of 8.6 mol % [3,2′-13C2]IAA and 5.4 mol % [3,3′-13C2]Trp was calculated. No significant enrichment of the [3-13C1]IAA isotopomer was observed. These results clearly show that IAA was synthesized from Trp without breakage of the 3,3′ bond.
Figure 2.
13C-NMR spectra of IAA (A) and proteinogenic Trp (B) isolated from kernels labeled with [3,3′-13C2]Trp (left) and of expansions of signals for C-3 and C-2′(IAA)/C-3′(Trp) (right). Coupling patterns are indicated.
Investigation of IAA Biosynthesis by Quantitative Assessment of Carbon Flux in Developing Maize Kernels: Studies with [U-13C6]Glc and [1,2-13C2]Acetate
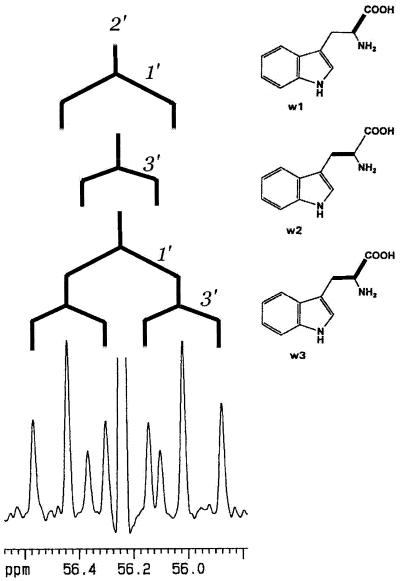
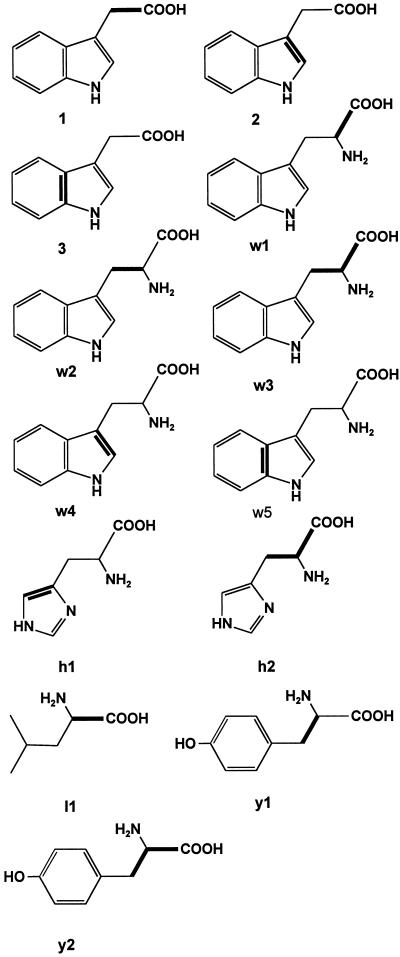
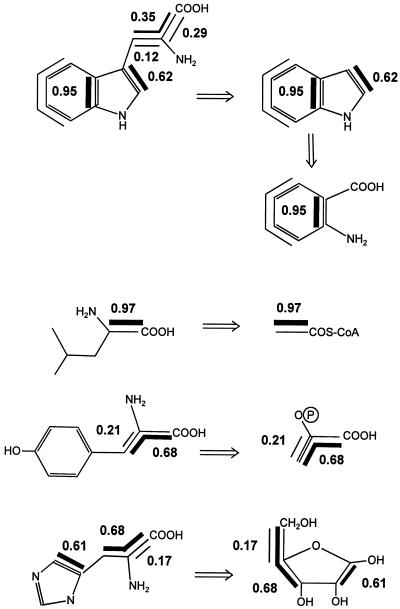
Experiments with l-[ring-2H5]Trp and d,l-[3,3′-13C2]Trp showed that the aromatic amino acid Trp serves as an efficient precursor of IAA in developing maize kernels. However this result cannot exclude alternative routes to IAA in this tissue. To assess the entire metabolic network leading to IAA, we performed experiments with the nonspecific precursors [U-13C6]Glc and [1,2-13C2]acetate followed by retrobiosynthetic NMR analysis. This technique allows a quantitative prediction of the general carbon flux in the biological system under study by a retrodictive analysis of 13C-labeling patterns (for review, see Bacher et al., 1999; Eisenreich and Bacher, 2000). Here, developing maize kernels were cultured on sterile medium supplied with 2 g [U-13C6]Glc L−1 or 3 g [1,2-13C2]acetate L−1. After 19 d of culture on labeling medium, IAA, Trp, Leu, Tyr, and His were isolated and analyzed by 13C-NMR spectroscopy. Isotopomers containing contiguous 13C atoms were quantified by analysis of 13C13C couplings. As an example, the 13C-2′ signal of Trp isolated from kernels labeled with [U-13C6]Glc is shown in Figure 3. Isotopomers with two contiguous 13C atoms result in a doublet signal due to 13C13C coupling. Isotopomers with three adjacent 13C atoms result in a double doublet reflecting two 13C couplings. The abundance of the isotopomers [1′,2′-13C2]Trp (w1), [2′,3′-13C2]Trp (w2), and [1′,2′,3′-13C3]Trp (w3) can be calculated from the relative signal intensities (Table II). The comparison of the isotopomer composition of the metabolite of interest (IAA) with the isotopomer composition (Table II) of several primary metabolites allows us to judge which metabolites are plausible precursors of IAA. Besides the biosynthesis from Trp (pathway 1) three alternative IAA biosynthetic pathways were tested for their quantitative relevance for IAA biosynthesis in vivo: the biosynthesis directly from IGP (pathway 4), the biosynthesis from indole and a C2 precursor that derives from acetyl-coenzyme A (CoA; pathway 2), and the biosynthesis from indole and a C3 unit that derives from phosphoenolpyruvate (PEP) or a triose-P (pathway 3). To compare the predicted and observed isotopomer distribution for these proposed biosynthetic pathways, it was necessary to obtain information about the isotopomer composition of the following metabolites: IAA, Trp, Rib-P, indole, anthranilate, acetyl-CoA, and PEP.
Figure 3.
13C signal of C-2′ of Trp isolated from kernels labeled with [U-13C6]Glc. 13C13C coupling patterns are indicated in italic numbers. From these data the abundance of the isotopomers [1′,2′-13C2]Trp (w1), [2′,3′-13C2]Trp (w2), and [1′,2′,3′-13C3]Trp (w3) can be calculated.
Table II.
Isotopomer analysis of multiple 13C-labeled IAA and amino acids isolated from developing maize kernels supplied with [U-13C6]Glc
| Compound | A | B | C | D | E |
|---|---|---|---|---|---|
| IAA | 1′,2′-13C2 | 1 | 0.54 | 0.22 | 0.39 |
| 2,3-13C2 | 2 | 1.00 | 0.40 | 0.70 | |
| 3a,7a-13C2 | 3 | 1.48 | 0.59 | 1.05 | |
| Trp | 1′,2′-13C2 | w1 | 0.30 | 0.14 | 0.29 |
| 2′,3′-13C2 | w2 | 0.13 | 0.06 | 0.12 | |
| 1′,2′,3′-13C3 | w3 | 0.36 | 0.17 | 0.35 | |
| 2,3-13C2 | w4 | 0.64 | 0.30 | 0.62 | |
| 3a,7a-13C2 | w5 | 0.98 | 0.64 | 0.95 | |
| His | 1,2-13C2 | h3 | 0.26 | 0.10 | 0.17 |
| 1,2,3-13C3 | h2 | 1.00 | 0.39 | 0.68 | |
| 4,5-13C2 | h1 | 0.90 | 0.35 | 0.61 | |
| Leu | 1,2-13C2 | l1 | 1.01 | 0.47 | 0.97 |
| Tyr | 2,3-13C2 | y1 | 0.32 | 0.12 | 0.21 |
| 1,2,3-13C3 | y2 | 1.05 | 0.40 | 0.68 |
A, Isotopomers analyzed. B, Abbreviation for the analyzed isotopomer (see Fig. 4). C, mol % of isotopomer carrying 13C nuclei at least at the designated positions. D, Fraction of multiple 13C-labeled isotopomers of all isotopomers carrying 13C at the designated positions (see “Materials and Methods”). E, Fraction of multiple 13C-labeled isotopomers of all isotopomers carrying 13C at the designated positions incorporated during the labeling (see “Materials and Methods”). This normalization enables comparison of metabolites with different de novo biosynthetic rate using 13C-enrichment data: IAA, 2.50% 13C; Trp, 2.14% 13C; Leu, 2.14% 13C; Tyr, 2.63% 13C; His, 2.57% 13C.
The isotopomer composition of Trp, Leu, Tyr, and His was determined (Table II; Fig. 4). Based on established mechanisms of amino acid biosynthesis, the isotopomer distribution of central metabolites were reconstructed from the labeling data of the amino acids (Fig. 5). Only a fraction of a given metabolite is synthesized de novo during the labeling period. To account for this fact the amount of each isotopomer was normalized to 100% de novo biosynthesis for each metabolite considered (see “Materials and Methods”). Specifically, the abundance of the [1,2-13C2]- acetate isotopomer could be inferred from the normalized abundance of [1,2-13C2]Leu (Fig. 4, l1) as the acetate moiety of acetyl-CoA is incorporated into C-1/C-2 of Leu (Oaks, 1965). The abundance of [2,3-13C2]PEP and [1,2,3-13C3]PEP was derived from the abundance of [2,3-13C2]Tyr (y1) and [1,2,3-13C3]Tyr (y2). As indole is the direct precursor of Trp, the abundance of [2,3-13C2]indole and [3a,7a-13C2]indole could be inferred from the abundance of [2,3-13C2]Trp (w4) and [3a,7a-13C2]Trp (w5), respectively. Anthranilic acid is a precursor of indole (for review, see Schmid and Amrhein, 1995). [1,2-13C2]Anthranilic acid could be inferred from [3a,7a-13C2]indole, the precursor of [3a,7a-13C2]Trp (w5; Fig. 5). His derives from phosphoribosyl pyrophosphate (Wiater et al., 1971) and therefore the Rib-labeling pattern can be determined from the His-labeling data: [1,2-13C2]Rib-P from [4,5-13C2]His (Fig. 4, h1) and [3,4,5-13C3]Rib-P from [1,2,3-13C3]His (Fig. 4, h2).
Figure 4.
Isotopomers analyzed to allow comparison of predictions for different hypothetical IAA biosynthetic pathways with the observed IAA data. The according isotopomer formula and the definition of the abbreviation is given in Table II. Contiguous 13C labeling is shown by bold chemical bonds.
Figure 5.
Incorporation of [U-13C6]Glc into developing maize kernels. Labeling patterns of metabolic intermediates indole, anthranilic acid, acetyl-CoA, PEP, and Rib can be reconstructed from observed labeling patterns of the isolated amino acids Trp, Leu, Tyr and His on the basis of established mechanisms. Contiguous 13C labeling is shown by bold lines. The number next to this line represents the normalized abundance of the isotopomer (Table II).
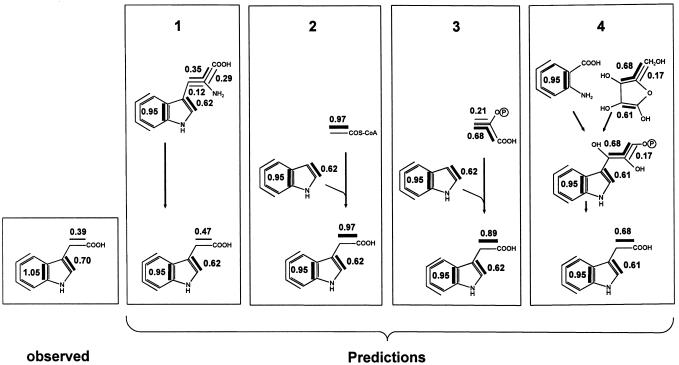
All metabolites isolated from kernels labeled with [U-13C6]Glc showed incorporation of label (Table II). From the absolute 13C enrichment of each metabolite the rate of de novo biosynthesis during the labeling period could be calculated. Using this enrichment data, the abundance of multiply labeled isotopomers could be normalized (see “Materials and Methods”). In all metabolites analyzed we observed incorporation of intact blocks that resulted in 13C2- and 13C3-isotopomers in a much larger amount than that expected from statistic incorporation of 13C. Normalized abundance of 0.39 for [1′,2′-13C2]IAA, 0.70 for [2,3-13C2]IAA, and 1.05 for [3a,7a-13C2]IAA was observed (Table II). The relative abundance of the analyzed isotopomers (Table II), normalized for 100% de novo biosynthesis, and the isotopomer compositions of the inferred metabolic intermediates are summarized in Figure 5. Using the isotopomer composition of these metabolic intermediates, the labeling pattern of IAA via hypothetical mechanisms could be predicted. More specifically, the normalized abundance of [1′,2′-13C2]IAA, [2,3-13C2]IAA, and [3a,7a-13C2]IAA was predicted for the Trp-dependent (pathway 1) and three hypothetical Trp-independent pathways described above (pathway 2–4; Fig. 6). The measured IAA data and the four predictions show similar normalized abundance of the [2,3-13C2]IAA and [3a,7a-13C2]IAA (0.95 for all predictions shown) isotopomer.
Figure 6.
Observed and predicted labeling pattern of IAA. Predictions are made on the basis of metabolites derived from kernels labeled with [U-13C6]Glc. 1, Prediction for a Trp-dependent IAA biosynthesis. 2, Prediction for a hypothetical pathway via indole and a C2 metabolite derived from acetyl-CoA. 3, Prediction for a hypothetical pathway via indole and a C3 metabolite derived from PEP or triose-P. 4, Prediction for a hypothetical IAA biosynthesis directly from IGP, the isotopomer composition of which can be deduced from anthranilate and Rib. Contiguous 13C labeling is shown by bold lines and the normalized abundance of the isotopomer is given (Tables II and III).
The ratio [2,3-13C2]IAA to [3a,7a-13C2]IAA could be more accurately determined as it is not influenced by the rate of de novo biosynthesis. In this case the predictions for pathways 1, 2, 3 (0.65), and 4 (0.64) matched well with the observed ratio (0.67) indicating that comparable isotopomer composition of the indole ring can be reconstructed using IAA, Trp, or Trp plus His primary data. This confirmed the origin of IAA from the shikimic acid pathway and it showed that, despite the complexity of the system, the technique of retrobiosynthetic analysis can be applied. On the other hand, the normalized abundance of the [1′,2′-13C2]IAA isotopomer for the acetic acid side chain (Table III; Fig. 6), and the [1′,2′-13C2]IAA to [2,3-13C2]IAA ratio (Table III) showed significant differences between the observed and predicted isotopomer ratios for Trp as a precursor of IAA. These differences can be explained by separate Trp pools for IAA and protein biosynthesis (see “Discussion”), which would differ in the biosynthetic origin of the Ser side chain of Trp. The observed isotopomer ratios clearly exclude, however, that IAA is synthesized from IGP or from indole plus a metabolite derived from acetyl-CoA or PEP. As comparable isotopomer compositions can be expected for PEP and triose-P, a triose-P can be also excluded as a precursor of the side chain of IAA.
Table III.
Observed and predicted labeling pattern of IAA
| Precursor | [1′,2′-13C2]IAA | [2,3-13C2]IAA | Ratio |
|---|---|---|---|
| Trp | 0.47 | 0.62 | 0.76 |
| IGP | 0.68 | 0.61 | 1.11 |
| Indole plus acetate-derived metabolite | 0.97 | 0.62 | 1.56 |
| Indole plus PEP-derived metabolite | 0.89 | 0.62 | 1.43 |
| Observed | 0.39 | 0.70 | 0.56 |
Predictions are made on the basis of metabolites derived from kernels labeled with [U-13C6]Glc. The normalized abundance of [1′,2′-13C2]IAA and [2,3-13C2]IAA and the ratio [1′,2′-13C2]IAA to [2,3-13C2]IAA is given. Four hypothetical IAA biosynthetic pathways were analyzed for their plausibility. (a) Biosynthesis from Trp: [1′,2′-13C2]IAA derives from [1′,2′,3′-13C3]Trp (w3) plus [2′,3′-13C2]Trp (w2). (b) Biosynthesis from IGP: [1′,2′-13C2]IAA derives from [3,4,5-13C3]Rib-P. (c) Biosynthesis from indole plus an acetate derived metabolite: [1′,2′-13C2]IAA derives from [1,2-13C2]acetyl-CoA. (d) Biosynthesis from indole plus a PEP or triose-P-derived metabolite: [1′,2′-13C2]IAA derives from [1,2,3-13C3]PEP plus [2,3-13C2]PEP. For a biosynthesis from IGP [2,3-13C2]IAA derives from [1,2-13C2]Rib-P, for the other hypothetical pathways [2,3-13C2]IAA derives from [2,3-13C2]indole.
Kernels were also cultured on a medium with [1,2-13C2]acetate. IAA and Trp isolated from this tissue did not show any significant enrichment of the [1′,2′-13C2]IAA, [2′,3′-13C2]Trp, [2,3-13C2]IAA, and [2,3-13C2]Trp isotopomer. On the other hand, 0.87% mol [1,2-13C2]Leu that derives from [1,2-13C2]acetyl CoA was observed. This gives additional evidence that a hypothetical IAA biosynthetic pathway via indole plus a C2 unit derived from acetyl CoA does not play a major role for IAA biosynthesis in vivo.
DISCUSSION
Plants can synthesize IAA by multiple pathways and several Trp-dependent pathways have been proposed. IAA synthesis can also occur independent of Trp via a Trp precursor, e.g. indole or IGP. This biosynthetic redundancy (Fig. 1; for review, see Bartel, 1997; Normanly and Bartel, 1999) has complicated the elucidation of IAA biosynthesis. Various pathways can exist in different tissues or at various developmental stages of one individual plant.
Relatively large amounts of IAA conjugates accumulate during development in the maize endosperm (Bandurski et al., 1998). IAA from this deposit is probably required for polar embryo development and for the development of the maize seedling. Seedlings that are not dissected from the kernel import IAA from the endosperm (Epstein et al., 1980). IAA de novo biosynthesis is initiated late in seedling development (Jensen and Bandurski, 1996) and is not detectable in the first week after germination. Therefore, biosynthesis, conjugation and mobilization of IAA can be studied in the maize kernel. The aim of our study was to elucidate IAA biosynthesis under physiological conditions in this model system.
Label from Trp Is Efficiently Incorporated into IAA in the Maize Endosperm
Crude enzyme preparations from maize endosperm converted both indole (Rekoslavskaya and Bandurski, 1994; Rekoslavskaya, 1995) and Trp (Östin et al., 1999) into IAA. We modified the kernel tissue culture system to obtain efficient labeling of IAA in a system that closely resembles the in vivo situation (Cobb and Hannah, 1983; Cully et al., 1984). Deuterated Trp and [13C2]Trp were used for the incorporation experiments. When 2 mm l-[2H5]Trp was used, the relative enrichment was 24% for IAA and 47% for Trp isolated from total protein. In contrast, with 20 mm l-[2H5]Trp 89% of IAA and 64% of Trp were labeled. These differences in specific enrichment could indicate a significantly higher de novo biosynthesis rate of IAA in the presence of high exogenous Trp concentrations. Unlabeled indole did not suppress the conversion of Trp to IAA. These findings suggest that Trp-dependent IAA biosynthesis is the predominant route in the developing maize endosperm. Similar results were obtained with liquid endosperm preparations (Östin et al., 1999).
NMR analysis of IAA that incorporated label from d,l-[13C2]Trp clearly showed that IAA is synthesized without breakage of the 3,3′ bond. This excludes a hypothetical pathway that would involve the breakdown of Trp to yield indole as an intermediate of IAA biosynthesis.
Retrobiosynthetic Analysis of IAA Biosynthesis
The Trp-labeling experiment with in vitro cultured kernels provided evidence for Trp-dependent IAA biosynthesis. However, it cannot be excluded that specific precursors can influence the biosynthetic route. We therefore evaluated the origin of the IAA carbon skeleton when specific precursors were omitted. The high concentration of IAA in the maize endosperm and the improvement of the kernel culture described in this manuscript allowed the isolation of IAA in quantities sufficient for retrobiosynthetic analysis based on 13C-NMR spectroscopy. Uniformly labeled Glc as a general precursor provided an assessment of the IAA biosynthetic route reflecting the natural situation. The results obtained excluded IAA synthesis from IGP because the abundance of the [1′,2′-13C2]IAA isotopomer was by far too low to originate from [2′,3′-13C2]IGP and [1′,2′,3′-13C3]IGP. Due to the low abundance of [1′,2′-13C2]IAA, IAA formation from indole and condensation with metabolites that derive from the PEP pool or from acetyl-CoA can also be excluded (Table III; Fig. 6). The exclusion of acetyl-CoA and acetate derived metabolites like glycolate, mevalonate, etc. as precursors of the side chain of IAA was confirmed by feeding with [1,2-13C2]acetate. No incorporation of acetate into the IAA side chain was observed, although acetate was efficiently metabolized, e.g. to Lys.
If IAA biosynthesis does occur entirely from Trp, then the normalized isotopomer composition for the indole ring and the side chain of IAA and Trp should be identical. We observed a good agreement for the indole ring, but the normalized isotopomer abundance of [1′,2′-13C2]IAA was 17% lower than expected (Tables II and III; Fig. 6). One possible explanation for this result is that a fraction of IAA is synthesized from a Trp precursor. We can exclude IAA formation from IGP or via the condensation of indole with metabolites that derive from PEP or acetyl-CoA (see above). In theory the data can be explained by the addition of C1 units to indole for the biosynthesis of a fraction of total IAA. Such a mechanism has not yet been associated with IAA biosynthesis. Therefore the involvement of a Trp-independent pathway remains purely speculative.
Alternatively, we postulate the existence of two different Trp pools. This hypothesis would predict that the side chains of Trp in the two pools would have different isotopomer patterns. The side chain of Trp is derived from Ser and there are two different Ser pathways in plants: a pathway via Gly and a pathway via 3-phosphoglycerate (Ho et al., 1998, 1999). The expected abundance of the [1′,2′,3′-13C3]Trp isolated from [U-13C6]Glc-labeled kernels should differ dramatically for these two Ser biosynthetic pathways: When Ser is synthesized from two molecules of Gly, no [1′,2′,3′-13C3]Trp should be detectable; in contrast, when Ser is synthesized from 3-phosphoglycerate that derives from PEP, the normalized abundance of [1′,2′,3′-13C3]Trp should be similar to [1,2,3-13C3]PEP. We observed a normalized abundance of 0.35 for [1′,2′,3′-13C3]Trp and 0.68 for [1′,2′,3′-13C3]PEP, deduced from [1′,2′,3′-13C3]Tyr (Table II). From these data we conclude that the Ser side chain of Trp that is incorporated into proteins is synthesized via both pathways and these pathways contribute approximately equal amounts to this Ser pool.
A similar ratio has been obtained for Ser biosynthesis in heterotrophic Beta vulgaris cell cultures (Werner, 1996). The Ser side chain of the Trp, from which IAA is synthesized, should derive to a larger extent from two molecules of Gly. This would indicate separate Trp pools for IAA and protein biosynthesis. A different experimental approach will be necessary to test this hypothesis, since the soluble Trp (Radwanski and Last, 1995) is only a few percent of total Trp. These low levels are not sufficient for detection of the isotopomer composition.
In summary, we conclude that IAA is synthesized from Trp in developing maize kernels. No evidence for a major contribution of a Trp-independent pathway was found. Several pathways for the conversion of Trp to IAA have been proposed (Fig. 1) and a few genes have been cloned encoding candidate genes of these pathways (for review, see Normanly and Bartel, 1999). These molecular “probes” can now be used to further dissect IAA metabolism in the maize endosperm.
MATERIALS AND METHODS
Materials
[U-13C6]Glc was purchased from Isotec (Miamisburg, OH). l-[Ring-2H5]Trp, [2-13C]indole, [3-13C]indole, and [13C]formaldehyde were purchased from Promochem (Wesel, Germany).
Synthesis of [3,3′-13C2]Trp
(1H-[3-13C]Indol-3-yl[13C]Methyl)-N,N-Dimethylamine (Compound a)
Glacial acetic acid (700 μL) was mixed with 700 μL of a 20% (w/v) aqueous solution of 13C-formaldehyde and 450 μL of an aqueous solution of 50% (w/v) dimethylamine-HCl. The mixture was cooled to 0°C. 3-13C-Indole (500 mg, 4.24 mmol) was added with stirring. After 3 h, 10 mL of 2 n NaOH was added to the clear solution. A precipitate of colorless crystals of compound a was formed immediately. The mixture was extracted twice with 20 mL of ethyl acetate. The organic layer was dried (Na2SO4) and evaporated. Yield: 723 mg (4.11 mmol, 97%).
2-Acetylamino-2-(1H-[3-13C]Indol-3-yl)[13C]Methyl-Malonic Acid Diethyl Ester (Compound b)
Ninety-five milligrams (4.13 mmol) of sodium was dissolved in 9 mL of absolute ethanol. Seven hundred and twenty milligrams (4.09 mmol) of compound a was added to the sodium ethanolate solution. Then 900 mg of acetamino diethyl malonate and 1,020 mg of dimethylsulfate were added. The solution was stirred for 12 h. Water (50 mL) was then added. The mixture was extracted twice with 20 mL of ethyl acetate and dried (NaSO4), and the solvent was removed. Yield: 1.388 g (97%).
2-Acetylamino-2-(1H-[3-13C]Indol-3-yl[13C]Methyl)-Malonic Acid (Compound c)
Compound b (1.370 g, 3.93 mmol) was dissolved in an aqueous solution of 3 g of LiOH in 25 mL of water. The solution was stirred by refluxing at 180°C for 12 h. Then the solution was cooled to room temperature and acidified with 2 n HCl taking care that the temperature did not exceed 25°C. The solution was extracted twice with 20 mL of ethyl acetate and dried (Na2SO4), and the solvent was removed under reduced pressure at 40°C. Yield: 951 mg (83%).
2-Acetylamino-2-(1H-[3-13C]Indolyl)[3′-13C]Propionic Acid (Compound d)
Compound c (930 mg) was suspended in 20 mL of water and the mixture was stirred at 100°C for 1 h. The mixture was cooled to room temperature and extracted three times with 20 mL of ethyl acetate. The organic layers were dried with Na2SO4 and the solvent was removed under reduced pressure at 40°C. Yield: 540 mg (79%).
2-Amino-2-(1H-[3-13C]Indol-3-yl)[3′-13C]Propionic Acid (d,l-[3,3′-13C2]Trp)
Compound d (520 mg) was stirred at 150°C in 2.0 mL of hydrazine in a sealed pressure tube for 15 h. The hydrazine was then evaporated under reduced pressure. The residue was dissolved in 5 mL of water and purified on an ion exchanger (Dowex 50 W X8, Sigma-Aldrich, Deisenhofen, Germany) by elution with aqueous ammonia (25% [w/v]). Yield: 195 mg (45%). M (calculated Mr) = 206.21; MS: m/z (%): 206 (3), 161 (2), 132 (100), 118 (5), 112 (26), 97 (29), 56 (31). 1H-NMR (600 MHz, dimethyl sulfoxide): d (doublet) = 3.01 (dm, 1JCH = 127.2 Hz, 1 H), 3.31 (dm, 1JCH = 129.3 Hz, 1 H), 3.50 (m, 1 H), 6.96 (dd, 3JHH = 7.5 Hz, 7.5 Hz, 1 H), 7.05 (dd, 3JHH = 7.5 Hz, 7.5 Hz, 1 H), 7.24 (d, 2JCH = 6.6 Hz, 1 H), 7.35 (d, 3JHH = 7.8 Hz, 1 H), 7.56 (d, 3JHH = 7.8 Hz, 1 H), 7.64 (s [singlet], 2 H, NH2), 11.05 (s, 1 H, NH). 13C-NMR (151 MHz, dimethyl sulfoxide): d = 27.17 (d, 1JCC = 49.0 Hz), 54.75 (d, 1JCC = 49.0 Hz), 109.54 (d, 1JCC = 49.0 Hz), 111.33, 118.20, 118.36, 120.80, 124.14 (d, 1JCC = 53.9 Hz), 127.30 (d, 1JCC = 53.9 Hz), 136.34, 170.51.
Tissue Culture of Developing Maize Kernels
Plants of the commercial hybrid Pioneer 3394 were grown in one field in Johnston, Iowa. Ear shoots were kept bagged until the plants were self-pollinated, 2 to 3 d after silk emergence. Ears were harvested 4 d after pollination, placed on ice, and processed within 4 h of harvesting, essentially following the method of Gengenbach (1977). In the field, protruding silks were cut off and the outer most husks removed. In the laboratory all steps were conducted under aseptic conditions. Holding ears by the shank, each ear was sprayed with alcohol and flamed before removing the outer husks. When only the most inner layer of husks was left, the ear was again lightly sprayed with alcohol and flamed. Longitudinal sections of the ear, two kernels wide and with cob tissue at least 3 mm deep, were removed from the middle one-third of the ear. The wedges were further sectioned to eight-kernel blocks and the two kernels at each end of the cob piece were carefully removed to leave four developing kernels per block. Care was taken during preparation to prevent the drying out of kernels and cob tissue. The culture trays consisted of disposable Phytatrays (P5929, Sigma, St. Louis) each with a sterile 1- to 1.5-cm-deep block of florist foam (Wet Foam Block, StyroFab, Waxahachie, TX). The tray lid (used as a base) was filled with 50 to 80 mL of culture medium, enough to completely soak the foam and leave 2 mL of free medium. Kernel blocks were pressed onto the wet foam block until level with the surface of the foam block. On average, each tray contained 12 four-kernel blocks. With a hot dissecting needle, four small holes were made on the sides of the tray lids to allow for gas exchange. Before assembling the culture tray foam blocks and liquid culture medium were sterilized by autoclaving for 30 min. The culture medium used was a modification of that described by Gengenbach and Jones (1994) and contained per liter 80 g of Suc or, in case of the [U-13C6]Glc-labeling experiment 80 g of Glc, 4.33 g of Murashige-Skoog salts, 2 g of l-Asn monohydrate, 400 μg of thiamine hydrochloride, and 10 mg streptomycin sulfate. The medium was adjusted to pH 5.8 with 4 n HCl before autoclaving for 35 min. Cultures were incubated at 24°C, in the dark, and inspected every 3 d for possible contamination. After 7 d of culture, kernel blocks were transferred to fresh trays containing labeled medium and incubated for an additional 19 d. Overall, kernels were harvested 30 d after pollination and frozen at −80°C.
Culturing kernels on florist foam made more efficient use of expensive labeled material than previously reported systems. Here, only 1 to 2 mL of medium was used per harvested kernel. Cully et al. (1984), plating on agar media, used an average of 8 mL of medium per kernel; Singletary and Below (1989), using liquid medium and a wire/paper platform, employed 23 to 31 mL of medium per kernel.
Isolation of IAA and Amino Acids
To allow the isolation of metabolites in quantities sufficient for NMR analysis IAA was isolated from IAA conjugates and amino acids were isolated from total protein. Frozen kernels were ground in liquid nitrogen and extracted three times with 70% (v/v) acetone in water and twice with n-hexane:acetone, 1:1 (v/v). The residue was divided equally for Trp isolation and isolation of the other amino acids. For purification of IAA the volume of the supernatant was reduced to 30%. The solution was adjusted to pH 14 by addition of 10 n NaOH, incubated for 1 h to allow hydrolysis of conjugates (Ueda and Bandurski, 1969), adjusted to pH 2 by addition of concentrated HCl and extracted three times with 0.7 volume of ethyl acetate:n-hexane (3:1, v/v). The solvent was removed and the residue was dissolved in 2 mL of methanol. IAA was purified by two sequential isocratic HPLC steps using a nucleosil RP18 column (10 × 250 mm, flow rate: 6 mL/min, Merck, Rahway, NJ). The effluent was monitored photometrically (278 nm). For the first HPLC step methanol:acetic acid:water (10:9:81, v/v) was used as solvent. IAA eluted at 25 min. For the second HPLC step methanol:acetic acid:water (40:6:54, v/v) was used as solvent. IAA eluted at 6 min. Fractions containing IAA were combined and extracted twice with ethyl acetate. The organic phase was evaporated to dryness under reduced pressure.
An aliquot of the solvent-extracted tissue was subjected to alkaline hydrolysis and Trp was isolated as described earlier (Eisenreich et al., 1991).
A second aliquot of biomass was boiled for 24 h under reflux in 6 n HCl containing 4% (v/v) thioglycolic acid. Amino acids were separated by ion-exchange chromatography as previously described (Eisenreich et al., 1991). Leu and Tyr were further purified by reversed-phase HPLC using an RP18 column (21 × 250 mm, Macherey-Nagel, Düren, Germany) with water as eluent. The retention volumes were 120 and 150 mL, respectively. His was purified by preparative thin-layer chromatography on silica plates that were developed with n-butanol:acetic acid:water (4:1:1, v/v). His was detected by spraying a small section of the plate with ninhydrin.
GC-MS Analysis
EI-MS, HR EI-MS, and GC-MS were performed on a MAT 90 double focussing mass spectrometer (Finnigan, Bremen, Germany), equipped with an EI ion source operated at 70 eV. To derivatize IAA to its methylester, 0.1 mg of IAA was dissolved in 100 μL of methanol, and 20 μL of trimethylchlorosilane was added. After 2 h at 25°C the solvent was blown off with nitrogen. For sample injection the residue was dissolved in 100 μL of methanol. Trp was derivatized to trimethylsilyl-Trp: 50 μL of N-methyl-N-(trimethylsilyl) trifluoroacetamide were added to 0.1 mg of Trp. The mixture was kept at 25°C for 2 h and then injected onto the GC column. For GC-MS a GC 3400 gas chromatograph (Varian, Palo Alto, CA) with a fused silica DB-5 ms capillary column (30 m × 0.25 mm, coated with a 0.1-μm layer of liquid phase) and helium as carrier gas was used for sample separation. The injector temperature was kept at 300°C and injection volumes were 0.2 to 0.4 μL of a 1% to 2% (w/v) solution. Temperature program: 2 min isothermal at 50°C, then 10°C/min up to 300°C, finally 15 min isothermal at 300°C.
NMR Spectroscopy
1H- and 13C-NMR spectra of isolated metabolites were recorded at 500.13 and 125.76 MHz, respectively, with a DRX 500 spectrometer (Bruker Instruments, Billerica, MA) equipped with a dual 1H/13C probe head. Synthetic compounds were analyzed using Bruker AMX 600 and ARX 300 spectrometers. 13C-NMR spectra were measured as follows: 45° pulse (3 μs); repetition time, 3.2 s; spectral width, 29 kHz; data set, 64 kilo-words; temperature, 10°C; zero-filling to 128 kilo-words, and gaussian apodization prior to Fourier transformation; 1H decoupling by WALTZ16 during acquisition and relaxation. The IAA signal assignments are based on two-dimensional homo- and heteronuclear NMR analysis (double-quantum filtered correlation spectroscopy, heteronuclear multiple-quantum correlation, and heteronuclear multiple-bond multiple-quantum correlation; data not shown). IAA was dissolved in deuterated methanol. Isolated Trp and Tyr were dissolved in 0.1 n NaOD. Leu and His were dissolved in 0.1 n DCl.
Analysis of 13C Enrichment
For each amino acid analyzed the 13C enrichment of one carbon carrying a proton atom was calculated after integration of 13C-induced satellite signals in the 1H spectrum. This 13C enrichment was set as standard to calculate the 13C enrichment of the other positions in the 13C spectrum after calibration with a natural abundance sample. The mean 13C enrichment of compounds isolated form kernels labeled with [U-13C6]Glc was calculated from the 13C enrichment of all proton carrying positions.
Analysis of Isotopomer Composition
In 1H decoupled 13C-NMR spectroscopy one 13C atom results in a singlet signal. The presence of a second neighboring 13C nucleus results in a doublet signal with a coupling constant of 30 to 70 Hz. If two neighboring 13C nuclei characterized by different coupling constants are incorporated, a doublet of a doublet is observed (Fig. 3). If the coupling constants are the same, a 1:2:1 signal is observed. For each carbon position the central singlet signal, the doublet satellite signals and the double doublet satellite signals were integrated and the fraction of the total integral was calculated. This fraction represents the fraction of multiply 13C-labeled isotopomers of all 13C containing isotopomers (Table II, D). This value multiplied by the 13C enrichment (e) yields the isotopomer content (c) in mol % (Table II, C).
Calculations for the Analysis of the [U-13C6]Glc-Labeling Experiment
To allow comparison of 13C-NMR data of metabolites with different rates of de novo biosynthesis the data were normalized to 100% de novo biosynthesis. The normalized fraction of multiply 13C-labeled isotopomer n, which we introduce here (Table II, E) represents the fraction of the multiply 13C-labeled isotopomer of all 13C-containing isotopomers that result from the incorporation of 13C nuclei that derive from [U-13C6]Glc. It can be calculated from the isotopomer content (c) and the 13C abundance (e) using the following formula: n = c × (e − 1.1%)−1, with the natural abundance of 13C being 1.1%.
ACKNOWLEDGMENTS
We thank Kim Johnson for technical assistance and Wolfgang Steglich and Meinhart Zenk for their advice and helpful discussions.
Footnotes
This research was supported by the Deutsche Forschungsgemeinschaft (grant nos. SPP 1067 and SFB 369) and by Fonds der Chemischen Industrie.
LITERATURE CITED
- Bacher A, Rieder C, Eichinger D, Arigoni D, Fuchs G, Eisenreich W. Elucidation of novel biosynthetic pathways and metabolite flux patterns by retrobiosynthetic analysis. FEMS Microbiol Rev. 1999;22:567–589. [Google Scholar]
- Bandurski RS, Cohen JD, Slovin J, Reinecke DM. Auxin biosynthesis and metabolism. In: Davies JP, editor. Plant Hormones: Physiology, Biochemistry, and Molecular Biology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 39–65. [Google Scholar]
- Bandurski RS, Kowalczyk S, Lezniki A, Mekhedov S, Momonoki Y, Oguri S. Metabolic targets for control of IAA levels in maize. Plant Growth Regulation Society of America, Proceedings of the 25th Annual Meeting. 1998;26:181–186. [Google Scholar]
- Bartel B. Auxin biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:51–66. doi: 10.1146/annurev.arplant.48.1.51. [DOI] [PubMed] [Google Scholar]
- Bartel B, Fink GR. Differential regulation of an auxin producing nitrilase gene family in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1994;91:6649–6653. doi: 10.1073/pnas.91.14.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartling D, Seedorf M, Schmidt RC, Weiler EW. Molecular characterization of two cloned nitrilases from Arabidopsis thaliana: key enzymes in the biosynthesis of the plant hormone indole-3-acetic acid. Proc Natl Acad Sci USA. 1994;91:6021–6025. doi: 10.1073/pnas.91.13.6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BG, Hannah LC. Development of wild type, shrunken-1 and shrunken-2 maize kernels grown in vitro. Theor Appl Genet. 1983;65:47–51. doi: 10.1007/BF00276261. [DOI] [PubMed] [Google Scholar]
- Cully DE, Gengenbach BG, Smith JA, Rubenstein I, Connelly JA, Park W. Endosperm protein synthesis and l-[35S]methionine incorporation in maize kernels cultured in vitro. Plant Physiol. 1984;74:389–394. doi: 10.1104/pp.74.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenreich W, Bacher A. Elucidation of biosynthetic pathways by retrodictive/predictive comparison of isotopomer patterns determined by NMR spectroscopy. In: Setlow JK, editor. Genetic Engineering, Principles and Methods. New York: Plenum Press; 2000. (in press) [DOI] [PubMed] [Google Scholar]
- Eisenreich W, Schwarzkopf B, Bacher A. Biosynthesis of nucleotides, flavins and deazaflavins in Methanobacterium thermoautotrophicum. J Biol Chem. 1991;266:9622–9631. [PubMed] [Google Scholar]
- Epstein E, Cohen JD, Bandurski RS. Concentration and metabolic turnover of indoles in germinating kernels of Zea mays. Plant Physiol. 1980;65:415–421. doi: 10.1104/pp.65.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengenbach BG. Development of maize caryopses resulting from in vitro pollination. Planta. 1977;134:91–93. doi: 10.1007/BF00390100. [DOI] [PubMed] [Google Scholar]
- Gengenbach BG, Jones RJ. In vitro culture of maize kernels. In: Freeling M, Walbot V, editors. The Maize Handbook. Berlin: Springer-Verlag; 1994. pp. 705–708. [Google Scholar]
- Hillebrand H, Bartling D, Weiler E. Structural analysis of the nit2/nit1/nit3 gene cluster encoding nitrilases, enzymes catalyzing the terminal activation step in indole-acetic acid biosynthesis in Arabidopsis thaliana. Plant Mol Biol. 1998;36:89–99. doi: 10.1023/a:1005998918418. [DOI] [PubMed] [Google Scholar]
- Ho CL, Noji M, Saito K. Plastidic pathway of serine biosynthesis: molecular cloning and expression of 3-phos-phoserine phosphatase from Arabidopsis thaliana. J Biol Chem. 1999;274:11007–11012. doi: 10.1074/jbc.274.16.11007. [DOI] [PubMed] [Google Scholar]
- Ho CL, Noji M, Saito M, Yamazaki M, Saito K. Molecular characterization of plastidic phosphoserine aminotransferase in serine biosynthesis from Arabidopsis. Plant J. 1998;16:443–452. doi: 10.1046/j.1365-313x.1998.00313.x. [DOI] [PubMed] [Google Scholar]
- Jensen PJ, Bandurski RS. Incorporation of deuterium into indole-3-acetic acid and tryptophan in Zea mays seedlings grown on 30% deuterium oxide. J Plant Physiol. 1996;147:697–702. [Google Scholar]
- Michalczuk L, Ribnicky DM, Cooke TJ, Cohen JD. Regulation of indole-3-acetic acid biosynthetic pathways in carrot cell cultures. Plant Physiol. 1992;100:1346–1353. doi: 10.1104/pp.100.3.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J. Auxin metabolism. Physiol Plant. 1997;100:431–442. [Google Scholar]
- Normanly J, Bartel B. Redundancy as a way of life: IAA metabolism. Curr Opin Plant Biol. 1999;2:207–213. doi: 10.1016/s1369-5266(99)80037-5. [DOI] [PubMed] [Google Scholar]
- Normanly J, Cohen JD, Fink GR. Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc Natl Acad Sci USA. 1993;93:10355–10359. doi: 10.1073/pnas.90.21.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oaks A. The synthesis of leucine in maize embryos. Biochim Biophys Acta. 1965;11:79–89. doi: 10.1016/0304-4165(65)90474-5. [DOI] [PubMed] [Google Scholar]
- Östin A, Ilic N, Cohen JD. An in vitro system from maize seedlings for tryptophan-independent IAA biosynthesis. Plant Physiol. 1999;119:173–178. doi: 10.1104/pp.119.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwanski ER, Last RL. Tryptophan biosynthesis and metabolism: biochemical and molecular genetics. Plant Cell. 1995;7:921–934. doi: 10.1105/tpc.7.7.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapparini F, Cohen JD, Slovin J. Indole-3-acetic acid biosynthesis in Lemna gibba studied using stable isotope labeled anthranilate and tryptophan. Plant Growth Reg. 1999;27:139–144. [Google Scholar]
- Rekoslavskaya NI. Biosynthesis pathways of IAA and tryptophan in the developing maize endosperm: in vitro study. Fiziologiya Rastenii (Moscow) 1995;42:165–174. [Google Scholar]
- Rekoslavskaya NI, Bandurski RS. Indole as a precursor of indole-3-acetic acid in Zea mays. Phytochemistry. 1994;35:905–909. [Google Scholar]
- Schmid J, Amrhein N. Molecular organization of the shikimate pathway in higher plants. Phytochemistry. 1995;39:737–749. [Google Scholar]
- Sekimoto H, Seo M, Kawakami N, Komano T, Desloire S, Liotenberg S, Marion-Poll A, Cabaoche M, Kamiya Y, Koshiba T. Molecular cloning of aldehyde oxidases in Arabidopsis thaliana. Plant Cell Physiol. 1998;39:433–442. doi: 10.1093/oxfordjournals.pcp.a029387. [DOI] [PubMed] [Google Scholar]
- Singletary GW, Below FE. Growth and composition of maize kernels cultured in vitro with varying supplies of carbon and nitrogen. Plant Physiol. 1989;89:341–346. doi: 10.1104/pp.89.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam YY, Normanly J. Determination of indole-3-pyruvic acid levels in Arabidopsis thaliana by gas chromatography selected ion monitoring mass spectroscopy. J Chromatography. 1998;800:101–108. doi: 10.1016/s0021-9673(97)01051-0. [DOI] [PubMed] [Google Scholar]
- Thimann KV. Hormone Action in the Whole Life of Plants. Amherst: University of Massachusetts Press; 1977. [Google Scholar]
- Ueda M, Bandurski RS. A quantitative estimation of alkali-labile indole-3-acetic acid compounds in dormant and germinating maize kernels. Plant Physiol. 1969;44:1175–1181. doi: 10.1104/pp.44.8.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner I. Biosynthese von Gallussäure und Aminosäuren in Pflanzen. PhD Thesis. Munich: Technische Universität; 1996. [Google Scholar]
- Wiater A, Krajewska-Grynkiewicz K, Klopotowski T. Histidine biosynthesis and its regulation in higher plants. Acta Biochim Pol. 1971;18:299–307. [PubMed] [Google Scholar]
- Wright AD, Sampson MB, Neuffer G, Michalczuk L, Slovin JP, Cohen JD. Indole-3-acetic acid biosynthesis in the mutant maize orange pericarp, a tryptophan auxotroph. Science. 1991;254:998–1000. doi: 10.1126/science.254.5034.998. [DOI] [PubMed] [Google Scholar]